Abstract
An iron-superoxide dismutase (FeSOD) with an unusual subcellular localization, VuFeSOD, has been purified from cowpea (Vigna unguiculata) nodules and leaves. The enzyme has two identical subunits of 27 kD that are not covalently bound. Comparison of its N-terminal sequence (NVAGINLL) with the cDNA-derived amino acid sequence showed that VuFeSOD is synthesized as a precursor with seven additional amino acids. The mature protein was overexpressed in Escherichia coli, and the recombinant enzyme was used to generate a polyclonal monospecific antibody. Phylogenetic and immunological data demonstrate that there are at least two types of FeSODs in plants. An enzyme homologous to VuFeSOD is present in soybean (Glycine max) and common bean (Phaseolus vulgaris) nodules but not in alfalfa (Medicago sativa) and pea (Pisum sativum) nodules. The latter two species also contain FeSODs in the leaves and nodules, but the enzymes are presumably localized to the chloroplasts and plastids. In contrast, immunoblots of the soluble nodule fraction and immunoelectron microscopy of cryo-processed nodule sections demonstrate that VuFeSOD is localized to the cytosol. Immunoblot analysis showed that the content of VuFeSOD protein increases in senescent nodules with active leghemoglobin degradation, suggesting a direct or indirect (free radical-mediated) role of the released Fe in enzyme induction. Therefore, contrary to the widely held view, FeSODs in plants are not restricted to the chloroplasts and may become an important defensive mechanism against the oxidative stress associated with senescence.
Superoxide dismutases (SODs; EC 1.15.1.1) are ubiquitous metalloenzymes that catalyze the dismutation of superoxide radicals and thus prevent oxidative damage in all organisms. Three classes of SODs, differing in the metals at their catalytic active site, are known in plants. The CuZnSODs are localized in the cytosol, chloroplasts, nucleus, and apoplast; the MnSODs in the mitochondria and peroxisomes; and the FeSODs in the chloroplasts (Sandalio et al., 1987; Van Camp et al., 1990; Bowler et al., 1994; Ogawa et al., 1996). There have been contradictory reports on the distribution of FeSODs in higher plants. Once thought to be confined to a few families of vascular plants (Bridges and Salin, 1981), it is now accepted that the FeSOD (sodB) genes are amply distributed in plants but only are expressed under certain, yet poorly defined, conditions (Van Camp et al., 1990; Bowler et al., 1994).
In an earlier study (Becana et al., 1989), we found FeSOD in nodules of cowpea (Vigna unguiculata) and, more recently, in nodules of other legumes (Rubio et al., 2001). The deduced sequence of the FeSOD of alfalfa (Medicago sativa) nodules (Rubio et al., 2001) contains a typical plastid signal peptide, in agreement with other plant FeSODs. The deduced sequence of the FeSOD from cultured soybean (Glycine max) cells was also proposed to have a plastid signal peptide (Crowell and Amasino, 1991), but a closer analysis of the N-terminal sequence shows this not to be the case (this work). In this study, we have investigated in detail the FeSOD of cowpea nodules (VuFeSOD). We have cloned the VuFeSOD cDNA in Escherichia coli and have used the recombinant enzyme to raise a monospecific polyclonal antibody. The mature enzyme was characterized and localized in the nodule cytosol. This most unusual subcellular localization, along with immunoblots of other legume nodules, indicates that there are at least two well-defined groups of FeSODs in plants. Expression analysis of VuFeSOD suggests an antioxidative role of the enzyme during senescence of cowpea nodules.
RESULTS
Characterization of VuFeSOD and Its Derived Protein
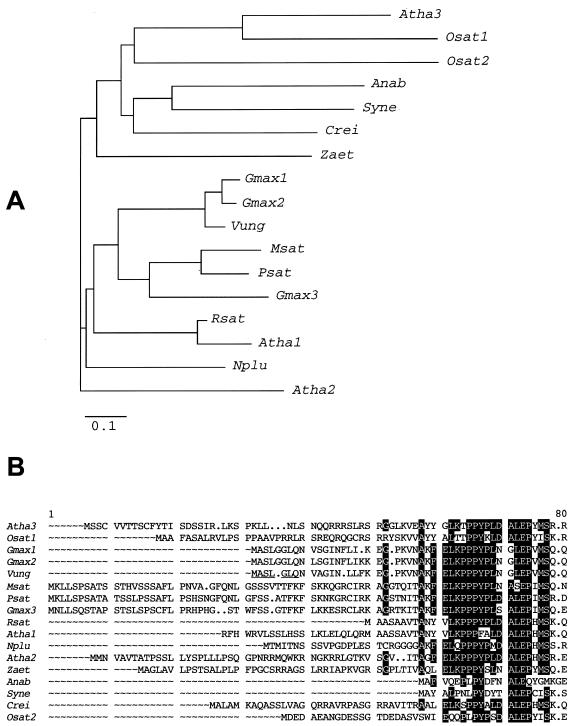
Primers were designed against conserved sequences of FeSODs from Chlamydomonas reinhardtii and higher plants and were used to isolate a clone, VuFeSOD, from a cowpea nodule cDNA library. The same primers enabled us to isolate cDNA clones encoding FeSODs from pea (Pisum sativum; accession no. AJ496175) and soybean (accession no. AF108084) nodules. The VuFeSOD sequence was 1,043 bp long and contained an open reading frame of 738 bp, which encodes a protein of 245 amino acids, with a molecular mass of 27,411 D and a pI of 5.31. The predicted protein contains the residues thought to be essential for FeSOD activity (Tyr-58, Trp-100, and Asn-185) and metal binding (His-54, His-102, Asp-202, and His-206), as well as the residues (Ala-97, Gln-98, Trp-100, and Ala-186) proposed as primary candidates to distinguish FeSODs from MnSODs (Van Camp et al., 1990; Bowler et al., 1994). The amino acid sequence of VuFeSOD was highly homologous (94% identity) with the soybean FeSOD (isozymes 1 and 2) but had lower homology (72%–81% identity) with soybean FeSOD (isozyme 3) and with the FeSODs of alfalfa (Rubio et al., 2001) and pea nodules (Fig. 1A).
Figure 1.
