Abstract
We analyzed the expression profile and cellular localization of the maize (Zea mays) Rpd3-type histone deacetylases genes ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 (indicated as ZmHDA101, ZmHDA102, and ZmHDA108 in the Plant Chromatin Database). This study shows that maize Rpd3 transcripts are present in all the organs and cellular domains analyzed, but we found that their amounts change during development, accumulating in the inner region of the endosperm, in vascular zones of the nucellus, in the tapetum, and in the tetrads. A similar expression profile and nucleus-cytoplasmic localization was observed for ZmRpd3 proteins. Glutathione S-transferase pull-down assays show that ZmRpd3 proteins can interact with the maize retinoblastoma-related (ZmRBR1) protein, an important regulator of cell cycle progression, and with the maize retinoblastoma-associated protein (ZmRbAp1). However, the three ZmRpd3 proteins do not mutually compete in the binding. These results suggest a general role of ZmRpd3 genes in the plant cell cycle and development. These observations also provide indications on possible mechanisms regulating their transcription and protein accumulation. Similarities in the gene expression profiles and protein interactions may indicate that functional redundancy among members of the ZmRpd3 gene family exists. However, a degree of functional divergence is also supported by our findings.
Plant development is a striking example of a highly orchestrated biological process. Recent advances demonstrate that this intricate process is accomplished by diverse mechanisms and networks that operate at distinct levels within the nucleus (Goodrich and Tweedie, 2002). A fundamental mechanism controlling the selectivity of gene expression is the ability of many transcription factors to access the genome of eukaryotes (Struhl, 1999). This is achieved by packaging genes into chromatin, which impedes the binding of any proteins to their target DNA sequences. The accessibility of DNA to protein interaction is regulated by different enzymatic complexes that modulate nucleosomal structure. In the past few years, it has been shown that posttranslational modifications of histones, including acetylation, methylation, phosphorylation, and ubiquitination play a key role in modulating dynamic changes in chromatin structure and gene activity (Wu and Grunstein, 2000). Distinct histone modification patterns, together with direct modifications of the DNA, such as DNA methylation, are believed to form an epigenetic code acting as epigenetic marks or docking elements specifically read by regulatory factors that, in turn, can alter chromatin structure and regulate transcription (Strahl and Allis, 2000; Schreiber and Bernstein, 2002; Turner, 2002).
Histone acetylation is the best-characterized type of histone modification (Cress and Seto, 2000; Roth et al., 2001). The enzymes responsible for maintaining the steady-state balance of histone acetylation are the histone acetyltransferases (HATs) and histone deacetylases (HDACs). Both enzymes are members of distinct gene families and exist as multiprotein complexes. Many of the recently identified HATs and HDACs turned out to be transcriptional co-activators and co-repressors, thus establishing a direct link between histone acetylation and regulation of gene transcription. Mechanisms and factors controlling gene activity by affecting chromatin structure are largely conserved in eukaryotes, including plants (Lusser 2002). However, the sessile nature of plants, which makes them more sensitive to environmental signals, and the relative plasticity of their cell fate suggest that specific features of the chromatin-mediated control of gene transcription exist in plants. Evidence has shown that the basic features of histone acetylation in plants resemble those of other eukaryotes, but marked differences, reflected by novel classes of HDACs not identified in other experimental systems, have been reported (Lusser et al., 2001).
In plants, different HDAC genes have been identified and classified into three distinct gene families (http://chromdb.biosci.arizona.edu; Pandey et al., 2002). The first family, named the HDA gene family, contains members related to the yeast sequences Rpd3 and Hda1 (Rundlett et al., 1996; Taunton et al., 1996; Rossi et al., 1998; Lechner et al., 2000). This family is further divided into three classes based on their degree of homology with Rpd3 (class I), Hda1 (class II), or a third group of sequences phylogenetically distinct from the first two classes. The members of the second family of plant HDACs, termed the SRT family, are related to yeast Sir2 (Imai et al., 2000). In contrast to other eukaryotes, plants contain a third family of enzymes, the nucleolar-phosphoproteins HD2 (HDT gene family), which appear to be plant-specific (Lusser et al., 1997; Dangl et al., 2001).
Recently, attempts have been made to characterize plant HDACs at their functional level, providing the first indication on the biological role of these enzymes. Treatment with the HDAC inhibitor trichostatin A, showed that histone acetylation is involved in silencing rRNA genes in allotetraploid Brassica sp. (Pikaard, 1999). Furthermore, it has been reported that a maize Rpd3-like enzyme (ZmRpd3I) is physically associated with the maize retinoblastoma-related (ZmRBR1) protein, a key regulator of cell cycle progression (Rossi and Varotto, 2002; Rossi et al., 2003). Moreover, these proteins cooperate in repressing gene transcription in plant cells, thus suggesting a role of ZmRpd3I in cell cycle control. Recent studies, using antisense-mediated down-regulation and overexpression of Rpd3 genes in plants, reveal a variety of pleiotropic effects that indicate the involvement of Rpd3-type HDACs in plant development (Tian and Chen, 2001; Wu et al., 2000; Jang et al., 2003). Furthermore, Rpd3 genes have been identified in screens for mutants displaying inhibition of transgene silencing and defects in RNA-directed DNA methylation (Murfett et al., 2001; Aufsatz et al., 2002). Despite this evidence, the mechanisms responsible for the Rpd3-mediated control of gene activity, knowledge about which genes are directly affected by these chromatin-modifier factors, and the specific role of HDACs in plant development remain elusive. Similarly, possible differences in functional activities among members of the same gene family also need to be clarified.
Maize represents one of the best-characterized systems for studies of HDACs because members of all three families described above have been identified and biochemically characterized (for review, see Lusser et al., 2001). To gain insights into the role played by maize Rpd3 genes (ZmRpd3) in plant development and to investigate peculiarities among members of this gene family, we analyzed their gene expression profile and cellular localization in various plant organs at different developmental stages. Glutathione S-transferase (GST) pull-down assays were also performed to assess the ability of different ZmRpd3 proteins to interact with ZmRBR1 and with a maize retinoblastoma-associated protein (ZmRbAp1), a histone-binding protein that participates in forming a ZmRBR1/ZmRpd3 complex (Rossi et al. 2001; Rossi et al., 2003). Our results suggest a general role of maize Rpd3-type HDACs during plant development. In addition, although our data suggest a functional redundancy among members of the ZmRpd3 gene family, they indicate that a certain degree of functional divergence is likely to exist.
RESULTS
Maize Rpd3 Sequences and Phylogenetic Analysis
To examine the relationship among maize Rpd3-type genes, we considered two ZmRpd3 cDNA sequences, termed ZmRpd3I/ZmHD1BI and ZmHD1BII, previously characterized at the molecular and biochemical level (Rossi et al., 1998; Lechner et al., 2000). In addition, several expressed sequence tag (EST) clones homologous to Rpd3-type HDACs have been identified in the maize genome database (http://www.zmdb.iastate.edu). Contig formation by EST assembly has identified three ZmRpd3 cDNA sequences (ZmHDA101, ZmHDA102, and ZmHDA108 in the Plant Chromatin Databases; http://chromdb.biosci.arizona.edu). ZmHDA101 and ZmHDA108 exhibit high sequence homology with ZmRpd3I/ZmHD1BI and ZmHD1BII, respectively, suggesting allelic polymorphism. Reverse transcriptase-PCR amplification was performed using total RNA extracted from seedlings of the maize B73 inbred line and primers designed to specific regions of ZmRpd3I/ZmHD1BI, ZmHD1BII, and ZmHDA102 sequences, respectively. Sequencing of the amplified fragments, corresponding to full-length cDNAs, revealed 100% homology with the sequences reported in the Plant Chromatin Database. We will therefore hereinafter refer to the maize Rpd3 genes as ZmRpd3/101 (formerly ZmRpd3I/ZmHD1BI or ZmHDA101), ZmRpd3/102 (formerly ZmHDA102), and ZmRpd3/108 (formerly ZmHD1BII or ZmHDA108).
