Abstract
Integrin-linked kinase (ILK) is a multidomain focal adhesion (FA) protein that functions as an important regulator of integrin-mediated processes. We report here the identification and characterization of a new calponin homology (CH) domain-containing ILK-binding protein (CH-ILKBP). CH-ILKBP is widely expressed and highly conserved among different organisms from nematodes to human. CH-ILKBP interacts with ILK in vitro and in vivo, and the ILK COOH-terminal domain and the CH-ILKBP CH2 domain mediate the interaction. CH-ILKBP, ILK, and PINCH, a FA protein that binds the NH2-terminal domain of ILK, form a complex in cells. Using multiple approaches (epitope-tagged CH-ILKBP, monoclonal anti–CH-ILKBP antibodies, and green fluorescent protein–CH-ILKBP), we demonstrate that CH-ILKBP localizes to FAs and associates with the cytoskeleton. Deletion of the ILK-binding CH2 domain abolished the ability of CH-ILKBP to localize to FAs. Furthermore, the CH2 domain alone is sufficient for FA targeting, and a point mutation that inhibits the ILK-binding impaired the FA localization of CH-ILKBP. Thus, the CH2 domain, through its interaction with ILK, mediates the FA localization of CH-ILKBP. Finally, we show that overexpression of the ILK-binding CH2 fragment or the ILK-binding defective point mutant inhibited cell adhesion and spreading. These findings reveal a novel CH-ILKBP–ILK–PINCH complex and provide important evidence for a crucial role of this complex in the regulation of cell adhesion and cytoskeleton organization.
Keywords: focal adhesion, integrin-linked kinase, calponin homology, integrins, cell adhesion
Introduction
Cell adhesion to the extracellular matrix (ECM) is mediated primarily by integrins (Hynes 1992), which together with soluble factors, control gene expression, differentiation, proliferation, and morphological changes (Clark and Brugge 1995; Schwartz et al. 1995; Dedhar and Hannigan 1996; Sastry and Horwitz 1996). Focal adhesions (FAs) are integrin-rich cell adhesion sites through which the ECM is physically linked to the actin cytoskeleton and signals are transduced into the intracellular compartment (for review see Jockusch et al. 1995; Burridge and Chrzanowska-Wodnicka 1996; Calderwood et al. 2000). A selective group of cytoplasmic proteins, which include catalytic proteins such as FA kinase (FAK) and noncatalytic proteins such as α-actinin, talin, and paxillin are recruited to FAs in response to cell adhesion (Parsons et al. 1994; Ilic et al. 1997; Turner 1998). Consistent with the fundamental roles of cell matrix adhesions in control of cell behavior, many components of the FAs are well conserved in different organisms ranging from nematodes and insects to vertebrates (Hynes and Zhao 2000).
Integrin-linked kinase (ILK) is an evolutionally conserved FA protein that is involved in the integrin-mediated processes (Hannigan et al. 1996; Dedhar et al. 1999; Li et al. 1999; Wu 1999). ILK participates in the regulation of cell adhesion, growth, gene expression, differentiation, cell shape change, and ECM assembly. Furthermore, recent genetic studies in Caenorhabditis elegans [Mackinnon, A.C., and B.D. Williams. 2000. 40th American Society for Cell Biology Annual Meeting. 2664. (Abstr.)] and Drosophila (Zervas et al. 2001) have demonstrated that lack of ILK expression results in phenotypes resembling that of integrin-null mutants, suggesting that ILK is indispensable for integrin function during embryonic development.
