Abstract
Background and purpose:
There is good evidence that agents interacting with the endocannabinoid system in the body can also interact with the peroxisome proliferator-activated receptor γ. The present study was designed to test whether the reverse is true, namely whether peroxisome proliferator-activated receptor γ ligands have direct effects upon the activity of the endocannabinoid metabolizing enzyme fatty acid amide hydrolase.
Experimental approach:
Fatty acid amide hydrolase activity was measured in rat brain homogenates, C6 glioma and RBL2H3 basophilic leukaemia cells. Cellular uptake of anandamide was also assessed in these cells.
Key results:
Peroxisome proliferator-activated receptor γ activators inhibited the metabolism of the endocannabinoid anandamide in rat brain homogenates with an order of potency MCC-555 > indomethacin ≈ ciglitazone ≈ 15-deoxy-Δ12,14-prostaglandin J2 ≈ pioglitazone > rosiglitazone > troglitazone. The antagonists BADGE, GW9662 and T0070907 were poor inhibitors of anandamide hydrolysis. The inhibition by ciglitazone was competitive and increased as the pH of the assay buffer was decreased; the Ki value at pH 6.0 was 17 μM. In intact C6 glioma cells assayed at pH 6.2, significant inhibition of anandamide hydrolysis was seen at 3 μM ciglitazone, whereas 100 μM was required to produce significant inhibition at pH 7.4. Ciglitazone also interacted with monoacylglycerol lipase as well as with cannabinoid CB1 and CB2 receptors.
Conclusions and implications:
Ciglitazone may be useful as a template for the design of novel dual action anti-inflammatory agents which are both inhibitors of fatty acid amide hydrolase and agonists at the peroxisome proliferator-activated receptor γ.
Keywords: endocannabinoid, anandamide, fatty acid amide hydrolase, cannabinoid receptors, peroxisome proliferator-activated receptor γ, ciglitazone
Introduction
The peroxisome proliferator-activated receptor (PPAR) family of nuclear receptors are ligand-activated transcription factors, and are of considerable current interest in relation to their use for the treatment of inflammatory conditions and cancer (for reviews, see Moraes et al., 2006; Wang et al., 2006). There are three classes of PPAR, α, γ and β/δ and a variety of both synthetic and naturally occurring ligands for these agents have been described. In the case of PPARγ, ligands include the thiazolidinediones such as rosiglitazone, used for the treatment of type II diabetes, as well as arachidonic acid derivatives such as 15-deoxy-Δ12,14-prostaglandin-J2 (Moraes et al., 2006).
One family of endogenous compounds that have been shown to interact with the PPARs is the N-acylethanolamines. The most well-studied of these compounds, anandamide (AEA, arachidonoylethanolamide), is primarily described in the literature as an endogenous agonist at cannabinoid (CB) receptors (Devane et al., 1992) but activates PPARγ receptors at micromolar concentrations (Bouaboula et al., 2005; see also, Gasperi et al., 2007) and acts synergistically with the PPARα receptor agonist in the formalin test of inflammatory pain, the combination being antagonized by the CB1 receptor antagonist/inverse agonist rimonabant (Russo et al., 2007). Palmitoylethanolamide and oleoylethanolamide have no direct effect upon CB receptors, but activate PPARα receptors, an action which has been suggested to contribute to their anti-inflammatory (palmitoylethanolamide) and satiety-producing (oleoylethanolamide) properties (Fu et al., 2003; Lo Verme et al., 2005).
N-acylethanolamines are primarily metabolized by the enzyme fatty acid amide hydrolase (FAAH), and treatment of rodents with the selective FAAH inhibitor 3′-carbamoyl-biphenyl-3-yl-cyclohexylcarbamate (URB597) produces beneficial effects in experimental models of inflammation and inflammatory pain that can be reduced by SR144528 (Holt et al., 2005; Jayamanne et al., 2006). SR144528 was originally described as a selective CB2 receptor antagonist/inverse agonist (Rinaldi-Carmona et al., 1998), but has recently been shown to block PPARα receptor-mediated responses (Lo Verme et al., 2006). Anandamide can also interact with cyclooxygenase-2 as a substrate to form prostaglandin ethanolamides, which as a class do not interact with CB receptors, but do have a range of biological actions, including effects that may involve the PPARγ pathway (Yu et al., 1997; Berglund et al., 1999; Rockwell and Kaminski, 2004). The other main endocannabinoid, 2-arachidonoylglycerol, is also a substrate for FAAH (Goparaju et al., 1998) (although in the brain, monoacylglycerol lipase (MGL) is probably more important, Dinh et al., 2002) as well as COX-2 (Kozak et al., 2000), and causes an activation of PPARγ (Rockwell et al., 2006).
The findings that AEA (and 2-AG) can interact with FAAH, cyclooxygenase-2 and PPARγ would suggest that there may be an overlap of the structural requirements for association with these three targets. In support of this, non-steroidal, anti-inflammatory agents such as indomethacin and ibuprofen, which have a primary action upon cyclooxygenase enzymes, can also interact directly with both PPARγ and FAAH (Paria et al., 1996; Fowler et al., 1997; Lehmann et al., 1997). These findings raise the possibility that agents with a primary action upon PPARγ, such as the thiazolidinediones, may also interact directly with FAAH. This possibility has been investigated in the present study.
Materials and methods
Preparation of rat brain homogenates
Brains (without cerebellum) from adult male Sprague–Dawley rats were used in this study. The frozen brains were thawed on ice and homogenized with a glass homogenizator in 20 ml of 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 1 mM MgCl2, pH 7.0. The homogenates were centrifuged at ∼35 000 g for 20 min (4°C). The supernatants were discarded, the pellets resuspended in 20 ml buffer and centrifuged again. The pellets were then resuspended in 10 ml buffer and incubated at 37°C for 15 min to remove all endogenous FAAH substrates which otherwise could interfere with the assay. After the incubation, the homogenates were centrifuged a final time at ∼35 000 g for 20 min (4°C). The supernatants were discarded and the pellets were resuspended in Tris-HCl buffer (50 mM Tris-HCl, 1 mM ethylenediaminetetraacetic acid (EDTA), 3 mM MgCl2, pH 7.4). The homogenates were frozen at −80°C in aliquots. Ethical permission for the study was obtained from the local ethical committee.
Culturing of cells
Rat C6 glioma cells (passage range 14–24) were obtained from the European Collection of Cell Cultures (Porton Down, Wiltshire, UK). F10-Ham nutrient mixture containing 25 mM HEPES, L-glutamine, 10% fetal bovine serum and 100 U ml −1 penicillin+100 μg ml−1 streptomycin was used as culture medium. Rat RBL-2H3 basophilic leukaemia cells (passage range 30–36) were obtained from the American Type Culture Collection (Manassas, VA, USA). Minimal essential medium containing Earl's salts, 2 mM L-glutamine, 15% fetal bovine serum and 100 U ml−1 penicillin+100 μg ml−1 streptomycin was used as culture media. The cells were grown in 75 cm2 culturing flasks at 37°C, 5% CO2 in humidified atmosphere at normal atmospheric pressure. The cell culture media was changed every 2–3 days and passage was performed two times a week.