A, Unrooted phylogenetic tree of FeSOD proteins from cyanobacteria, green algae, and higher plants. The tree was constructed with the neighbor-joining method of the Clustal W suite of programs (Thompson et al., 1994). The bar represents percentages of 1,000 “bootstraps,” and the bar represents 0.1 substitution per site. Abbreviations and DNA data bank of Japan/EMBL/GenBank accession nos. for the protein sequences are: Atha3, Arabidopsis isozyme 3 (NP_197722); Osat1, rice (Oryza sativa) isozyme 1 (JG0179); Osat2, rice isozyme 2 (BAA92737); Syn, Synechocystis sp. (NP_441397); Ana, Anabaena sp. (AAD51417); Crei, Chlamydomonas reinhardtii (JC4611); Zaet, Zantedeschia aethiopica (AAC633778); Atha2, Arabidopsis isozyme 2 (NP_199923); Msat, alfalfa (AAL32441); Psat, pea partial FeSOD (AJ496175); Gmax1, soybean isozyme 1 (P28759); Gmax2, soybean isozyme 2 (AF108084); Gmax3, soybean isozyme 3 (The Institute for Genomic Research database; http://www.tigr.org/tdb/tgi/plant.shtml; TC125105); Vung, cowpea (AAF28773); Rsat, radish (Raphanus sativus; AAC15842); Atha1, Arabidopsis isozyme 1 (P21276); and Nplu, wild tobacco (Nicotiana plumbaginifolia; A39267). B, Comparison of deduced N-terminal sequences. Conserved residues on eight or more sequences are displayed as white letters on a black background. The cleavage site of the VuFeSOD precursor is underlined.
Prediction programs such as PSORT and ChloroP indicated that VuFeSOD lacks a plastid signal peptide (Fig. 1B) and suggested a cytosolic localization. To confirm this, the enzyme was purified from cowpea nodules and leaves by anion-exchange or affinity chromatography. Two independent enzyme preparations from each plant organ were N-terminally sequenced producing identical results. Comparison of the N-terminal sequence (NVAGINLL) with the derived sequence of the precursor protein revealed the presence of seven extra amino acids (MASLGLQ; Fig. 1B). The same prediction programs suggested that soybean FeSOD (isozymes 1 and 2) are cytosolic, whereas soybean FeSOD (isozyme 3) and pea nodule FeSOD have plastid signal peptides (Fig. 1B), in agreement with the homology data mentioned above.
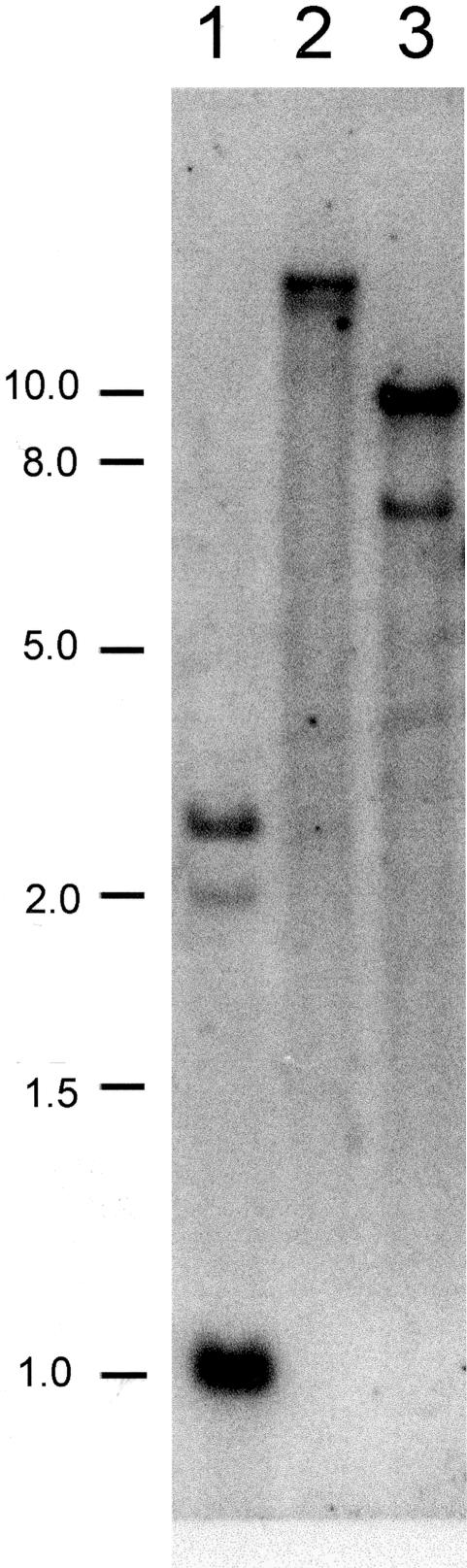
Southern-blot analysis of cowpea genomic DNA was used to estimate the number of gene copies with a probe comprising the entire open reading frame of VuFeSOD. Two hybridization bands were observed with restriction enzymes that do not cut inside the gene (EcoRI and EcoRV) and one additional band of smaller size when using an enzyme that cuts inside the gene (HindIII). Hence, two copies of the sodB gene appear to be present in the cowpea genome (Fig. 2).
Figure 2.
Southern-blot analysis of VuFeSOD. Genomic DNA (15 μg per lane) was digested with HindIII (1), EcoRV (2), or EcoRI (3). The probe was the complete open reading frame (735 bp) of VuFeSOD. DNA size markers in kilobase pairs are shown to the left.
Overproduction and Purification of Recombinant VuFeSOD
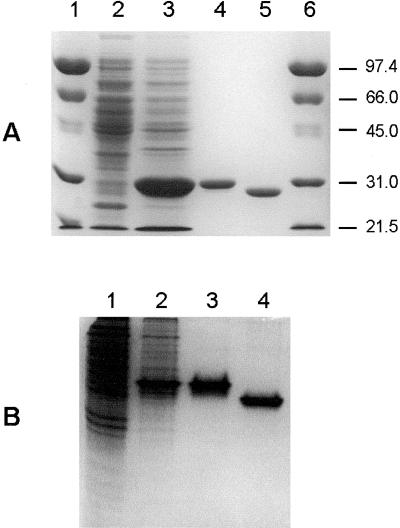
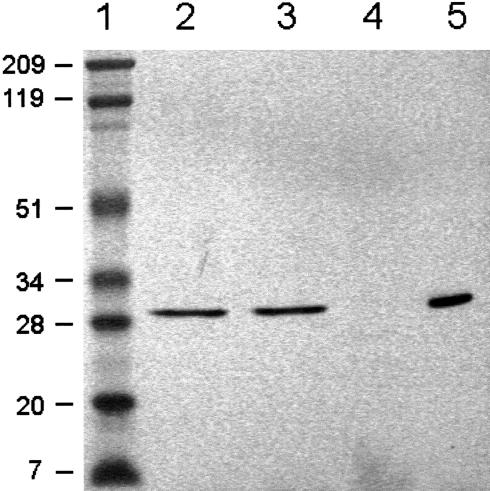
A 733-bp fragment coding for the mature protein was cloned into the NdeI and NotI sites of the expression vector pET-28a(+) and was used to transform E. coli BL21. Analysis of cell extracts by SDS-PAGE revealed the overproduction of a protein of approximately 30 kD (Fig. 3A). Recombinant VuFeSOD was purified to homogeneity, as judged by SDS-PAGE (Fig. 3A) and native-PAGE (Fig. 3B), by a single step of affinity chromatography using a HiTrap metal-chelating column, which selectively bound the (His)6-tagged protein. The molecular mass (29.7 kD) of the purified recombinant protein on SDS-PAGE agreed with the molecular mass calculated from its amino acid sequence and included 21 amino acids from the cloning vector at the N terminus. Thrombin removed 17 of those amino acids, including the (His)6 tag, as confirmed by SDS-PAGE (Fig. 3A) and native-PAGE (Fig. 3B). The affinity purification step was able to remove the FeSOD and other SODs of E. coli from the recombinant VuFeSOD preparation, as can be seen in Figure 4 (compare lanes 1 and 2).