Although it is clear that ILK is an important component of FAs, how ILK functions in FA is not completely understood. At the molecular level, ILK consists of three structurally distinct motifs. At the NH2 terminus of ILK lie four ankyrin (ANK) repeats, which are responsible for binding to PINCH (Tu et al. 1999). The PINCH binding is required for FA localization of ILK (Li et al. 1999). In addition, it connects ILK to other proteins including Nck-2, a SH2/3-containing adaptor protein involved in the growth factor and small GTPase signaling (Tu et al. 1998). The importance of the ILK–PINCH interaction is underscored by recent genetic studies in C. elegans (Hobert et al. 1999), showing that lack of PINCH causes a phenotype that is identical to that of null mutation of β-integrin/pat-3 or ILK/pat-4. COOH-terminal to the ANK repeat domain is a pleckstrin homology-like motif that likely binds PtdIns(3,4,5)P3 and participates in the regulation of the kinase activity (Delcommenne et al. 1998). ILK COOH-terminal domain exhibits significant homology to other kinase catalytic domains (Dedhar et al. 1999). ILK phosphorylates several key proteins including PKB/Akt and glycogen synthase kinase 3 and regulates their activation (Delcommenne et al. 1998). In addition to catalyzing serine/threonine phosphorylation, the multidomain structure and FA localization of ILK suggest that it could potentially function as a scaffolding protein mediating multiple protein–protein interactions. To test this hypothesis, we have carried out studies aimed at identifying additional ILK-binding proteins. We report here the identification of a new calponin homology (CH) domain-containing ILK-binding protein (CH-ILKBP) that is widely expressed and highly conserved among different organisms. Furthermore, we have characterized the interaction between ILK and CH-ILKBP and show that CH-ILKBP, ILK, and PINCH form a complex in cells. Additionally, we demonstrate that CH-ILKBP localizes to FAs and the ILK-binding activity is required for the FA localization. Finally, we provide functional evidence showing that CH-ILKBP is critically involved in the regulation of cell adhesion and spreading.
Materials and Methods
Yeast Two-Hybrid Assays
A cDNA fragment encoding human ILK residues 136–452 was inserted into the EcoRI/XhoI sites of a pLexA vector (CLONTECH Laboratories, Inc.). The resulting construct (pLexA/ILK136–452) was used as a bait to screen a human heart MATCHMAKER LexA cDNA library (>3 × 106–independent clones) (CLONTECH Laboratories, Inc.) as described (Tu et al. 1998, Tu et al. 1999). 48 positive colonies were obtained and analyzed by PCR. A majority (39 out of the 48) of them contain cDNA inserts with an identical size (1.5 kb). Four clones from this major positive group were selected, and the cDNA inserts were sequenced. They contain an identical cDNA fragment encoding CH-ILKBP residues 29–372. A cDNA encoding full-length CH-ILKBP was isolated from a human heart cDNA library (CLONTECH Laboratories, Inc.) by PCR using the following primers: 5′-GGTCCATATGGCCACCTCCCCGCAGAAG-3′ and 5′-CTGCTCGAGTCACTCCACGTTACGGTAC-3′). Additionally, we performed yeast two-hybrid binding assays to determine the interactions between specific CH-ILKBP and ILK sequences. The interactions were quantified by measuring the β-galactosidase activity as described (Tu et al. 1998, Tu et al. 1999).
Northern Blot
A 32P-labeled CH-ILKBP cDNA probe was prepared by labeling a human CH-ILKBP cDNA fragment (encoding residues 1–229) using a random-primed DNA labeling kit (Boehringer). A blot containing equal amount (2 μg /lane) of polyA+ RNA from different human tissues (CLONTECH Laboratories, Inc.) was hybridized with the 32P-labeled CH-ILKBP probe following the manufacturer's protocol.
Site-directed Mutagenesis
A QuickChange™ site-directed mutagenesis system (Stratagene) was used to change F at position 271 to D. The point mutation was confirmed by DNA sequencing.
Generation of Glutathione-S-Transferase– and Maltose-binding Protein–CH-ILKBP Fusion Proteins
DNA fragments encoding CH-ILKBP sequences were prepared by PCR and inserted into the EcoRI/XhoI sites of a pGEX-5x-1 vector (Amersham Pharmacia Biotech) or the EcoRI/SalI sites of a pMAL-C2 vector (New England BioLabs, Inc.). The recombinant vectors were used to transform Escherichia coli cells. The expression of the glutathione S-transferase (GST) and maltose-binding protein (MBP) fusion proteins was induced with IPTG, and they were purified by affinity chromatography using glutathione-Sepharose 4B and amylose-agarose (Tu et al. 1999), respectively.