FAAH activity assay in rat brain homogenates
The assay was carried out essentially as described previously (Boldrup et al., 2004). Briefly, test compounds (10 μl, in ethanol, except for rosiglitazone, troglitazone and 2-chloro-5-nitro-N-4-pyridinyl-benzamide (T0070907) (in dimethyl sulphoxide (DMSO)) and pioglitazone (in DMSO:ethanol 1:1 v/v)), 165 μl of homogenates (1.5 μg protein per assay unless otherwise stated) diluted in 10 mM Tris-HCl, 1 mM EDTA, pH 7.4 (unless otherwise stated) and [Et-3H]AEA (that is, AEA labelled in the ethanolamine part of the molecule, 25 μl in 10 mM Tris-HCl, 1 mM EDTA, pH 7.4 containing 1% w/v fatty acid-free bovine serum albumin), assay concentration 2 μM (unless otherwise stated), were added to the tubes. In some cases, a preincubation phase was used between test compound and homogenate before addition of AEA; this is indicated in the figure legends. The tubes were incubated for 5–10 min at 37°C, as indicated. Reactions were stopped by putting the tubes on ice and adding 400 μl of activated charcoal mixture (80 μl active charcoal, 320 μl 0.5 M HCl). The samples were mixed three times and left at room temperature for ∼30 min. To sediment the charcoal, the tubes were centrifuged at ∼700 g for 10 min. An aliquot (200 μl) of the supernatant was transferred from each tube to scintillation vial for analysis of tritium content by liquid scintillation spectroscopy with quench correction. Blank values were for assays conducted in the absence of homogenate.
AEA uptake in C6 and RBL-2H3 cells
The assay was that of Rakhshan et al. (2000) as modified by Sandberg and Fowler (2005). Briefly, C6 or RBL-2H3 cells were plated with an initial density of 2 × 105 cells per well. The plates were incubated overnight at 37°C in humidified atmosphere with 5% CO2. Cells were then washed once with Krebs–Ringer HEPES buffer (120 mM NaCl, 4.7 mM KCl, 2.2 mM CaCl2, 10 mM HEPES, 0.12 mM KH2PO4, 0.12 mM MgSO4, pH 7.4 or pH 6.2, as appropriate) containing 1% bovine serum albumin and once with buffer without bovine serum albumin. Krebs–Ringer HEPES buffer containing 0.1% fatty acid-free bovine serum albumin (330 μl) was added to each well followed by addition of 10 μl URB597 (to a final assay concentration of 0.1 μM (C6 cells) or 1 μM (RBL2H3 cells)) or vehicle. The plate was incubated at 37°C for 10 min. Aliquots (10 μl) of ciglitazone or rosiglitazone (containing 2 μl ethanol to ensure sufficient solubility) were added followed by a further 10 min of incubation at 37°C. An aliquot (50 μl) of [Ara-3H]AEA (that is, AEA labelled in the arachidonoyl part of the molecule, final concentration of 0.1 μM (C6 cells) or 0.2 μM (RBL2H3 cells)) was added and uptake was allowed for 5 min at 37°C. The uptake was stopped by placing the plates on ice and washing the cells three times with cold Krebs–Ringer HEPES buffer containing 1% bovine serum albumin (500 μl). NaOH (0.2 M, 500 μl) was added to each well and the plate was incubated 15 min at 75°C. Aliquots of 300 μl were taken from each well and transferred to scintillation vials for analysis of tritium content by liquid scintillation spectroscopy with quench correction. The assay was also run on plates without cells, which were treated exactly the same as for plates containing cells.
FAAH activity in C6 and RBL-2H3 cells
The assay of Paylor et al. (2006) was used. Briefly, C6 or RBL-2H3 cells were plated in 24-well plates, incubated overnight and washed as described above. Krebs–Ringer HEPES buffer containing 0.1% fatty acid-free bovine serum albumin (340 μl) was added to each well followed by addition of 10 μl of ciglitazone or rosiglitazone (containing 2 μl ethanol to ensure sufficient solubility) and the plate was preincubated for 10 min at 37°C. Then, 50 μl of [Et-3H]AEA (final concentration of 0.1 μM (C6 cells) or 0.2 μM (RBL2H3 cells)) was added and incubation continued for another 20 min. To terminate the reaction, the plate was placed on ice and 400 μl of cold methanol was added. The cells were collected by scraping the wells and aliquots of 400 μl were transferred to glass tubes. Chloroform (200 μl) was added to each tube and the samples were mixed two times. Phases were separated by centrifugation and aliquots (200 μl) of the aqueous phase were transferred to scintillation vials for analysis of tritium content by liquid scintillation spectroscopy with quench correction. Blank values were for wells alone.
Inhibition of 2-oleoylglycerol hydrolysis
The assay of Brengdahl and Fowler (2006) was used. Briefly, cytosol preparations of cerebellum from adult male Sprague–Dawley rats (available at the Department) or commercially available recombinant MGL were used. The assay was performed in 96-well flat-bottomed microplates. Test compound (5 μl, diluted in ethanol) were added to each well of a 96-well flat-bottomed microplate (5 μl pure EtOH to controls and blanks). Aliquots (35 μl) of rat cerebellar cytosolic fractions (2.5 μg protein per well) or recombinant human MGL (0.03 μl per well) diluted in assay buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.4) were added to each well. The plate was preincubated at 25°C for 10 min. [3H]2-oleoylglycerol (2-OG) (10 μl, in 10 mM Tris-HCl, 1 mM EDTA, pH 7.4 containing 1% w/v fatty acid-free bovine serum albumin) was added to the wells to give an assay concentration of 2 μM. The plate was incubated for 2 h (cytosol) or 1 h (recombinant MGL) at 25°C. The reaction was stopped by placing the plate on ice and adding 100 μl phenyl sepharose solution (20 μl phenyl sepharose, 80 μl 1.5 M NaCl, 0.5 M HCl). The plate was left on ice for approximately 30 min to allow the phenyl sepharose to settle, and 30 μl of the aqueous phase was transferred from each well to scintillation vials for analysis of tritium content by liquid scintillation spectroscopy with quench correction. Blank values were for wells not containing cytosolic fractions or MGL.