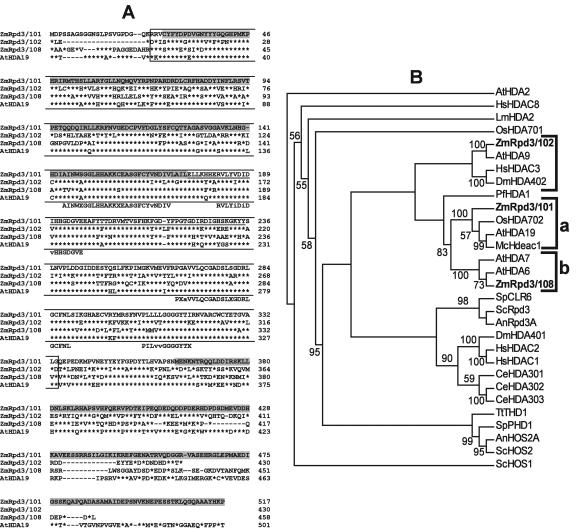
Sequence comparison among the three maize Rpd3-type and one Arabidopsis Rpd3-type proteins revealed that ZmRpd3/101 shares 49.5%, 55%, and 75% amino acid sequence identity with ZmRpd3/102, ZmRpd3/108, and AtHDA19, respectively (Fig. 1A). Particularly, the degree of homology among these sequences was higher in the HDAC catalytic domain (PF00805 in the Protein Family “Pfam” database) than in the N- and C-terminal regions, which appeared to be specific to each ZmRpd3 protein. Phylogenetic analysis, performed using the HDAC domains of 30 Rpd3 sequences from different eukaryotes identified in the GenBank database (Pandey et al., 2002) revealed that the three maize proteins group into three distinct branches (Fig. 1B). Specifically, ZmRpd3/101 clusters with AtHDA19 and OsHDA702 (cluster a), ZmRpd3/108 appears to be related to AtHDA6 and AtHDA7 (cluster b), whereas ZmRpd3/102 and AtHDA9 form a distinct branch, more divergent with respect to the other two. Similar results were obtained when a phylogenetic tree was produced using the N- and C-terminal regions of the 30 Rpd3 proteins (data not shown). These results indicate that the three ZmRpd3-type HDACs, along with other plant proteins, are included in three phylogenetically separate groups that may represent different orthologs.
Figure 1.
Amino acid sequences alignment and phylogenetic analysis of maize Rpd3-like proteins. A, ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 sequences from maize and AtHDA19 sequence from Arabidopsis were aligned using ClustalW software (Thompson et al., 1994) and were subsequently edited with Genedoc software (http://www.psc.edu/biomed/genedoc/). The number of amino acids is indicated at the right site. A dash indicates gaps in the alignment, whereas asterisks represent amino acid identity. The white box indicates the HDAC domain (PF00805 in Pfam database) present in different Rpd3 proteins. Shaded boxes represent the N- and C-terminal regions of ZmRpd3/101 involved in binding of ZmRBR1 and ZmRbAp1; an internal domain that enhances ZmRpd3/101-ZmRbAp1interaction is also underlined (Rossi et al., 2003). Below the multiple sequence alignment, consensus motifs found in all Rpd3 class I HDACs are shown. These motifs represent highly conserved sequence positions in Rpd3-like sequences from 30 different eukaryotes (see below) identified by generating a logo sequence (http://www.bio.cam.ac.uk/cgi-bin/seqlogo/logo.cgi). Upper- and lowercase letters represent positions 98% and 60% conserved within class I HDACs, respectively; X indicates variable positions. B, Unrooted neighbor-joining tree of the HDAC domain of 30 Rpd3 class I HDAC proteins was obtained using ClustalW and Phylip software packages (Felsenstein, 1989; Thompson et al., 1994). Nomenclature of the proteins is referred to Pandey et al. (2002). Proteins and their accession numbers follow: AnRpd3A (AAF80489), AnHOS2A (AAF80490), AtHDA2 (AAD40129), AtHDA6 (BAB10553), AtHDA7 (BAB09994), AtHDA9 (CAB72470), AtHDA19 (AAB66486), CeHDA301 (CAB03984), CeHDA302 (Q09440), CeHDA303 (CAB03224), DmHDA401 (AAC61494), DmHDA402 (AAC83649), HsHDAC1 (Q13547), HsHDAC2 (NP_001518), HsHDAC3 (NP_003874), HsHDAC8 (NP_060956), LmHDA2 (CAC14522), McHdeac1 (AAF82385), OsHDA701 (BAB61857), OsHDA702 (AAK01712), PfHDA1 (AAD22407), ScRpd3 (P32561), ScHOS1 (Q12214), ScHOS2 (P53096), SpCLR6 (CAA19053), SpPHD1 (O13298), TtTHD1 (AAG00980), ZmRpd3/101 (AAK67142), ZmRpd3/102 (AAL33655), and ZmRpd3/108 (AAD10139). Abbreviations for species are: Aspergillus nidulans (An), Arabidopsis (At), Caenorhabditis elegans (Ce), fruit fly (Drosophila melanogaster; Dm), human (Homo sapiens; Hs), Leishmania major (Lm), common ice plant (Mesembryanthemum crystallinum; Mc), rice (Oryza sativa; Os), Plasmodium falciparum (Pf), Brewer's yeast (Saccharomyces cerevisiae; Sc), fission yeast (Schizosaccharomyces pombe; Sp), Tetrahymena thermophila (Tt), and maize (Zm). The class III HDACs AtHDA2 was used as out-group. Bootstrap values (100 replicates) are indicated at the nodes of the tree and are expressed as percentages; a 50% cut-off was applied. The ZmRpd3 proteins are highlighted in bold. Brackets indicate the three clusters containing ZmRpd3 sequences.
Expression Profiles of ZmRpd3 Transcripts
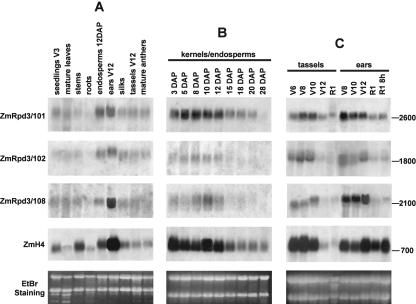
The expression levels of the ZmRpd3 transcripts were analyzed in northern-blot experiments using total RNA prepared from different maize organs. The results revealed that transcripts hybridizing with DNA probes specific for ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 and corresponding to the predicted length of the three ZmRpd3 cDNAs were present in all the organs analyzed (Fig. 2A). Transcript abundance of each of the three ZmRpd3 genes was variable across different organs, with a higher than average abundance in endosperms and ears.
Figure 2.
Northern-blot analysis of ZmRpd3 transcripts. Total RNA (20 μg) extracted from different maize organs (A), kernels/endosperms (B), and ears and tassels (C) harvested at different developmental stages was fractionated through an agarose/formaldehyde denaturing gel and blotted onto nylon membrane. The membranes were hybridized with DNA probes specific for each ZmRpd3 gene and with a probe specific for the histone H4 transcript. Ethidium bromide staining of the gel is shown as input control. The length of the transcripts (number of nucleotides) is reported on the right of each panel. The organs or developmental stages are reported according to indications from the Iowa State University of Science and Technology (1989; see “Materials and Methods”).
The expression patterns of ZmRpd3 genes were also assessed at different stages of endosperm, ear, and tassel development. These organs were chosen because they represent useful model systems to investigate the correlation between ZmRpd3 gene expression and development in maize (Cheng et al., 1983; Slocombe et al., 1999; McSteen et al., 2000). Northern analysis showed that the amount of each ZmRpd3 transcript changes during development of endosperm, ear, and tassel (Fig. 2, B and C). In these organs, the ZmRpd3 genes displayed similar expression profiles; in particular, ZmRpd3 transcripts were more abundant during the first phases of development and then decreased during later developmental stages. High amounts of ZmRpd3 RNAs appeared to be correlated with high hybridization signals of the DNA replication marker histone H4 (Meshi et al., 2000), indicating that these developmental stages are characterized by a high DNA replication activity (Cheng et al., 1983; Slocombe et al., 1999) and suggesting a possible coupling of ZmRpd3 function and DNA replication events.