GST Fusion Protein Pull-Down Assays
Mouse C2C12 cells were lysed with 1% Triton X-100 in 20 mM Tris HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1 mM Na3VO4, 2 mM AEBSF, 5 μg/ml pepstatin A, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. The lysates were precleared and then incubated with equal amounts of GST–CH-ILKBP fusion proteins or GST for 2 h at 4°C. GST and GST fusion proteins were precipitated with glutathione-Sepharose beads and ILK was detected by Western blotting with anti-ILK antibody 65.1 (5 μg/ml).
Generation of Monoclonal Anti–CH-ILKBP Antibodies
Mouse monoclonal anti–CH-ILKBP antibodies were prepared using GST fusion protein containing CH-ILKBP residues 29–372 as an antigen based on a previously described method (Tu et al. 1998). Hybridoma supernatants were initially screened for anti–CH-ILKBP activities by ELISA and Western blotting using MBP–CH-ILKBP29–372. Antibodies that recognize MBP–CH-ILKBP29–372 were selected and further tested by Western blotting using GST–CH-ILKBP fusion proteins and mammalian cell lysates.
Expression of FLAG– and Green Fluorescent Protein–CH-ILKBP Fusion Proteins in Mammalian Cells
DNA fragments encoding CH-ILKBP sequences were cloned into the EcoRI/KpnI sites of the p3XFLAG–CMV-14 vector (Sigma-Aldrich) or those of the pEGFP-N1 vector (CLONTECH Laboratories, Inc.). C2C12 cells, rat mesangial cells, and CHO K1 cells were transfected with the vectors using LipofectAmine PLUS (Life Technologies) (Huang et al. 2000).
Coimmunoprecipitation Assays
To immunoprecipitate endogenous CH-ILKBP, C2C12 cells were lysed with 1% Triton X-100 in the Hepes buffer (50 mM Hepes, pH 7.1, 150 mM NaCl, 10 mM Na4P2O7, 2 mM Na3VO4, 100 mM NaF, 10 mM EDTA) containing protease inhibitors. The cell lysates (750 μg) were incubated with 750 μl of hybridoma culture supernatant containing anti–CH-ILKBP antibody 1D4 or 750 μl of unconditioned medium as a control for 2 h. The samples were then mixed with 50 μl of UltraLink immobilized protein G (Pierce Chemical Co.). After incubation for 2 h, the beads were washed four times, and the proteins bound were released from the beads by boiling in 75 μl of SDS-PAGE sample buffer for 5 min. The samples (10 μl/lane) were analyzed by Western blotting with anti–CH-ILKBP antibody 3B5, anti-ILK antibody 65.1, antipaxillin antibody (clone 349; Transduction Laboratories), and rabbit polyclonal anti-PINCH antibodies, respectively.
To immunoprecipitate FLAG-tagged, wild-type, or mutant forms of CH-ILKBP, lysates (prepared as described above) of the C2C12 transfectants (500 μg lysates) or the CHO transfectants (750 μg lysates) were mixed with 70 μl agarose beads conjugated with anti-FLAG antibody M2 (Sigma-Aldrich). The precipitated proteins were released from the beads by boiling in 50 μl of SDS-PAGE sample buffer for 5 min and analyzed by Western blotting (10 μl/lane).
To immunoprecipitate FLAG-PINCH, C2C12 cells were transfected with pFLAG–CMV-2/PINCH, which was generated by inserting the full-length PINCH cDNA into the BglII/SalI sites of the pFLAG–CMV-2 vector (Sigma-Aldrich) using LipofectAmine2000 (Life Technologies). 24 h after transfection, the cells were lysed with 1% Triton X-100 in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 2 mM Na3VO4, and 100 mM NaF containing protease inhibitors. The lysates (1 mg) were mixed with 30 μl agarose beads conjugated with anti-FLAG antibody M2. The precipitated proteins were released from the beads by boiling in 60 μl of SDS-PAGE sample buffer for 5 min and analyzed by Western blotting (10 μl/lane).