Figure 3.
Overproduction and purification of recombinant VuFeSOD. A, SDS gel (10% [w/v] acrylamide) stained with Coomassie. Lanes 1 and 6, Prestained molecular mass markers in kilodaltons. Lane 2, Wild-type BL21 cells (60 μg of protein). Lane 3, Transformed cells 1 h after isopropylthio-β-galactoside (IPTG) induction (60 μg of protein). Lane 4, Recombinant VuFeSOD after affinity-chromatography purification (5 μg of protein). Lane 5, Same as lane 4 treated with thrombin (5 μg of protein). B, Native gel (15% [w/v] acrylamide) stained with Coomassie. Lane 1, Wild-type BL21 cells (60 μg of protein). Lane 2, Transformed cells after 1-h induction with IPTG (60 μg of protein). Lane 3, Purified recombinant VuFeSOD (5 μg of protein). Lane 4, Same as lane 3 treated with thrombin (5 μg of protein).
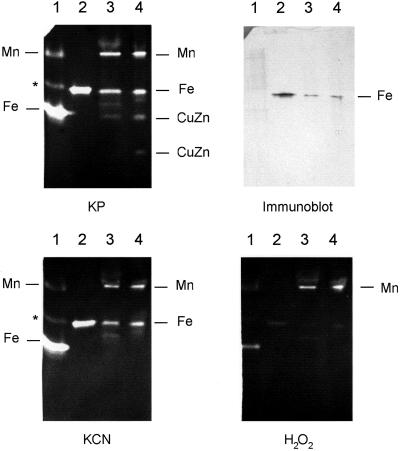
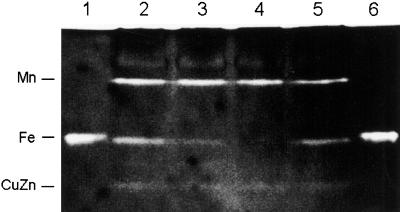
Figure 4.
Inhibitor studies of recombinant VuFeSOD and SOD isozymes of cowpea. Four native gels (15% [w/v] acrylamide) were run with identical samples. Three of them were incubated, before SOD activity staining, with potassium phosphate buffer alone, buffer plus KCN, and buffer plus H2O2, respectively. The fourth gel (without inhibitors) was used for the immunoblot. Lane 1, BL21 cells (60 μg). Lane 2, Recombinant VuFeSOD (0.5 μg) treated with thrombin. Lane 3, Cowpea nodule extract (60 μg). Lane 4, Cowpea leaf extract (60 μg). Lane 1 shows the MnSOD and FeSOD, as well as the putative hybrid Mn/FeSOD (asterisk), of E. coli (Clare et al., 1984). Lane 4 shows the MnSOD and FeSOD as well as the cytosol (upper band) and plastid (lower band) CuZnSODs of cowpea leaves.
Characterization of VuFeSOD Antibody
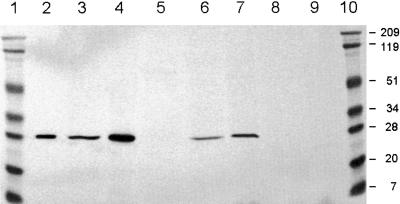
Pure recombinant VuFeSOD was used to raise a monospecific polyclonal antibody. This recognized a single protein band of approximately 27 kD, which corresponds to the subunit size of VuFeSOD, in preparations of affinity-purified recombinant VuFeSOD and in extracts of cowpea nodules and leaves (Fig. 5). Immunoblots showed that recombinant VuFeSOD treated with thrombin migrated to the same position as the enzyme from nodules or leaves (Fig. 5). The presence of FeSODs in bacteroids and nodule extracts from other legumes was also investigated using the antibody. The immunoreactive protein of approximately 27 kD was absent in cowpea bacteroids or in alfalfa and pea nodules, but was clearly detectable in soybean and common bean nodules (Fig. 5). Because alfalfa and pea nodules do contain FeSOD (Rubio et al., 2001), we conclude that the enzymes from soybean and common bean are more antigenically related to VuFeSOD than are the other FeSODs.
Figure 5.
Immunoblot analysis of recombinant VuFeSOD in legume nodule extracts. Samples were subjected to 10% (w/v) SDS-PAGE and proven with anti-VuFeSOD antibody. Lanes 1 and 10, Prestained molecular mass markers in kilodaltons. Lane 2, Purified recombinant VuFeSOD treated with thrombin. Lane 3, Cowpea nodules (60 μg). Lane 4, Cowpea leaves. Lane 5, Cowpea bacteroids. Lane 6, Soybean nodules. Lane 7, Common bean (Phaseolus vulgaris) nodules. Lane 8, Alfalfa nodules. Lane 9, Pea nodules. All lanes were loaded with 60 μg of protein, except lane 2, which was loaded with 0.2 μg of protein.
Native-PAGE in the absence (control) or presence of inhibitors (Fig. 4) showed that the SOD isozyme composition of cowpea is similar to that of other legumes (Rubio et al., 2001). Thus, nodules and leaves contain mitochondrial MnSOD (resistant to KCN and H2O2), FeSOD (resistant to KCN and inhibited by H2O2), and cytosolic CuZnSOD (inhibited by KCN and H2O2). A plastid CuZnSOD (fast-migrating isozyme) was also clearly observed in the leaves. SOD activity gels also revealed that recombinant VuFeSOD is highly active, resistant to KCN but inhibited by H2O2, and migrates at the same position as endogenous FeSOD. Immunoblots of native gels showed that the antibody specifically recognized the FeSOD isozyme (Fig. 4).
The specificity of the antibody was further tested by immunoprecipitation and subsequent analysis of SOD isozymes on native gels (Fig. 6). The FeSOD activity was greatly reduced after immunoprecipitation with 1 μg of antibody and was completely suppressed with 4 μg of antibody, whereas the MnSOD and CuZnSOD activities were unaffected.
Figure 6.
