Abstract
Expression of Cah1, encoding a periplasmic carbonic anhydrase in Chlamydomonas reinhardtii Dangeard, is activated when cells are exposed to low-CO2 conditions (0.04% [v/v]) in light. By using an arylsulfatase reporter gene, a regulatory region essential for the transcriptional activation of Cah1 was delimited to a 63-bp fragment between –293 and –231 relative to the transcription start site. Linker-scan analysis of the 63-bp region identified two enhancer elements, EE-1 (AGATTTTCACCGGTTGGAAGGAGGT) and EE-2 (CGACTTACGAA). Gel mobility shift assays indicated that nuclear extracts purified from cells grown under low-CO2 conditions in light contained DNA-binding proteins specifically interacting with EE-1 and EE-2. Gel mobility shift assays using mutant oligonucleotide probes revealed that the protein binding to EE-1 preferentially recognized a 9-bp sequence stretch (AGATTTTCA) of EE-1, containing a conserved sequence motif named EEC, GANTTNC, which is also present in EE-2. The EE-1- and EE-2-binding proteins interacted with the EECs contained in both of the two enhancer elements in vitro. Four EECs in the 5′-upstream region from –651 to –231 of Cah1 played a central role in the transcriptional activation of Cah1 under low-CO2 conditions. These EEC-binding proteins were present even in cells grown under high-CO2 conditions (5% [v/v]) or in the dark when Cah1 is not activated. On the basis of these results, the relationship between the transcriptional regulation of Cah1 and protein-binding to the enhancer elements in the 5′-upstream region of Cah1 is discussed.
Aquatic photosynthetic organisms, such as the green alga Chlamydomonas reinhardtii, acclimate to CO2-limiting stress by inducing a carbon-concentrating mechanism (Aizawa and Miyachi, 1986; Badger and Price, 1994; Kaplan and Reinhold, 1999). This mechanism involves active uptake of inorganic carbon into the cells mostly in the form of HCO3 – and catalyzed conversion of HCO3 – to CO2, which is the substrate of Rubisco. Although a number of genes regulated in response to changes in the CO2 concentration have been identified in C. reinhardtii (Fukuzawa et al., 1990; Villand et al., 1997; Spalding, 1998), cis-acting elements and transcription factors functioning in the gene regulation are still poorly understood. The expression of Cah1 is repressed when cells are maintained under high-CO2 (5% [v/v]) conditions, whereas the expression is activated within 1 h after transfer to low-CO2 (0.04% [v/v]) conditions (Fukuzawa et al., 1990). This activation does not occur when cells are cultured in the dark or in the presence of 3-(3,4-dichlorophenyl)-1,1-dimethylurea, suggesting that photosynthetic electron flow is required (Dionisio-Sese et al., 1990). By using a reporter gene encoding arylsulfatase (Ars) driven by the β2-tubulin minimal promoter, transcription of Cah1 has been shown to be controlled by two regulatory regions: a silencer region, which represses the transcription under high-CO2 conditions or in the dark, and an enhancer region, which activates it under low-CO2 conditions in light (Kucho et al., 1999). However, the detailed nucleotide sequences of cis-acting elements and presence of DNA-binding proteins related to Cah1 regulation have not been identified.
In this paper, we describe a conserved DNA sequence in the two enhancer elements involved in the transcriptional activation of Cah1 under low-CO2 conditions and the presence of DNA-binding proteins that interact with the enhancer elements in nuclear extracts.
RESULTS
A 63-bp Region between –293 and 231 Is Essential for Transcriptional Activation of Cah1 under Low-CO2 Conditions in Light
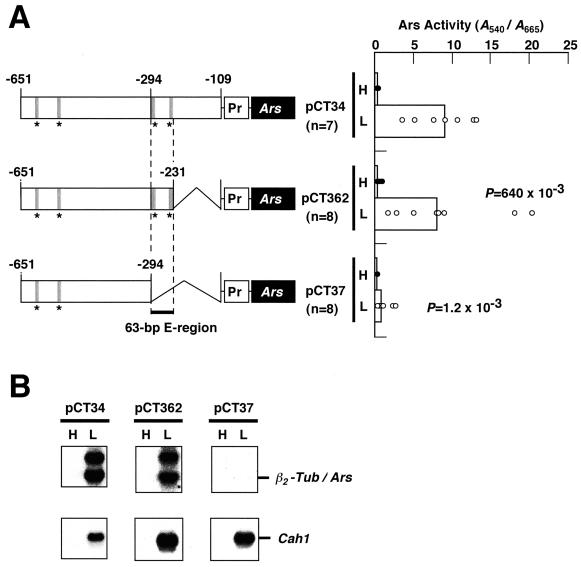
Previously, we showed that a 543-bp Cah1 upstream region between –651 and –109 was sufficient to confer CO2 responsiveness on the β2-tubulin minimal promoter, and a 185-bp region between –293 and –109 was essential to activate the transcription under low-CO2 conditions (Kucho et al., 1999). To delimit the region essential for activation, the 3′ end of the 543-bp fragment was truncated to position –231 and fused to the Ars reporter gene driven by the β2-tubulin minimal promoter (Davies and Grossman, 1994; Fig. 1A, pCT362). Then, the chimeric construct was transformed into C. reinhardtii cells, and the Ars activity of the transformants was measured under high- and low-CO2 conditions. Chimeric constructs, pCT34 and pCT37, were used as positive and negative controls for the activation of Ars expression under low-CO2 conditions, respectively (Kucho et al., 1999). In C. reinhardtii, individual transformants carrying the same chimeric construct have been reported to show different levels of expression possibly dependent on the positions where the constructs are integrated into the genome (Quinn and Merchant, 1995; Kucho et al., 1999). Therefore, for each chimeric construct, at least seven independent transformants containing a single transgene were assayed for Ars activity, and the significance of differences in the activities among transgenic lines with different constructs was evaluated statistically by the nonparametric Mann-Whitney U test. We regarded differences showing P values less than 25 × 10–3 as statistically significant. As shown in Figure 1A, transformants harboring pCT362 exhibited comparable Ars activities to cells harboring pCT34 under low-CO2 conditions (P = 640 × 10–3). On the other hand, cells carrying pCT37 showed significantly lower expression under low-CO2 conditions than that of pCT34. Northern-blot analyses of representative transformants with pCT362 and pCT37 revealed that a significant amount of Ars mRNA accumulated in cells with pCT362 but not in those with pCT37 (Fig. 1B). An additional band observed above the β2-Tub/Ars transcript corresponds to be a premature transcript containing Ars introns (Kucho et al., 1999). These results indicate that the 63-bp region between –293 and –231 is essential for transcriptional activation of Cah1 under low-CO2 conditions in light. Therefore, the 63-bp region was named the E-region because it contains enhancer element(s) functioning in transcriptional activation under these conditions.