CB receptor binding
The assay was carried out in 96-well flat-bottomed microplates. [3H]CP-55,940 (final concentration 0.4 nM) in assay buffer (50 mM Tris-HCl pH 7.4, 1 mM EDTA, 3 mM MgCl2, 0.5% bovine serum albumin (w/v)) was added to each well in an aliquot of 50 μl followed by 50 μl of test compound dissolved in a combination of assay buffer/ethanol (ciglitazone, [[6-[(2-fluorophenyl)methoxy]-2-napthalenyl]methyl]-2,4- thiazolidinedione (MCC-555)) or assay buffer/DMSO (HU 210). Aliquots (150 μl) of either rat brain membranes (the same as used for FAAH assays, 15 μg per well for CB1 receptors) or membranes expressing human CB2 receptors, 0.44 μg per well, were added to the wells. The plates were incubated at 37°C for 60 min and tilted four times during this time. The samples were collected on a Whatman GF/C filter pre-wet with 0.2% (v/v) polyethylenimine and washed with 50 mM Tris-HCl pH 7.4, containing 0.1% (w/v) bovine serum albumin using a cell harvester. The resulting filter spots were transferred to scintillation vials and analysed for radioactivity by liquid scintillation spectroscopy with quench correction. Non-specific binding was determined using 1 μM HU 210.
Statistical analyses
For curves when the observed maximal inhibition was greater than 50%, pI50 and IC50 values were obtained using the built-in programme sigmoidal dose–response curve, variable slope of the GraphPad Prism computer programme (GraphPad Software Inc., San Diego, CA, USA), with top (uninhibited) and bottom (minimum activity remaining) values set to 100 and 0, respectively. Statistical evaluations (as shown in the text) were undertaken using the same computer programme. Kmapp values were calculated from pooled data using the direct linear plot (Eisenthal and Cornish-Bowden, 1974) analysis and the Enzyme Kinetics v1.4 computer programme (Trinity Software, Campton, NH, USA).
Materials
Anandamide [ethanolamine-1-3H] (specific activity 2.22 TBq mmol−1), anandamide [arachidonyl 5,6,8,9,11,12,14,15-3H] (specific activity 7.4 TBq mmol−1) and 2-OG (glycerol-1,2,3-3H) (specific activity 0.74 TBqmmol−1) were purchased from American Radiolabeled Chemicals Inc., St Louis, MO, USA CP-55,940 [side chain-2,3,4(N)-3H] (specific activity 5.94 TBq mmol−1) was obtained from Perkin Elmer, Boston, MA, USA. Human CB2 receptor membrane preparations were purchased from Bio-Xtal, Mundolsheim, France via the Axxora platform (Swedish Distributor In vitro Sweden AB, Stockholm, Sweden). Ciglitazone, 15-deoxy-Δ12,14-prostaglandin-J2, GW9662 (2-chloro-5-nitrobenzanilide), MCC-555, pioglitazone, URB597, methyl arachidonyl fluorophosphonate (MAFP), non-radioactive AEA and recombinant human MGL were obtained from the Cayman Chemical Co, Ann Arbor, MI, USA. 2,2′-[(1-methylethylidene) bis(4,1-phenyleneoxy-methylene)]bisoxirane (BADGE) and T0070907 were purchased from Biomol international, Plymouth Meeting, PA, USA. HU 210 ((6aR)-trans-3-(1,1-dimethylheptyl)-6a,7,10,10a-tetrahydro-1-hydroxy-6,6-dimethyl-6H-dibenzo[b,d]pyran-9-methanol) was purchased from Tocris Bioscience, Ellisville, MO, USA. Indomethacin, 3-(2-aminoethyl)-5-((4-ethoxyphenyl)methylene)-2,4-thiazolidinedione, activated charcoal, 2-OG and fatty acid-free bovine serum albumin were obtained from Sigma Aldrich, St Louis, MO, USA. Phenyl sepharose was obtained from Amersham Bioscience, Uppsala, Sweden.
Results
Inhibition by PPARγ-ligands of FAAH in rat brain homogenates
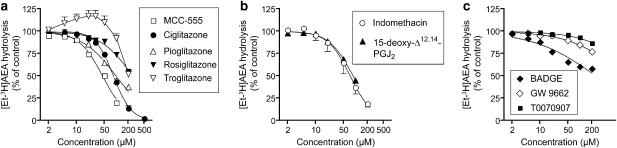
The effects of a series of 10 PPARγ ligands upon the ability of rat brain membrane preparations to hydrolyse the FAAH substrate AEA (2 μM) are shown in Figure 1. At the outset, it should be pointed out for the sake of clarity that the effects studied are direct effects of the compounds upon the enzyme activity rather than effects secondary to transcriptional activation of PPARγ. Among the thiazolidinedione class of compounds, MCC-555 (pI50 value 4.35±0.02; IC50 value 45 μM) was the most potent inhibitor, followed by ciglitazone (pI50 value 4.08±0.01, IC50 value 84 μM), pioglitazone (pI50 value 4.01±0.04, IC50 value 97 μM), rosiglitazone (26 and 45% inhibition at 100 and 200 μM, respectively), and troglitazone (8 and 48% inhibition at 100 and 200 μM, respectively) (Figure 1a). The curve for troglitazone showed an apparent stimulation of activity. However, with the exception of the highest concentration tested (200 μM), the 95% confidence limits of the data points straddled 100%. Indomethacin and 15-deoxy-Δ12,14-prostaglandin J2 were roughly equivalent in potency to ciglitazone, with pI50 values (IC50 values in brackets) of 4.14±0.05 (73 μM) and 4.06±0.02 (87 μM), respectively (Figure 1b). 3-(2-Aminoethyl)-5-((4-ethoxyphenyl)methylene)-2,4-thiazolidinedione, an extracellular signal-regulated kinase docking domain inhibitor, was also tested, but produced <15% inhibition of the hydrolysis of 0.5 μM AEA at the highest concentration tested (100 μM, data not shown). The PPARγ antagonists BADGE, GW9662 and T0070907 were poor inhibitors of AEA hydrolysis (Figure 1c). A PPARβ/δ agonist, GW501516, was also tested, and found to inhibit AEA hydrolysis with a pI50 value of 4.52±0.02, corresponding to an IC50 value of 30 μM (data not shown).
Figure 1.
Interaction of PPARγ ligands with FAAH in rat brain membrane fractions. (a) thiazolidinediones; (b) other PPARγ activators; (c) PPARγ antagonists. The compounds were preincubated with the homogenates for 10 min before addition of 2 μM [3H-Et]AEA and incubation for a further 10 min (assay pH 7.4). Data are means±s.e.m. (when not enclosed by the symbols), n=3. AEA, anandamide (arachidonoylethanolamide); FAAH, fatty acid amide hydrolase; MCC-555, [[6-[(2-fluorophenyl)methoxy]-2-napthalenyl]methyl]-2,4-thiazolidinediones; PPAR, peroxisome proliferator-activated receptor; T0070907, 2-chloro-5-nitro-N-4-pyridinyl-benzamide.