Despite similarities in the transcription profiles, the relative amounts of ZmRpd3 transcripts differed in the organs and in the different stages of organ development analyzed. For example, ZmRpd3/108 RNA was particularly abundant in the ear at V12 stage with respect to the other two transcripts (Fig. 2A), whereas the level of ZmRpd3/101 RNA was higher during endosperm development compared with ZmRpd3/102 and ZmRpd3/108 transcripts (Fig. 2B).
Characterization of Antibodies and Immunodetection of ZmRpd3 Proteins
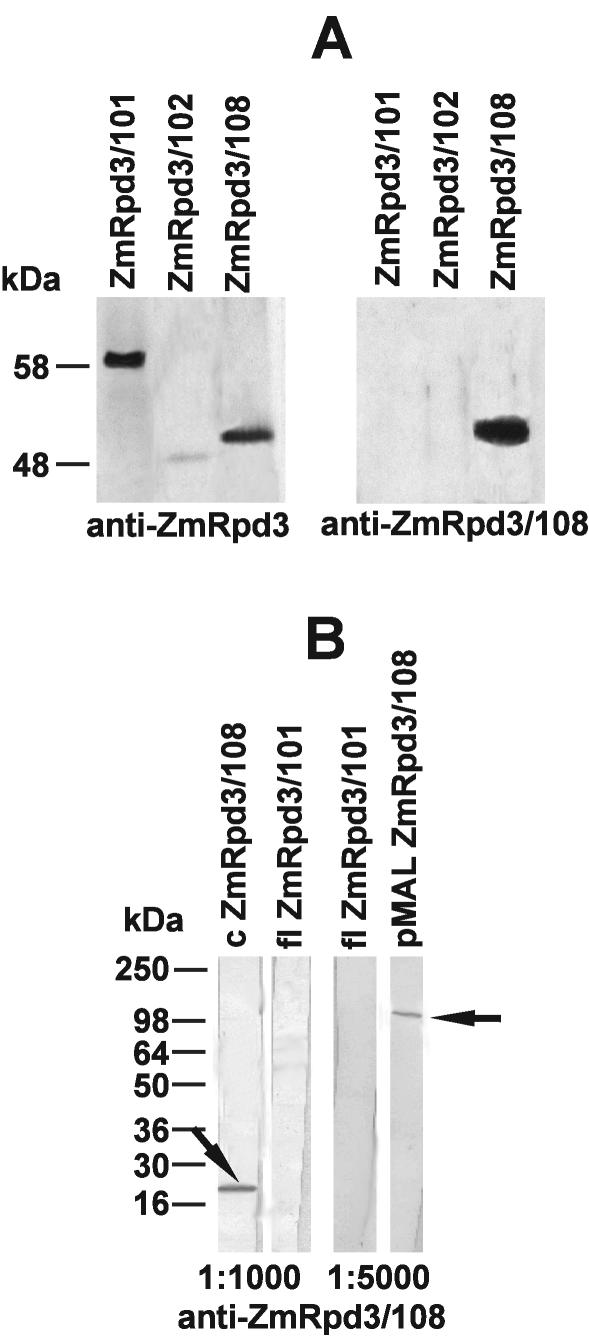
Antibodies raised against the full-length ZmRpd3/101 (anti-ZmRpd3; Rossi et al., 2003) and the C-terminal domain of ZmRpd3/108 (anti-ZmRpd3/108) were tested for their ability to specifically recognize ZmRpd3 proteins. Western-blot analysis of the in vitro-translated (IVT) ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 products revealed that the anti-ZmRpd3 antibody detected three polypeptides corresponding to the predicted molecular masses of the ZmRpd3 proteins (ZmRpd3/101, 58 kD; ZmRpd3/102, 49 kD; and ZmRpd3/108, 51 kD), because this antibody is likely to recognize the HDAC domain conserved throughout different ZmRpd3 proteins (Fig. 3A, left panel). Conversely, the anti-ZmRpd3/108 antibody detected only the 51-kD polypeptide, corresponding to ZmRpd3/108 protein (Fig. 3A, right panel). Accordingly, the anti-ZmRpd3/108 antibody specifically recognized only the full-length ZmRpd3/108 recombinant protein and its C-terminal part, whereas no signal was observed with the complete sequence of ZmRpd3/101 (Fig. 3B). These results indicate that the anti-ZmRpd3 antibody can recognize different ZmRpd3 proteins, whereas anti-ZmRpd3/108 antibody is specific to ZmRpd3/108.
Figure 3.
Characterization of antibodies. A, IVT ZmRpd3 proteins were fractionated by SDS-PAGE, blotted, and immunodetected with anti-ZmRpd3 antibody (left panel) and anti-ZmRpd3/108 antibody (right panel). B, Recombinant full-length ZmRpd3/108 fused to the maltose-binding domain (pMAL ZmRpd3/108), C-terminal part of ZmRpd3/108 (c ZmRpd3/108), and full-length ZmRpd3/101(fl ZmRpd3/101) proteins were expressed in Escherichia coli, and the protein extracts were fractionated by SDS-PAGE followed by blotting onto nylon membranes and immunodetection with different dilutions of anti-ZmRpd3/108 antibody. Arrows mark the band specifically recognized by the antibody. The size of molecular markers is reported.
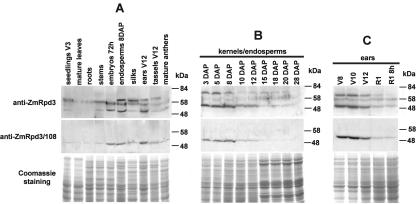
To investigate the pattern of ZmRpd3 protein accumulation, western-blot analyses were performed using crude protein extracts prepared from various maize organs and from endosperm and ear tissues at different developmental stages. Three immunoreactive bands were detected using the anti-ZmRpd3 antibody (Fig. 4, A–C, top panel). The first one, a polypeptide with a molecular mass of approximately 58 kD, may represent ZmRpd3/101 because it corresponds to the predicted size of this protein. A 51-kD polypeptide was also detected, and it represents the ZmRpd3/108 protein because a corresponding band is specifically recognized by the anti-ZmRpd3/108 antibody (Fig. 4, A–C, middle panel). A polypeptide corresponding to the predicted molecular mass of ZmRpd3/102 protein (49 kD) seems not to be detected by these two antibodies, probably for its low amount in maize crude protein extracts. However, an additional polypeptide of 64 kD was detected by the anti-ZmRpd3 antibody (Fig. 4, A–C, top panel) in the maize crude protein extracts. The 64-kD polypeptide is likely to represent one ZmRpd3 protein because it was specifically immunoprecipitated, together with significant HDAC activity, by the anti-ZmRpd3 antibody from endosperm crude extracts (Rossi et al., 2003; our unpublished data). This polypeptide may represent an additional Rpd3 protein not yet identified in maize, or alternatively, it might correspond to a protein variant of one of the three ZmRpd3 described in this work, migrating at this position due to posttranslational modifications.
Figure 4.
Western-blot analysis of ZmRpd3 protein. One hundred micrograms of crude protein extracts from various maize organs (A), from different developmental stages of kernel/endosperm (B), and ear (C) was fractionated by SDS-PAGE, blotted, and immunodetected with anti-ZmRpd3 antibody (top panels). The filters were subsequently stripped and reprobed with anti-ZmRpd3/108 antibody (middle panels). The size of molecular markers is indicated. Coomassie staining of SDS-PAGEs loaded with the protein extracts are shown as loading control (bottom panels).