Immunoprecipitation of VuFeSOD in cowpea nodules. Lanes 1 and 6, Thrombin-treated recombinant VuFeSOD (0.3 μg). Lane 2, Nodule extract (60 μg of protein) plus 0.2 μg of protein antibody. Lane 3, Nodule extract plus 1 μg of antibody. Lane 4, Nodule extract plus 4 μg of antibody. Lane 5, Nodule extract with no antibody but with 4 μg of preimmune serum (control).
Expression of VuFeSOD in Nodules
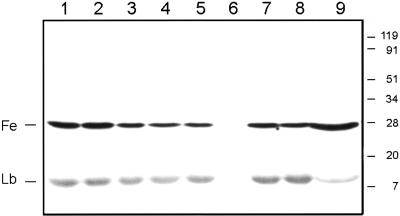
Immunoblots were used to study the effects of stressful conditions and natural senescence (aging) on the nodule content of VuFeSOD (Fig. 7). There were either no changes, or very minor decreases, in VuFeSOD protein levels in nodules of plants exposed to salt or water stress, continuous darkness, or excess nitrate. In contrast, nodule aging had clear effects on VuFeSOD content. The protein was not detectable in very young nodules (25-d-old plants) but was relatively abundant in mature nodules (46-d-old plants) and in senescent nodules (82-d-old plants). In the latter plants, two nodule populations could be distinguished by their color (“red-brown” and “green”), which reflected increased leghemoglobin (Lb) degradation (Arredondo-Peter et al., 1997). The VuFeSOD content of green nodules was clearly greater than that of red-brown nodules. The decrease in Lb content of green nodules was confirmed by incubating the same membrane with anti-Lb antibody (Fig. 7). Exposure of immunoblot membranes for longer times resulted in a very faint signal for Lb in the youngest nodules (25-d-old plants), but there was still no detectable signal for VuFeSOD (data not shown).
Figure 7.
Effect of stress and aging on VuFeSOD and Lb protein levels in cowpea nodules. Lane 1, Untreated (control) nodules; lane 2, salt stress; lane 3, dark stress; lane 4, drought; lane 5, nitrate excess; lane 6, young nodules (25-d-old plants); lane 7, mature nodules (46-d-old plants); lane 8, senescent red-brown nodules (82-d-old plants); lane 9, senescent green nodules (82-d-old plants). Control and stressed nodules were harvested from 46-d-old plants. Prestained molecular mass markers are shown in kilodaltons. All lanes were loaded with 60 μg of protein. The same membrane was challenged with the anti-VuFeSOD and anti-Lb antibodies.
Localization of VuFeSOD in Cowpea Nodules
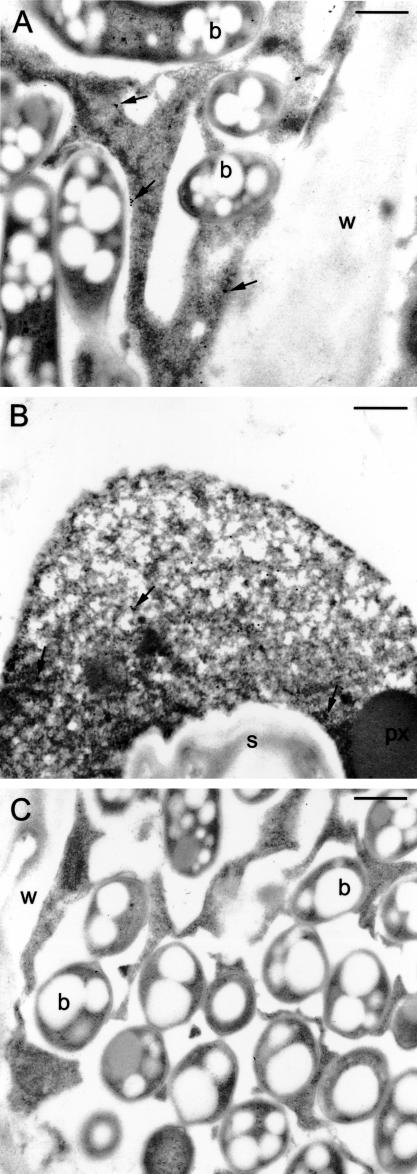
The cytosolic localization predicted from the VuFeSOD sequence was investigated with immunoblots and immunoelectron microscopy. Cowpea nodules and leaves were extracted in iso-osmotic medium and centrifuged at 15,000g for 20 min and at 100,000g for 1 h. Immunoblots indicated that VuFeSOD was in the soluble fraction (“cytosol”) and not in the membrane pellet (Fig. 8). This was confirmed by immunoelectron microscopy of high-pressure/freeze-substituted nodules, which showed significant labeling in the cytosol of both infected and uninfected (“interstitial”) cells (Fig. 9). Gold particles in infected cells were counted and statistically analyzed (Table I). The low labeling (3.9 gold particles per 25 μm2) in bacteroids and negligible labeling (<0.8 gold particles per 25 μm2) in the mitochondria and peroxisomes were not significantly different from sections treated with non-immune serum. Similar counting results were obtained for uninfected cells. Additional controls, in which the antibody (1:100, v/v) was pre-adsorbed with increasing concentrations (35–700 μg protein mL–1) of VuFeSOD protein, showed a drastic (70%) reduction in labeling of infected cells (data not shown).
Figure 8.
Immunoblots showing localization of VuFeSOD in the soluble fraction of nodules (“cytosol”). Lane 1, Prestained molecular mass markers in kilodaltons. Lane 2, Cowpea nodule extract. Lane 3, Cowpea nodule cytosol fraction. Lane 4, Cowpea nodule membrane fraction. Lane 5, Purified recombinant VuFeSOD treated with thrombin. All lanes were loaded with 60 μg of protein, except lane 5, which was loaded with 0.2 μg of protein.
Figure 9.
Immunogold localization of VuFeSOD. A, Labeling in the cytosol of infected cells (arrows). There is some minor nonspecific labeling of bacteroids. B, Labeling in the cytosol of uninfected cells (arrows). C, Serial section to A that was incubated in non-immune serum. Note the absence of significant labeling. b, Bacteroid; w, cell wall; s, starch; px, peroxisome. Bars = 500 nm.
Table I.
Number of gold particles in the cytosol and bacteroids of infected cells from cowpea nodules
| Treatment | Cytosol | Bacteroids |
|---|---|---|
| number of gold particles per 25 μm2 | ||
| FeSOD antibody | 6.0 ± 0.5* | 3.9 ± 0.4 |
| Non-immune serum | 1.2 ± 0.1 | 3.4 ± 0.4 |
Values are means ± se of 32 micrographs (each micrograph 25 μm2) taken from sections of three nodules incubated with FeSOD antibody or non-immune serum. Means denoted with an asterisk are significantly different (P < 0.01) from the corresponding non-immune serum according to the Fisher's F test.