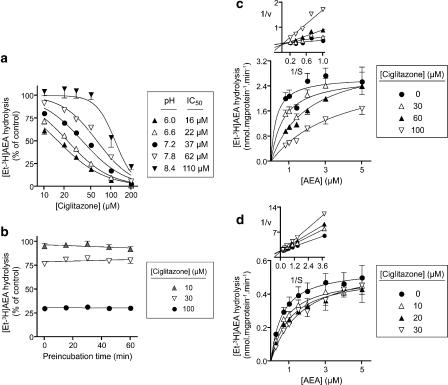
The interaction of ciglitazone with FAAH was investigated in more detail. The inhibition was sensitive to the assay pH, with IC50 values for inhibition of 0.5 μM AEA hydrolysis ranging from 16 μM at pH 6.0 to 110 μM at pH 8.4 (Figure 2a). Preliminary experiments also found the potency of rosiglitazone to increase as the assay pH was lowered, although the effect of pH was more modest than seen with ciglitazone. Thus, at a concentration of 50 μM, the % of control hydrolysis of 0.5 μM AEA was 70, 75, 78 and 82 at pH 6.0, 6.6, 7.2 and 7.8, respectively (means, n=2). The corresponding values at 100 μM rosiglitazone were 53, 55, 57 and 64%, respectively.
Figure 2.
Interaction of ciglitazone with FAAH in rat brain membrane fractions. (a) Effect of ciglitazone upon the hydrolysis of 0.5 μM [3H-Et]AEA over the pH range 6.0–8.4. The protein contents were 3.0, 2.1, 1.5, 1.1 and 0.8 μg per assay at pH 6.0, 6.6, 7.2, 7.8 and 8.4, respectively, the differences reflecting the pH optimum of the enzyme (Schmid et al., 1985). Preincubation times were 10 min. In (b), ciglitazone was preincubated for different times before addition of 2 μM [3H-Et]AEA (assay pH 7.4). In (c) and (d), no preincubation phase was used and the pH of the assay buffer was either 7.4 ((c) 1.5 μg protein per assay) or 6.0 ((d) 2 μg protein per assay). In (a–c), the incubation times with [3H-Et]AEA were 5 min, and in (d) it was 10 min. Data are means±s.e.m., n=3. The insets to (c and d) illustrate the competitive nature of the inhibition. AEA, anandamide (arachidonoylethanolamide); FAAH, fatty acid amide hydrolase.
At an assay pH of 7.4, the inhibition of FAAH by ciglitazone showed no time dependency (Figure 2b) and was competitive in nature (Figure 2c). A competitive interaction was also seen with MCC-555 (data not shown). In these cases, the Km values in the absence of inhibitor were lower than the lowest substrate concentration used, precluding determination of Ki values. However, when the experiments with ciglitazone were run at pH 6, the competitive nature of the inhibition was again seen (Figure 2d), and a Ki value of 17 μM could be determined from a secondary replot of the Kmapp values vs inhibitor concentration. For a competitive inhibitor, a pH dependency would be seen at a given substrate concentration if the Km value for the substrate is pH dependent. To assess this possibility, two membrane fractions were assayed at five substrate concentrations (0.2, 0.4, 0.6, 0.8 and 1.0 μM) at three pH values, 6.0, 7.2 and 8.4. The Km values calculated for the pooled data by direct-linear plot were 0.73, 0.43 and 0.35 μM at pH 6.0, 7.2 and 8.4, respectively. Using these values and the Cheng–Prusoff relation, the Ki values for the data shown in Figure 2a can be estimated as 9, 17 and 45 μM at pH 6, 7.2 and 8.4, respectively. Thus, the pH dependency shown in Figure 2a is due both to a decreased affinity of the enzyme for the substrate and an increased inhibitory potency of ciglitazone as the assay pH is lowered.
Inhibition of AEA accumulation and hydrolysis in RBL2H3 cells by ciglitazone
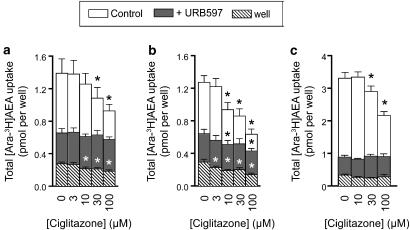
To determine whether or not ciglitazone could affect AEA metabolism in intact cells, the rate of [3H-Ara]AEA uptake and [3H-Et]AEA hydrolysis was determined using C6 glioma and RBL2H3 basophilic leukaemia cells. With respect to uptake, the data are somewhat obfuscated by the ability of ciglitazone to affect the retention of AEA by the wells alone (Figures 3a and b). However, statistically significant reductions in the total uptake of AEA into C6 cells were seen at 30 and 100 μM ciglitazone at an assay pH of 7.4 (Figure 3a), and at 10, 30 and 100 μM ciglitazone at an assay pH of 6.2 (Figure 3b). RBL2H3 cells were also tested at pH 7.4 and found to be sensitive to 30 and 100 μM ciglitazone (Figure 3c). In these cell lines, FAAH has been shown to regulate the rate of AEA uptake (Rakhshan et al., 2000; Kaczocha et al., 2006; Thors et al., 2007), and, as expected, the selective FAAH inhibitor URB597 greatly reduced the rate of uptake. However, ciglitazone was without obvious effect upon the uptake for URB597-treated cells (Figures 3a–c), indicating that the effect of ciglitazone upon the accumulation of AEA is secondary to inhibition of FAAH. Rosiglitazone was also investigated at concentrations of 30 and 100 μM in the C6 cells in these experiments and found to produce effects for both control and URB597-treated cells. Thus, at pH 6.2, for example, the total uptake was 1.28±0.08, 0.67±0.03 and 0.45±0.03 (control) and 0.65±0.06, 0.51±0.05 and 0.43±0.04 (URB597-treated) pmol per well for 0, 30 and 100 μM rosiglitazone, respectively (means±s.e.m., n=6, where the zero values are the same as those in Figure 3b). However, the compound also reduced the retention of AEA by the wells alone at these concentrations, rendering interpretation of these data somewhat difficult.
Figure 3.
Effects of ciglitazone upon the uptake of [3H]AEA. The plates containing either C6 cells at pH 7.4 (a) C6 cells at pH 6.2 (b) or RBL2H3 cells at pH 7.4 (c) or wells alone (all three panels) were preincubated with either vehicle or URB597 for 10 min followed by addition of ciglitazone and 10 min incubation. [Ara-3H]AEA was added and uptake was allowed for 5 min. URB597 concentrations were 0.1 μM for (a and b), and 1 μM for (c). [Ara-3H]AEA concentrations were 0.1 μM for (a and b), and 0.2 μM for (c). In each case, the total size of the column represents the uptake. Values shown are means±s.e.m., n=6. *P<0.05 vs the respective control value, Dunnett's multiple comparison test following significant one-way analysis of variance (ANOVA) for repeated measures for the compound. AEA, anandamide (arachidonoylethanolamide); URB597, 3′-carbamoyl-biphenyl-3-yl-cyclohexylcarbamate.