Western-blot analysis indicated that the 58-kD protein was detectable in all maize organs analyzed and its amount did not change across endosperm and ear development (Fig. 4, A–C, top panel). Conversely, the 64-kD polypeptide was detected by anti-ZmRpd3 antibody only in endosperm, silk, ear, and tassel (Fig. 4A, top panel) and was more abundant during the first stages of endosperm and ear development and then decreased during later stages (Fig. 4, B and C, top panel). The ZmRpd3/108-specific 51 kD polypeptide was only slightly detectable in mature leaves, stems, and roots, and its expression pattern resembled that of the 64-kD polypeptide during endosperm and ear development (Fig. 4, A–C, top and middle panel).
Comparison of the transcript and protein expression patterns of ZmRpd3 genes revealed differences in the timing of protein accumulation with respect to RNA during endosperm development. For example, ZmRpd3/108 protein is abundant in kernels at 3, 5, and 8 d after pollination (DAP) and then starts to decrease, whereas RNA levels gradually increase in the first stages of development and reach their maximum level in 10/12 DAP endosperms (Figs. 2B and 4B). The finding that ZmRpd3 proteins start to decrease before their corresponding RNAs reach their maximum level suggests that, at least in a specific period during endosperm development (from 8 to 12 DAP), the abundance of ZmRpd3 proteins is regulated by mechanisms operating either at the translational (i.e. reduction of the RNA availability for protein synthesis) or at the posttranslational levels (i.e. increase of protein degradation and/or reduction of protein stability).
Cellular Localization of Maize Rpd3 Transcripts and Proteins
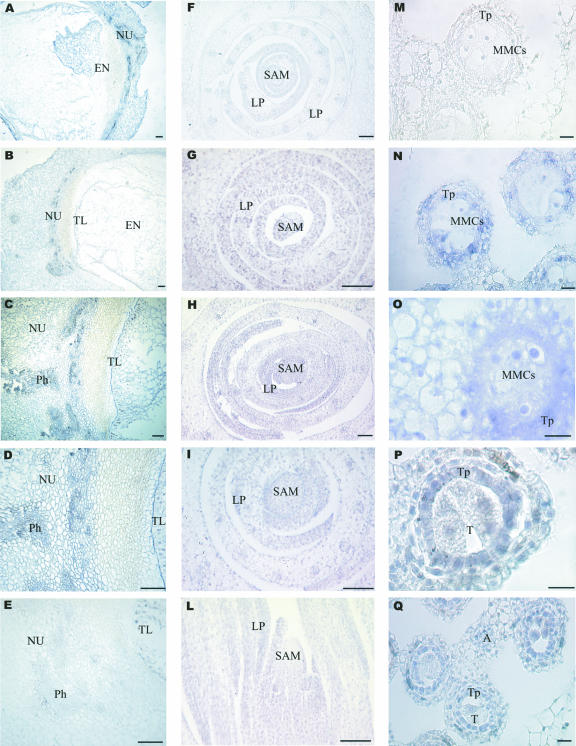
In situ hybridization experiments were carried out to analyze the cellular localization of ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 transcripts during development of the maize seed and shoot apical meristem (SAM) and during microsporogenesis at premeiotic and tetrad stages. Results indicated that the three ZmRpd3 transcripts showed a similar pattern of cellular localization in the organs and tissues investigated (Fig. 5). In particular, during seed development each ZmRpd3 gene was expressed in all cellular domains of the kernel: maternal tissue, embryo, and endosperm (Fig. 5, A–E; data not shown). ZmRpd3 transcripts started to be detectable at 3 DAP and over cellularization and endoreduplication stages accumulated in the endosperm (Fig. 5, A and B), showing an abundance gradient as they were more localized in the inner region of the endosperm than in the transfer cell domain facing the maternal tissue (data not shown; Rossi et al., 2003). At these developmental stages, the hybridizing signals were also detected in the vascular zone of the nucellar parenchyma (Fig. 5, C–E).
Figure 5.
Localization of ZmRpd3 transcripts in maize tissues. A through E, Longitudinal sections through a maize kernel at 6 (A) and 10 (B–E) DAP were hybridized with antisense probes specific for ZmRpd3/101 (A, B, and D) and ZmRpd3/102 (C) transcripts. E, Hybridization with ZmRpd3/101 sense probe is reported as an example of negative control. EN, Endosperm; NU, nucellus; TL, transfer layer; Ph, phloem. F through L, Cross (F–I) and longitudinal (L) sections through the shoot apex were hybridized with antisense probes specific for ZmRpd3/101 (G and L), ZmRpd3/102 (H), and ZmRpd3/108 (I). Hybridization with sense ZmRpd3/101 is shown as negative control (F). LP, Leaf primordium. M through Q, Cross sections of anthers during microsporogenesis at premeiotic (M–O) and tetrad stages (P and Q) were hybridized with sense (M) or antisense (N) probes specific for the histone H4 transcript and with antisense probes specific for ZmRpd3/101 (O), ZmRpd3/102 (P), and ZmRpd3/108 (Q). A, Anthers; T, tetrad; Tp, tapetum. The purple/blue staining represents the hybridization signal. Bars = 100 μm.
Cellular localization of ZmRpd3 transcripts was analyzed in shoot apexes at two different developmental stages: in 8-d-old seedlings and at the V2 plant stage. In both stages, the three genes were constitutively expressed in all cell types (Fig. 5, F–L). The signals were more pronounced in the small and round cells of the SAM and of leaf primordia compared with the older cells of leaf primordia. This is probably due to the fact that the first cell type is rich in cytoplasm, whereas vacuoles occupy most of the cell volume in the second cell type. ZmRpd3 transcripts were also localized in all anther tissues, with a higher level of expression in tapetal cells and microspore mother cells (MMCs; Fig. 5, O–Q). In the tapetum, ZmRpd3 transcripts were present from the onset of meiosis up to the early microspore stage, thus decreasing when tapetal cells start to degenerate at the tasseling stage. Furthermore, the ZmRpd3 genes were expressed in MMCs in tetrads, whereas in the early uni-nucleate microspore, the transcript level decreased.
Cellular localization of the histone H4 transcript was detected in the same maize tissues (Fig. 5, M and N; data not shown) revealing remarkable differences with respect to the localization of ZmRpd3 transcripts. In fact, as expected for a DNA replication marker (Meshi et al., 2000), histone H4 mRNAs specifically localized in active proliferating cells, whereas ZmRpd3 transcripts are present in all cellular domains.
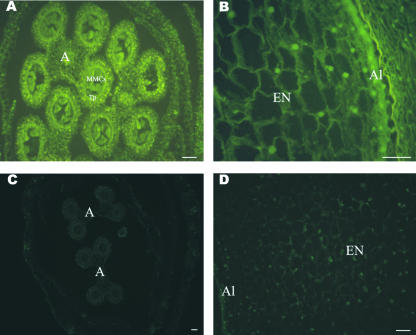
Detection of ZmRpd3 proteins was performed using the anti-ZmRpd3 antibody, which, as mentioned above, recognizes different ZmRpd3-related polypeptides and the anti-ZmRpd3/108 antibody specific for this member of the ZmRpd3 protein family. Incubation of kernel sections, obtained from seeds harvested at 10 DAP (endoreduplication phase) and 16 DAP (starch accumulation phase), with anti-ZmRpd3 antibody revealed a localization of ZmRpd3 proteins in all the cellular domains of the kernel. In addition, it was evident that the antibody reacted both in cytoplasm and nucleus (data not shown). Specific localization of ZmRpd3/108 protein was assessed in anther sections during microsporogenesis, in endosperm at 10 DAP, and in 8-d-old shoot apexes (Fig. 6; data not shown). In anthers and endosperm, the antibody reacted both in cell cytoplasm and nucleus (Fig. 6, A and B). A higher signal was observed in tapetum and MMCs of anthers (Fig. 6A) and in the aleurone of the endosperm (Fig. 6B). In these cases, the signal was particularly high in the cell cytoplasm. In shoot apexes, the signal was detected mainly in cell cytoplasm with a less evident staining of the nucleus (data not shown).