To immunoprecipitate paxillin, C2C12 lysates (500 μg) were mixed with 2 μg of antipaxillin antibody or 2 μg of irrelevant mouse IgG. The immune complexes were precipitated with 50 μl of UltraLink immobilized protein G, and the proteins bound were released from the beads by boiling in 50 μl of SDS-PAGE sample buffer for 5 min. The samples (10 μl/lane) were analyzed by Western blotting.
Immunofluorescence Staining of Cells
Immunofluorescence staining was performed as described (Li et al. 1999). In brief, cells were plated on fibronectin-coated coverslips, fixed, and stained with mouse anti–CH-ILKBP antibody 1D4 (hybridoma supernatant), mouse anti-FLAG antibody M2 (4 μg/ml), rabbit anti-FAK antibody (4 μg/ml) (Santa Cruz Biotechnology, Inc.), and/or rabbit anti–β-catenin antiserum (1:2,000 dilution) (Sigma-Aldrich). The mouse and rabbit antibodies were detected with a fluorescein-conjugated anti–mouse IgG antibody (28 μg/ml) and a Rhodamine redTX-conjugated anti–rabbit IgG antibody (10 μg/ml), respectively. In control experiment, no specific staining was observed with unconditioned medium or with ID4 supernatants that were preincubated with 1 μM GST–CH-ILKBP for 1 h. For staining of cells expressing green fluorescent protein (GFP)–CH-ILKBP fusion proteins, anti-ILK and FAK antibodies were detected with Rhodamine redTX-conjugated anti–mouse and anti–rabbit IgG antibodies, respectively. Actin stress fibers were visualized by staining the cells with rhodamine-labeled phalloidin.
Isolation of Cytoskeleton Fractions
Cytoskeleton fractions were isolated from C2C12 cells as described (Yang et al. 1994). In brief, C2C12 cell monalayers were extracted with 0.7 ml of cold PBS containing 1% Trion X-100 and protease inhibitors. The Triton X-100–insoluble material (cytoskeletal fractions) remaining on the dishes was extracted with equal volume (0.7 ml) of PBS containing 1% SDS and protease inhibitors. The distributions of CH-ILKBP and other FA proteins were analyzed by Western blotting (30 μl/lane).
Cell Adhesion and Spreading Assays
For assays using CHO cells, the cells were transfected with FLAG vectors encoding CH-ILKBP, CH2 (residues 222–372), CH-ILKBP F271→D point mutant, CH2 deletion mutant (residues 1–229), or a FLAG vector lacking CH-ILKBP sequence using LipofectAmine PLUS. Cell adhesion assays were performed as described (Wu et al. 1995). In brief, the transfectants were harvested with trypsin 1 d after transfection and washed twice with α-MEM containing 10% FBS, twice with serum-free Opti-MEM (Life Technologies), and kept in suspension for 30 min. The cells (3 × 104/well) were seeded in collage IV–coated 96-well plates (Becton Dickinson). After incubation at 37°C under a 5% CO2–95% air atmosphere for 90 min, the wells were washed three times with PBS. The numbers of adhered cells were quantified by measuring N-acetyl-β-d-hexosaminidase activity (Landegren 1984).
For assays using C2C12 cells, the cells were transfected with FLAG vectors encoding CH-ILKBP (FLAG–CH-ILKBP), CH2 (residues 222–372) (FLAG-CH2), CH2 deletion mutant (residues 1–229) (FLAG-ΔCH2), or a FLAG control vector using LipofectAmine PLUS. C2C12 cells stably expressing the FLAG-tagged proteins were selected with 1 mg/ml of G418 (Life Technologies). Two independent clones expressing each form of CH-ILKBP were isolated. C2C12 cells expressing FLAG-CHILKBP (clones A28 and A31), FLAG-CH2 (clones B20 and B45), FLAG-ΔCH2 (clones C31 and C46), and the vector control cells were harvested with trypsin. The cells were washed twice with DME containing 10% FBS and twice with DME containing 1% BSA. The cells were kept in suspension for 30 min and then seeded in 12-well plates coated with collagen I (Becton Dickinson). The cells were allowed to adhere at 37°C for 30 min, and four randomly selected fields were photographed. The plates were then washed twice with PBS, and four randomly selected fields were photographed after the wash. The numbers of the total cells (before wash) and the adhered cells (after wash) from the four randomly selected fields were counted manually. The percentage of cell adhesion is presented as the number of adhered cells divided by the number of total cells. In addition to manual counting, we also measured cell adhesion using the hexosaminidase method and obtained similar results.
