Abstract
Nonhomologous end joining (NHEJ) directly rejoins DNA double-strand breaks (DSBs) when recombination is not possible. In Saccharomyces cerevisiae, the DNA polymerase Pol4 is required for gap filling when a short 3’ overhang must prime DNA synthesis. Here, we examined further end variations to test specific hypotheses regarding Pol4 usage in NHEJ in vivo. Surprisingly, Pol4 dependence at 3’ overhangs was reduced when a nonhomologous 5’ flap nucleotide was present across from the gap, even though the mismatched nucleotide was resynthesized, not incorporated. In contrast, a gap with a 5’ deoxyribosephosphate (dRP) was as Pol4-dependent as a gap with a 5’ phosphate, demonstrating the importance of the downstream base in relaxing the Pol4 requirement. Combined with prior observations of Pol4-independent NHEJ of nicks with 5’ hydroxyls, we suggest that base stacking interactions across the broken strands can stabilize a joint, allowing another polymerase to substitute for Pol4. This model predicts that a unique function of Pol4 is to actively stabilize template strands that lack stacking continuity. We also explored whether NHEJ end processing can occur via short and long patch pathways analogous to base excision repair. Results demonstrated that 5’ dRPs could be removed in the absence of Pol4 lyase activity. The 5’ flap endonuclease Rad27 was not required for repair in this or any situation tested, indicating that still other NHEJ 5’ nucleases must exist.
Keywords: DNA repair, nonhomologous end joining, DNA polymerase, double-strand break, nuclease
INTRODUCTION
In the absence of a homologous template, the nonhomologous end joining (NHEJ) pathway repairs DNA double-strand breaks (DSBs) by directly ligating the two ends [1]. The Ku heterodimer (Yku70 and Yku80 in yeast) threads onto the ends to initiate NHEJ [2], and the termini are thought to be held in proximity, at least in yeast, by the Mre11/Rad50/Xrs2 complex [3]. DNA ligase IV (Dnl4/Lif1 in yeast) ligates the break, restoring the duplex [4]. These proteins are sufficient for “simple religation” NHEJ of breaks with compatible termini. Joining of damaged termini is not as well understood, but is important because DNA damaging agents like ionizing radiation and reactive oxygen species typically create DSBs with “dirty” ends [5-8].
Pol4, the only Pol X polymerase in S. cerevisiae, is dispensable for simple religation NHEJ [9], but is strictly required when a gap in the joint must be filled using an unstable primer-template pair, as is the case with 3’ overhangs [10]. Other polymerases can compensate for the loss of Pol4 when a stably paired primer is available, for example at 5’ overhangs [10]. However, Pol4 is surprisingly not required for joining of nicked 3’ overhang DSBs with 5’ hydroxyls [10], even though yeast lack a 5’ kinase and thus demand resynthesis of the damaged 5’ nucleotide [11]. Unknown factors must contribute to the stability of primer-template pairing and therefore the specific need for Pol4.
The end processing mechanisms used in base excision repair (BER) are biochemically analogous to those required in NHEJ. BER is initiated when a glycosylase and an abasic endonuclease cleave a damaged base, leaving a 1-nt gap with 3’ hydroxyl and 5’ deoxyribosephosphate (dRP) termini. Two sub-pathways of mammalian BER function redundantly to complete repair (Figure 1A). In short patch BER, the Pol X family polymerase Pol β fills the 1-nt gap and removes the 5’ dRP with its lyase activity [12,13]. In the long patch pathway, either Pol δ or Pol β performs displacement synthesis [14,15], yielding a 5’ flap, which is cleaved by FEN-1 (Rad27 in yeast) [16]. Compellingly, Pol4 and Pol β are each non-processive gap-filling polymerases, and both have 5’ dRP lyase activity [17-19]. Also, Rad27 physically interacts with Dnl4 and Pol4, can process 5’ flaps at DSBs in vitro [20], and has been implicated in NHEJ of ends with 5’ flaps in vivo [21]. Based on these considerations, we hypothesized that Pol4 might participate in both short and long patch sub-pathways of NHEJ (Figure 1B).
Figure 1.
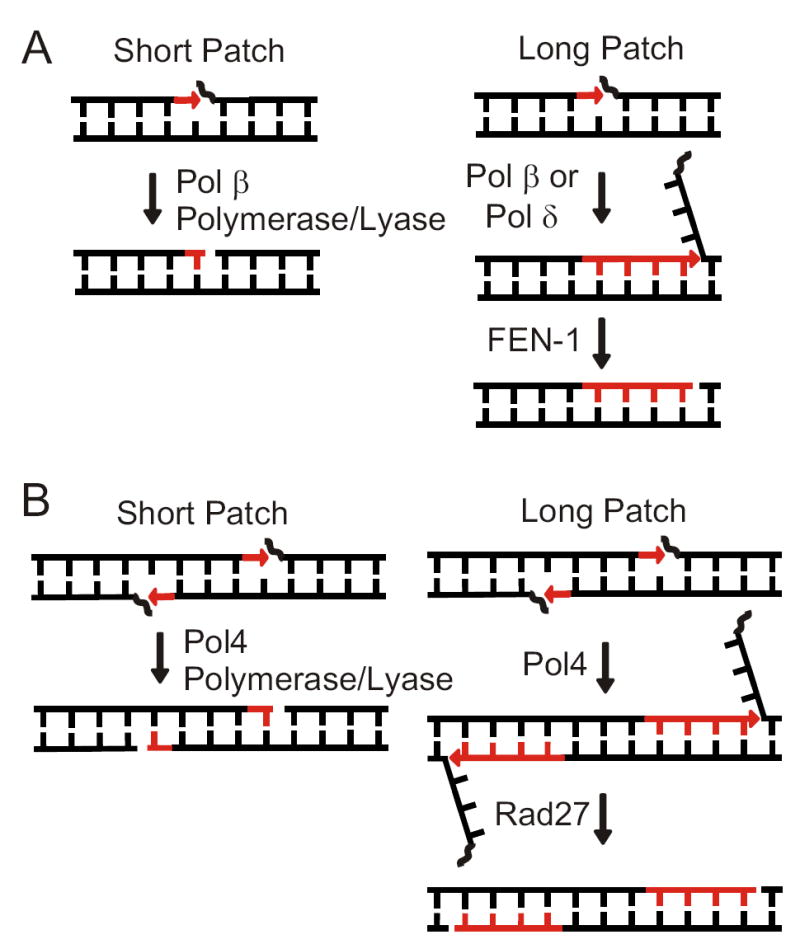
(A) Short and long patch pathways of mammalian BER. (B) Analogous modes of 5’ processing in putative short and long patch pathways of NHEJ.