Figure 6.
Localization of ZmRpd3/108 protein in maize tissues. A and B, Sections were incubated with anti-ZmRpd3/108 antibody. A, Cross section through a maize floret at a premeiotic stage during microsporogenesis. A, Anther; Tp, tapetum. B, Longitudinal section through maize endosperm at 10 DAP. EN, Endosperm; Al, aleurone. C and D, Sections were not treated with the secondary antibody. C, Cross section through a maize floret at a premeiotic stage. D, Longitudinal section through maize endosperm at 10 DAP. Bars = 100 μm.
Analysis of Interactions of Different ZmRpd3 Proteins with ZmRBR1 and ZmRbAp1
The binding of ZmRpd3/101 with the maize retinoblastoma-related (ZmRBR1) and maize retinoblastoma-associated (ZmRbAp1) proteins has been reported previously (Rossi et al., 2003). To verify whether different members of maize Rpd3-type HDACs possess different capabilities or different mechanisms of interaction with regulatory protein partners, we tested whether ZmRpd3/102 and ZmRpd3/108 were able to associate to ZmRBR1 and ZmRbAp1.
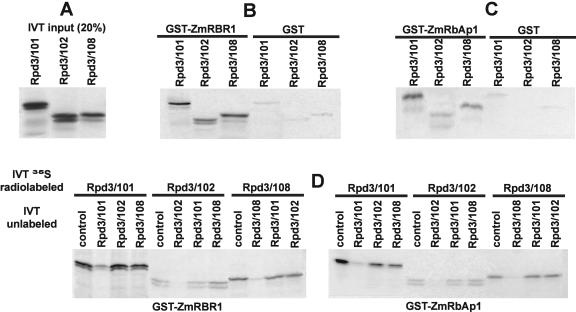
GST pull-down experiments were carried out using recombinant GST-ZmRBR1 and GST-ZmRbAp1 proteins (Rossi et al., 2003) and IVT ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 proteins. To allow a comparison of binding abilities, equimolar amounts of IVT ZmRpd3 products were used in the experiments (Fig. 7A). The results showed that the three ZmRpd3 proteins were retained by GST-ZmRBR1 (Fig. 7B). IVT ZmRpd3 proteins were also able to interact with recombinant GST-ZmRbAp1 (Fig. 7C). The different amount of IVT products retained by GST-ZmRbAp1-coated beads might suggest differences in the competence of each ZmRpd3 protein to bind ZmRbAp1; however, further investigations are needed to verify whether this occurs during in vivo formation of the ZmRpd3s/ZmRbAp1 complex.
Figure 7.
Analysis of protein interactions among ZmRpd3s, ZmRBR1, and ZmRbAp1. A, Autoradiography of an SDS polyacrylamide gel loaded with 10% of the IVT ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 proteins used as input in GST pull-downs; equimolar amounts of the IVT products were used in the experiments. In vitro GST pull-down assays were performed using beads carrying recombinant GST-ZmRBR1 (B) or GST-ZmRbAp1 (C) incubated with IVT products of the different ZmRpd3 proteins. Beads with GST alone were used as negative control. D, GST pull-down assays were carried out as in B and C, but beads carrying either GST-ZmRBR1 (left panel) or GST-ZmRbAp1 (right panel) were first incubated with unlabeled IVT ZmRpd3 proteins and then IVT 35S radiolabeled products (35S IVT) were added (5:1 ratio unlabeled: radiolabeled) to the reaction mix for further incubation. All possible combinations of unlabeled and radiolabeled ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 were tested. Control indicates that only radiolabeled IVT and no unlabeled products were added to the reaction mix.
To test whether the ZmRpd3 proteins mutually compete in binding ZmRBR1 and ZmRbAp1, radiolabeled IVT ZmRpd3 proteins were incubated with glutathione Sepharose beads, carrying GST-fused recombinant ZmRBR1 or ZmRbAp1 proteins, in the presence of the unlabeled ZmRpd3 products. The results showed that each unlabeled ZmRpd3 protein competed with its radiolabeled counterpart, whereas addition of an unlabeled ZmRpd3 protein, different from the radiolabeled one, did not affect the binding (Fig. 7D). These experiments indicate that the three ZmRpd3 proteins are able to interact with both ZmRBR1 and ZmRbAp1; however, because they do not compete with each other in the interaction, different regions of ZmRBR1 and ZmRbAp1 may be involved in the binding with each ZmRpd3 protein.
DISCUSSION
In this study, a detailed analysis was performed of the expression and cellular localization profiles of three Rpd3-type HDACs from maize. Our results indicate that the three ZmRpd3 genes identified so far and termed ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 are ubiquitously expressed in all maize organs analyzed. In addition, their products localize in all cellular domains during different stages of kernel, shoot, and anther development. This is consistent with a general requirement of ZmRpd3-type HDACs and suggests their involvement in various regulatory networks, acting in distinct physiological processes. In this context, an overall activity of Rpd3 has recently emerged through transcriptional profiling of the yeast Rpd3 mutant and by an extensive analysis of Rpd3 binding and deacetylation activities throughout the complete genome (Bernstein et al., 2000; Kurdistani et al., 2002; Robyr et al., 2002). Among the several classes of genes regulated by Rpd3 activity, it was shown that Rpd3 function is primarily related to cell cycle and anabolic genes. In accordance with a role of ZmRpd3 genes in the control of the cell cycle progression, we report that the three ZmRpd3 proteins are able to interact with the maize retinoblastoma-related protein ZmRBR1, a key regulator of the G1/S transition (Rossi and Varotto, 2002), and with the maize retinoblastoma-associated protein ZmRbAp1, a histone-binding protein believed to be involved in nucleosome assembly (Rossi et al., 2001). In addition, Lechner et al. (2000) proposed a possible direct link between ZmRpd3 activities and DNA replication. In the present study, although high amounts of ZmRpd3 RNAs appear to be correlated with expression of the DNA replication-associated marker histone H4, we also observe marked differences in their localization profiles. In fact, the histone H4 RNA specifically localized in regions containing active proliferating cells, whereas ZmRpd3 is widespread in all cellular domains. This observation indicates that the possible involvement in DNA replication is just one of the multiple aspects of ZmRpd3 functions. The expression profile of ZmRpd3 genes suggests a more general association with intense metabolic activity. ZmRpd3 transcripts and proteins accumulate during the initial stages of kernel, ear, and tassel development, which are characterized by elevated metabolic activity, cell division, and DNA replication rate (Cheng et al., 1983; Irish and Nelson, 1991; Slocombe et al., 1999). Furthermore, ZmRpd3 RNAs and proteins exhibit a gradient of abundance in endosperm domains, because they are more localized in the inner region than in the transfer cell domain (Rossi et al., 2003) and accumulate more in tapetal cells and tetrads than in other tissues during anther maturation. Hence, the level of ZmRpd3 expression is high in cellular domains that exhibit elevated metabolic activities to provide nutrients for either embryo or pollen development (Piffanelli and Murphy, 1998; Slocombe et al., 1999).