For cell spreading, C2C12 cells expressing different forms of CH-ILKBP were prepared as described above and seeded in 12-well plates coated with collagen I. The plates were incubated at 37°C under a 5% CO2-95% air atmosphere for different periods of time. The cell morphology (phase–contrast image) was recorded with a DVC-1310C Magnafire™ digital camera (Optronics). Unspread cells were defined as round phase–bright cells, whereas spread cells were defined as cells with extended processes, lacking a rounded morphology and not phase–bright (Komoriya et al. 1991; Richardson et al. 1997). The percentage of cells adopting spread morphology was quantified by analyzing ≥300 cells from three randomly selected fields (>100 cells/field).
Online Supplemental Material
Figure S1: Association of FLAG–CH-ILKBP, FLAG-F271D, and FLAG-ΔCH2 with Triton X-100–insoluble cytoskeleton fractions. Figure S2: CH-ILKBP binds to ILK but not paxillin in rat embryo fibroblasts (REF-52). Figure S3: Immunofluorescence staining of GFP–CH-ILKBP–expressing cells with monoclonal antipaxillin antibody. Supplemental figures available at http://www.jcb.org/cgi/content/full/153/3/585/DC1.
Results
Identification of a Novel ILK-binding Protein by Yeast Two-Hybrid Screens
To identify proteins that interact with the COOH-terminal region of ILK, we screened a human heart LexA cDNA library with a bait construct encoding ILK residues 136–452. 48 positive clones were obtained. PCR analyses showed that 39 out of the 48 positive clones contained cDNA inserts with an identical size (1.5 kb). Four clones (clones 10, 12, 35, and 36) from this major positive group were selected and sequenced. The results showed that they contained an identical cDNA insert. BLAST searches of cDNA database revealed that this insert overlaps with several EST clones and a cDNA encoding an “Unnamed Protein Product” (sequence data available from Genbank/EMBL/DDBJ under accession numbers AA057458, AA316121, and AK001655). A cDNA encoding the full-length ILKBP was isolated from the heart cDNA library by PCR. It encodes a protein of 372 residues comprising two CH domains at the COOH-terminal region (Fig. 1 A) and was therefore designated as CH domain–containing ILK-binding protein or CH-ILKBP. The CH domains of CH-ILKBP share significant homology with those of α-actinin, filamin, and other actin-binding proteins. Proteins that are structurally closely related are present in other organisms including C. elegans (Fig. 1 A), suggesting that CH-ILKBP, like ILK and PINCH, represents an ancient protein.
Figure 1.