To test these ideas, we added various requirements for 5’ processing to 3’ overhang joints that we previously showed to depend on Pol4 for gap filling [10]. Strikingly, adding an unpaired flap nucleotide to the 5’ end adjacent to a gap relieved the requirement for Pol4, even though the mismatched nucleotide was not incorporated during NHEJ. In contrast, point mutations revealed that the polymerase activity of Pol4 was required for resynthesizing a base that had been excised to form a 5’ dRP. These results suggest a model in which the unique action of Pol4 is to overcome the lack of continuity of base-stacking in the primer-template pair. Pol4 lyase activity was dispensable for rejoining DSBs with 5’ dRP termini. Additional mutation of rad27 did not impair joining of 5’ dRPs, or numerous 5’ flap structures, indicating that yeast have other redundant mechanisms for processing 5’ termini that are more complex than a simple model of short and long patch repair. We also attempted to ask whether Pol4 is required to resynthesize a damaged 3’ nucleotide. A 3’ dideoxynucleotide was chosen as a model but proved to greatly reduce the NHEJ efficiency, indicating that the requisite NHEJ 3’ nuclease depends on a 3’ hydroxyl.
MATERIALS AND METHODS
Yeast strains
S. cerevisiae strains used for plasmid transformation were isogenic derivatives of the previously described wild-type strain YW389 (MATα ade2D0 his3D200 leu2 lys2-801 trp1D63 ura3D0). YW438 (pol4Δ∷MET15), [10], and YW514 (pol4-D367E) [9] were previously described. YW1831 (pol4-KK247∷247RR) was constructed by inserting URA3 into pol4 and then replacing URA3 using a tailed PCR product containing the mutant allele. The mutation was confirmed by sequencing. YW1807 was created by transforming the RNH35 overexpression plasmid pTW572 into YW389. YW1866 (pol4-KK247∷247RR rad27Δ∷kanMX4) and YW1899 (pol4Δ∷TRP1 rad27Δ∷kanMX4) were created via the one-step gene replacement technique followed by transformation with pTW572.
Construction of a simple religation NHEJ control plasmid
pTW571 was designed to be co-transformed into yeast as a simple relegation NHEJ control with pTW423-based OMPs. To construct pTW571, LEU2 was amplified from pRS315 with oligonucleotides tailed with NotI and SalI sites, and the PCR product was ligated into the polylinker of pRS411. pTW571 was digested with ClaI, which makes a DSB within LEU2. Neither pTW571 nor the OMP can serve as a recombination template for the other.
RNH35 overexpression plasmid
To create pTW572, RNH35 was amplified from yeast genomic DNA with oligonucleotides that add tails to target the PCR product into SmaI-cut pTW436 [22]. The resulting HIS3-marked 2μ plasmid drives RNH35 expression from the ADH1 promoter. Insertion of RNH35 was verified by PCR.
Oligonucleotide-modified plasmids for NHEJ assays
OMPs were created as previously described. Briefly, two pairs of annealed oligonucleotides designed to restore the ADE2 coding sequence were ligated onto the ends of pTW423 digested with BglII and XhoI. Plasmids were purified by agarose gel electrophoresis and the concentration quantified by UV spectrometry. Because the DSB side of the oligonucleotides initially bore 5’ hydroxyls to prevent ligation of the DSB in vitro, OMPs were treated with T4 polynucleotide kinase (NEB) followed by a second round of purification. Oligonucleotide ligation was monitored by primer extension, and 70-90% of the plasmid was typically ligated (data not shown). Because ligation efficiency varies slightly, small NHEJ differences between OMPs are not significant. Inter-strain differences with a given OMP are independent of this effect.
Construction of DSBs with 5’ dRP termini
Oligonucleotides were synthesized with deoxyuracil residues in the desired dRP position. After ligation, OMPs were treated with T4 PNK (NEB) to add 5’ phosphates, and then UDG (NEB) to remove the uracil bases, leaving 5’ dRPs. UDG treatment was inferred to be effective because it rendered joints Pol4-dependent when a gap was created (Figure 3C), and was also verified by primer extension (data not shown).
Figure 3.
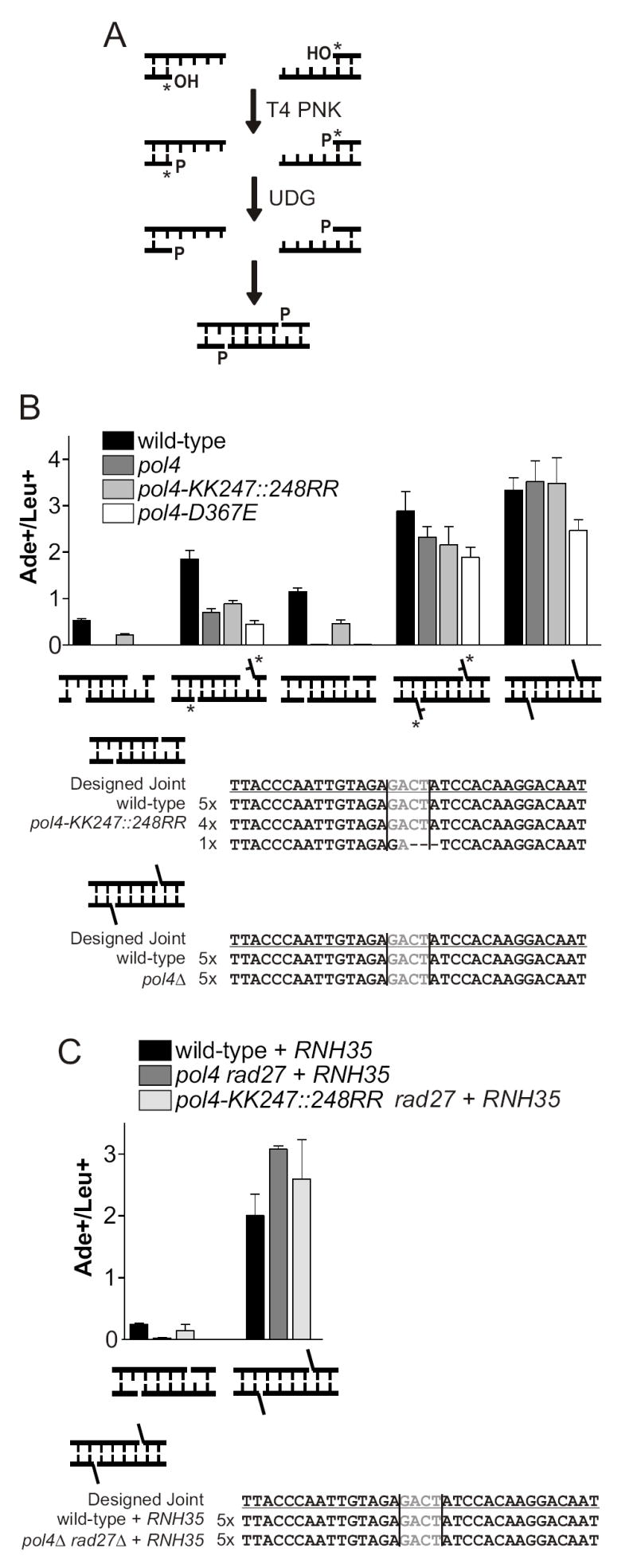
5’ dRP lesions demand gap filling by Pol4, but do not require the Pol4 lyase activity or Rad27. (A) Schematic for construction of OMPs with 5’ dRP termini. Deoxyuracil residues are indicated in gray. Treatment with T4 PNK followed by UDG forms a 5’ dRP. (B) Polymerase domain point mutant pol4-D367E eliminates repair of 5’ dRP-containing DSBs only when a missing base must be synthesized. Lyase domain mutant pol4-KK247∷248RR does not impair rejoining when 5’ dRPs are present. (C) Additional mutation of rad27 does not impair joining. Data are expressed as in Figure 2.
pBX2 NHEJ assay
PCR was done on 16 Ade+ colonies per strain and acrylamide gel electrophoresis was used to separate the products by size. 86% of the joints in wild-type and 100% in rad27Δ cells were flap joints.