Figure 1.
Schematic drawings of the chimeric constructs and Ars expression of their corresponding transgenic lines. A, The 3′-nested deletions of a Cah1 upstream region were fused to the Ars reporter gene (Ars) driven by the β2-tubulin minimal promoter (Pr). Asterisks represent positions of consensus sequences in the enhancer element (EEC, see text). The 63-bp enhancer region (E-region) between –293 and –231 essential for the transcriptional activation of Cah1 under low-CO2 conditions in light is highlighted. These chimeric constructs were transformed into C. reinhardtii cells, and the Ars enzyme activity in each transformant was measured under high-CO2 (H) and low-CO2 (L) conditions in light. Black and white circles represent the Ars activities in independent transformants under high- and low-CO2 conditions, respectively. Bars indicate the median values. The number of transformants used for the Ars activity measurements is represented by n. The Ars activities in transformants carrying pCT362 and pCT37 under low-CO2 conditions were compared with those in transformants carrying pCT34 by the Mann-Whitney U test, and the resulting P values are indicated. B, Northern-blot analyses of representative strains harboring the indicated chimeric constructs using 32P-labeled Ars- or Cah1-specific probes.
Linker-Scan Analysis of the E-Region Identifies Two Putative Enhancer Elements
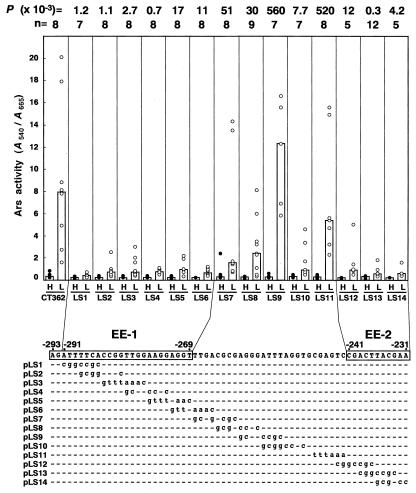
To evaluate which nucleotides within the E-region are critical for CO2-responsive activation of Cah1 expression, linker-scan analysis was conducted. A consecutive series of nucleotide substitutions were introduced into the E-region of pCT362, generating 14 linker-scan constructs (Fig. 2, pLS1–pLS14). These constructs were transformed into C. reinhardtii cells, and the Ars activities exhibited by the transgenic cell lines (LS1 to LS14) under low-CO2 conditions were compared statistically to those exhibited by a control strain, CT362, harboring pCT362. At least five independent transformants harboring a single-copy chimeric construct were selected by Southern-blot analyses (data not shown), and the Ars activities were tested for each transformant. Transgenic lines LS1 to LS6 showed significant decreases in the Ars activity under low-CO2 conditions compared with CT362 (P value < 25 × 10–3). The median values of the Ars activity in these strains were at least one-tenth lower than that in CT362. However, nucleotide substitution from (–268)-TTGA-(–265) to AAAC in LS6 was not responsible for the reduction in Ars activity, because substitution of the TTGA sequence with GCGG in LS7 did not affect the activity significantly (P = 51 × 10–3). These results suggest that the nucleotide sequence from –291 to –269 plays an important role in the transcriptional activation of Cah1 under low-CO2 conditions. Because the A and G nucleotides at positions –293 and –292 were essential for binding of a nuclear protein to EE-1 (described below), a 25-bp nucleotide sequence stretch from –293 to –269, AGATTTTCACCGGTTGGAAGGAGGT, was assumed to be a CO2-responsive enhancer element and designated EE-1. In addition, transgenic lines LS12, LS13, and LS14 exhibited significantly lower Ars activities than the control CT362 (P < 25 × 10–3), whereas strain LS11 did not show such low activity (P = 520 × 10–3), suggesting that an 11-bp nucleotide sequence stretch from –241 to –231, CGACTTACGAA, designated EE-2, also could be another CO2-responsive enhancer element.
Figure 2.
Linker-scan analyses of the E-region harboring two putative enhancer elements, EE-1 and EE-2. A series of nucleotide substitutions were introduced into the E-region of the control construct pCT362. The nucleotide substitutions are indicated by lowercase letters, and nucleotides identical with the wild-type sequence are represented by dashes. The Ars activities of linker-scan constructs were measured under high-CO2 (H) or low-CO2 (L) conditions in light. Black and white circles represent the Ars activities in independent transformants under high- and low-CO2 conditions, respectively. Bars indicate the median values. The number of transformants tested for the activity is represented by n. The Ars activity of each linker-scan construct under low-CO2 conditions was compared with that of the control construct pCT362 by the Mann-Whitney U test, and the resulting P values are indicated above the graph.