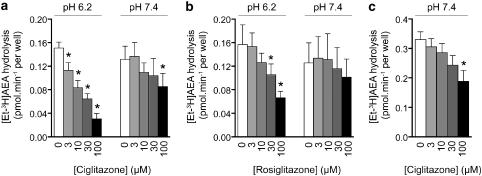
The hydrolysis of [3H-Et]AEA by intact C6 and RBL2H3 cells and its inhibition by ciglitazone was more straightforward to interpret. At an assay pH of 7.4, ciglitazone was a weak inhibitor of the hydrolysis of AEA by either cell line, producing significant effects only at the highest concentration tested (Figures 4a and c), whereas at pH 6.2, the compound significantly reduced the hydrolysis at all concentrations tested (Figure 4a). Consistent with the data in the brain homogenates, rosiglitazone was a weaker inhibitor of anandamide hydrolysis than ciglitazone, but again showed a pH sensitivity (Figure 4b). The effect of MCC-555 upon the uptake and hydrolysis of AEA was also tested, but the ethanol concentrations required to produce sufficient solubilization of this compound were outside the acceptable range for the assays.
Figure 4.
Effect of ciglitazone upon the hydrolysis of [3H]AEA. The plates were preincubated with ciglitazone (a for C6 cells, c for RBL2H3 cells) or rosiglitazone (b, C6 cells) for 10 min followed by addition of [Et-3H]AEA (0.1 (a and b) or 0.2 μM (c)) and 20 min incubation. Values shown are means±s.e.m., n=3–6. *P<0.05 vs the respective control value, Dunnett's multiple comparison test following significant one-way ANOVA for repeated measures for the compound. AEA, anandamide (arachidonoylethanolamide); ANOVA, analysis of variance.
Interaction of ciglitazone and MCC-555 with other components of the CB system
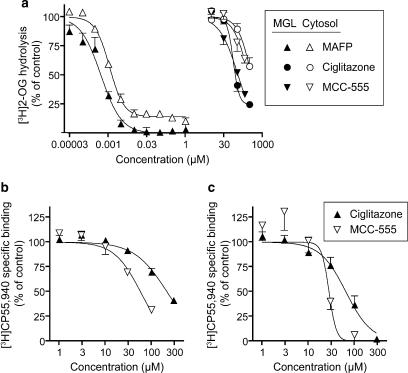
To determine whether MCC-555 and ciglitazone interact with other components of the CB system, their effects upon the activity of MGL and the binding of [3H]CP-55,940 to CB1 and CB2 receptors was assessed. The effects of ciglitazone and MCC-555 upon the hydrolysis of the MGL substrate 2-OG were determined for two enzyme sources: rat brain cytosol and human recombinant MGL (Figure 5a). Both ciglitazone and MCC-555 showed a concentration-dependent inhibition of 2-OG hydrolysis by rat brain cytosolic fractions although less than 50% inhibition had been attained at the highest concentration tested (300 and 200 μM for ciglitazone and MCC-555, respectively). Inhibition tests were also carried out on human recombinant MGL. Both ciglitazone and MCC-555 inhibited 2-OG hydrolysis by the recombinant MGL with pI50 values of 3.99±0.06 and 3.96±0.03, respectively, corresponding to IC50 values of 100 and 110 μM, respectively. MAFP, a potent non-selective serine hydrolase inhibitor known to inhibit MGL (Dinh et al., 2002) was included in the study as a positive control and produced the expected inhibition of 2-OG hydrolysis, although a small residual (∼10%) hydrolytic activity was seen for the cytosolic fractions (Figure 5a).
Figure 5.
(a) Inhibition by ciglitazone, MCC-555 and MAFP of [3H]2-OG hydrolysis by rat brain cytosolic fractions and by recombinant human MGL. The enzyme sources were preincubated with test compounds for 10 min at room temperature followed by addition of 2 μM [3H]2-OG. Incubation was carried out at room temperature for 2 h with cytosol preparations and 1 h with recombinant human MGL preparations. Values shown are mean±s.e.m., n=3–6. (b and c) Inhibition by ciglitazone and MCC-555 of the specific binding of 0.4 nM [3H]CP-55,940 to rat brain CB1 (b) and human recombinant CB2 receptors. (c) Values shown are means±s.e.m., n=3. CB, cannabinoid; MAFP, methyl arachidonyl fluorophosphonate; MCC-555, 5-[[6-[(2-fluorophenyl)methoxy]-2-napthalenyl]methyl]-2,4-thiazolidinedione; MGL, monoacylglycerol lipase; 2-OG, 2-oleoylglycerol.
The effects of MCC-555 and ciglitazone upon the binding of [3H]CP-55,940 to rat brain CB1 and human recombinant CB2 receptors are shown in Figures 5b and c, respectively. Both compounds inhibited the binding to CB1 receptors, with pI50 values of 4.24±0.05 and 3.67±0.04 for MCC-555 and ciglitazone, respectively, corresponding to IC50 values of 57 and 210 μM, respectively. The compounds were more potent towards CB2 receptors, with pI50 values of 4.55±0.19 and 4.19±0.07 for MCC-555 and ciglitazone, respectively, corresponding to IC50 values of 28 and 65 μM, respectively.
Discussion
In view of recent reports that compounds interacting with the endocannabinoid system of the body can also activate PPARγ, the present study was designed with a view to determine whether the reverse is true, that is whether PPARγ-ligands have direct effects upon FAAH, the key enzyme metabolizing AEA. Our data indicate that this is the case, and both MCC-555 and ciglitazone were competitive inhibitors of FAAH. It is rather difficult to compare potencies for lipophilic compounds in different assays, particularly when cell-free experiments are used. However, comparison of rank order of potencies within a given assay are valid. For the FAAH inhibition, the rank order of potency was MCC-555>ciglitazone≈pioglitazone>rosiglitazone>troglitazone. This differs from their rank order of potency towards PPARγ, where the order of potency is rosiglitazone (0.06)>pioglitazone (0.69)≈troglitazone (0.78)≈MCC-555 (∼1)>ciglitazone (3) (numbers in brackets indicate the EC50 values in μM for the compounds in transactivation assays (Reginato et al., 1998; Willson et al., 1996, 2000). This means that of the compounds tested, ciglitazone has the greatest FAAH inhibitory potency relative to its ability to activate PPARγ.
Both ciglitazone and MCC-555 were also found to interact with MGL and with CB1 and CB2 receptors. Once again, comparison of potencies between assays is difficult, but the data can be compared with values for indomethacin obtained using the same assays: over the concentration range tested (10–300 μM), this compound did not inhibit the 2-OG hydrolysing activity of either the rat cytosolic enzyme or the recombinant MGL, but it inhibited the binding of [3H]CP-55,940 to rat brain CB1 and human recombinant CB2 receptors with pI50 values of 3.58±0.19 and 3.89±0.03, respectively, corresponding to IC50 values of 260 and 130 μM, respectively (data from figures 6c–e of Holt et al., 2007). Thus, the relative potencies of ciglitazone and indomethacin for FAAH vs CB receptors are rather similar, whereas indomethacin is more selective than ciglitazone for FAAH vs MGL.