Construction of plasmids with 3’ dideoxy termini
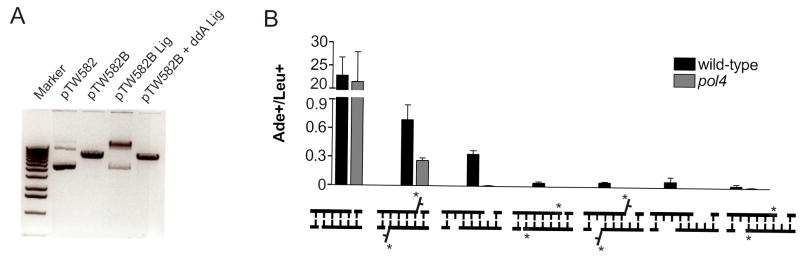
Pre-existing BstXI sites in pES16 [4] were silently mutated by ligating PCR products created with tailed primers into the vector, creating pTW578. Recognition sequences for BstXI, which cleaves at variable sites allowing for construction of DSBs with desired overhangs, were inserted between codons 2 and 3 of ADE2 by ligating two PCR products into pTW578 cut with BglII and NotI. The resulting plasmids contain a BstXI site on each side of stop codons in each reading frame. BstXI digestion and joining of the overhangs in the target configuration places ADE2 in-frame. pTW582 (flap), pTW583 (compatible) and pTW584 (gap) were digested with BstXI and incubated with 40 U of TdT (NEB) in a 100 μl reaction containing 0.25 mM CoCl2, 0.15 mM ddATP (pTW582 and pTW583) or ddTTP (pTW583 and 584), 7.5 μg plasmid, and NEBuffer 4 for 2 hours at 37°C. TdT was inactivated by incubation at 70°C for 10 minutes, and the plasmid purified with GeneClean (QBioGene) and quantified by UV spectrometry. To verify 3’ dideoxynucleotide addition, 500 ng of pTW582 cut with BstXI was incubated with T4 DNA Ligase (NEB) in a 10 μl reaction for 2 hours at ~16°C before and after TdT treatment. Ligations were run out on a 0.8% agarose gel, and loss of ligation following TdT treatment was noted (Figure 5A).
Figure 5.
3’ dideoxynucleotide termini inhibit NHEJ. (A) Addition of the 3’ dideoxynucleotide was verified using pTW582, which forms compatible overhangs upon BstXI digestion. The linearized plasmid was incubated with T4 DNA ligase before (lane 4) and after TdT treatment (lane 5). (B) BstXI-digested plasmids were transformed into wild-type and pol4Δ yeast before and after TdT treatment. Data are expressed as in Figure 2.
Yeast transformation
Plasmids were transformed into yeast using a high efficiency lithium acetate method as previously described. Strains containing the HIS3-marked RNH35 expression plasmid were grown overnight in synthetic defined glucose media lacking histidine to maintain selection for the plasmid. Other strains were grown in rich YPAD media. All strains were diluted into YPAD for outgrowth. 100 ng of linear OMP (marked with URA3) was co-transformed with 100 ng of pTW571 cut within LEU2 with ClaI as a simple religation NHEJ control. Cells were plated in parallel to medium lacking either uracil or leucine and grown at 30°C for 3 days. The relative repair efficiency for a strain-plasmid combination is expressed as the ratio of Ade+ (white) colonies on plates lacking uracil to colonies on plates lacking leucine. Note that ADE2 restoration effectively selects for the target joints, but alternative Ade+ joints can also occur and will be represented in the graphs. A minimum of 150, and more typically 500-1200, Ade+ colonies were counted for each joint in wild-type yeast, except where noted. Similar colony numbers were counted for mutant strains when possible. Individual data panels represent results collected in parallel with a single preparation of plasmid and carrier DNAs.
RESULTS
Unpaired 5’ nucleotides relieve the requirement for Pol4 at 3’ overhangs with gaps
We previously showed that Pol4 is required for gap filling during NHEJ of 3’ overhangs where the primer-template pair adjacent to the gap is unstable [10]. Stabilization of the primer terminus, either by increasing the overhang length or changing the overhang polarity, reduced or eliminated dependence on Pol4 [10]. To gain insight into other parameters that affect whether Pol4 is required for NHEJ, we first used oligonucleotide-modified plasmids (OMPs) [23] to compare joints with gaps to those with mismatched nucleotides. In this system, annealed oligonucleotides are ligated onto restriction site ends in an essential region of the ADE2 gene to allow selection for a specific NHEJ event. As a control, the OMP is co-transformed with a plasmid containing a ClaI DSB in LEU2, which requires NHEJ but not end processing. Therefore, a reduction in the ratio of Ade+ to Leu+ colonies indicates a defect in end processing. Consistent with previous results, a joint with 3’ overhangs, a 1-nt gap and a 1-nt nonhomologous 3’ flap was highly dependent on Pol4 for rejoining, showing a 22-fold defect in pol4Δ cells (Figure 2A, second joint). Surprisingly, moving the flaps to the 5’ termini reduced the pol4Δ defect to only 2-fold (Figure 2A, third joint), approximately the same defect seen at 5’ overhangs with no flaps [10]. To validate this finding, we created a similar joint with shorter overhangs (Figure 2B, second joint). This joint was slightly less efficient than a comparable joint with no gaps (Figure 2B, first joint) and in fact showed no defect in pol4Δ cells (Figure 2B, second joint).
Figure 2.