DISCUSSION
In this paper, we have studied in detail an FeSOD from cowpea. Comparison of the N-terminal sequence of the mature protein with the cDNA-derived sequence indicates that the precursor protein contains seven additional amino acids. Prediction programs for subcellular localization suggested that VuFeSOD is located to the cytosol. This was subsequently demonstrated by immunoblot analysis and by immunogold labeling using a monospecific antibody. Phylogenetic and immunological analyses showed that at least two types of FeSOD may exist in legumes, which are localized in the plastids (alfalfa and pea) or the cytosol (cowpea and common bean). However, soybean nodules probably contain two putative cytosolic FeSODs (isoenzymes 1 and 2, with no apparent signal peptide) and one putative plastid FeSOD (isoenzyme 3). In fact, more than three FeSOD isozymes can be detected in activity gels of soybean nodule extracts (J.F. Moran, M.C. Rubio, and M. Becana, unpublished data).
The subcellular localization of VuFeSOD is reminiscent of cyanobacteria, which contain an FeSOD in the cytosol and a MnSOD in the thylakoids (Obinger et al., 1998), as opposed to eukaryotic algae and higher plants, which have a chloroplastic FeSOD (Van Camp et al., 1990; Kliebenstein et al., 1998). In the stroma, FeSODs eliminate the superoxide radicals generated by electron leakage to oxygen from photosystems and ferredoxin (Tsang et al., 1991). There are some reports localizing FeSOD in peroxisomes or mitochondria (Droillard and Paulin, 1990; Srivalli and Khanna-Chopra, 2001), but these organelle localizations have not been substantiated by immunoelectron microscopy or by isolating cDNA clones with the required targeting sequences. To our knowledge, there is only one other case where the N terminus of a plant FeSOD has been sequenced. The enzyme of wild tobacco is synthesized as a precursor protein with a plastid signal peptide that correctly targets it to the chloroplasts (Van Camp et al., 1990; Kurepa et al., 1997). Although VuFeSOD has a putative cleavage site region (KFELKPPPYP) that is highly homologous to the N terminus of the wild tobacco enzyme (KFELQPPPYP), we were unable to detect an equivalent protein in cowpea by either N-terminal sequencing of purified enzyme preparations or SDS-gel analysis of crude extracts.
Native VuFeSOD has an apparent molecular mass of 57 kD (Becana et al., 1989), and the subunit molecular mass in the presence or absence of β-mercaptoethanol is approximately 27 kD. Hence, the enzyme of cowpea nodules, and probably of other legumes (judging from the SDS-gel analysis), is a dimer with two equal subunits that are not joined covalently, as also occurs with other FeSODs (Almansa et al., 1994; Bowler et al., 1994).
Immunoblot analysis showed that there were important changes in VuFeSOD protein levels during nodule aging. The VuFeSOD protein was not detectable in young nodules but became abundant in mature and old nodules. This may be one reason why FeSOD was not previously detected in leaves of young cowpea plants (Corpas et al., 1991). The VuFeSOD protein was particularly abundant in nodules that had turned green due to Lb breakdown. We propose that the increase in VuFeSOD protein is, at least in part, due to the release of Fe from Lb. Free Fe can catalyze the generation of highly damaging hydroxyl radicals, which may be a contributing factor in nodule senescence (Becana and Klucas, 1992). This hypothesis would explain why the stress treatments, which appear not to induce Lb degradation under our experimental conditions, did not induce VuFeSOD expression. Thus, our results suggest that free Fe (or heme) may induce VuFeSOD, directly or indirectly, as a protective mechanism against oxidative stress. A direct link between Fe supply and FeSOD activity has been demonstrated in tobacco (Nicotiana tabacum), where Fe deficiency decreases FeSOD protein and activity as a result of a lower transcription rate (Kurepa et al., 1997). However, VuFeSOD was also detected in the interstitial cells of nodules. Although these cells are known to express Lb at lower levels than infected cells (VandenBosch and Newcomb, 1988), we cannot rule out the possibility that factors other than free Fe induce VuFeSOD in cowpea nodules and leaves.
In summary, this is the first conclusive report of the presence of an extra-plastidial FeSOD in higher plants. Phylogenetic and immunological analyses confirm the distinction between cytosolic and chloroplastic FeSODs. The VuFeSOD protein is more abundant in nodules with active Lb degradation, suggesting that the enzyme is induced by Fe and plays an antioxidative role with advancing senescence.
MATERIALS AND METHODS
Plant Material and Treatments
Nodulated cowpea (Vigna unguiculata Walp. cv California blackeye no. 5 × Bradyrhizobium sp. [Vigna] strain 32H1) plants were grown for 46 d (vigorous vegetative stage) in pots containing 2:1 (v/v) perlite:vermiculite (three plants per pot) under controlled environment conditions (Matamoros et al., 1999). To study the effect of stress on VuFeSOD expression, plants were subjected to one of the following treatments: 0.1 m NaCl, 10 mm KNO3, continuous darkness, or water stress (by withholding irrigation). All treatments were applied for 5 d in the growth chamber. To study the effect of age on VuFeSOD expression, plants were harvested at 25, 46, or 82 d from germination.
Cloning and Characterization of VuFeSOD cDNA
A cowpea nodule λgt11 cDNA library (Luan et al., 2000) was PCR screened using two degenerated primers (5′-GC[A/T]TTCAACAA[T/C]GC[T/A]GC[T/A]CAGG-3′ and 5′-TC[A/C]AG[G/A]TAGTAAGCATGCTCCCA-3′) based on plant FeSOD sequences deposited in the databases. The same primers were combined with λ forward and reverse primers to obtain the 5′- and 3′-untranslated regions by PCR. The PCR mixture contained 0.5 μm primers, 200 μm deoxynucleoside triphosphates, 1.5 mm MgCl2, and 2 units of Taq polymerase (Invitrogen, Carlsbad, CA) in a final volume of 25 μL of the PCR buffer (20 mm Tris-HCl, pH 8.4, and 50 mm KCl). PCR conditions were as follows: an initial cycle at 95°C for 3 min; 45 cycles of 55°C for 1 min, 72°C for 0.5 min (internal fragments), or 72°C for 1 min (5′ and 3′ fragments), and 95°C for 1 min; and a final cycle at 72°C for 5 min. The resulting PCR products were size fractionated on agarose gels, purified with the Geneclean kit (Bio 101, Vista, CA), and subcloned into the linearized vectors pCR2.1 or pCRII (Invitrogen). The same primers and similar procedures were used to isolate cDNA clones encoding soybean and pea nodule FeSODs from the corresponding cDNA libraries.