One interesting property of the inhibition of brain [3H]AEA hydrolysis by ciglitazone was its sensitivity to the assay pH used. The increase in potency as the pH is reduced could in theory be due to an increasing contribution of the hydrolytic enzyme N-acylethanolamine-hydrolysing acid amidase, which is found in the brain and shows a pH optimum of 5 (Ueda et al., 2001). However, in membrane preparations of the type used here, we have not been able to demonstrate measurable activity of this enzyme (Holt et al., 2007). A more important determinant of the pH sensitivity may be the ionization state of the molecule (the pKa value of ciglitazone was estimated to be 7.65 by Lipinski et al. (1991) although a simpler and more soluble thiazolidinedione molecule had a pKa (H2O) value of 6.40). A similar pH dependency has been seen for the inhibition of FAAH by the acidic non-steroidal anti-inflammatory drugs indomethacin, flurbiprofen and ibuprofen, the latter two showing a similar pH shift in intact cells when the pH of the extracellular medium is lowered (Holt et al., 2001; Fowler et al., 2003; Holt and Fowler, 2003). In contrast, URB597 is less effective at a lower pH (Paylor et al., 2006). The C6 cells used here can buffer, to a certain extent, the reduction of the extracellular pH, so that the change in the intracellular pH is only about 0.4 U (Holt and Fowler, 2003). The difference in potency of ciglitazone, and even rosiglitazone, for the C6 cells seen at assay pH values of 7.4 and 6.2 is thus larger than would have been predicted from the data using rat brain homogenates. This would suggest that the main effect of the pH shift in the C6 cells is to allow an increased permeability of the compounds into the cells.
The concentrations of ciglitazone producing inhibition of AEA hydrolysis in intact cells are in the range of those often used to assess the effects of this agent upon cellular function, particularly in experiments investigating PPARγ-independent effects of this compound (see Okuyama et al., 2005; Weng et al., 2006; Soller et al., 2007 and references therein). With respect to C6 glioma cells, Zander et al. (2002) utilized incubations for 1–8 days with 30–100 μM ciglitazone to assess the effects of this compound upon C6 glioma cell viability. They found that the decreased viability was blocked by BADGE, which would argue in favour of a PPARγ-mediated effect. In contrast, Pérez-Ortiz et al. (2004) found that a 48 h incubation of C6 cells with 20 μM ciglitazone decreased cell viability in a manner only partially blocked by GW9662. This PPARγ-independent action is unlikely to be mediated by FAAH inhibition, since in our hands URB597 has no effect upon C6 glioma cell viability (De Lago et al., 2006), whereas ciglitazone produces a rapid loss of cell viability when assayed under the same conditions (IC50 values of 94, 68 and 23 μM for incubations of 3, 6 and 24 h, respectively, using calcein fluorescence to assess cell viability; Lenman A and Fowler CJ, unpublished data) – indeed, an ability of ciglitazone to affect glutathione content was suggested to underlie its effects in this cell line (Pérez-Ortiz et al., 2004). Nonetheless, these experiments underline both the importance of utilizing PPARγ antagonists when assessing the cellular effects of thiazolidinedione compounds in general and ciglitazone in particular and of recognizing the possibility that the endocannabinoid system can contribute to PPARγ-independent actions of ciglitazone.
The present study naturally raises the question as to whether the thiazolidinediones produce effects upon the endocannabinoid system in man. A peak concentration of rosiglitazone of 598±117 ng ml−1 has been reported for a single 8 mg oral dose (Thummel et al., 2006), corresponding to ∼1.7 μM. For pioglitazone, the situation is complicated by the presence of two major metabolites, but the peak concentration of the compound itself following a 45 mg oral dose given once daily for 10 days is 1.6±0.2 μg ml−1 (Thummel et al., 2006), corresponding to ∼4.5 μM. It thus seems unlikely that these two compounds will directly affect FAAH at normal dosing in man. It is of course possible that indirect effects upon FAAH activity secondary to PPARγ activation can occur, and future studies should investigate this possibility, although it may in some cases be difficult to determine whether such effects are the result of changes in cell viability (see above) or differentiation state (see Gasperi et al., 2007, for a study on the endocannabinoid system in undifferentiated and differentiated 3T3-L1 cells).
In conclusion, the present study has demonstrated that PPARγ agonists are capable of interacting directly with the endocannabinoid system and that ciglitazone can reduce the activity of FAAH in intact cells, particularly when the extracellular pH is reduced (such as is seen in conditions of inflammation, Häbler, 1929). Given that both FAAH inhibitors and PPARγ agonists are of current interest as potential anti-inflammatory agents (see Holt et al., 2005; Moraes et al., 2006), the present study would suggest that ciglitazone may be a useful template for the design of compounds with potent actions upon both these targets.
Acknowledgments
We thank the Swedish Research Council (Grant no. 12158, medicine) and the Research Funds of the Medical Faculty, Umeå University for financial support.
Abbreviations
- AEA
anandamide (arachidonoylethanolamide)
- BADGE
2,2′-[(1-methylethylidene) bis(4,1-phenyleneoxy-methylene)]bisoxirane)
- CB
cannabinoid
- CP-55940
(−)-cis-3-[2-hydroxy-4-(1,1-dimethylheptyl) phenyl]-trans-4-(3-hydroxypropyl) cyclohexanol)
- FAAH
fatty acid amide hydrolase
- GW9662
2-chloro-5-nitrobenzanilide
- HU 210
(6aR)-trans-3-(1,1-dimethylheptyl)-6a,7,10,10a-tetrahydro-1-hydroxy-6,6 dimethyl-6H-dibenzo[b,d]pyran-9-methanol
- KRH buffer
Krebs–Henseleit-bicarbonate buffer
- MAFP
methyl arachidonyl fluorophosphonate
- MCC-555
5-[[6-[(2-fluorophenyl)methoxy]-2-napthalenyl]methyl]-2,4-thiazolidinedione
- MGL
monoacylglycerol lipase
- 2-OG
2-oleoylglycerol
- PPAR
peroxisome proliferator-activated receptor
- T0070907
2-chloro-5-nitro-N-4-pyridinyl-benzamide
- URB597
3′-carbamoyl-biphenyl-3-yl-cyclohexylcarbamate
Conflict of interest
The authors state no conflict of interest.