The analysis of the expression and localization patterns of ZmRpd3 transcripts and proteins reported in this study indicates possible mechanisms regulating the expression and accumulation of these enzymes. The finding that the amount of ZmRpd3 mRNAs changes during the development of various maize organs suggests a control of their transcription rate. Nevertheless, the accumulation of ZmRpd3 mRNAs during endosperm development is delayed with respect to the corresponding proteins, suggesting mechanisms regulating protein abundance at the posttranscriptional and posttranslational levels. In this context, it has been proposed that posttranslational modifications of animal HDACs such as phosphorylation and sumoylation, besides their function in modulating HDAC activity and complex formation, might act as signals to increase or decrease protein stability and the degree of protein degradation (Pflum et al., 2001; David et al., 2002). Immunolocalization experiments suggest further mechanisms of regulation of ZmRpd3 protein abundance by cellular compartmentalization. In accordance with previous observations on the subcellular localization of maize HDAC activities (Grabher et al., 1994), our results indicate that the ZmRpd3/108 enzyme is localized in both nucleus and cytoplasm of endosperm, anther, and SAM cells. The cytoplasmic localization of ZmRpd3 proteins may be related to their requirement for deacetylation of newly synthesized acetylated histones, which need specific steps of posttranslational modifications to be properly assembled into chromatin (Mello and Almouzni, 2001). However, cytoplasmic localization of Rpd3-type class I HDACs might also be due to nucleus-cytoplasmic shuttling of these enzymes: a mechanism believed to be crucial both for the control of HDAC activity in the nucleus and for ensuring HDAC activity on cytoplasmic nonhistone proteins (for review, see Khochbin et al., 2001). Taken together these results indicate that diverse mechanisms are involved in the control of ZmRpd3 abundance during plant development. Interestingly, it has been recently reported that down-regulation of the maize Gcn5-type HAT determined histone hypoacetylation and a concomitant reduction of ZmRpd3/101 and ZmRpd3/108 expression, whereas treatment of maize cells with the HDAC inhibitor trichostatin A provoked hyperacetylation and decreased the level of ZmGcn5 transcription (Bhat et al., 2003). In accordance with the scenario proposed by these authors, it may be therefore speculated that a number of distinct mechanisms operate coordinately to regulate the transcription rate, the protein abundance, and the activity of HATs and HDACs. These mechanisms may act in response to alteration of the global level of histone acetylation within the cell with the aim to balance these changes (hyperacetylation or hypoacetylation) by directly regulating abundance and activity of HATs and HDACs.
Our results obtained from the analysis of ZmRpd3 expression patterns and protein interactions provide at least a partial answer to whether functional redundancy rather than distinct activity occurs among different members of the ZmRpd3 gene family. Functional redundancy of these enzymes is indicated by the observations that the three ZmRpd3 genes display an overlapping expression and localization pattern as well as that they are able to interact with ZmRBR1 and ZmRbAp1. Functional redundancy is not surprising in maize, an allotetraploid species that evolutionarily originated by genomic duplication (Gaut and Doebley, 1997), and can be explained by considering that the maintenance of the histone acetylation levels could be so critical to cell growth that multiple, functionally redundant HDACs are required. However, this is not the case for yeast or mammals, where different members of the Rpd3 family exhibit only a small degree of functional overlap (Bernstein et al., 2000; Lagger et al., 2002; Robyr et al., 2002). On the other hand, the present study also provides lines of evidence supporting functional differences among ZmRpd3 enzymes. The expression pattern of the three ZmRpd3 transcripts is not completely identical because differences in the level of each ZmRpd3 RNA in different organs and during organ development have been observed. In addition, sequence comparison and phylogenetic analysis indicate that the three ZmRpd3 proteins are included in three separate groups; each of these clusters may represent different orthologs, with possible functional diversification. Furthermore, N- and C-terminal regions of ZmRpd3 proteins display a low degree of sequence homology and appear to be specific to each ZmRpd3 protein. Interestingly, it was shown that these regions are implicated in the interaction of ZmRpd3/101 with ZmRBR1 and ZmRbAp1 (Fig. 1A; Rossi et al., 2003) and might be related to specific interaction capabilities/mechanisms among different members of the ZmRpd3 family. Finally, competition assays showed that ZmRpd3 proteins do not compete with each other in binding both ZmRBR1 and ZmRbAp1, suggesting that different regions of ZmRBR1 and ZmRbAp1 may be involved in the interaction with distinct ZmRpd3 proteins. Similar to mammals (for review, see Cress and Seto, 2000), each ZmRpd3 may concomitantly participate in forming multiprotein complexes that require distinct HDAC enzymatic properties to execute its regulatory function. Unfortunately, attempts to test whether the three ZmRpd3 proteins possess different enzymatic properties have so far been inconclusive because, as previously reported (for review, see de Ruijter et al., 2003), production of recombinant proteins of this class of HDACs usually originates inactive forms. The analysis of maize-specific mutants, none of which has so far been described, is an unavoidable goal to elucidate the functions fulfilled by each member of ZmRpd3 gene family.
MATERIALS AND METHODS
Plant Material
All preparations described in this paper were obtained from greenhouse- and field-grown seedlings and kernels of the maize (Zea mays) B73 inbred line. Identification of the developmental stages of maize organs was made according to indications from the Iowa State University of Science and Technology (1989), dividing plant development into vegetative (V) and reproductive (R) stages and in which each V stage is designated numerically to indicate the number of leaves. The R1 stage represents silking, whereas R1 8 h indicates ears harvested 8 h after pollination. RNA and protein were extracted from all parts of the kernel in seeds harvested at 3, 5, and 8 DAP, whereas endosperm tissue was obtained through microdissection of seeds harvested at 10, 12, 18, 20, and 28 DAP. Similarly, ears and tassels were obtained through microdissection of the reproductive apexes at the indicated developmental stages.
Isolation of ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 cDNAs
Total RNA was extracted from seedlings of the B73 maize inbred line and first-strand cDNA was synthesized using a 17-bp oligo(dT) primer and Superscript reverse transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Subsequently, the full-length ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 cDNAs were obtained by PCR amplification. Specifically, a 1,857-bp ZmRpd3/101 fragment, a 1,424-bp ZmRpd3/102 fragment, and a 1,551-bp ZmRpd3/108 fragment were isolated using the following primer combinations: HDA101-5 (5′-GGCGATGGACCCGTCATCG-3′) and HDA101-3 (5′-GACTGTACAACTGAGATCATGGG-3′) located at nucleotide positions –4 and 1,853 with respect to the ATG starting codon of ZmRpd3/101; HDA102-5 (5′-GTGAAGCGATGCTGGAGAAAGACCG-3′) and HDA102-3 (5′-TGACCTGCTAAATTAGATCCCTGCTG-3′) located at nucleotide positions –8 and 1,416 with respect to the ATG starting codon of ZmRpd3/102; and HDA108-5 (5′-ATGGCGGCTTCTGGTGAGG-3′) and HDA108-3 (5′-TCTAGTCTAGTTACCAACACGCAGG-3′) located at nucleotide positions 1 and 1,551 with respect to the ATG starting codon of ZmRpd3/108. All identified cDNAs were sequenced on both strands to avoid ambiguity.
Phylogenetic Analysis
Rpd3-type HDACs from different eukaryotes were identified through a BLAST search of the public GenBank database (http://www.ncbi.nlm.nih.gov) using maize ZmRpd3/101 as a query sequence. Twenty-seven of these sequences, plus the three maize sequences described in this work, were selected according to the indication of Pandey et al. (2002). The catalytic HDAC domains within these amino acid sequences were identified by screening the Protein Family (Pfam) database (http://pfam.wustl.edu/). Complete protein sequences, HDAC domains, and N- plus C-terminal regions were aligned using ClustalW software (Thompson et al., 1994) and edited with the Genedoc program (http://www.psc.edu/biomed/genedoc/). Unrooted phylogenetic trees were constructed by the distance method using the neighbor-joining algorithm implemented in the Phylip software package (Felsenstein, 1989) followed by bootstrap analysis to assess the branch patterns.