Nuclear Extract from Cells Grown under Low-CO2 Conditions in Light Contains Proteins That Interact with Enhancer Elements, EE-1 and EE-2
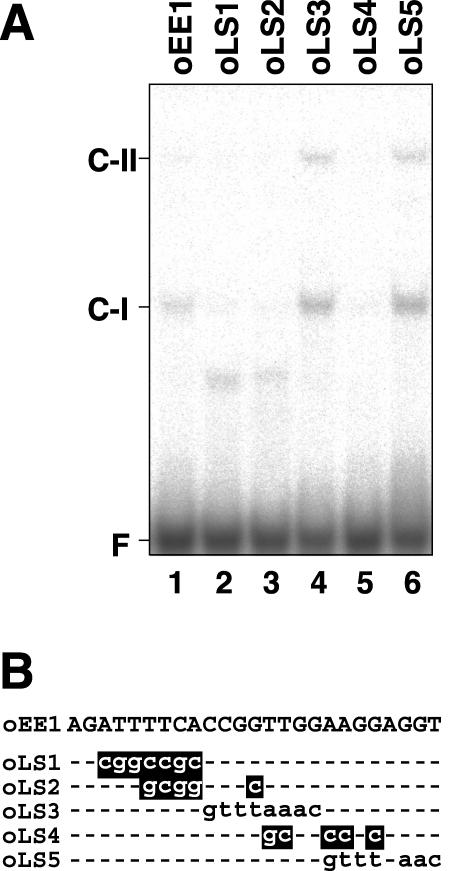
Because the two putative enhancer elements, EE-1 and EE-2, were assumed to function in transcriptional activation under low-CO2 conditions in light, it was possible that DNA-binding proteins interacting with these elements could be present in nuclear extracts from cells grown under these conditions. To determine whether these elements were involved in DNA-protein interaction, gel mobility shift assays were carried out. When a 32P-labeled double-stranded oligonucleotide probe of EE-1 (oEE1) was used, a retarded DNA-protein complex (C-I) was detected (Fig. 3A, lane 1), indicating that a protein which binds to EE-1 was present in the nuclear extracts. When modified oligonucleotides, oLS1, oLS2, and oLS4, that carry identical nucleotide substitutions to those in constructs pLS1, pLS2, and pLS4, respectively, were used as probes (Fig. 3B), the shifted C-I band was not detected (Fig. 3A, lanes 2, 3, and 5). These results showed good correlation with the fact that these substitutions exhibited significantly reduced inducibility of Ars activities under low-CO2 conditions in vivo (Fig. 2). On the contrary, probes oLS3 and oLS5 formed the C-I band (Fig. 3A, lanes 4 and 6), although cells harboring linker-scan constructs pLS3 and pLS5, containing identical nucleotide substitutions to probes oLS3 and oLS5, respectively, were defective in CO2-responsive activation of Ars expression (Fig. 2). These probes detected an additional DNA-protein complex, C-II, which was not observed with the wild-type EE-1 probe, oEE1 (Fig. 3A, lanes 4 and 6). Because both oLS3 and oLS5 probes contained an 8-bp sequence stretch of GTTTAAAC as a consequence of nucleotide substitutions (Fig. 3B), it is possible that the additional shifted bands were generated by the artificial 8-bp sequence stretch.
Figure 3.
Gel mobility shift assay using the EE-1 probe and its derivatives. A, 32P-labeled double-stranded oligonucleotide of EE-1 (lane 1) and linker-scan oligonucleotides (lanes 2–6) were incubated with 2.5 μg of nuclear proteins purified from cells grown under low-CO2 conditions in light. Probes oLS1 to oLS5 carry identical nucleotide substitutions to those in constructs pLS1 to pLS5, respectively (Fig. 2). F indicates the free probe. C-I and C-II indicate the shifted bands. B, Nucleotide sequence of EE-1 and linker-scan oligonucleotides. Mutations that abolished C-I complex formation are highlighted.
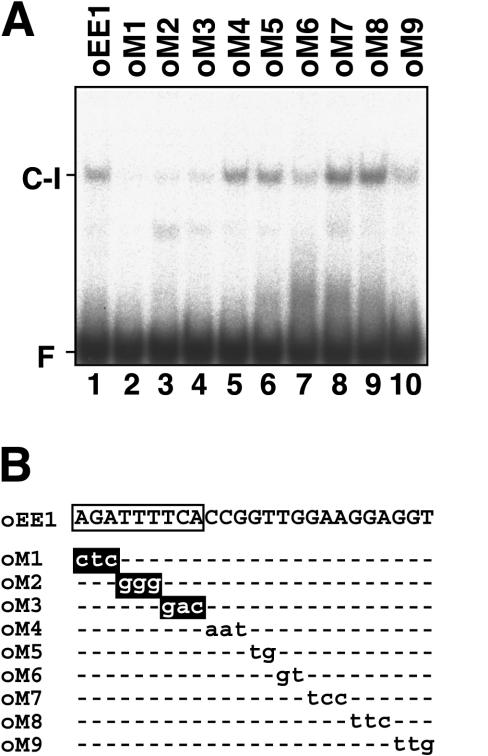
To decrease the artificial effects of nucleotide substitution, mutant oligonucleotide probes oM1 to oM9 carrying consecutive 2- or 3-bp nucleotide substitutions were tested by the gel mobility shift assay (Fig. 4). Mutations in the first 9 bp of EE-1 (AGATTTTCA) caused drastic decreases in the formation of C-I (Fig. 4A, lanes 2–4), whereas those in the remaining 3′ portion of EE-1 had lesser effects (Fig. 4A, lanes 5–10). On the basis of these results, the 9-bp AGATTTTCA sequence stretch was assumed to be a core sequence of EE-1, which is critical for interaction with the DNA-binding protein contained in the C-I complex.
Figure 4.
Gel mobility shift assay using DNA fragments of EE-1 carrying consecutive 2- or 3-bp nucleotide substitutions. A, 32P-labeled double-stranded oligonucleotide probes of EE-1 (lane 1) and modified probes carrying consecutive 2- or 3-bp nucleotide substitutions as shown in B (lanes 2–10) were incubated with 2.5 μg of nuclear proteins purified from cells grown under low-CO2 conditions in light. F indicates the free probe. B, Nucleotide substitutions introduced into EE-1 are shown by lowercase letters. Nucleotide substitutions that abolished C-I complex formation are highlighted. A core sequence of EE-1 critical for interaction with the DNA-binding protein is boxed.
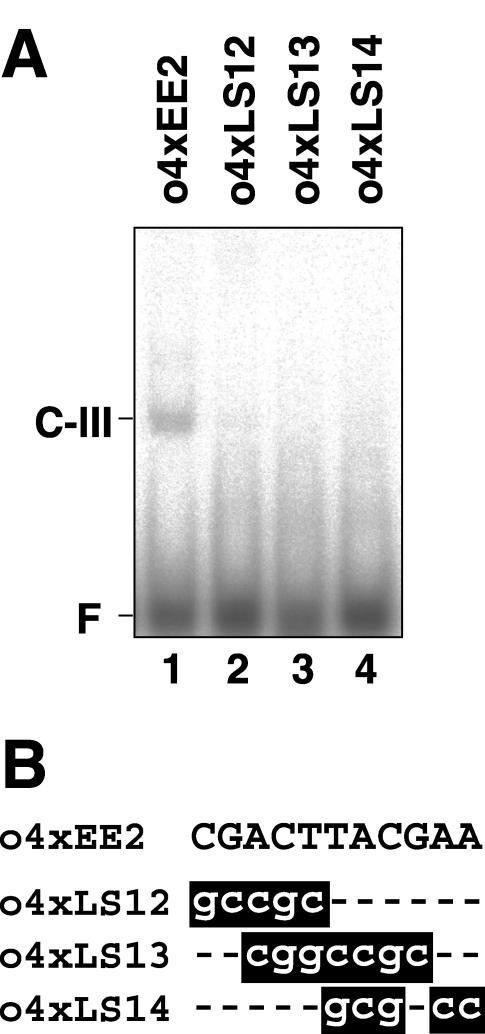
On the other hand, a 32P-labeled double-stranded oligonucleotide probe containing four tandem copies of EE-2 (o4xEE2) produced another retarded DNA-protein complex, C-III (Fig. 5A, lane 1). This indicates the presence of a protein that binds to EE-2 in the nuclear extracts. When modified probes o4xLS12, o4xLS13, and o4xLS14, carrying nucleotide substitutions identical to those in constructs pLS12, pLS13, and pLS14, respectively, were used (Fig. 5B), the C-III complex was not detected (Fig. 5A, lanes 2–4). These results coincide with the in vivo observations that cells harboring these constructs exhibited significantly reduced Ars activities under low-CO2 conditions (Fig. 2).