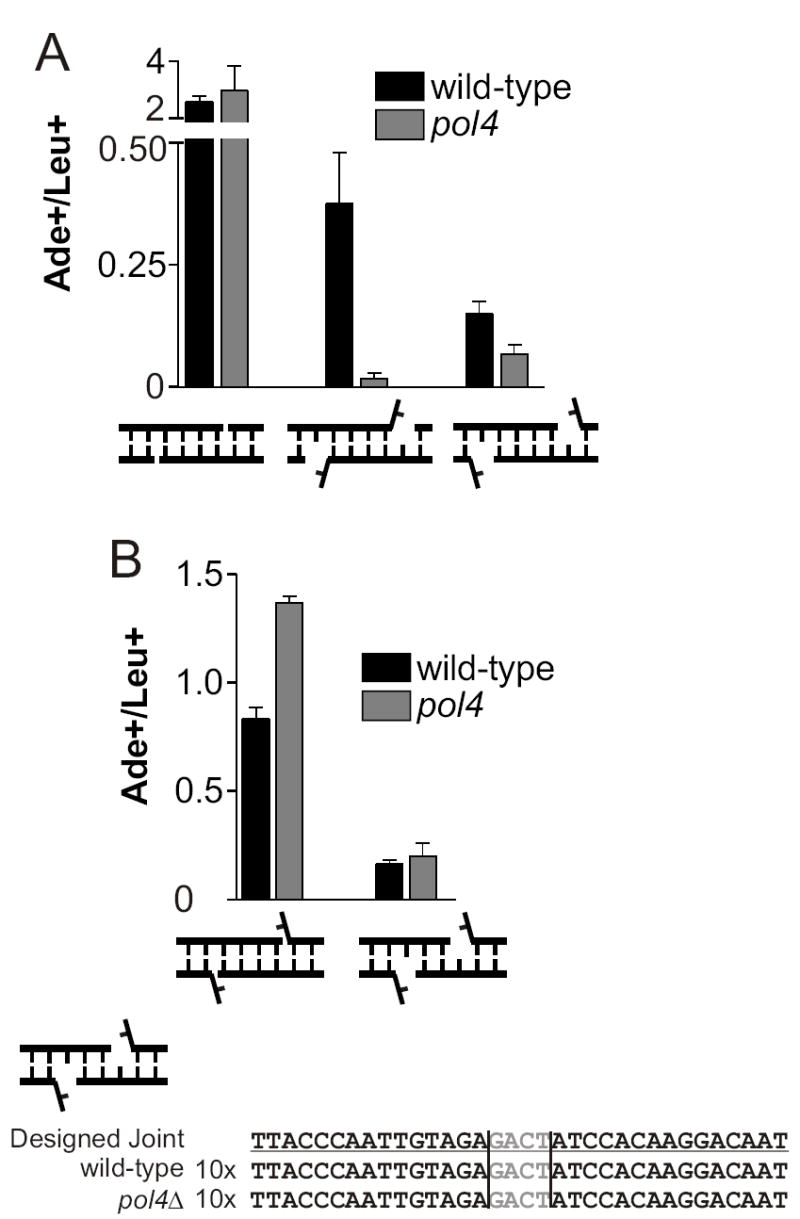
The requirement of Pol4 for gap filling is relaxed in the presence of 5’ flaps. (A) and (B) OMPs were used to create DSBs with the indicated structures. Repair is expressed as the ratio of Ade+ colonies to Leu+ colonies. Ade+ colonies arise from accurate repair of the OMP, whereas Leu+ colonies result from a simple religation NHEJ event (see Materials and Methods). A reduction in this ratio indicates a defect in end processing. Each point represents the mean ± standard deviation from three independent transformations. Vertical lines on sequences denote the overhang position, and annealed nucleotides are shown in gray.
To explain this unexpected Pol4-independent joining, we first reasoned that the mismatched 5’ nucleotide might be incorporated during NHEJ, avoiding the need for polymerization. If this were the case, 50% of the completed joints would be expected to have a sequence that included a flap nucleotide, assuming mismatch correction without strand bias. However, sequencing of 10 Ade+ colonies each from wild-type and pol4Δ yeast revealed that the flap sequence was never present in the rejoined plasmid (Figure 2B), suggesting that the nucleotide was indeed excised and resynthesized by a polymerase other than Pol4.
Pol4 polymerase activity is required at a DSB with 5’ dRP termini and 1-nt gaps
To further explore the influence of 5’ status on the requirement for Pol4, we next asked whether Pol4 participates in repair of DSBs with 5’ dRPs, which differ from nick and 5’ flap substrates in that they have a 5’ sugar but lack a 5’ base. DSBs with 5’ dRPs could be formed in vivo as a direct consequence of the DNA damaging agent or if BER enzymes process damaged bases on opposite strands. 5’ dRP termini were again added to 4-base 3’ overhangs because DSBs with this configuration are repaired efficiently and show strong dependence on Pol4 for gap filling [10]. Oligonucleotides were synthesized with deoxyuracil residues where dRPs were desired, ligated onto the plasmid, and subsequently treated with T4 PNK and UDG to phosphorylate the 5’ termini and remove the uracil bases, respectively (Figure 3A).
These plasmids were transformed into wild-type and pol4 yeast both before and after UDG treatment. We first examined a joint in which removal of the uracil yields 1-nt gaps adjacent to 5’ dRPs (Figure 3B, second and third joints). The pol4Δ strain showed about a 2-fold defect at rejoining the uracil-containing DSB (Figure 3B, second joint). This mild defect is likely due to a low level of uracil cleavage in the cell by Ung1, the yeast homolog of UDG, which would create gaps predicted to require Pol4. Indeed, when the uracil was pre-excised in vitro by UDG, leaving a 5’ dRP across from an unpaired nucleotide, the joining efficiency in wild-type cells decreased and repair became strictly Pol4-dependent (Figure 3B, third joint). This demonstrates both that UDG treatment was successful and that repair of the dRP substrate required Pol4.
Because Pol4 dependence of the above dRP joint could be due to either the Pol4 polymerase or lyase activities, or both, we next tested activity-specific point mutations. Considering polymerization first, pol4-D367E mutates a catalytic aspartate in the Pol4 polymerase domain and abolishes polymerase activity in vitro and in vivo [9], but is not expected to impair dRP lyase activity via the distinct 8 kDa domain. pol4-D367E cells proved as deficient as pol4Δ cells at rejoining the break with dRPs and 1-base gaps, demonstrating that at least the Pol4 polymerization activity is required. This requirement is in marked contrast to the 5’ mismatch substrates above and shows that the base of the downstream nucleotide is critical for enabling synthesis by another polymerase. The sugar-phosphate backbone still present in the dRP was not sufficient.
Pol4 lyase activity is not required at DSBs with 5’ dRP termini
The above data are potentially consistent with repair occurring by either a short patch or a long patch NHEJ pathway. Our assay cannot determine the number of bases synthesized during repair, but short and long patch pathways might be distinguished by their mechanism of 5’ dRP removal, by analogy to BER (Figure 1). We therefore asked whether the Pol4 lyase can participate in 5’ dRP processing during NHEJ (Figure 1B). pol4-KK247∷248RR mutation eliminates two lysines predicted to catalyze the lyase reaction based on alignments with Pol β, although this mutation does not completely disable the lyase activity in vitro [17]. As previously observed [9], pol4-KK247∷248RR cells showed a mild defect at rejoining a DSB containing simple Pol4-dependent gaps (Figure 3A, first joint). Since this joint does not require lyase activity, this is likely due to destabilizing effects of the mutation on the 8 kDa domain, which binds the downstream 5’ terminus in Pol X polymerases [24]. Most importantly, the pol4-KK247∷248RR strain showed only a similar minor defect at the joint with a gap and 5’ dRP (Figure 3B, first joint), unlike the severe defect seen in the polymerase mutant. Sequencing revealed that the Ade+ colonies produced by pol4-KK247∷248RR cells were indeed the target joint (Figure 3B).