RNA Preparation and Northern-Blot Analysis
Total RNA was extracted with Trizol reagent (Invitrogen). For northern analysis, total RNA (20 μg) was subjected to electrophoresis through 1.2% (w/v) agarose/1.1% (v/v) formaldehyde gels, blotted onto Hybond-N membrane (Amersham Biosciences, Uppsala) and hybridized with 32P-radiolabeled DNA probes. Probes specific for ZmRpd3/101, 102, and 108 transcripts were identified in the 3′ ends of the cDNA sequences, which exhibit only a very low degree of homology (Fig. 1A). Probes were obtained by PCR amplification of ZmRpd3/101, 102, and 108 clones. Specifically, a 530-bp ZmRpd3/101 fragment (bp 1,290–1,820 from ATG) was obtained using primers HDA101/2-5 (5′-AGGCTGTGGAAGAGTCATC-3′) and HDA101-3 (5′-GACTGTACAACTGAGATCATGGG-3′); a 511-bp ZmRpd3/102 fragment (bp 904–1,415 from ATG) was obtained using primers HDA102/2-5 (5′-AGGAGAATGTAGCACGGTGTTGG-3′) and HDA102-3 (5′-TGACCTGCTAAATTAGATCCCTGCTG-3′); and a 598-bp ZmRpd3/108 fragment (bp 953–1,551 from ATG) was obtained using primers HDA108/2-5 (5′-TCAGAAATGTTGCACGCTGCTGG-3′) and HDA108-3 (5′-TCTAGTCTAGTTACCAACACGCAGG-3′). The DNA fragments were cloned into pBluescript SK (Stratagene, La Jolla, CA) and tested to verify that no cross-hybridization occurred among the three probes. A 240-bp fragment, corresponding to the cDNA sequence of the maize histone H4 (bp 78–318 from ATG; GenBank accession no. BE512572) was also used as a probe and was obtained by PCR amplification using the primers H4-5 (5′-CATCCAGGGCATCACGAAGCC-3′) and H4-3 (5′-CGAAGCCGTAGAGCGTGCG-3′). Hybridization and washes were carried out as previously described (Rossi et al., 1998).
Antibody Production, Protein Extraction, and Western-Blot Analysis
For anti-ZmRpd3/108 antibody production, the 3′ end of the ZmRpd3/108 cDNA (cZmRpd3/108, covering amino acids 359–458) was PCR amplified using primers UnVor1 (5′-CGAGCTCTCCAACCAAAAAGTGTTG-3′) and UnRev2 (5′-CAAGCTTCTACAGATCATCTTTTGG-3′) and cloned into pQE-32 vector (Qiagen USA, Valencia, CA) after restriction with SacI and HindIII. Overexpression of ZmRpd3/108 was carried out in Escherichia coli strain M15 [pREP4] (Qiagen USA), and the recombinant protein with the 6×-His-Tag fused to the N-terminal part was purified by nickelnitrilotriacetic acid agarose affinity chromatography (Qiagen USA) under denaturing conditions (6 m urea) according to the manufacturer's instructions. The purified peptide was used to generate rabbit polyclonal antiserum against the C-terminal part of ZmRpd3/108. IgGs were purified from serum by affinity chromatography using Protein G Sepharose chromatography (Amersham Biosciences). Production of the anti ZmRpd3 antibody (against the entire ZmRpd3/101 polypeptide) has previously been described (Rossi et al., 2003).
Crude protein extracts from different maize organs were prepared as previously described (Rossi et al., 2001). Protein quality and concentration were determined by Coomassie staining of SDS-polyacrylamide gels and using Bradford assays. For immunoblotting, 100 μg of the extracts was separated by SDS-PAGE, blotted onto Immobilon-P membranes (Millipore, Bedford, MA), and incubated with the indicated antibody. ZmRpd3 antibody was used more concentrated than in previous experiments (1:500 versus 1:1,500 [v/v]; Rossi et al., 2003) to allow detection of additional immunoreactive bands; a 1:2,500 (v/v) dilution of the ZmRpd3/108 antibody was used for western-blot analysis except where otherwise stated. Horseradish peroxidase-conjugated goat anti-rabbit IgG (Sigma-Aldrich, St. Louis) was used as a secondary antibody; the ECL system (Amersham Biosciences) was used for detection.
In Situ Hybridization and Immunolocalization
Preparation of sections from various plant materials (kernels at 5, 8, 12, 14, and 16 DAP; apices at 8 and 15 d after seed germination; anthers from the MMCs to uninucleate microspore stage of development) and in situ hybridization experiments were carried out as previously described (Rossi et al., 2001). To obtain DIG-UTP (Roche Diagnostics, Mannheim, Germany)-labeled antisense RNA probes, the same histone H4 and ZmRpd3-specific DNA fragments employed in northern-blot analysis (see above) were used as template for in vitro transcription. The corresponding sense transcripts were used as negative controls. Sections were incubated with antisense or sense probes in 50% (v/v) formamide at 45°C to 48°C overnight. After hybridization, the slides were extensively washed in 2× SSC at 45°C to 48°C and treated with 20 μg mL–1 RNaseA (Roche Diagnostics). DIG detection and signal visualization were done using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate (Roche Diagnostics) following the manufacturer's instructions.
Preparation of sections from the same plant materials and immunolocalization experiments were performed as previously described (Rossi et al., 2001). Anti-ZmRpd3 and anti-ZmRpd3/108 were used as primary antibodies at a 1:250 (v/v) dilution. Slides were incubated for 2 h at room temperature with the primary antibody and for 1 h at room temperature with an anti-rabbit secondary antibody. Secondary antibodies conjugated with both alkaline phosphatase (A2556, Sigma-Aldrich) and fluorescein isothiocyanate were used. Images were acquired using a DC 300F camera (Leica, Wetzlar, Germany).
GST Pull-Down Assays
Production of GST-fused ZmRBR1 (ZmRBR1 A/B pocket plus C-terminal domain) and ZmRbAp1 (full-length protein) was described previously (Rossi et al., 2003). For in vitro transcription/translation, the entire coding sequences of ZmRpd3/101, ZmRpd3/102, and ZmRpd3/108 were introduced into pGEM 3Zf(+) (Promega, Madison, WI) under T7 promoter regulation. In vitro GST pull-down experiments were performed as previously described (Rossi et al., 2003). The amounts of GST-fusion proteins were quantified by Bradford assays and Coomassie staining of SDS-polyacrylamide gels. Equimolar amounts of beads coated with GST domain were used as a negative control in the pull-downs. IVT proteins were quantified by autoradiography of SDS-polyacrylamide gels, and equimolar amounts were used in pull-down assays. For competition experiments, glutathione Sepharose beads (Amersham Biosciences) coated with GST-ZmRBR1 were incubated for 2 h at 4°C with unlabeled IVT proteins in NETN buffer (20 mm Tris-HCl, pH 8, 100 mm NaCl, 1 mm EDTA, and 0.5% [v/v] NP-40), washed twice with the same buffer, and then incubated with [35S]Met-radiolabeled IVT products in NETN buffer for 4 h at 4°C. For experiments with GST-ZmRbAp1, the incubation and washes were in Z′ buffer (25 mm HEPES-NaOH, pH 7.5, 12.5 mm MgCl2, 150 mm KCl, 20% [v/v] glycerol, and 0.1% [v/v] NP-40). A 5:1 ratio between unlabeled and radiolabeled IVT proteins was used.
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes.
Acknowledgments
We are grateful to Hans Hartings for help in the sequence and phylogenetic analysis and for critical reading of the manuscript. We thank Massi-miliano Lauria, Alexandra Lusser, and Monica Sturaro for critical reading of the manuscript and Mayra Bernardi for help in manuscript preparation.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.025403.
This work was supported by the Framework Program V of the European Community (Zeastar project no. QLRT–2000–00020), by Ministero delle Politiche Agricole (Roma, Italy), and by the Austrian Science Foundation (project no. P14528).
