Abstract
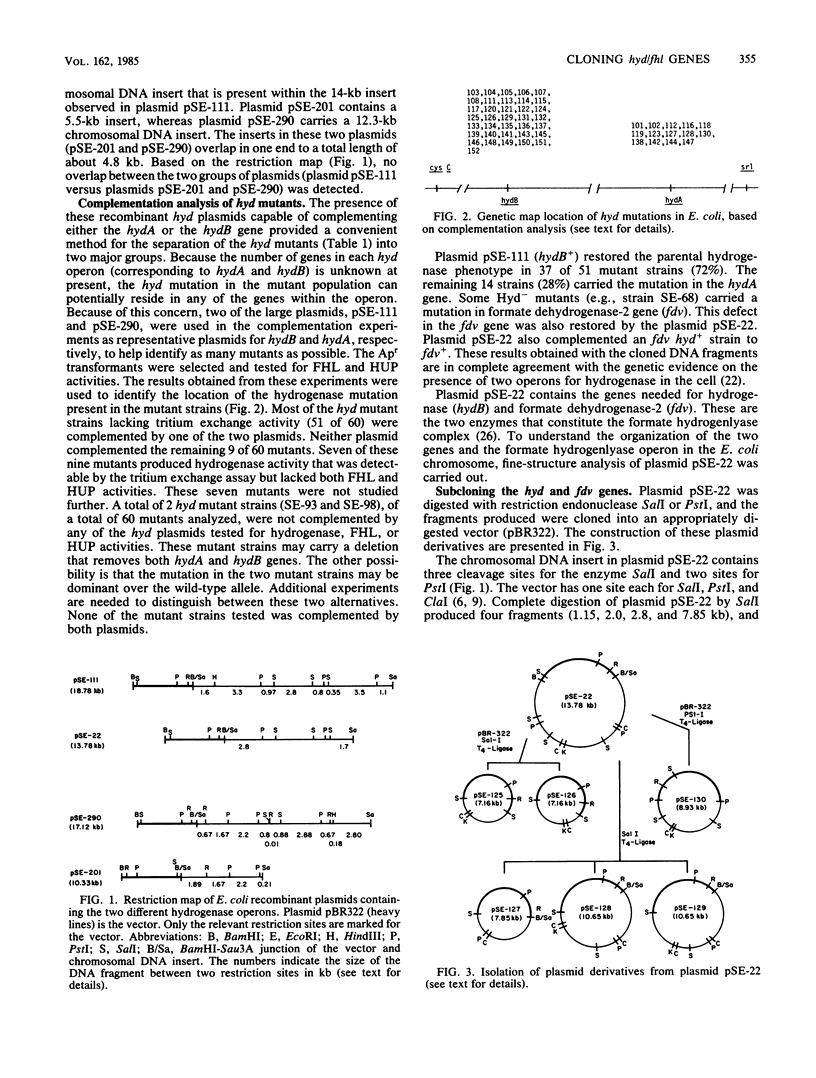
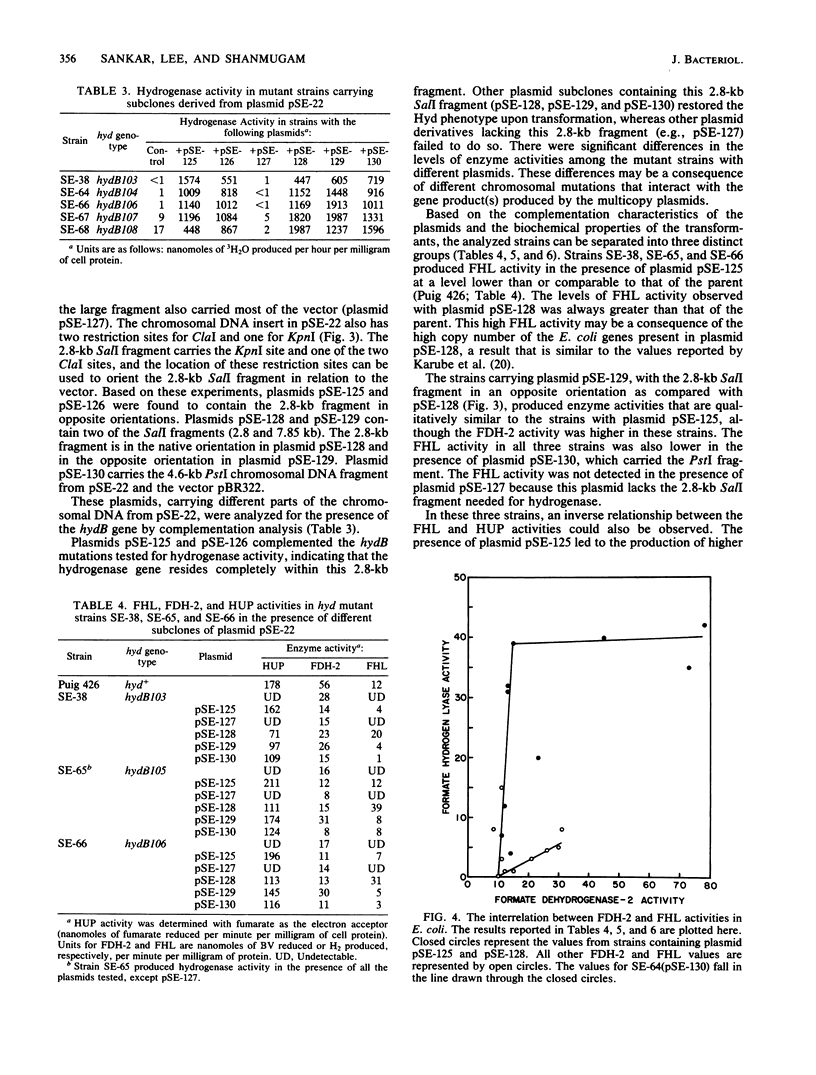
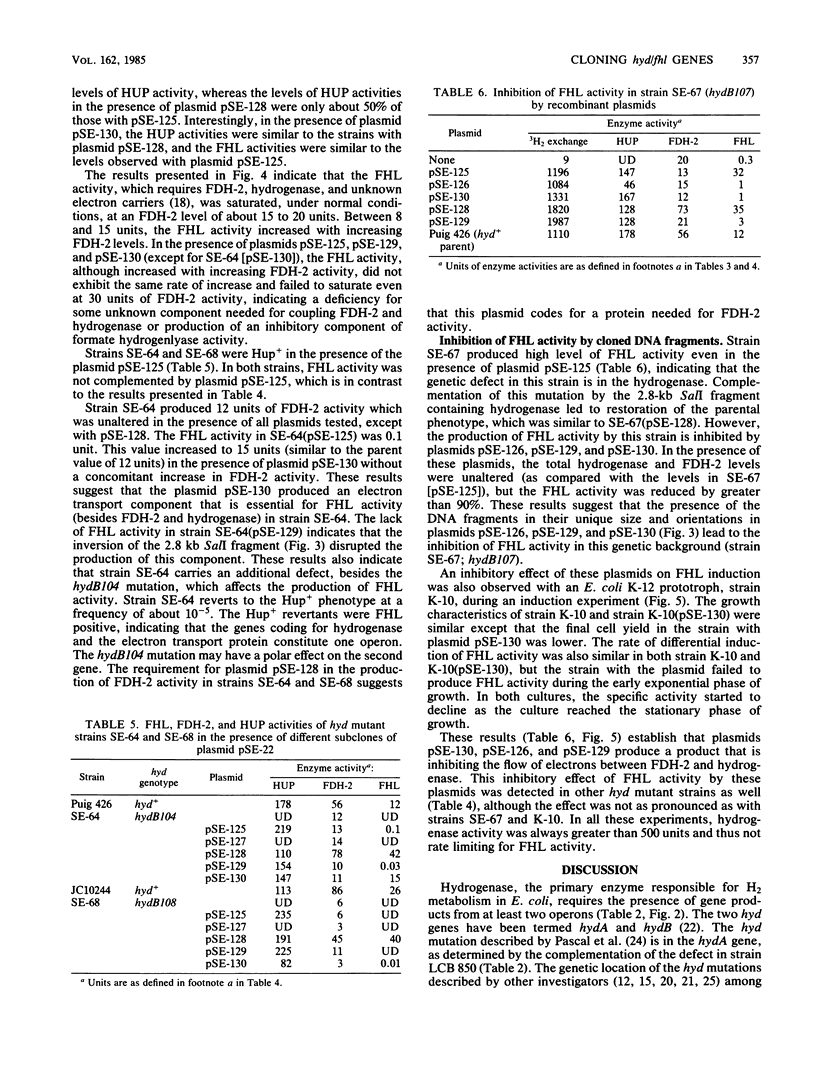
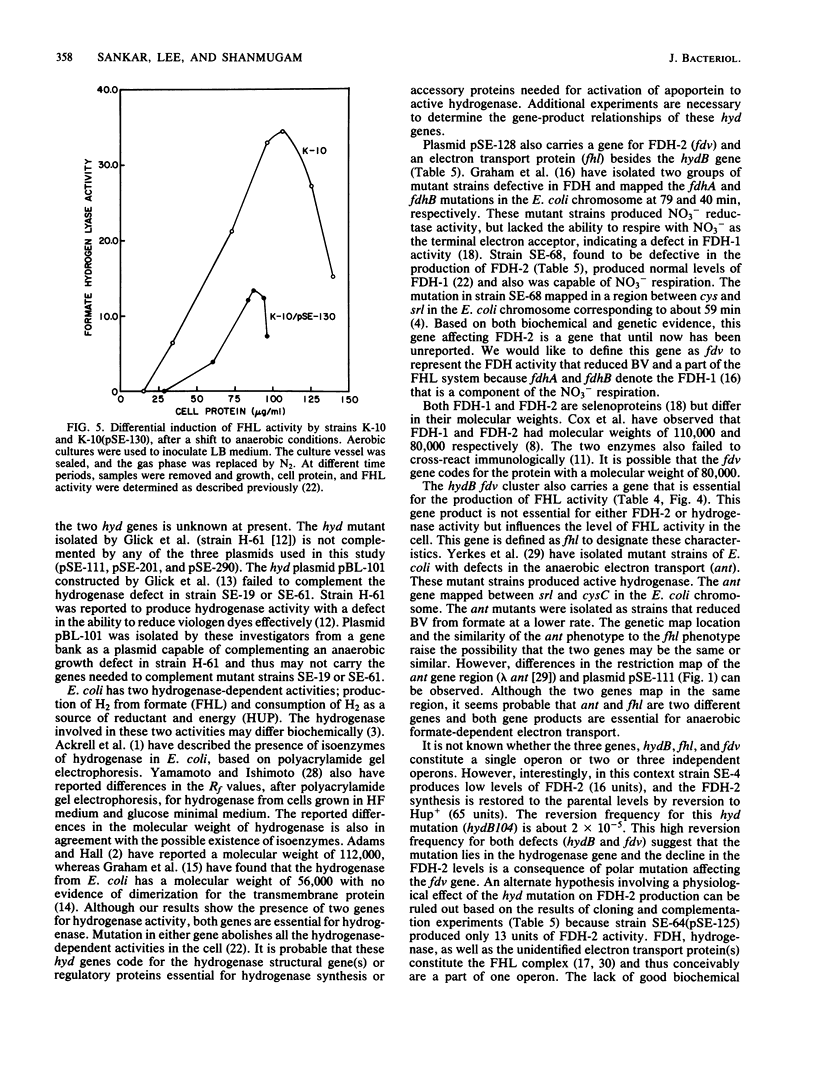
Escherichia coli has two unlinked genes that code for hydrogenase synthesis and activity. The DNA fragments containing the two genes (hydA and hydB) were cloned into a plasmid vector, pBR322. The plasmids containing the hyd genes (pSE-290 and pSE-111 carrying the hydA and hydB genes, respectively) were used to genetically map a total of 51 mutant strains with defects in hydrogenase activity. A total of 37 mutants carried a mutation in the hydB gene, whereas the remaining 14 hyd were hydA. This complementation analysis also established the presence of two new genes, so far unidentified, one coding for formate dehydrogenase-2 (fdv) and another producing an electron transport protein (fhl) coupling formate dehydrogenase-2 to hydrogenase. Three of the four genes, hydB, fhl, and fdv, may constitute a single operon, and all three genes are carried by a 5.6-kilobase-pair chromosomal DNA insert in plasmid pSE-128. Plasmids carrying a part of this 5.6-kilobase-pair DNA (pSE-130) or fragments derived from this DNA in different orientations (pSE-126 and pSE-129) inhibited the production of active formate hydrogenlyase. This inhibition occurred even in a prototrophic E. coli, strain K-10, but only during an early induction period. These results, based on complementation analysis with cloned DNA fragments, show that both hydA and hydB genes are essential for the production of active hydrogenase. For the expression of active formate hydrogenlyase, two other gene products, fhl and fdv are also needed. All four genes map between 58 and 59 min in the E. coli chromosome.