Figure 5.
Gel mobility shift assay using DNA fragments containing four tandem copies of EE-2 (o4xEE2) and its derivatives. A, 32P-labeled double-stranded oligonucleotide of o4xEE2 (lane 1) and modified oligonucleotides carrying linker-scan mutations (lanes 2–4) were incubated with 2.5 μg of nuclear proteins purified from cells grown under low-CO2 conditions in light. Probes o4xLS12 to o4xLS14 carry identical mutations to those in pLS12 to pLS14, respectively (Fig. 2). F indicates the free probe. B, Nucleotide sequences of o4xEE2 and modified oligonucleotides including mutations. Mutations that abolished C-III complex formation are highlighted.
EE-1- and EE-2-Binding Proteins Interact with Both of the Two Enhancer Elements
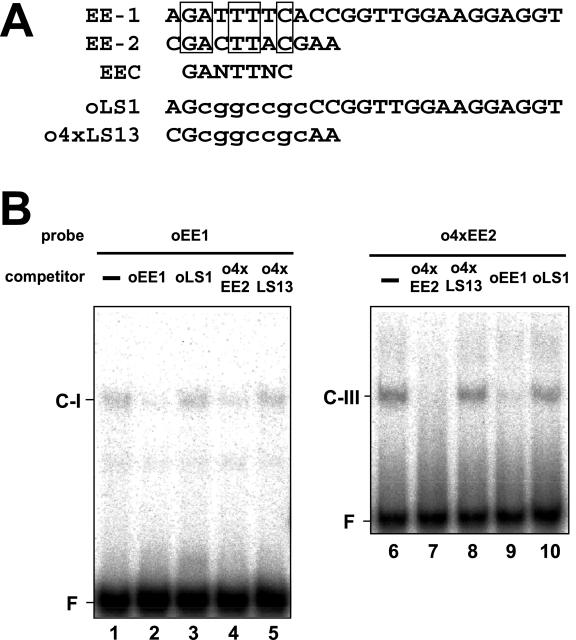
The two enhancer elements, EE-1 and EE-2, contain a 7-bp consensus motif named EEC, GANTTNC (Fig. 6A). If the EE-1- and EE-2-binding proteins interact with their target sequences by recognizing EEC, it is possible that the EE-1-binding protein could bind to EE-2 and that the EE-2-binding protein could bind to EE-1. To examine these possibilities, cross-competition assays were carried out (Fig. 6B). First, when oEE1 was used as a probe, formation of the C-I complex was competed by the presence of an excess of unlabeled o4xEE2 as well as by that of oEE1 itself (Fig. 6B, lanes 2 and 4). In contrast, addition of o4xLS13, in which the consensus motif in o4xEE2 was mutated (Fig. 6A), did not decrease the signal intensity of C-I (Fig. 6B, lane 5). These results strongly suggest that the EE-1-binding protein interacts with EE-2 by recognizing EEC. Second, when o4xEE2 was used as a probe, formation of the C-III complex was competed by the presence of oEE1 as well as by that of o4xEE2 itself (Fig. 6B, lanes 7 and 9). Addition of oLS1, carrying nucleotide substitutions in the consensus motif in oEE1 (Fig. 6A), did not cause the competition (Fig. 6B, lane 10). These results indicate that the EE-2-binding protein also interacts with EE-1 by recognizing EEC.
Figure 6.
Comparison of the nucleotide sequences of EE-1 and EE-2, and cross-competition assays using oEE1 and o4xEE2 probes. A, Conserved nucleotides between EE-1 and EE-2 are boxed. In oLS1 and o4xLS13, nucleotide substitutions are shown by lowercase letters. B, 32P-labeled oEE1 (lanes 1–5) and 32P-labeled-o4xEE2 (lanes 6–10) probes were incubated with 600-fold molar excess amounts of unlabeled competitors as indicated above. F indicates the free probe.
DNA-Binding Proteins Are Constitutively Present in Nuclear Extracts Irrespective of Changes in CO2 Levels and Light Illumination
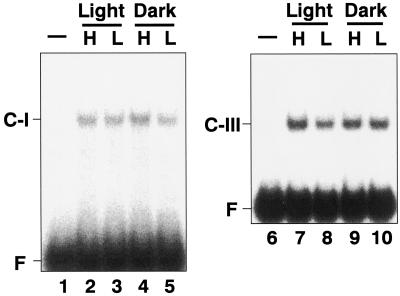
To test the effects of CO2 concentration and light illumination on the DNA-protein complex formations, nuclear extracts were isolated from cells grown under high- and low-CO2 conditions in the light or the dark and then gel mobility shift assays were performed with the oEE1 and o4xEE2 probes (Fig. 7). The retarded bands, C-I and C-III, which were observed with the nuclear extracts from cells grown under low-CO2 conditions in light were also detected with those from the cells grown under high-CO2 conditions (Fig. 7, lanes 2, 4, 7, and 9) or in the dark (Fig. 7, lanes 4, 5, 9, and 10) when expression of Cah1 is not activated. These results indicate that the DNA-binding proteins that bind to the two enhancer elements are present in the nuclear extracts irrespective of changes in the CO2 concentration and light illumination.
Figure 7.
Gel mobility shift assay using nuclear extracts purified from cells grown under high-CO2 conditions or in the dark. Nuclear extracts were prepared from cells grown under high-CO2 (H; lanes 2, 4, 7, and 9) or low-CO2 (L; lanes 3, 5, 8, and 10) conditions and in the light (lanes 2, 3, 7, and 8) or the dark (lanes 4, 5, 9, and 10), and then incubated with 32P-labeled probes, oEE1 (lanes 1–5) or o4xEE2 (lanes 6–10). F indicates the free probe.