References
- Aufsatz W, Mette MF, Van Der Winden J, Matzke M, Matzke AJ (2002) HDA6, a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J 21: 6832–6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Tong JK, Schreiber SL (2000) Genome-wide studies of histone deacetylase function in yeast. Proc Natl Acad Sci USA 97: 13708–13713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RA, Riehl M, Santandrea G, Velasco R, Slocombe S, Donn G, Steinbiss HH, Thompson RD, Becker HA (2003) Alteration of GCN5 levels in maize reveals dynamic responses to manipulating histone acetylation. Plant J 33: 455–469 [DOI] [PubMed] [Google Scholar]
- Cheng PC, Greyson RI, Walden DB (1983) Organ initiation and the development of unisexual flowers in the tassel and ear of Zea mays. Am J Bot 70: 450–462 [Google Scholar]
- Cress WD, Seto E (2000) Histone deacetylases, transcriptional control, and cancer. J Cell Physiol 184: 1–16 [DOI] [PubMed] [Google Scholar]
- Dangl M, Brosch G, Haas H, Loidl P, Lusser A (2001) Comparative analysis of HD2 type histone deacetylases in higher plants. Planta 213: 280–285 [DOI] [PubMed] [Google Scholar]
- David G, Neptune MA, DePinho RA (2002) SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J Biol Chem 277: 23658–23663 [DOI] [PubMed] [Google Scholar]
- de Ruijter AJ, Van Gennip AH, Caron HN, Kemp S, Van Kuilenburg AB (2003) Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 370: 737–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP: phylogeny inference package version. Cladistics 5: 164–166 [Google Scholar]
- Gaut BS, Doebley JF (1997) DNA sequence evidence for the segmental allotetraploid origin of maize. Proc Natl Acad Sci USA 94: 6809–6814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich J, Tweedie S (2002) Remembrance of things past: chromatin remodeling in plant development. Annu Rev Cell Dev Biol 18: 707–746 [DOI] [PubMed] [Google Scholar]
- Grabher A, Brosch G, Sendra R, Lechner T, Eberharter A, Georgieva EI, Lopez-Rodas G, Franco L, Dietrich H, Loidl P (1994) Subcellular location of enzymes involved in core histone acetylation. Biochemistry 33: 14887–14895 [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403: 795–800 [DOI] [PubMed] [Google Scholar]
- Iowa State University of Science and Technology (1989) How a Corn Plant Develops. Special Report Number 48. Cooperative Extension Service, Ames, IA
- Irish EE, Nelson TM (1991) Identification of multiple stages in the conversion of maize meristems from vegetative to floral development. Development 112: 891–898 [Google Scholar]
- Jang IC, Pahk YM, Song SI, Kwon HJ, Nahm BH, Kim JK (2003) Structure and expression of the rice class-I type histone deacetylase genes OsHDAC1-3: OsHDAC1 overexpression in transgenic plants leads to increased growth rate and altered architecture. Plant J 33: 531–541 [DOI] [PubMed] [Google Scholar]
- Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D (2001) Functional significance of histone deacetylase diversity. Curr Opin Genet Dev 11: 162–166 [DOI] [PubMed] [Google Scholar]
- Kurdistani SK, Robyr D, Tavazoie S, Grunstein M (2002) Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet 31: 248–254 [DOI] [PubMed] [Google Scholar]
- Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T et al. (2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J 21: 2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner T, Lusser A, Pipal A, Brosch G, Loidl A, Goralik-Schramel M, Sendra R, Wegener S, Walton JD, Loidl P (2000) RPD3-type histone deacetylase in maize embryos. Biochemistry 39: 1683–1692 [DOI] [PubMed] [Google Scholar]
- Lusser A (2002) Acetylated, methylated, remodeled: chromatin states for gene regulation. Curr Opin Plant Biol 5: 437–443 [DOI] [PubMed] [Google Scholar]
- Lusser A, Brosch G, Loidl A, Haas H, Loidl P (1997) Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science 277: 88–91 [DOI] [PubMed] [Google Scholar]
- Lusser A, Kolle D, Loidl P (2001) Histone acetylation: lesson from the plant kingdom. Trends Plant Sci 6: 59–65 [DOI] [PubMed] [Google Scholar]
- McSteen P, Laudencia-Chingcuanco D, Colasanti J (2000) A floret by any other name: control of meristem identity in maize. Trends Plant Sci 5: 61–66 [DOI] [PubMed] [Google Scholar]
- Mello JA, Almouzni G (2001) The ins and outs of nucleosome assembly. Curr Opin Genet Dev 11: 136–141 [DOI] [PubMed] [Google Scholar]
- Meshi T, Taoka KI, Iwabuchi M (2000) Regulation of histone gene expression during the cell cycle. Plant Mol Biol 43: 643–657 [DOI] [PubMed] [Google Scholar]
- Murfett J, Wang XJ, Hagen G, Guilfoyle TJ (2001) Identification of Arabidopsis histone deacetylase HDA6 mutants that affect transgene expression. Plant Cell 13: 1047–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Slinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA (2002) Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res 30: 5036–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piffanelli P, Murphy DJ (1998) Novel organelles and targeting mechanisms in the anther tapetum. Trends Plant Sci 3: 250–253 [Google Scholar]
- Pikaard CS (1999) Nucleolar dominance and silencing of transcription. Trends Plant Sci 4: 478–483 [DOI] [PubMed] [Google Scholar]
- Pflum MK, Tong JK, Lane WS, Schreiber SL (2001) Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J Biol Chem 276: 47733–47741 [DOI] [PubMed] [Google Scholar]
- Robyr D, Suka Y, Xenarios I, Kurdistani SK, Wang A, Suka N, Grunstein M (2002) Microarray deacetylation maps determine genome-wide functions for yeast histone deacetylases. Cell 109: 437–446 [DOI] [PubMed] [Google Scholar]
- Rossi V, Hartings H, Motto M (1998) Identification and characterisation of an RPD3 homologue from maize (Zea mays L.) that is able to complement an rpd3 null mutant of Saccharomyces cerevisiae. Mol Gen Genet 258: 288–296 [DOI] [PubMed] [Google Scholar]
- Rossi V, Locatelli S, Lanzanova C, Boniotti B, Varotto S, Pipal A, Goralik-Schramel M, Lusser A, Gatz C, Gutierrez C et al. (2003) A maize histone deacetylase and retinoblastoma-related protein physically interact and cooperate in repressing gene transcription. Plant Mol Biol 51: 401–413 [DOI] [PubMed] [Google Scholar]
- Rossi V, Varotto S (2002) Insights into the G1/S transition in plants. Planta 215: 345–356 [DOI] [PubMed] [Google Scholar]
- Rossi V, Varotto S, Locatelli S, Lanzanova C, Lauria M, Zanotti E, Hartings H, Motto M (2001) The maize WD-repeat gene ZmRbAp1 encodes a member of the MSI/RbAp sub-family and is differentially expressed during endosperm development. Mol Genet Genom 265: 576–584 [DOI] [PubMed] [Google Scholar]
- Roth SY, Denu JM, Allis CD (2001) Histone acetyltransferases. Annu Rev Biochem 70: 81–120 [DOI] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M (1996) HDA1 and RPD3 are members of distinct histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci USA 93: 14503–14508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber SL, Bernstein BE (2002) Signaling network model of chromatin. Cell 111: 771–778 [DOI] [PubMed] [Google Scholar]
- Slocombe S, Maitz M, Hueros G, Becker HA, Yan G, Mueller M, Varotto S, Santandrea G, Thompson RD (1999) Genetic control of endosperm development. In VEA Russo, DJ Cove, LG Edgar, R Jaenisch, F Salamini, eds, Development Genetics, Epigenetics and Environmental Regulation. Springer-Verlag, New York, pp 185–197
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Struhl K (1999) Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell 98: 1–4 [DOI] [PubMed] [Google Scholar]
- Taunton J, Hassing CA, Schreiber SL (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272: 408–411 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Chen ZJ (2001) Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proc Natl Acad Sci USA 98: 200–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM (2002) Cellular memory and the histone code. Cell 111: 285–291 [DOI] [PubMed] [Google Scholar]
- Wu J, Grunstein M (2000) 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci 25: 619–623 [DOI] [PubMed] [Google Scholar]
- Wu K, Mali K, Tian L, Brow D, Miki B (2000) Functional analysis of a RPD3 histone deacetylase homologue in Arabidopsis thaliana. Plant Mol Biol 44: 167–176 [DOI] [PubMed] [Google Scholar]