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackrell B. A., Asato R. N., Mower H. F. Multiple forms of bacterial hydrogenases. J Bacteriol. 1966 Oct;92(4):828–838. doi: 10.1128/jb.92.4.828-838.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. W., Hall D. O. Purification of the membrane-bound hydrogenase of Escherichia coli. Biochem J. 1979 Oct 1;183(1):11–22. doi: 10.1042/bj1830011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams M. W., Mortenson L. E., Chen J. S. Hydrogenase. Biochim Biophys Acta. 1980 Dec;594(2-3):105–176. doi: 10.1016/0304-4173(80)90007-5. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Chopra I., Howe T. G. Bacterial resistance to the tetracyclines. Microbiol Rev. 1978 Dec;42(4):707–724. doi: 10.1128/mr.42.4.707-724.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J. C., Edwards E. S., DeMoss J. A. Resolution of distinct selenium-containing formate dehydrogenases from Escherichia coli. J Bacteriol. 1981 Mar;145(3):1317–1324. doi: 10.1128/jb.145.3.1317-1324.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GRAY C. T., GEST H. BIOLOGICAL FORMATION OF MOLECULAR HYDROGEN. Science. 1965 Apr 9;148(3667):186–192. doi: 10.1126/science.148.3667.186. [DOI] [PubMed] [Google Scholar]