DISCUSSION
Although the physiological aspects of acclimation of photosynthetic microalgae to CO2-limiting stress have been extensively studied, regulatory components such as cis-acting elements and transcription factors that control the expression of genes involved in the acclimation have not been extensively described in eukaryotic microalgae. In a cyanobacterium, Synechocystis sp. PCC6803, a consensus CO2-responsive sequence motif, TCAATG-(N10)-ATCAAT, has been described based on deletion analyses of the promoter region of the ndh3 operon encoding NAD(P)H dehydrogenase subunits (Figge et al., 2001). Two LysR family transcriptional regulators have been identified, cmpR regulating the cmp operon encoding a low-CO2-inducible ABC-type bicarbonate transporter (Omata et al., 2001), and ndhR regulating the ndh3 operon (Figge et al., 2001). In the photosynthetic eukaryote C. reinhardtii, 5′-upstream regions of the three CO2-responsive genes have been analyzed to date. An identical 194-bp sequence in the 5′-upstream region shared by the two mitochondrial carbonic anhydrase genes, Mca1 and Mca2, is sufficient to confer CO2-responsiveness on the promoterless Ars reporter gene (Villand et al., 1997).
We previously reported that the 185-bp 5′-upstream region from –293 to –109 relative to the transcription start site was required for transcriptional activation of Cah1 encoding a periplasmic carbonic anhydrase under low-CO2 conditions in light (Kucho et al., 1999). Here, further deletion analysis of this region revealed that a 63-bp region from –293 to –231 (E-region) was essential for the CO2-responsive transcriptional activation. Furthermore, linker-scan analysis identified two putative enhancer elements, EE-1 (AGATTTTCACCGGTTGGAAGGAGGT; –293 to –269) and EE-2 (CGACTTACGAA; –241 to –231), within the E-region (Fig. 2). These two elements shared a core sequence motif named EEC, GANTTNC. Gel mobility shift assays revealed that DNA-binding proteins that interact with EE-1 and EE-2 are present in C. reinhardtii nuclear extracts. Three types of nucleotide substitutions within EE-2 that caused a decrease in Ars induction under low-CO2 conditions (Fig. 2) abolished the interaction of the DNA-binding protein with EE-2 in vitro (Fig. 5). This good correlation between the in vivo and in vitro data suggests that the EE-2-binding protein could be one of the transcription factors involved in the CO2-responsive activation of Cah1 expression.
Although the in vivo linker scan analysis using transgenic cell lines showed that the whole EE-1 region was essential for activating Cah1 expression (Fig. 2), gel shift assays using mutated oligonucleotide probes (oM1–oM9) demonstrated that only 9 bp in the 5′ part of EE-1 was recognized by the DNA-binding protein (Fig. 4). One possible explanation for this discrepancy is to assume that the reduction in Ars activity under low-CO2 conditions by nucleotide substitutions LS3 to LS6 is caused by other undetectable DNA-binding protein(s), which binds to 16 bp in the 3′ part of EE-1.
From the cross-competition assays (Fig. 6), it was strongly suggested that there is one EEC-binding protein in the nuclear extracts, and this binds to both EE-1 and EE-2. In total, four EECs are found in the 5′-upstream region of Cah1 from –651 to –231 (–615 to –607, GATTTGC; –550 to –542, GAATTCC; –293 to –285, GATTTTC [in EE-1]; –241 to –233, GACTTAC [in EE-2]; Fig. 1). Deletion of the former two EECs caused a severe decrease in transcriptional activation under low-CO2 conditions, and deletion of all four EECs completely abolished the activation (Kucho et al., 1999; Fig. 1). Furthermore, loss of the latter two EECs also caused a severe decrease in activation (Fig. 1, pCT37 [L]). These results strongly suggest that EECs play a central role in the transcriptional activation of Cah1 under low-CO2 conditions. EEC was also found within the upstream region of another low-CO2-inducible gene, Mca1, which encodes mitochondrial carbonic anhydrase in C. reinhardtii, at a position between –47 and –41 relative to the transcription start site (Villand et al., 1997). Because the upstream region up to –194 was shown to be sufficient for the CO2-responsive transcriptional activation, it is possible that EEC could act as a common CO2-responsive enhancer element in C. reinhardtii.
A sequence similarity search of EEC in the motif database PLACE for plant cis-acting regulatory DNA elements (Higo et al., 1999) revealed a striking similarity with box III, a light-responsive element from the rbcS-3A gene in pea (Pisum sativum; Kuhlemeier et al., 1987; Table I). This similarity corresponds with the function of EEC because transcriptional activation by the E-region containing two EECs occurs in the light. Box III was shown to interact with a nuclear regulatory protein, GT-1 (Green et al., 1987). Molecular dissection of cloned GT-1 indicated that the trihelix (helix-helix-turn-helix) motif was essential for DNA binding, and that it bound to the target DNA as a dimer (Hiratsuka et al., 1994). GT-1 not only binds to box III, but also to an additional five cis-acting elements upstream of the rbcS-3A gene with the consensus sequence G(A/G)(A/T)AA(A/T) (Green et al., 1988). Recently, detailed amino acid sequence comparisons revealed that GT-1 is closely related to Myb proteins that function as transcription activators in diverse organisms ranging from animals to plants (Nagano, 2000). EEC also shows sequence similarities with the E1A enhancer element of the adenovirus type 5 (Hearing and Shenk, 1983) and the human β-interferon enhancer (Goodbourn et al., 1985; Table I), which is bound by a nuclear factor, IRF-1 (Miyamoto et al., 1988), that contains a Myb-type DNA-binding domain. Therefore, it is possible that Mybrelated proteins could bind to EEC and regulate the CO2-responsive expression of Cah1 and Mca1 in C. reinhardtii.
Table I.
Cis-acting elements showing sequence similarities with the EEC
| Organism | Gene | Element | Sequencea | Reference |
|---|---|---|---|---|
| C. reinhardtii | Cah1 | EE-1 | aagattttcaccggttggaaggaggt | This report |
| C. reinhardtii | Cah1 | EE-2 | ccgacttacgaa | This report |
| C. reinhardtii | Mca1 | — | ccgaatttct | Villand et al. (1997) |
| Pea | rbcS-3A | Box III | atcATTTTCact | Kuhlemeier et al. (1987) |
| Human | β-IFN | cactttcacttc | Goodbourn et al. (1985) | |
| Adenovirus type 5 | E1A | — | attttcacttcc | Hearing and Shenk (1983) |
a Nucleotides of the EEC and its corresponding nucleotides are underlined. Consensus sequence of GT-1 binding sites is capitalized.