The 5′-RACE procedure was used to verify the start codon for cowpea and soybean FeSOD cDNAs. This was found not to be necessary for the plastid FeSOD of pea nodules. RNA was extracted from cowpea nodules as described by Verwoerd et al. (1989) and reverse transcribed according to the manufacturer's protocol (Invitrogen). For the PCR reactions, each of two primers (5′-TC[A/C]AG[G/A]TAGTAAGCATGCTCCCA-3′ and 5′-AGGTTCCATGTAAGTTCTGGG-3′) was used in combination with an abridged anchor primer using 5 μL of purified cDNA as template. The PCR fragments amplified in both reactions were found to be identical.
DNA sequencing was performed by the dideoxy method using an ABI Prism 377 automated sequencer and AmpliTaqDNA Polymerase (Applied Biosystems, Foster City, CA). Homology searches were performed with BLAST (Altschul et al., 1997). Sequence alignments and analyses were performed with the Gap and PileUp programs of the Genetics Computer Group package (Madison, WI). Predictions of subcellular localization and signal peptide analyses were made using MitoProtII (Claros, 1995), PSORT (Nakai and Kanehisa, 1992), and ChloroP and TransitP (Center for Biological Sequence Analysis, Department of Biotechnology, Technical University of Denmark, Copenhagen).
Heterologous Expression of VuFeSOD
Primers (5′-GCATATGAATGTGGCTGGCATCA-3′ and 5′-CGTCGACTATCATGCACTTAAAGCC-′3) containing, respectively, NdeI and SalI restriction sites (underlined), were designed to amplify by PCR a 700-bp fragment (encoding the mature protein) using the nodule cDNA library as a template. The PCR mix contained 0.2 μm primers, 240 μm dNTPs, 1.5 mm MgCl2, 0.05% (v/v) W-1 detergent (Invitrogen), and 1.25 units of Taq polymerase in a final volume of 25 μL of the PCR buffer (20 mm Tris-HCl, pH 8.4, and 50 mm KCl). PCR conditions were as follows: 95°C for 3 min; 35 cycles of 61°C for 45 s, 72°C for 1 min, and 94°C for 45 s; and a final extension at 72°C for 5 min. The PCR product was cloned into the linearized pCRII vector (Invitrogen), and the pCRII::VuFeSOD construct was digested with NdeI and SalI restriction enzymes and separated on agarose gels. The 700-bp fragment was gel extracted using the Geneclean kit and was directionally subcloned in the pET-28a(+) vector (Novagen, Madison, WI) into the NdeI and SalI sites. Cells of Escherichia coli DH5α (BD Biosciences Clontech, Palo Alto, CA) and BL21(DE3) (Invitrogen) were transformed with the resultant pET-28a::VuFeSOD construct. Positive colonies were selected with kanamycin (100 μg mL–1) in Luria-Bertani plates.
In-Gel SOD Activity
SOD isozymes were separated in 15% (w/v) native-PAGE gels and activity bands were visualized by the method of Beauchamp and Fridovich (1971). This is based on SOD inhibition of nitroblue tetrazolium reduction by photochemically generated superoxide radicals. The identification of the three types of isozymes was based on the differential inhibition of SOD activity on gels preincubated with 3 mm KCN or 5 mm H2O2 for 1 h (Rubio et al., 2001).
Purification of Recombinant VuFeSOD
Transformed E. coli BL21(DE3) cells, containing the pET-28a(+)::VuFeSOD construct, were grown at 37°C in Luria-Bertani broth containing kanamycin (100 μg mL–1) until the A600 reached 0.5. IPTG was then added at a final concentration of 1 mm. Cells were further incubated for 4 h, pelleted by centrifugation, and stored at –80°C until used. The cell paste was thawed on ice and resuspended in 10 mL of buffer containing 20 mm sodium phosphate (pH 7.5), 500 mm NaCl, 0.1% (v/v) Triton X-100, DNase I (25 μg mL–1), RNase (50 μg mL–1), pepstatin (1.25 μg mL–1), and leupeptin (1.25 μg mL–1). The cell suspension was incubated with lysozyme (2 μg mL–1) at 4°C for 15 min with gentle stirring, sonicated, and cleared by centrifugation at 48,000g at 4°C for 15 min. The resulting supernatant was chromatographed on a 5-mL HiTrap Chelating column (Amersham Biosciences, Uppsala), loaded with Ni, and equilibrated with 20 mm sodium phosphate (pH 7.5), 500 mm NaCl, and 0.1% (v/v) Triton X-100. The column was successively washed with 30 mL each of equilibrating buffer containing 0, 10, and 50 mm imidazole. A linear gradient from 50 to 500 mm imidazole (120 mL in total) was applied, and recombinant VuFeSOD was eluted at 160 mm imidazole. This was removed with Centricon C-10 filters, and fractions were directly stored at 4°C or made up to 0.5 mm EDTA and 50% (v/v) glycerol and stored at –80°C.
The fused recombinant VuFeSOD containing a (His)6-tag was digested overnight at 4°C with thrombin (Sigma-Aldrich, St. Louis). Protein purification was monitored on native-PAGE gels (stained for SOD activity) and SDS-PAGE gels (stained with Coomassie Brilliant Blue R-250). Protein was determined by a dye-binding assay (Bio-Rad Laboratories, Hercules, CA) using bovine serum albumin as a standard.
Purification of Native VuFeSOD
For purification of native VuFeSOD, 60 g of cowpea leaves was homogenized at 4°C using an Omnimixer (Sorvall, Wilmington, DE) with 400 mL of an optimized SOD extraction buffer consisting of 50 mm potassium phosphate (pH 7.8), 0.1 mm EDTA, 0.1% (v/v) Triton X-100, and 1% (w/v) polyvinylpyrrolidone-10. The extract was filtered through cheesecloth and centrifuged at 12,000g at 4°C for 20 min to remove cell debris. Proteins were precipitated at 4°C between 35% and 65% (w/v) ammonium sulfate and resuspended in 25 mL of 10 mm potassium phosphate (pH 7.8). The suspension was dialyzed overnight at 4°C against the same buffer, centrifuged to remove precipitated proteins, and chromatographed on a DEAE cellulose column (2.5 × 8.5 cm; DE52, Whatman, Maidstone, Kent, UK) equilibrated with the same buffer. After loading the sample and washing the column with equilibrating buffer, the proteins were eluted with a linear gradient (400 mL in total) of 10 to 100 mm potassium phosphate (pH 7.8) at 1.2 mL min–1. Fractions containing most FeSOD activity (monitored by gel SOD staining as described above) were pooled, concentrated, and loaded on a preparative (2-mm thick) 15% (w/v) native-PAGE gel. The FeSOD activity band was cut out, and the protein was eluted, concentrated, and loaded on a 10% (w/v) SDS-PAGE gel. The protein was blotted onto a polyvinylidene difluoride membrane (Immobilon-Psq, Millipore, Bedford, MA) at 15 mV for 20 min with a Milliblot II graphite electroblotter (Millipore), and the 27-kD band was N-terminal sequenced.