- Glick B. R., Wang P. Y., Schneider H., Martin W. G. Identification and partial characterization of an Escherichia coli mutant with altered hydrogenase activity. Can J Biochem. 1980 Apr;58(4):361–367. doi: 10.1139/o80-047. [DOI] [PubMed] [Google Scholar]
- Glick B. R., Zeisler J., Banaszuk A. M., Friesen J. D., Martin W. G. The identification and partial characterization of a plasmid containing the gene for the membrane-associated hydrogenase from E. coli. Gene. 1981 Nov;15(2-3):201–206. doi: 10.1016/0378-1119(81)90129-3. [DOI] [PubMed] [Google Scholar]
- Graham A., Boxer D. H., Haddock B. A., Mandrand-Berthelot A. M., Jones R. W. Immunochemical analysis of the membrane-bound hydrogenase of Escherichia coli. FEBS Lett. 1980 May 5;113(2):167–172. doi: 10.1016/0014-5793(80)80584-9. [DOI] [PubMed] [Google Scholar]
- Graham A. The organization of hydrogenase in the cytoplasmic membrane of Escherichia coli. Biochem J. 1981 Aug 1;197(2):283–291. doi: 10.1042/bj1970283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddock B. A., Jones C. W. Bacterial respiration. Bacteriol Rev. 1977 Mar;41(1):47–99. doi: 10.1128/br.41.1.47-99.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R. W. The role of the membrane-bound hydrogenase in the energy-conserving oxidation of molecular hydrogen by Escherichia coli. Biochem J. 1980 May 15;188(2):345–350. doi: 10.1042/bj1880345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karube I., Urano N., Yamada T., Hirochika H., Sakaguchi K. Cloning and expression of the hydrogenase gene from Clostridium butyricum in Escherichia coli. FEBS Lett. 1983 Jul 11;158(1):119–122. doi: 10.1016/0014-5793(83)80689-9. [DOI] [PubMed] [Google Scholar]