The formations of DNA-protein complexes with oEE1 and o4xEE2 probes were not affected by the CO2 concentration or light illumination (Fig. 7), suggesting that interactions of these DNA-binding proteins with EEC would occur in cells grown under high-CO2 conditions or in the dark when Cah1 is not activated. Coupled with the data from linker-scan experiments, these observations suggest that the EEC sites may be important cis-acting elements that constitutively bind one or more proteins that serve to assist in the regulated transcription of the Cah1 gene. Therefore, it is possible that there may be the production of another protein under low-CO2 that may bind to the EEC-binding protein complex and that this EEC-binding protein-activator complex could stimulate transcriptional activity.
Previously, we have reported that the upstream silencer region of Cah1 from –651 to –294 is necessary to suppress the Cah1 expression under high-CO2 or in the dark (Kucho et al., 1999). This silencer region contributes tight regulation of the Cah1 transcription in response to changes in CO2 levels and light illumination. Further analyses of the DNA-binding protein specifically interacting with the silencer region could clarify the importance of the negative regulation of Cah1 gene. Because it is reported that posttranslational modifications of transcription factors, such as phosphorylation (Hunter and Karin, 1992) and/or recruitment of coactivators (Singh, 1998), play a central role in transcriptional activation, it is possible that posttranslational modifications of the EEC-binding proteins could be involved in the CO2-responsive transcriptional activation of Cah1. It is noteworthy that binding of GT-1 to a light-responsive cis-acting element is also considered to be necessary for regulation of the rbcS-3A gene in pea (Green et al., 1988). However, it has been suggested that modulation of the binding activity by calcium-dependent phosphorylation plays a role in the regulation (Maréchal et al., 1999).
Recently, a regulatory gene, Ccm1, encoding a zinc-finger protein, was identified from C. reinhardtii high-CO2-requiring mutants C16 and cia5 (Fukuzawa et al., 2001). Because these mutants are defective in the induction of several CO2-responsive genes, including Cah1, and CCM1 protein is constitutively expressed in the WT strain, it is possible that CCM1 may bind to CO2-responsive cis-acting elements, including EEC. The functional relationships between CCM1 protein and these regulatory cis-acting elements or DNA-binding protein remain to be elucidated. On the basis of the observation that there are EEC-binding proteins in the nuclear extracts regardless of changes in the CO2 concentration and light illumination, it is more likely that other cofactors that interact with the EEC-binding proteins, possible repressor proteins, and/or posttranslational modifications of the EEC-binding proteins would be crucial to the regulation of Cah1 expression in response to the CO2 concentration and light illumination. It is important to isolate other factors from regulatory mutants that are defective in the induction of Cah1 expression, to elucidate the regulatory mechanisms of CO2-responsive genes.
MATERIALS AND METHODS
Culture Conditions
For high-CO2 conditions, Chlamydomonas reinhardtii Dangeard cells were cultured in high salt (HS) medium (Sueoka, 1960) supplemented with 20 mm MOPS (pH 7.2) with aeration by air containing 5% (v/v) CO2 at 28°C. For low-CO2 conditions, cultures were bubbled with ordinary air containing 0.04% (v/v) CO2. Cultures were illuminated at an intensity of 150 μmol m–2 s–1. For Ars assays, cells were cultured in HS medium supplemented with 0.4 mm magnesium sulfate and 20 mm MOPS (pH 7.2; HSM+S; Kucho et al., 1999).
Chimeric Constructs
To generate pCT362, a DNA fragment containing the Cah1 upstream sequence from position –651 to –231 relative to the transcription start site was amplified by PCR. The amplified fragment was inserted into a bluntended KpnI site of pJD100, containing the Ars reporter gene driven by the β2-tubulin minimal promoter (Davies and Grossman, 1994). Chimeric constructs, pCT34 and pCT37, were generated as described previously (Kucho et al., 1999). Linker-scan mutations were introduced by a modified inverse PCR method with pCT362 as the template DNA. Primer sets were designed so as to generate a NotI (GCGGCCGC) or PmeI (GTTTAAAC) restriction site when the inverse PCR products were self-ligated (Table II). These primers were phosphorylated at the 5′ end by T4 polynucleotide kinase. Using the primers LS1-f to LS14-f in combination with LS1-r to LS14-r, inverse PCR was performed with LA Taq polymerase (TaKaRa, Kyoto). The amplified fragments were blunt-ended by T4 DNA polymerase, self-ligated, and transformed into Escherichia coli DH5α.
Table II.