At later stages of the work, VuFeSOD was purified from 8 to 10 g of nodules by ammonium sulfate fractionation and anion-exchange chromatography, as described above. The protein, concentrated to 2.4 mL of 100 mm Tris-HCl (pH 7.8), was loaded on an affinity column (Actigel ALD, Sterogene, Carlsbad, CA), prepared as follows. Approximately 1.5 mg of anti-VuFeSOD antibody was coupled to 2 mL of resin in 100 mm potassium phosphate (pH 7.8). The coupling reagent was 100 mm NaCNBH3 and the reaction was left to proceed for 2 h at room temperature. The coupled resin was placed in a column and equilibrated with 100 mm Tris-HCl (pH 7.8) containing 1 m NaCl. After loading the sample, the column was washed with three column volumes of the same buffer. The protein was eluted with 2.5 mL of 200 mm Gly (pH 3.0), immediately buffered with 500 mm Tris-HCl (pH 8.0), concentrated, loaded on 10% (w/v) SDS-PAGE, and transferred to Immobilon-Psq using a mini-transblot unit (Bio-Rad Laboratories) for subsequent N-terminal sequencing.
Antibody Production and Characterization
Affinity-purified recombinant VuFeSOD (>3 mg) was used to raise polyclonal monospecific antibodies in rabbits following conventional immunization protocols (BioGenes, Berlin). The specificity of the anti-VuFeSOD antibody was assessed by immunoprecipitation (Miller et al., 1987). Cowpea nodules were thoroughly ground at 4°C in SOD extraction buffer (see above). Immunoprecipitations were carried out in four aliquots (150 μg of protein) of the nodule soluble fraction incubated at 4°C for 90 min with 0 (control), 0.2, 1, or 4 μg of antibody, respectively. Then, 30 μg of goat anti-rabbit IgG was added to each aliquot, and the sample was further incubated for 90 min at 4°C. Solutions were cleared by centrifugation and electrophoresed on native gels to determine the isozyme pattern as described above.
Southern-Blot Analysis
Genomic DNA was isolated from cowpea leaves using the cetyltrimethyl ammonium bromide method (Saghai-Maroof et al., 1984). DNA (16 μg) was digested with the appropriate restriction enzymes (Fig. 2), electrophoresed on 0.7% (w/v) agarose gels using 1.5× Tris-acetate-EDTA buffer, and capillary transferred to Hybond-N+ membranes following standard procedures (Sambrook et al., 1989). The probe, consisting of the complete open reading frame, was labeled with [32P]dCTP using the Megaprime II labeling kit (Amersham Biosciences). Hybridization and washings were performed at 65°C (Sambrook et al., 1989). Membranes were exposed for 1 to 4 d (Imaging screen K, Eastman Kodak, Rochester, NY), and the screen was scanned with a Molecular Imager FX (Bio-Rad Laboratories).
Immunoblots
Proteins were extracted in SOD medium, separated in 10% (w/v) SDS-PAGE, and electroblotted to polyvinylidene difluoride membranes. In some cases, 15% (w/v) native-PAGE gels were used for immunoblots. Membranes were blocked with 5% (w/v) skim milk in 20 mm Tris-buffered saline, at 4°C overnight. The primary antibody was anti-VuFeSOD (1:1,000, v/v) or anti-Lb (1:1,500, v/v). The secondary antibody was goat anti-rabbit IgG alkaline phosphatase conjugate (1:30,000, v/v; Sigma-Aldrich) or horseradish peroxidase conjugate (1:20,000, v/v; Sigma-Aldrich). Immunoreactive bands were visualized with the nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl-phosphate system (Sigma-Aldrich) or with a chemiluminescent substrate (Pierce Chemical, Rockford, IL), respectively. Controls were run with rabbit serum to check for nonspecific adsorption of primary antibody.
Localization of VuFeSOD in Nodules
To determine whether VuFeSOD is in the soluble fraction or associated with membranes, cowpea nodules (1 g) were homogenized with a mortar and pestle at 4°C with 5 mL of an iso-osmotic medium consisting of 0.3 m mannitol, 1 mm EDTA, and 20 mm potassium phosphate (pH 7.8). The extract was filtered through Miracloth and centrifuged successively at 15,000g for 20 min (“extract”), and at 100,000g for 1 h (“cytosol”). The pellet (“membranes”) was washed with iso-osmotic medium and was resuspended in 20 mm potassium phosphate (pH 7.8), 1 mm EDTA, and 0.1% (v/v) Triton X-100. Immunoblot analysis of the three nodule fractions was performed after 10% (w/v) SDS-PAGE as described above.
For immunogold localization of VuFeSOD by electron microscopy, mature cowpea nodules were fixed in 2.5% (v/v) glutaraldehyde in 100 mm sodium cacodylate buffer (pH 7.0). Nodule slices (200 μm) were taken on a Vibratome 1000 (Agar Scientific, Stansted, UK). One-millimeter-diameter discs were punched from the slices, and air was extracted from the discs under mild vacuum while they were immersed in 1-hexadecene (Leica, Wetzlar, Germany) The discs were then high-pressure frozen according to Studer et al. (2001) using an EM-PACT (Leica). The frozen slices were freeze-substituted in anhydrous acetone containing 0.5% (w/v) uranyl acetate using a Leica EM AFS at –90, –65, and –45°C over a period of 68.5 h, before being embedded and polymerized in Lowicryl HM23 (Polysciences, Warrington, PA) at –45°C (24 h) and 25°C (48 h). Ultrathin sections (80 nm) were cut on an Ultracut E microtome (Leica) and collected on pioloform/carbon-coated nickel grids. Immediately after sectioning, and before the immunolabeling procedure, the grids containing the sections were placed on drops of iso-butanol (2-methylpropan-1-ol)-saturated water for 1 h. This procedure “softens” the resin and allows for increased labeling density (Roberts, 2002). The sections were then immediately immunogold labeled according to the procedures of James et al. (1996). They were first placed for 1 h on a blocking/diluting buffer containing 1% (v/v) Tween 20 and 1% (w/v) bovine serum albumin in Tris-buffered saline (10 mm Tris-HCl [pH 7.5], 150 mm NaCl, 0.5 g L–1 polyethyleneglycol-20 K, and 14 mm Na3N) and then incubated for 1 h in a 1:100 dilution (in buffer) of the primary antibody (rabbit anti-VuFeSOD). After washing, the grids were incubated in a 1:100 dilution of goat anti-rabbit antibodies conjugated to 15-nm gold particles (BBInternational, Cardiff, UK) for 1 h. The sections were viewed and photographed using a transmission electron microscope (1200 EX, JEOL, Tokyo).