References
- Berglund BA, Boring DL, Howlett AC. Investigation of structural analogs of prostaglandin amides for binding to and activation of CB1 and CB2 cannabinoid receptors in rat brain and human tonsils. Adv Exp Med Biol. 1999;469:527–533. doi: 10.1007/978-1-4615-4793-8_77. [DOI] [PubMed] [Google Scholar]
- Boldrup L, Wilson SJ, Barbier AJ, Fowler CJ. A simple stopped assay for fatty acid amide hydrolase avoiding the use of a chloroform extraction phase. J Biochem Biophys Methods. 2004;60:171–177. doi: 10.1016/j.jbbm.2004.04.020. [DOI] [PubMed] [Google Scholar]
- Bouaboula M, Hilairet S, Marchand F, Fajas L, Le Fur G, Casellas P. Anandamide induced PPARγ transcriptional activation and 3T3-L1 preadipocyte differentiation. Eur J Pharmacol. 2005;517:174–181. doi: 10.1016/j.ejphar.2005.05.032. [DOI] [PubMed] [Google Scholar]
- Brengdahl J, Fowler CJ. A novel assay for monoacylglycerol hydrolysis suitable for high-throughput screening. Anal Biochem. 2006;359:40–44. doi: 10.1016/j.ab.2006.07.004. [DOI] [PubMed] [Google Scholar]
- De Lago E, Gustafsson SB, Fernández-Ruiz J, Nilsson J, Jacobsson SOP, Fowler CJ. Acyl-based anandamide uptake inhibitors cause rapid toxicity to C6 glioma cells at pharmacologically relevant concentrations. J Neurochem. 2006;99:677–688. doi: 10.1111/j.1471-4159.2006.04104.x. [DOI] [PubMed] [Google Scholar]
- Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–1949. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- Dinh TP, Carpenter D, Leslie FM, Freund TF, Katona I, Sensi SL, et al. Brain monoglyceride lipase participating in endocannabinoid inactivation. Proc Natl Acad Sci USA. 2002;99:10819–10824. doi: 10.1073/pnas.152334899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenthal R, Cornish-Bowden A. The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem J. 1974;139:715–720. doi: 10.1042/bj1390715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CJ, Holt S, Tiger G. Acidic nonsteroidal anti-inflammatory drugs inhibit rat brain fatty acid amide hydrolase in a pH-dependent manner. J Enzyme Inhib Med Chem. 2003;18:55–58. doi: 10.1080/1475636021000049726. [DOI] [PubMed] [Google Scholar]
- Fowler CJ, Tiger G, Stenström A. Ibuprofen inhibits rat brain deamidation of anandamide at pharmacologically relevant concentrations. Mode of inhibition and structure–activity relationship. J Pharmacol Exp Ther. 1997;283:729–734. [PubMed] [Google Scholar]
- Fu J, Gaetani S, Oveisi F, Lo Verme J, Serrano A, Rodríguez de Fonseca F, et al. Oleylethanolamide regulates feeding and body weight through activation of the nuclear receptor PPAR-α. Nature. 2003;425:90–93. doi: 10.1038/nature01921. [DOI] [PubMed] [Google Scholar]
- Gasperi V, Fezza F, Pasquariello N, Bari M, Oddi S, Finazzi Agrò A, et al. Endocannabinoids in adipocytes during differentation and their role in glucose uptake. Cell Mol Life Sci. 2007;64:219–229. doi: 10.1007/s00018-006-6445-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goparaju SK, Ueda N, Yamaguchi H, Yamamoto S. Anandamide amidohydrolase reacting with 2-arachidonoylglycerol, another cannabinoid receptor ligand. FEBS Lett. 1998;422:69–73. doi: 10.1016/s0014-5793(97)01603-7. [DOI] [PubMed] [Google Scholar]
- Häbler C. Über den K- und Ca-gehalt von eiter und exsudaten und seine beziehungen zum entzündungsschmerz. Klin Wochenschr. 1929;8:1569–1572. [Google Scholar]
- Holt S, Fowler CJ. Anandamide metabolism by fatty acid amide hydrolase in intact C6 glioma cells. Increased sensitivity to inhibition by ibuprofen and flurbiprofen upon reduction of extra- but not intracellular pH. Naunyn-Schmiedeberg's Arch Pharmacol. 2003;367:237–244. doi: 10.1007/s00210-002-0686-z. [DOI] [PubMed] [Google Scholar]
- Holt S, Comelli F, Costa B, Fowler CJ. Inhibitors of fatty acid amide hydrolase reduce carrageenan-induced hind paw inflammation in pentobarbital-treated mice: comparison with indomethacin and possible involvement of cannabinoid receptors. Br J Pharmacol. 2005;146:467–476. doi: 10.1038/sj.bjp.0706348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S, Nilsson J, Omeir R, Tiger G, Fowler CJ. Effects of pH on the inhibition of fatty acid amidohydrolase by ibuprofen. Br J Pharmacol. 2001;133:513–520. doi: 10.1038/sj.bjp.0704113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S, Paylor B, Boldrup L, Alajakku K, Vandevoorde S, Sundström A, et al. Inhibition of fatty acid amide hydrolase, a key endocannabinoid metabolizing enzyme, by analogues of ibuprofen and indomethacin. Eur J Pharmacol. 2007;565:26–36. doi: 10.1016/j.ejphar.2007.02.051. [DOI] [PubMed] [Google Scholar]
- Jayamanne A, Greenwood R, Mitchell VA, Aslan S, Piomelli D, Vaughan CW. Actions of the FAAH inhibitor URB597 in neuropathic and inflammatory chronic pain models. Br J Pharmacol. 2006;147:281–288. doi: 10.1038/sj.bjp.0706510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczocha M, Hermann A, Glaser ST, Bojesen IN, Deutsch DG. Anandamide uptake is consistent with rate-limited diffusion and is regulated by the degree of its hydrolysis by fatty acid amide hydrolase. J Biol Chem. 2006;281:9066–9075. doi: 10.1074/jbc.M509721200. [DOI] [PubMed] [Google Scholar]
- Kozak KR, Rowlinson SW, Marnett LJ. Oxygenation of the endocannabinoid, 2-arachidonoylglycerol, to glyceryl prostaglandins by cyclooxygenase-2. J Biol Chem. 2000;275:33744–33749. doi: 10.1074/jbc.M007088200. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, Lenhard JM, Oliver BB, Ringold GM, Kliewer SA. Peroxisome proliferator-activated receptors α and γ are activated by indomethacin and other non-steroidal anti-inflammatory drugs. J Biol Chem. 1997;272:3406–3410. doi: 10.1074/jbc.272.6.3406. [DOI] [PubMed] [Google Scholar]
- Lipinski CA, Fiese EF, Korst RJ. pKa, log P and MedChem CLOGP fragment values of acidic heterocyclic potential bioisosteres. Quant Struct Act Relat. 1991;10:109–117. [Google Scholar]
- Lo Verme J, Fu J, Astarita G, La Rana G, Russo R, Calignano A, et al. The nuclear receptor PPAR-α mediates the antiinflammatory actions of palmitoylethanolamide. Mol Pharmacol. 2005;67:15–19. doi: 10.1124/mol.104.006353. [DOI] [PubMed] [Google Scholar]