Primers used in making linker-scan constructs
| Primera | Sequence (5′-3′)b | Position (5′-3′)c | Construct |
|---|---|---|---|
| LS1-f | CCGCCCGGTTGGAAGGAGGTTTGACGCGAG | -284 to -259 | pCT362LS1 |
| LS2-f | CCGCTTGGAAGGAGGTTTGACGCGAGGGAT | -280 to -255 | pCT362LS2 |
| LS3-f | AAACAAGGAGGTTTGACGCGAGGGATTTAG | -276 to -251 | pCT362LS3 |
| LS4-f | CCGCAGGTTTGACGCGAGGGATTTAGGTGC | -272 to -247 | pCT362LS4 |
| LS5-f | AAACTTGACGCGAGGGATTTAGGTGCGAGT | -268 to -243 | pCT362LS5 |
| LS6-f | AAACCGCGAGGGATTTAGGTGCGAGTCCGA | -264 to -239 | pCT362LS6 |
| LS7-f | CCGCAGGGATTTAGGTGCGAGTCCGACTTA | -260 to -235 | pCT362LS7 |
| LS8-f | CCGCATTTAGGTGCGAGTCCGACTTACGAA | -256 to -231 | pCT362LS8 |
| LS9-f | CCGCAGGTGCGAGTCCGACTTACGAAGGTA | -252 to -227 | pCT362LS9 |
| LS10-f | CCGCGCGAGTCCGACTTACGAAGGTACCAT | -248 to -223 | pCT362LS10 |
| LS11-f | AAACGACTTACGAAGGTACCATCGGGCCCC | -240 to -215 | pCT362LS11 |
| LS12-f | CCGCTACGAAGGTACCATCGGGCCCCCCCT | -236 to -211 | pCT362LS12 |
| LS13-f | CCGCAAGGTACCATCGGGCCCCCCCTCGAG | -232 to -207 | pCT362LS13 |
| LS14-f | CCGCTACCATCGGGCCCCCCCTCGAGGTCG | -228 to -203 | pCT362LS14 |
| LS1-r | CCGCTACAACGCTGCCAACGTGGTGGCGCG | -293 to -317 | pCT362LS1 |
| LS2-r | CCGCAATCTACAACGCTGCCAACGTGGTGG | -289 to -314 | pCT362LS2 |
| LS3-r | AAACTGAAAATCTACAACGCTGCCAACGTG | -285 to -310 | pCT362LS3 |
| LS4-r | CCGCCCGGTGAAAATCTACAACGCTGCCAA | -281 to -306 | pCT362LS4 |
| LS5-r | AAACCCAACCGGTGAAAATCTACAACGCTG | -277 to -302 | pCT362LS5 |
| LS6-r | AAACCCTTCCAACCGGTGAAAATCTACAAC | -273 to -298 | pCT362LS6 |
| LS7-r | CCGCACCTCCTTCCAACCGGTGAAAATCTA | -269 to -294 | pCT362LS7 |
| LS8-r | CCGCTCAAACCTCCTTCCAACCGGTGAAAA | -265 to -290 | pCT362LS8 |
| LS9-r | CCGCCGCGTCAAACCTCCTTCCAACCGGTG | -261 to -286 | pCT362LS9 |
| LS10-r | CCGCCCCTCGCGTCAAACCTCCTTCCAACC | -257 to -282 | pCT362LS10 |
| LS11-r | AAACACCTAAATCCCTCGCGTCAAACCTCC | -249 to -274 | pCT362LS11 |
| LS12-r | CCGCTCGCACCTAAATCCCTCGCGTCAAAC | -245 to -270 | pCT362LS12 |
| LS13-r | CCGCGGACTCGCACCTAAATCCCTCGCGTC | -241 to -266 | pCT362LS13 |
| LS14-r | CCGCAGTCGGACTCGCACCTAAATCCCTCG | -237 to -262 | pCT362LS14 |
a Forward primers (LSn-f) are designed along with Ars coding strand, and reverse primers (LSn-r) are designed along with complementary strand of Ars. b Underlined nucleotides indicate introduced mutations. c Mutated sequences in primers are not included in the numbers of positions.
Transformation
The host C. reinhardtii strain 5D (nit1-305, cw-15; Tam and Lefebvre, 1993) was cotransformed with chimeric constructs and pMN24E, which was generated by eliminating a 3-kb EcoRI fragment from pMN24 (Fernandez et al., 1989), using the glass beads method as described previously (Kucho et al., 1999).
Ars Enzyme Assay
Ars activity was assayed quantitatively as described previously (Kucho et al., 1999) with modifications as follows. One hundred and fifty microliters of cell suspensions was transferred to 96-well microtiterplates and incubated under high- or low-CO2 conditions under illumination at 150 μmol m–2 s–1 for 4 h. After the incubation, the A665 of the cell suspensions was determined with a microplate reader (Bio-Rad Laboratories, Hercules, CA) to evaluate the cell density. The cell suspensions were reacted with 1 mm α-naphthylsulfate (Sigma-Aldrich, St. Louis) at 37°C for 2 h, followed by centrifugation of the cells at 1,000g for 10 min. One hundred microliters of the supernatants was transferred to another set of microtiterplates, and 100 μL of a solution containing 4% (w/v) SDS and 0.4 m Na acetate (pH 4.8) was added to stop the reaction. The A540 was measured using the microplate reader immediately after the addition of 20 μL of 10 mg mL–1 tetrazotized o-dianisidine (Sigma-Aldrich). Ars activity was calculated by dividing the A540 by the A665. To evaluate the significance of differences in the Ars activity between transformants containing various chimeric constructs, the nonparametric Mann-Whitney U test was performed using the statistical program JSTAT (http://www.vector.co.jp/soft/win95/business/se030917.html).
Northern-Blot Analysis
Northern-blot analysis was performed as described by Kucho et al. (1999) with the oligonucleotide probe p-CA1–5′ specific to the Cah1 transcript and 0.8- and 1.2-kb BamHI fragments of Ars cDNA.
Nuclear Extracts Preparation
Nuclei were isolated from C. reinhardtii strain cw-15 in a solution containing 0.5% (v/v) NP-40, 25 mm HEPES-NaOH (pH 7.9), 20 mm KCl, 0.6 mm Suc, 10% (v/v) glycerol, and 5 mm dithiothreitol (DTT), as described previously (Keller et al., 1984). Nuclear protein fractions were extracted in a solution containing 20 mm HEPES-NaOH (pH 7.9), 1.5 mm MgCl2, 500 mm KCl, 0.2 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride, 0.5 mm DTT, and 25% (v/v) glycerol and then dialyzed against a solution containing 20 mm HEPES-NaOH (pH 7.9), 100 mm KCl, 0.2 mm EDTA, 0.2 mm phenylmethylsulfonyl fluoride, 0.5 mm DTT, and 20% (v/v) glycerol (Abmayr and Workman, 1997). The protein concentration of the nuclear extracts was determined using a protein assay reagent kit (Bio-Rad Laboratories).
Gel Mobility Shift Assay
To generate probes for gel mobility shift assays, complementary pairs of synthetic oligonucleotides were 5′ end labeled with [γ-32P]ATP, boiled for 5 min, and annealed by cooling to room temperature. The labeled probes were purified by non-denaturing PAGE. Binding reactions were carried out by incubating 0.5 to 1 ng of probe (20,000 cpm) with 2.5 μg of nuclear proteins in 15 μL of reaction buffer containing 4 μg of poly(dI-dC).poly(dI-dC), 4% (v/v) glycerol, 10 mm Tris-HCl (pH 7.5), 50 mm NaCl, 2.5 mm MgCl2, and 0.5 mm DTT at 22°C for 30 min. The reaction mixtures were electrophoresed in an 8% (w/v) non-denaturing polyacrylamide gel for the oEE1 probe or a 5% (w/v) non-denaturing polyacrylamide gel for the o4xEE2 probe in 0.5× Tris-borate/EDTA buffer at 30 mA (Chodosh, 1997). Oligonucleotides used as competitors were phosphorylated at the 5′ ends with cold ATP.