Distribution of Materials
Upon request, all novel materials described in this publication will be made available in a timely manner, and in limited amounts, for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Acknowledgments
We thank Carroll Vance for kindly supplying the pea nodule cDNA library and two anonymous reviewers and Frank Minchin for helpful comments and English correction.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.023010.
This work was supported by the Dirección General de Investigación (Ministerio de Ciencia y Tecnología, Spain; grant nos. PB98–0522 and AGL2002–02876). J.F.M. and M.C.R. were the recipients, respectively, of a postdoctoral contract and a postdoctoral fellowship funded by the Ministerio de Ciencia y Tecnología-European Union (I3P program).
In memoriam of Prof. Robert V. Klucas, our mentor and friend.
References
- Almansa MS, del Río LA, Sevilla F (1994) Characterization of an iron-containing superoxide dismutase from a higher plant, Citrus limonum. Physiol Plant 90: 339–347 [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arredondo-Peter R, Moran JF, Sarath G, Klucas RV (1997) Analysis of Vigna unguiculata leghemoglobin genes. R Bras Fisiol Veg 9: 143–149 [Google Scholar]
- Beauchamp C, Fridovich I (1971) Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem 44: 276–287 [DOI] [PubMed] [Google Scholar]
- Becana M, Klucas RV (1992) Transition metals in legume root nodules: iron-dependent free radical production increases during nodule senescence. Proc Natl Acad Sci USA 89: 8958–8962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becana M, Paris FJ, Sandalio LM, del Río LA (1989) Isoenzymes of superoxide dismutase in nodules of Phaseolus vulgaris L., Pisum sativum L., and Vigna unguiculata (L) Walp. Plant Physiol 90: 1286–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Van Camp W, Van Montagu M, Inzé D (1994) Superoxide dismutase in plants. Crit Rev Plant Sci 13: 199–218 [Google Scholar]
- Bridges SM, Salin ML (1981) Distribution of iron-containing superoxide dismutase in vascular plants. Plant Physiol 68: 275–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clare DA, Blum J, Fridovich I (1984) A hybrid superoxide dismutase containing both functional iron and manganese. J Biol Chem 259: 5932–5936 [PubMed] [Google Scholar]
- Claros MG (1995) MitoProt, a Macintosh application for studying mitochondrial proteins. Comput Appl Biosci 11: 441–447 [DOI] [PubMed] [Google Scholar]
- Corpas FJ, Sandalio LM, Palma JM, Leidi EO, Hernández JA, Sevilla F, del Río LA (1991) Subcellular distribution of superoxide dismutase in leaves of ureide-producing leguminous plants. Physiol Plant 82: 285–291 [Google Scholar]
- Crowell DN, Amasino RM (1991) Induction of specific mRNAs in cultured soybean cells during cytokinin or auxin starvation. Plant Physiol 95: 711–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droillard MJ, Paulin A (1990) Isozymes of superoxide dismutase in mitochondria and peroxisomes isolated from petals of carnation (Dianthus caryophyllus) during senescence. Plant Physiol 94: 1187–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James EK, Iannetta PPM, Nixon PJ, Whiston AJ, Peat L, Crawford RMM, Sprent JI, Brewin NJ (1996) Photosystem II and oxygen regulation in Sesbania rostrata stem nodules. Plant Cell Environ 19: 895–910 [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepa J, Bueno P, Kampfenkel K, Van Montagu M, Van den Bulcke, Inzé D (1997) Effects of iron deficiency on iron superoxide dismutase expression in Nicotiana tabacum. Plant Physiol Biochem 35: 467–474 [Google Scholar]
- Luan P, Aréchaga-Ocampo E, Sarath G, Arredondo-Peter R, Klucas RV (2000) Analysis of a ferric leghemoglobin reductase from cowpea (Vigna unguiculata) root nodules. Plant Sci 154: 161–170 [DOI] [PubMed] [Google Scholar]
- Matamoros MA, Moran JF, Iturbe-Ormaetxe I, Rubio MC, Becana M (1999) Glutathione and homoglutathione synthesis in legume root nodules. Plant Physiol 121: 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SS, Boylan KLM, Vance CP (1987) Alfalfa root nodule carbon dioxide fixation: III. Immunological studies of nodule phosphoenolpyruvate carboxylase. Plant Physiol 84: 501–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obinger C, Regelsberger G, Pircher A, Strasser G, Peschek GA (1998) Scavenging of superoxide and hydrogen peroxide in blue-green algae (cyanobacteria). Physiol Plant 104: 693–698 [Google Scholar]
- Ogawa KI, Kanematsu S, Asada K (1996) Intra- and extra-cellular localization of “cytosolic” CuZn-superoxide dismutase in spinach leaf and hypocotyl. Plant Cell Physiol 37: 790–799 [Google Scholar]
- Roberts IM (2002) Iso-butanol saturated water: a simple procedure for increasing staining intensity of resin sections for light and electron microscopy. J Microsc 207: 97–107 [DOI] [PubMed] [Google Scholar]
- Rubio MC, Ramos J, Webb KJ, Minchin FR, Gonzalez E, Arrese-Igor C, Becana M (2001) Expression studies of superoxide dismutases in nodules and leaves of transgenic alfalfa reveal abundance of iron-containing isozymes, posttranslational regulation, and compesation of isozymes activities. Mol Plant-Microbe Interact 14: 1178–1188 [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Nat Acad Sci USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sandalio LM, Palma JM, del Río LA (1987) Localization of manganese-superoxide dismutase in peroxisomes isolated from Pisum sativum L. Plant Sci 51: 1–8 [Google Scholar]
- Srivalli B, Khanna-Chopra R (2001) Induction of new isoforms of superoxide dismutase and catalase enzymes in the flag leaf of wheat during monocarpic senescence. Biochem Biophys Res Commun 288: 1037–1042 [DOI] [PubMed] [Google Scholar]
- Studer D, Graber W, Al-Amoudi A, Eggli P (2001) A new approach for cryofixation by high pressure freezing. J Microsc 203: 285–294 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang EWT, Bowler C, Hérouart D, Van Camp W, Villarroel R, Genetello C, Van Montagu M, Inzé D (1991) Differential regulation of superoxide dismutases in plants exposed to environmental stress. Plant Cell 3: 783–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Camp W, Bowler C, Villarroel R, Tsang EWT, Van Montagu M, Inzé D (1990) Characterization of iron superoxide dismutase cDNAs from plants obtained by genetic complementation in Escherichia coli. Proc Natl Acad Sci USA 87: 9903–9907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch KA, Newcomb EH (1988) The occurrence of leghemoglobin protein in the uninfected interstitial cells of soybean root nodules. Planta 175: 442–451 [DOI] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A (1989) A small scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]