The above experiment demonstrates that the polymerase activity of Pol4 is required to resynthesize a base corresponding to a 5’ dRP, but suggests that the lyase activity is dispensable for removing the 5’ dRP itself. However, the minor defect in pol4-KK247∷248RR cells could also be due to residual Pol4 lyase activity as observed in vitro [17]. As an alternate way to address 5’ dRP removal, we constructed a DSB with 5’ dRP termini that lacks gaps. Because this joint does not require DNA synthesis, it could be assayed in pol4Δ cells, circumventing the concern over residual Pol4 lyase activity. However, none of the pol4 mutant strains showed a defect at rejoining this break, either before or after UDG treatment (Figure 3B, fourth and fifth joints). Four of five Ade+ colonies sequenced from pol4Δ cells represented the target joint, with the remaining joint showing a 3-base deletion that placed ADE2 in frame (Figure 3B). While these experiments do not rule out the possibility that the lyase domain of Pol4 may remove 5’ dRPs during a short patch pathway of NHEJ, other mechanisms for 5’ dRP removal must exist.
Yeast can rejoin 5’ dRP-containing DSBs in the absence of both Pol4 and Rad27
Since Rad27 cleaves 5’ flaps in long patch BER [25] and has been implicated in NHEJ of ends with 5’ flaps both in vivo and in vitro [20,21], we hypothesized that this nuclease might catalyze 5’ dRP removal in the absence of the Pol4 lyase (Figure 1B). We sought to test rejoining of 5’ dRP-containing DSBs in pol4Δ rad27Δ double mutants, which would lack both the putative short and long patch NHEJ pathways. Because Rad27 cleaves 5’ flaps during Okazaki fragment processing, rad27Δ strains grow slowly [26] and are exquisitely sensitive to the yeast transformation procedure (data not shown). Overexpression of RNase H (RNH35), which catalyzes an alternate pathway of Okazaki fragment processing, suppresses the rad27Δ growth defects [27] and also reduces the toxicity of transformation (data not shown) without affecting the ratio of processed to simple religation NHEJ (see Figure 4B below). We therefore added an RNH35 overexpression plasmid to our strains. Somewhat surprisingly, deletion of rad27 on top of pol4-KK247∷248RR or pol4Δ did not affect rejoining of either of the 5’ dRP-containing DSBs (Figure 3C).
Figure 4.
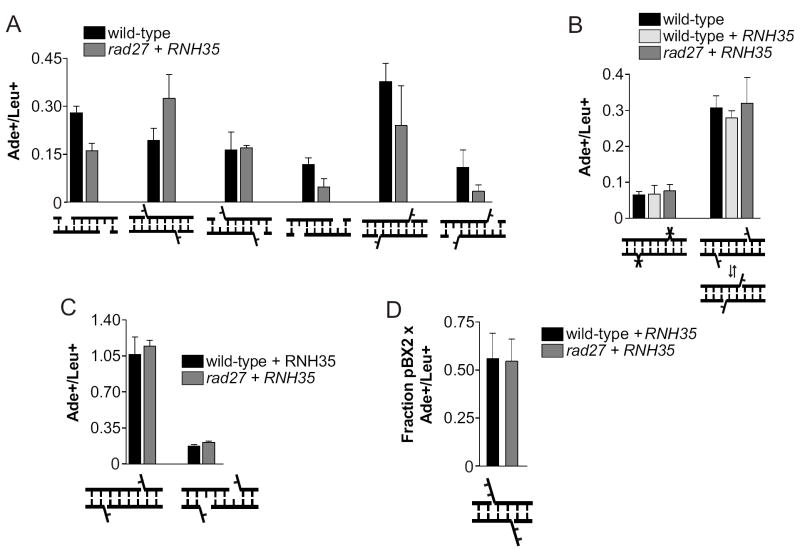
Rad27 is not required for rejoining of DSBs with gaps, flaps, and complex end structures. (A), (B) and (C) Joints with the indicated structures were created in OMPs, and data are expressed as in Figure 2. (D) pBX2 was cut with BglII and XhoI, forming the joint pictured within the ADE2 gene. PCR on 16 Ade+ colonies was used to differentiate flap joining from blunt joining, and data were corrected to show only the efficiency of flap joints in each strain.
Rad27 is not required for rejoining DSBs with flaps, gaps, and complex overhang structures
The lack of a 5’ dRP processing defect in rad27Δ cells could be due to the absence of displacement synthesis in NHEJ, which would be required to form the type of 5’ flap structure that Rad27 recognizes. We therefore used OMPs to generate such flaps directly and ask whether Rad27 is required for their processing. First, a panel of DSBs were generated in which the 4 annealed bases were kept constant, but each combination of 1-base gaps and flaps were added to both 3’ and 5’ overhangs (Figure 4A). rad27Δ yeast overexpressing RNH35 showed no obvious defect in any joint compared to wild-type yeast (Figure 4A).
Rad27 is much more efficient at cleaving dual flap structures than single flaps in vitro [28]. We therefore added 1-base unpaired flaps to both the 3’ and 5’ termini of the joint (Figure 4B, first joint). This structure could not be generated in a chromosome because it contains mismatches within the duplex, but it provides a model for a gap that has been overfilled and slipped back, as in long-patch BER [28]. rad27Δ cells repaired this DSB at wild-type levels (Figure 4B, first joint). We next constructed a more physiologically relevant version of this joint in which the flap is complementary, allowing it to anneal in either of two configurations (Figure 4B, second joint), but again, rad27Δ mutation had no effect on joining.
Because Rad27 interacts with Pol4 to coordinate flap cleavage and gap filling in vitro [20], we considered that Rad27 might only be required at joints where Pol4 is utilized. Pol4 is required only when the joint contains 3’ overhangs and gaps on both strands [10]. Addition of 5’ flaps to this type of joint relaxes the requirement for Pol4 (Figure 2), but Pol4 might normally perform gap filling at this type of joint when it is present. We therefore re-tested the joints used in Figure 2 in rad27Δ strains, but observed no reduction in joining (Figure 4C). Perhaps more informative is the observation that a joint with a 1-nt gap adjacent to a 5’ dRP could be repaired in pol4-KK247∷248RR rad27Δ cells (Figure 3C, first joint), indicating that Rad27 is not required for 5’ processing in the context of a Pol4-dependent joint.
The lack of a Rad27 requirement for any of the joints examined above contrasts with a previously reported 4.4-fold defect on a particular 5’ flap-containing joint [21]. To explore this discrepancy, we finally tested that substrate, pBX2, in our rad27Δ strain. The ADE2 reading frame can be restored in Bam HI/XhoI-cut pBX2 by a flap-containing joint (Figure 4D) or by rejoining of blunted ends. PCR with flanking primers was utilized to differentiate these two outcomes and adjust the Ade+/Leu+ ratio (data not shown). In our strains, rad27Δ cells formed the flap joint as efficiently as wild-type cells (Figure 4D).
Dideoxynucleotides greatly reduce the efficiency of NHEJ
In addition to damaged 5’ termini, Pol4 might be required to resynthesize damaged 3’ nucleotides. As a model to test this, we added dideoxynucleotides to the 3’ termini of plasmid-based DSBs. Although this lesion is unlikely to occur in nature, cells might treat these breaks similar to other forms of 3’ damage that cannot be directly reversed. The restriction enzyme BstXI was used to create DSBs with 3’ overhangs that form a compatible joint, a 1-nt gap, and a 1-nt flap. Dideoxynucleotides were added with terminal deoxynucleotidyl transferase (TdT), and ddNTP addition was verified by loss of ligation (Figure 5A). In all configurations tested, addition of dideoxynucleotides greatly reduced the rejoining efficiency in wild-type yeast (Figure 5B). Due to this low level of repair we were unable to reliably assess whether Pol4 is required to resynthesize a damaged 3’ terminal base, but these data show that the requisite but as yet uncharacterized NHEJ 3’ nuclease(s) require a 3’ hydroxyl.