Acknowledgments
We are grateful to John P. Davies and Paul A. Lefebvre for providing pJD100 and pMN24, respectively. We also thank Dr. Aaron Kaplan for his valuable suggestions.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.026492.
This work was supported by Grants-in-Aid from the Japanese Ministry of Education, Science and Culture and by the Japan Society for the Promotion of Science (grant no. JSPS–RFTF97R16001 to H.F.).
References
- Abmayr SM, Workman JL (1997) Preparation of nuclear and cytoplasmic extracts from mammalian cells. In FM Ausubel, R Brent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology. Current Protocols, New York, pp 12.1.1.1–12.1.9
- Aizawa K, Miyachi S (1986) Carbonic anhydrase and CO2 concentrating mechanisms in microalgae and cyanobacteria. FEMS Microbiol Rev 39: 215–233 [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K (1997) Current Protocols in Molecular Biology. Greene Publishing Associates and Wiley-Interscience, New York, pp 12.1.1–12.2.10
- Badger MR, Price GD (1994) The role of carbonic anhydrase in photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 45: 369–392 [Google Scholar]
- Chodosh LA (1997) Mobility shift DNA-binding assay using gell electrophoresis. In FM Ausubel, R Brent, RE Kingston, DD Moore, JG Seidman, JA Smith, K Struhl, eds, Current Protocols in Molecular Biology. Current Protocols, New York, pp 12.2.1–12.2.10
- Davies JP, Grossman AR (1994) Sequences controlling transcription of the Chlamydomonas reinhardtii β2-tubulin gene after deflagellation and during the cell cycle. Mol Cell Biol 14: 5165–5174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisio-Sese ML, Fukuzawa H, Miyachi S (1990) Light-induced carbonic anhydrase expression in Chlamydomonas reinhardtii. Plant Physiol 94: 1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez E, Schnell R, Ranum LPW, Hussey SC, Silflow CD, Lefebvre PA (1989) Isolation and characterization of the nitrate reductase structural gene of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 86: 6449–6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Cassier-Chauvat C, Chauvat F, Cerff R (2001) Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol Microbiol 39: 455–468 [DOI] [PubMed] [Google Scholar]
- Fukuzawa H, Fujiwara S, Yamamoto Y, Dionisio-Sese ML, Miyachi S (1990) cDNA cloning, sequence, and expression of carbonic anhydrase in Chlamydomonas reinhardtii: regulation by environmental CO2 concentration. Proc Natl Acad Sci USA 87: 4383–4387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuzawa H, Miura K, Ishizaki K, Kucho K, Saito T, Kohinata T, Ohyama K (2001) Ccm1, a regulatory gene controlling the induction of a carbon-concentrating mechanism in Chlamydomonas reinhardtii by sensing CO2 availability. Proc Natl Acad Sci USA 98: 5347–5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodbourn S, Zinn K, Maniatis T (1985) Human β-interferon gene expression is regulated by an inducible enhancer element. Cell 41: 509–520 [DOI] [PubMed] [Google Scholar]
- Green PJ, Kay SA, Chua N-H (1987) Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J 6: 2543–2549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green PJ, Yong M-H, Cuozzo M, Kano-Murakami Y, Silverstein P, Chua N-H (1988) Binding site requirements for pea nuclear protein factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A gene. EMBO J 7: 4035–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearing P, Shenk T (1983) The adenovirus type 5 E1A transcriptional control region contains a duplicated enhancer element. Cell 33: 695–704 [DOI] [PubMed] [Google Scholar]
- Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res 27: 297–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka K, Wu X, Fukuzawa H, Chua N-H (1994) Molecular dissection of GT-1 from Arabidopsis. Plant Cell 6: 1805–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Karin M (1992) The regulation of transcription by phosphorylation. Cell 70: 375–387 [DOI] [PubMed] [Google Scholar]
- Kaplan A, Reinhold L (1999) CO2 concentrating mechanisms in photosynthetic microorganisms. Annu Rev Plant Physiol Plant Mol Biol 50: 539–570 [DOI] [PubMed] [Google Scholar]
- Keller LR, Schloss JA, Silflow CD, Rosenbaum JL (1984) Transcription of α- and β-tubulin genes in vitro in isolated Chlamydomonas reinhardtii nuclei. J Cell Biol 98: 1138–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucho K, Ohyama K, Fukuzawa H (1999) CO2-responsive transcriptional regulation of CAH1 encoding carbonic anhydrase is mediated by enhancer and silencer regions in Chlamydomonas reinhardtii. Plant Physiol 121: 1329–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlemeier C, Fluhr R, Green PJ, Chua N-H (1987) Sequences in the pea rbcS-3A gene have homology to constitutive mammalian enhancers but function as negative regulatory elements. Genes Dev 1: 247–255 [DOI] [PubMed] [Google Scholar]
- Maréchal E, Hiratsuka K, Delgado J, Nairn A, Qin J, Chait BT, Chua N-H (1999) Modulation of GT-1 DNA-binding activity by calcium-dependent phosphorylation. Plant Mol Biol 40: 373–386 [DOI] [PubMed] [Google Scholar]
- Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T (1988) Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to INF-β gene regulatory elements. Cell 54: 903–914 [DOI] [PubMed] [Google Scholar]
- Nagano Y (2000) Several features of the GT-factor trihelix domain resemble those of the Myb DNA-binding domain. Plant Physiol 124: 491–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omata T, Gohta S, Takahashi Y, Hirano Y, Maeda S-I (2001) Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol 183: 1891–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn JM, Merchant S (1995) Two copper-responsive elements associated with the Chlamydomonas cyc6 gene function as targets for transcriptional activators. Plant Cell 7: 623–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KB (1998) Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiol 118: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MH (1998) CO2 acquisition: acclimation to changing carbon availability. In JD Rochaix, M Goldschmidt-Clermont, S Merchant, eds, The Molecular Biology of Chloroplasts and Mitochondria in Chlamydomonas. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 529–547
- Sueoka N (1960) Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 46: 83–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam L-W, Lefebvre PA (1993) Cloning of flagellar genes in Chlamydomonas reinhardtii by DNA insertional mutagenesis. Genetics 135: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villand P, Eriksson M, Samuelsson G (1997) Carbon dioxide and light regulation of promoters controlling the expression of mitochondrial carbonic anhydrase in Chlamydomonas reinhardtii. Biochem J 327: 51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]