- Krasna A. I. Mutants of Escherichia coli with altered hydrogenase activity. J Gen Microbiol. 1984 Apr;130(4):779–787. doi: 10.1099/00221287-130-4-779. [DOI] [PubMed] [Google Scholar]
- Macy J., Kulla H., Gottschalk G. H2-dependent anaerobic growth of Escherichia coli on L-malate: succinate formation. J Bacteriol. 1976 Feb;125(2):423–428. doi: 10.1128/jb.125.2.423-428.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PECK H. D., Jr, GEST H. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J Bacteriol. 1957 Jun;73(6):706–721. doi: 10.1128/jb.73.6.706-721.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal M. C., Casse F., Chippaux M., Lepelletier M. Genetic analysis of mutants of Escherichia coli K12 and Salmonella typhimurium LT2 deficient in hydrogenase activity. Mol Gen Genet. 1975 Nov 24;141(2):173–179. doi: 10.1007/BF00267682. [DOI] [PubMed] [Google Scholar]
- Pecher A., Zinoni F., Jatisatienr C., Wirth R., Hennecke H., Böck A. On the redox control of synthesis of anaerobically induced enzymes in enterobacteriaceae. Arch Microbiol. 1983 Nov;136(2):131–136. doi: 10.1007/BF00404787. [DOI] [PubMed] [Google Scholar]
- Yerkes J. H., Casson L. P., Honkanen A. K., Walker G. C. Anaerobiosis induces expression of ant, a new Escherichia coli locus with a role in anaerobic electron transport. J Bacteriol. 1984 Apr;158(1):180–186. doi: 10.1128/jb.158.1.180-186.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]