DISCUSSION
We previously showed that Pol4 is required for filling nucleotide gaps in NHEJ joints only at 3’ overhangs [10]. This unique ability of Pol4 suggests that it has acquired properties which allow it to deal with the limiting and unstable primer-template pairings inherent to 3’ overhangs. This interpretation is supported by in vitro analyses of Pol4 and the related mammalian Pol X NHEJ polymerases, Pol μ and Pol λ, including extensive structural data, which have revealed several unique features of the Pol X polymerases [17,24,29-34]. For example, the Pol X NHEJ polymerases make fewer template strand contacts [24] and have a propensity toward strand slippage [17,30,31], mismatch extension [32] and lesion bypass [33], observations consistent with a reduced dependence on a stable primer-template pairing. Indeed, a “gradient” of polymerase properties has been described in which Pol μ has the remarkable ability to catalyze template-dependent synthesis without any initial primer-template pairing [34].
Despite this general agreement between in vivo and in vitro observations, seemingly inconsistent results have also been observed. In particular, we unexpectedly showed that Pol4 is not required for NHEJ of fully compatible DSBs bearing 5’ hydroxyl termini [10] even though the lack of polynucleotide kinase in S. cerevisiae demands removal and resynthesis of the damaged 5’ nucleotide [11]. Such synthesis would occur from 3’ overhangs with the same number of primer-template base-pairs as Pol4-dependent joints, and yet can be catalyzed by another polymerase. This indicated that factors other than overhang polarity and length must influence the requirement for Pol4. Here, we demonstrate that dependence on Pol4 was relaxed when mismatched flap nucleotides were added to the 5’ termini of gap-containing joints, even though the mismatched nucleotides were not incorporated during repair and therefore polymerization must have occurred at the joint (Figure 2). In contrast, comparable joints with 5’ dRPs strictly required Pol4 for gap filling (Figure 3). These observations clearly point to the base on the 5’ terminal strand as a key parameter in determining Pol4 dependence, with the presence of a base allowing utilization of another polymerase even when the base cannot form a Watson-Crick base pair (Figure 2) or participate in ligation [10].
We suggest that base stacking interactions between the 5’ terminal base and an adjacent overhang base can stabilize a joint enough that another polymerase can substitute for Pol4. These interactions appear to be at least as important to promoting joint stability, and therefore polymerase promiscuity, as base pairing between the overhangs. In this model, which of the two strands might be stabilized by the 5’ terminal base must be considered. It is difficult to imagine the primer strand being involved, since a stacked 5’ base would necessarily occupy the polymerase active site and be incompatible with catalysis. In contrast, base stacking in the template strand would act to promote its continuity and might allow it to be productively engaged by another polymerase (Figure 6A). These ideas also provide an alternative explanation for our observation that Pol4 is not required when a gap exists on only one strand of a DSB joint [10]. Ligation might heal such a DSB primarily, but current results suggest that the nicked strand might also be stable enough to act as template for a non-Pol4 polymerase to fill the gapped strand. Genetic assays cannot differentiate these possibilities, but biochemical studies could provide further insight into the site of synthesis in joints stabilized by base stacking. Importantly, the relevant polymerase for such studies is not Pol4, but the currently unknown and apparently replicative polymerase that can substitute for it [10].
Figure 6.
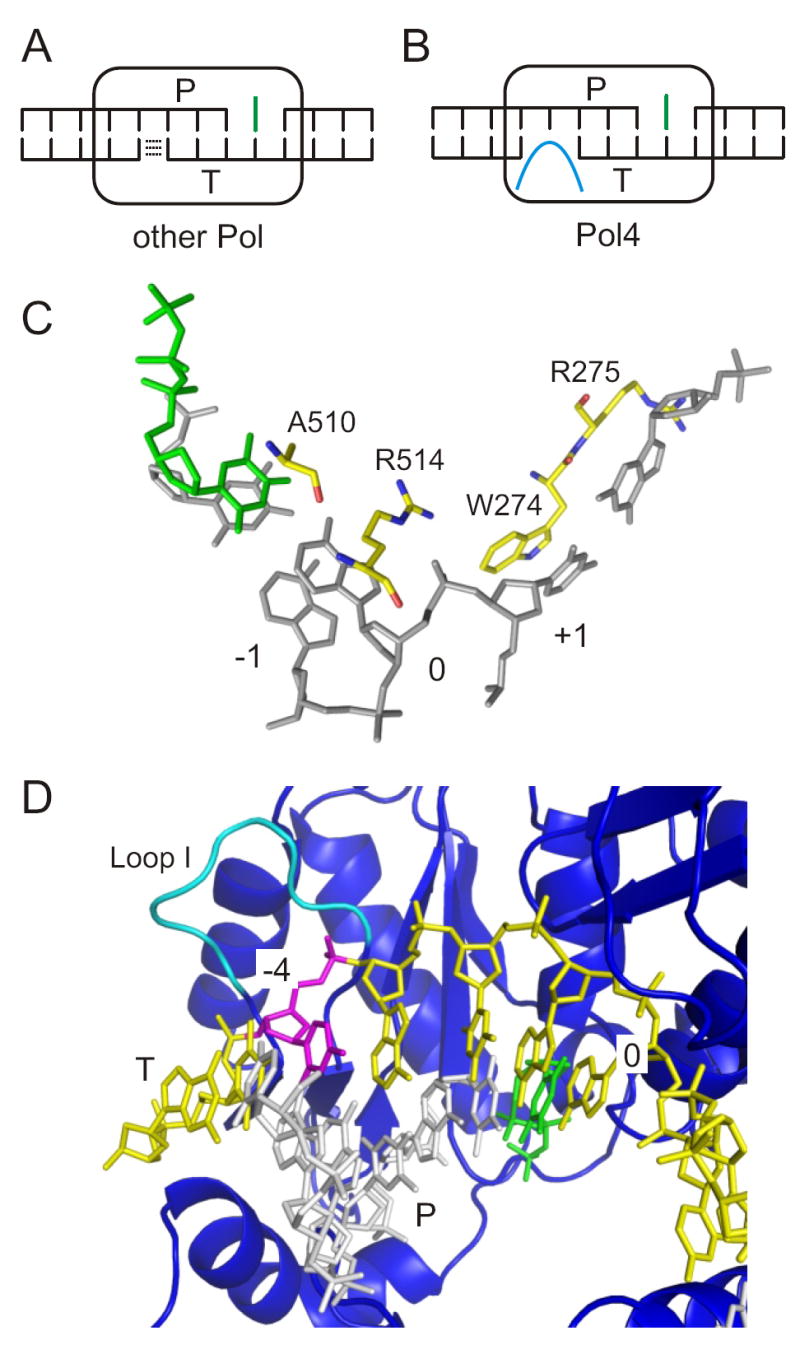
A model for Pol X polymerase action in NHEJ. (A) When a base exists on the 5’ terminal position of a template strand nick or mismatched 5’ flap, stacking interactions (dashed lines) are hypothesized to stabilize that strand sufficiently for its use by a polymerase other than Pol4. P = primer strand, T = template strand, green = incoming dNTP. (B) When gaps with no potential for base stacking exist on both strands, proteins motifs in Pol4 (cyan colored loop) are hypothesized to provide critical bridging contacts in the template strand gap. (C) Depiction of the extensive base stacking observed in a crystal structure of Pol λ (PDB 1XSN, [35]). The 0 (template nucleotide), -1 and +1 template strand positions are labeled. Pol λ residues are labeled and colored by element (yellow = carbon, blue = nitrogen, red = oxygen). Grey = DNA, green = incoming dNTP. (D) A different view of PDB 1XSN to highlight the position of Loop I (colored in cyan) relative to the expected position of a template strand gap in NHEJ (the -4 nucleotide colored in magenta, which might be absent in an NHEJ joint). The template strand (T) is otherwise yellow, the primer strand (P) white, the incoming dNTP green, and the protein in blue cartoon diagram.