- Lo Verme J, Russo R, La Rana G, Fu J, Farthing J, Mattace-Raso G, et al. Rapid broad-spectrum analgesia through activation of peroxisome proliferator-activated receptor-α. J Pharmacol Exp Ther. 2006;319:1051–1061. doi: 10.1124/jpet.106.111385. [DOI] [PubMed] [Google Scholar]
- Moraes LA, Piqueras L, Bishop-Bailey D. Peroxisome proliferator-activated receptors and inflammation. Pharmacol Ther. 2006;110:371–385. doi: 10.1016/j.pharmthera.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Okuyama K, Yamashita M, Kitabatake Y, Kawamura S, Takayanagi M, Ohno I. Ciglitazone inhibits the antigen-induced leukotrienes production independently of PPARγ in RBL-2H3 mast cells. Eur J Pharmacol. 2005;521:21–28. doi: 10.1016/j.ejphar.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Paria BC, Deutsch DD, Dey SK. The uterus is a potential site for anandamide synthesis and hydrolysis: differential profiles of anandamide synthase and hydrolase activities in the mouse uterus during the periimplantation period. Mol Reprod Dev. 1996;45:183–192. doi: 10.1002/(SICI)1098-2795(199610)45:2<183::AID-MRD11>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Paylor B, Holt S, Fowler CJ.The potency of the fatty acid amide hydrolase inhibitor URB597 is dependent upon the assay pH Pharmacol Res 200654481–485.Erratum published in Pharmacol Res55: 80 (2007) [DOI] [PubMed] [Google Scholar]
- Pérez-Ortiz JM, Tranque P, Vaqueor CF, Domingo B, Molina F, Calvo S, et al. Glitazones differentially regulate primary astrocyte and glioma cell survival. Involvement of reactive oxygen species and peroxisome proliferator-activated receptor-gamma. J Biol Chem. 2004;279:8976–8985. doi: 10.1074/jbc.M308518200. [DOI] [PubMed] [Google Scholar]
- Rakhshan F, Day TA, Blakely RD, Barker EL. Carrier-mediated uptake of the endogenous cannabinoid anandamide in RBL2H3 cells. J Pharmacol Exp Ther. 2000;292:960–967. [PubMed] [Google Scholar]
- Reginato MJ, Bailey ST, Krakow SL, Minami C, Ishii S, Tanaka H, et al. A potent antidiabetic thiazolidinedione with unique peroxisome proliferator-activated receptor γ-activating properties. J Biol Chem. 1998;273:32679–32684. doi: 10.1074/jbc.273.49.32679. [DOI] [PubMed] [Google Scholar]
- Rinaldi-Carmona M, Barth F, Millan J, Derocq J-M, Casellas P, Congy C, et al. SR 144528, the first potent and selective antagonist of the CB2 cannabinoid receptor. J Pharmacol Exp Ther. 1998;284:644–650. [PubMed] [Google Scholar]
- Rockwell CE, Kaminski NE. A cyclooxygenase metabolite of anandamide causes inhibition of interleukin-2 secretion in murine splenocytes. J Pharmacol Exp Ther. 2004;311:683–690. doi: 10.1124/jpet.104.065524. [DOI] [PubMed] [Google Scholar]
- Rockwell CE, Snider NT, Thompson JT, Heuvel JPV, Kaminski NE. Interleukin-2 suppression by 2-arachidonoyl glycerol is mediated through peroxisome proliferator-activated receptor γ independently of cannabinoid receptors 1 and 2. Mol Pharmacol. 2006;70:101–111. doi: 10.1124/mol.105.019117. [DOI] [PubMed] [Google Scholar]
- Russo R, LoVerme J, La Rana G, D'Agostino G, Sasso O, Calignano A, et al. Synergistic antinociception by the cannabinoid receptor agonist anandamide and the PPAR-α receptor agonist GW7647. Eur J Pharmacol. 2007;566:117–119. doi: 10.1016/j.ejphar.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg A, Fowler CJ. Measurement of saturable and non-saturable components of anandamide uptake into P19 embryonic carcinoma cells in the presence of fatty acid-free bovine serum albumin. Chem Phys Lipids. 2005;134:131–139. doi: 10.1016/j.chemphyslip.2004.12.010. [DOI] [PubMed] [Google Scholar]
- Schmid PC, Zuzarte-Augustin M, Schmid HHO. Properties of rat liver N-acylethanolamine amidohydrolase. J Biol Chem. 1985;260:14145–14149. [PubMed] [Google Scholar]
- Soller M, Dröse S, Brandt U, Brüne B, von Knethen A. Mechanism of thiazolidinedione-dependent cell death in Jurkat T cells. Mol Pharmacol. 2007;71:1535–1544. doi: 10.1124/mol.107.034371. [DOI] [PubMed] [Google Scholar]
- Thors L, Alajakku K, Fowler CJ. The ‘specific' tyrosine kinase inhibitor genistein inhibits the enzymic hydrolysis of anandamide. Implications for anandamide uptake. Br J Pharmacol. 2007;150:951–960. doi: 10.1038/sj.bjp.0707172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel KE, Shen DD, Isoherranen N, Smith HE.Design and optimization of dosage regimens: pharmacokinetic data Goodman & Gilman's The Pharmacological Basis of Therapeutics 2006Mcgraw-Hill: New York; 1787–1888.In: Brunton LL, Lazo JS, Parker KL (eds)11 ednMisc: pp [Google Scholar]
- Ueda N, Yamanaka K, Yamamoto S. Purification and characterization of an acid amidase selective for N-palmitoylethanolamine, a putative endogenous anti-inflammatory substance. J Biol Chem. 2001;276:35552–35557. doi: 10.1074/jbc.M106261200. [DOI] [PubMed] [Google Scholar]
- Wang T, Xu J, Yu X, Yang R, Han ZC. Peroxisome proliferator-activated receptor γ in malignant diseases. Crit Rev Oncol Hematol. 2006;58:1–14. doi: 10.1016/j.critrevonc.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Weng J-R, Chen C-Y, Pinzone JJ, Ringel MD, Chen C-S. Beyond peroxisome proliferator-activated receptor γ signaling: the multi-facets of the antitumor effect of thiazolidinediones. Endoc Relat Cancer. 2006;13:401–413. doi: 10.1677/erc.1.01182. [DOI] [PubMed] [Google Scholar]
- Willson TM, Brown PJ, Sternbach DD, Henke BR. The PPARs: from orphan receptors to drug discovery. J Med Chem. 2000;43:527–550. doi: 10.1021/jm990554g. [DOI] [PubMed] [Google Scholar]
- Willson TM, Cobb JE, Cowan DJ, Wiethe RW, Correa ID, Prakash SR, et al. The structure–activity relationship between peroxisome proliferator-activated receptor γ agonism and the antihyperglycemic activity of thiazolidinediones. J Med Chem. 1996;39:665–668. doi: 10.1021/jm950395a. [DOI] [PubMed] [Google Scholar]
- Yu M, Ives D, Ramesha CS. Synthesis of prostaglandin E2 ethanolamide from anandamide by cyclooxygenase-2. J Biol Chem. 1997;272:21181–21186. doi: 10.1074/jbc.272.34.21181. [DOI] [PubMed] [Google Scholar]
- Zander T, Kraus JA, Grommes C, Schlegel U, Feinstein D, Klockgether T, et al. Induction of apoptosis in human and rat glioma by agonists of the nuclear receptor PPARγ. J Neurochem. 2002;81:1052–1060. doi: 10.1046/j.1471-4159.2002.00899.x. [DOI] [PubMed] [Google Scholar]