A corollary of this model is that the unique function of Pol4, and presumably other Pol X NHEJ polymerases, is to stabilize joints via protein-DNA contacts when the stacking continuity of both DSB strands is disrupted. Pol X polymerases might also promote base pairing between the primer and template strands, but this energy gain would be the same for various Pol4-independent joints described here and previously [10] and cannot explain polymerase specificity. Thus, we infer that Pol4 achieves a productive catalytic complex by bridging the two sides of one or both DSB strands to overcome the missing contribution of base stacking to duplex stability (Figure 6B). Because of the extent of DNA bound by the polymerase [24,29], it is highly unlikely that NHEJ structural components such as Ku or MRX can provide this level of bridging; it must come from the polymerase itself. Crystal structures of Pol λ, the mammalian polymerase most related to Pol4, provide a first insight into the interactions that likely facilitate this bridging, which, not surprisingly, include extensive base stacking. In particular, residues R514, W274, the peptide backbone of R275 and A510 stack the bases of the template nucleotide (i.e. position 0), the +1 template strand nucleotide, the 5’ terminal base of the gap being filled, and the incoming nucleotide, respectively [35] (Figure 6C). Importantly, though, these interactions are with the primer strand gap. Our model suggests the further importance of interactions with the template strand gap. The previously described Loop I, which partially determines polymerase template dependence [36], is positioned to possibly provide such interactions [35] (Figure 6D), but current crystals are inconclusive in this regard because they contain continuous templates. Although a technical challenge, structures with discontinuous templates will be of paramount importance to evaluating the hypotheses forwarded here.
It is noteworthy that Pol4 dependence was not alleviated by the presence of 3’ mismatched flaps (Figure 2A, second joint). The bases on 3’ flaps would seem equally capable of promoting base stacking and therefore joint stability. However, these flaps demand nucleolytic processing prior to the action of a polymerase because they would interfere with use of the 3’ strand as a primer terminus. In contrast, the reduced Pol4 dependence of 5’ flap joints strongly suggests that polymerization does typically precede 5’ nucleolysis. Thus, these experiments tend to imply an inherent order to NHEJ processing steps, although further data will be required to describe this order in detail and what mechanisms enforce it.
The other issue addressed in this report is an explicit test of the short patch/long patch model of 5’ processing in NHEJ (Figure 1), suggested by the presence of 5’ dRP lyase activity in Pol4 [17] and the previously proposed role of Rad27 in NHEJ [20,21]. We were unable to provide clear support for either model, since ends with 5’ dRP termini could be rejoined in the absence of both the lyase activity of Pol4 and Rad27 (Figure 3). It is clear that yeast must have still other mechanisms for 5’ dRP removal during NHEJ, likely nucleases. Thus, our experiments do not rule out putative short-and long-patch pathways of NHEJ that depend on the Pol4 lyase or Rad27, respectively, but do reveal additional layers of complexity via multiple redundant mechanisms of 5’ processing. As these mechanisms are defined, the roles of Pol4 and Rad27 in short and long patch repair should be revisited.
Importantly, the lack of an NHEJ defect in our rad27Δ strains (Figure 4) contrasts with a previous report [21]. Several variables might contribute to this discrepancy. Rescue of the rad27Δ replication defects by overexpression of RNH35 allowed us to count many more colonies and ensured that we were not studying rare events in the small fraction of surviving rad27Δ cells. Additionally, yeast take up linear and supercoiled plasmids with different efficiencies depending on the growth state of the cells. In the previous report, a supercoiled plasmid was used as a control, and rad27Δ cells grew slower and divided fewer times in log phase than wild-type cells. Therefore, the perceived joining deficiency may have been affected by changes in the ratio of linear to supercoiled plasmid uptake, in contrast to our use of a linearized control plasmid. Importantly, the absence of a requirement for Rad27 in our genetic assays does not rule out a role for Rad27 in NHEJ. Given the biochemical evidence for its involvement [20], it is more likely that Rad27 is simply one of several mechanisms of 5’ end processing.
Acknowledgments
We would like to thank the Wilson lab for their support and critical readings of the manuscript. This work was supported by Public Health Service grant CA102563 (T.E.W.) and the University of Michigan Rackham Predoctoral Fellowship (J.M.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- 2.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 3.Chen L, Trujillo K, Ramos W, Sung P, Tomkinson AE. Promotion of Dnl4-catalyzed DNA end-joining by the Rad50/Mre11/Xrs2 and Hdf1/Hdf2 complexes. Mol Cell. 2001;8:1105–1115. doi: 10.1016/s1097-2765(01)00388-4. [DOI] [PubMed] [Google Scholar]
- 4.Wilson TE, Grawunder U, Lieber MR. Yeast DNA ligase IV mediates nonhomologous DNA end joining. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 5.Coquerelle T, Bopp A, Kessler B, Hagen U. Strand breaks and K’ end-groups in DNA of irradiated thymocytes. Int J Radiat Biol Relat Stud Phys Chem Med. 1973;24:397–404. doi: 10.1080/09553007314551251. [DOI] [PubMed] [Google Scholar]
- 6.Henner WD, Grunberg SM, Haseltine WA. Sites and structure of gamma radiation-induced DNA strand breaks. J Biol Chem. 1982;257:11750–11754. [PubMed] [Google Scholar]
- 7.K W, Huttermann J, Teoule R, editors. Effects of Ionizing Radiation on DNA. Springer-Verlag; New York: 1978. [Google Scholar]
- 8.Pastwa E, Neumann RD, Mezhevaya K, Winters TA. Repair of radiation-induced DNA double-strand breaks is dependent upon radiation quality and the structural complexity of double-strand breaks. Radiat Res. 2003;159:251–261. doi: 10.1667/0033-7587(2003)159[0251:roridd]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 9.Wilson TE, Lieber MR. Efficient processing of DNA ends during yeast nonhomologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway. J Biol Chem. 1999;274:23599–23609. doi: 10.1074/jbc.274.33.23599. [DOI] [PubMed] [Google Scholar]
- 10.Daley JM, Laan RL, Suresh A, Wilson TE. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J Biol Chem. 2005;280:29030–29037. doi: 10.1074/jbc.M505277200. [DOI] [PubMed] [Google Scholar]
- 11.Vance JR, Wilson TE. Uncoupling of 3’-phosphatase and 5’-kinase functions in budding yeast. Characterization of Saccharomyces cerevisiae DNA 3’-phosphatase (TPP1) J Biol Chem. 2001;276:15073–15081. doi: 10.1074/jbc.M011075200. [DOI] [PubMed] [Google Scholar]
- 12.Fortini P, Pascucci B, Parlanti E, D’Errico M, Simonelli V, Dogliotti E. The base excision repair: mechanisms and its relevance for cancer susceptibility. Biochimie. 2003;85:1053–1071. doi: 10.1016/j.biochi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Sobol RW, Prasad R, Evenski A, Baker A, Yang XP, Horton JK, Wilson SH. The lyase activity of the DNA repair protein beta-polymerase protects from DNA-damage-induced cytotoxicity. Nature. 2000;405:807–810. doi: 10.1038/35015598. [DOI] [PubMed] [Google Scholar]
- 14.Stucki M, Pascucci B, Parlanti E, Fortini P, Wilson SH, Hubscher U, Dogliotti E. Mammalian base excision repair by DNA polymerases delta and epsilon. Oncogene. 1998;17:835–843. doi: 10.1038/sj.onc.1202001. [DOI] [PubMed] [Google Scholar]
- 15.Prasad R, Dianov GL, Bohr VA, Wilson SH. FEN1 stimulation of DNA polymerase beta mediates an excision step in mammalian long patch base excision repair. J Biol Chem. 2000;275:4460–4466. doi: 10.1074/jbc.275.6.4460. [DOI] [PubMed] [Google Scholar]
- 16.Kim K, Biade S, Matsumoto Y. Involvement of flap endonuclease 1 in base excision DNA repair. J Biol Chem. 1998;273:8842–8848. doi: 10.1074/jbc.273.15.8842. [DOI] [PubMed] [Google Scholar]
- 17.Bebenek K, Garcia-Diaz M, Patishall SR, Kunkel TA. Biochemical properties of Saccharomyces cerevisiae DNA polymerase IV. J Biol Chem. 2005;280:20051–20058. doi: 10.1074/jbc.M501981200. [DOI] [PubMed] [Google Scholar]
- 18.Matsumoto Y, Kim K. Excision of deoxyribose phosphate residues by DNA polymerase beta during DNA repair. Science. 1995;269:699–702. doi: 10.1126/science.7624801. [DOI] [PubMed] [Google Scholar]
- 19.Kunkel TA. The mutational specificity of DNA polymerase-beta during in vitro DNA synthesis. Production of frameshift, base substitution, and deletion mutations. J Biol Chem. 1985;260:5787–5796. [PubMed] [Google Scholar]
- 20.Tseng HM, Tomkinson AE. Processing and joining of DNA ends coordinated by interactions among Dnl4/Lif1, Pol4, and FEN-1. J Biol Chem. 2004;279:47580–47588. doi: 10.1074/jbc.M404492200. [DOI] [PubMed] [Google Scholar]
- 21.Wu X, Wilson TE, Lieber MR. A role for FEN-1 in nonhomologous DNA end joining: the order of strand annealing and nucleolytic processing events. Proc Natl Acad Sci U S A. 1999;96:1303–1308. doi: 10.1073/pnas.96.4.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Della M, Palmbos PL, Tseng HM, Tonkin LM, Daley JM, Topper LM, Pitcher RS, Tomkinson AE, Wilson TE, Doherty AJ. Mycobacterial Ku and ligase proteins constitute a two-component NHEJ repair machine. Science. 2004;306:683–685. doi: 10.1126/science.1099824. [DOI] [PubMed] [Google Scholar]
- 23.Daley JM, Wilson TE. Rejoining of DNA double-strand breaks as a function of overhang length. Mol Cell Biol. 2005;25:896–906. doi: 10.1128/MCB.25.3.896-906.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Diaz M, Bebenek K, Krahn JM, Blanco L, Kunkel TA, Pedersen LC. A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology. Mol Cell. 2004;13:561–572. doi: 10.1016/s1097-2765(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 25.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) Embo J. 1997;16:3341–3348. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu J, Qian Y, Frank P, Wintersberger U, Shen B. Saccharomyces cerevisiae RNase H(35) functions in RNA primer removal during lagging-strand DNA synthesis, most efficiently in cooperation with Rad27 nuclease. Mol Cell Biol. 1999;19:8361–8371. doi: 10.1128/mcb.19.12.8361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kao HI, Henricksen LA, Liu Y, Bambara RA. Cleavage specificity of Saccharomyces cerevisiae flap endonuclease 1 suggests a double-flap structure as the cellular substrate. J Biol Chem. 2002;277:14379–14389. doi: 10.1074/jbc.M110662200. [DOI] [PubMed] [Google Scholar]
- 29.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA Polymerase mu. Nat Struct Mol Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 30.Bebenek K, Garcia-Diaz M, Blanco L, Kunkel TA. The frameshift infidelity of human DNA polymerase lambda. Implications for function. J Biol Chem. 2003;278:34685–34690. doi: 10.1074/jbc.M305705200. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Diaz M, Bebenek K, Krahn JM, Pedersen LC, Kunkel TA. Structural analysis of strand misalignment during DNA synthesis by a human DNA polymerase. Cell. 2006;124:331–342. doi: 10.1016/j.cell.2005.10.039. [DOI] [PubMed] [Google Scholar]
- 32.Picher AJ, Garcia-Diaz M, Bebenek K, Pedersen LC, Kunkel TA, Blanco L. Promiscuous mismatch extension by human DNA polymerase lambda. Nucleic Acids Res. 2006;34:3259–3266. doi: 10.1093/nar/gkl377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Y, Wu X, Guo D, Rechkoblit O, Taylor JS, Geacintov NE, Wang Z. Lesion bypass activities of human DNA polymerase mu. J Biol Chem. 2002;277:44582–44587. doi: 10.1074/jbc.M207297200. [DOI] [PubMed] [Google Scholar]
- 34.Nick McElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family X polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 35.Garcia-Diaz M, Bebenek K, Krahn JM, Kunkel TA, Pedersen LC. A closed conformation for the Pol lambda catalytic cycle. Nat Struct Mol Biol. 2005;12:97–98. doi: 10.1038/nsmb876. [DOI] [PubMed] [Google Scholar]
- 36.Juarez R, Ruiz JF, McElhinny SA, Ramsden D, Blanco L. A specific loop in human DNA polymerase mu allows switching between creative and DNA-instructed synthesis. Nucleic Acids Res. 2006;34:4572–4582. doi: 10.1093/nar/gkl457. [DOI] [PMC free article] [PubMed] [Google Scholar]