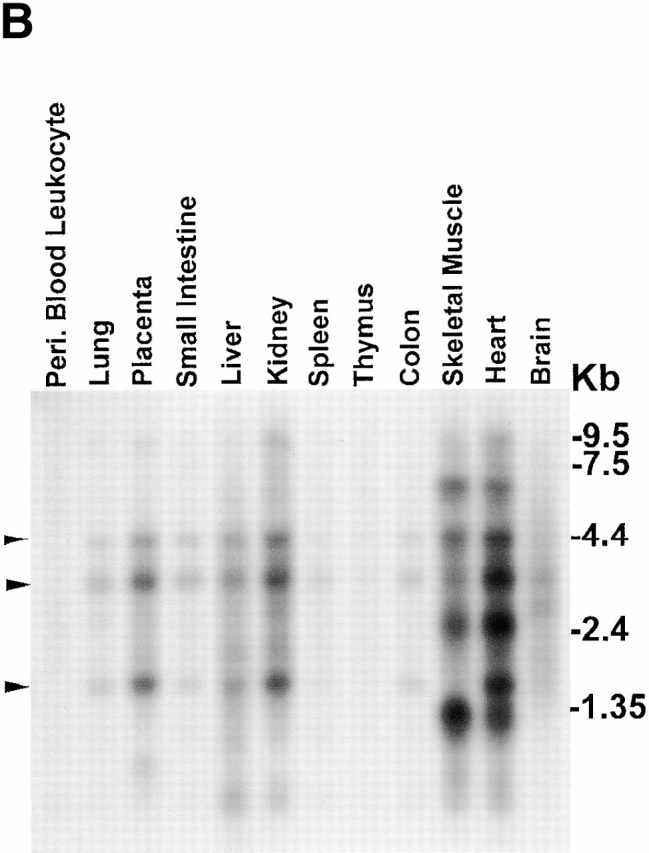
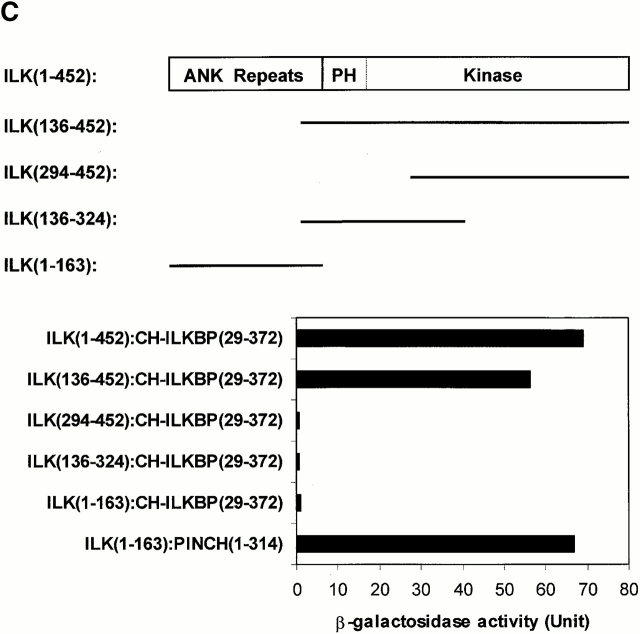
Identification of a new CH-containing ILK-binding protein. (A) Sequence alignment of human CH-ILKBP (sequence data available from GenBank/EMBL/DDBJ under accession number AF325830) with C. elegans T21D12.4 protein (sequence data available from GenBank/EMBL/DDBJ under accession no. AAC48090). Identical residues are indicated by gray background. The CH domains are underlined. (B) Northern blotting. Two micrograms of polyA+ RNA from human tissues were hybridized with a CH-ILKBP cDNA probe as described in Materials and Methods. Arrowheads indicate the positions of three transcripts (4.4, 3.5, and 1.7 kb). (C) ILK residues involved in the CH-ILKBP binding. The binding was determined by yeast two- hybrid binding assays as described in Materials and Methods. The numbers in parentheses indicate ILK, CH-ILKBP, and PINCH residues.
Northern blot analyses with a 32P-labeled CH-ILKBP cDNA probe showed three positive bands (4.4, 3.5, and 1.7 kb) in almost all human tissues tested, with strong signals detected in the heart, skeletal muscle, and kidney (Fig. 1 B). Additional bands, which likely represent tissue-specific CH-ILKBP isoforms or other related transcripts, were also detected in the heart and the skeletal muscle. The presence of multiple bands suggests that there likely exists a family of structurally related proteins. To confirm the interaction between ILK and CH-ILKBP, we cotransformed yeast cells with purified ILK and CH-ILKBP expression vectors and found that they readily bind to each other (Table ). In control experiments, replacement of either binding partner sequence with those of other proteins (for example, PINCH and lamin C) abolished the binding (Table ), confirming the specificity of the assay.
Table 1.
The ILK COOH-terminal Domain Specifically Interacts with CH-ILKBP in Yeast
| pLexA construct | pB42AD construct | Reporter gene | |
|---|---|---|---|
| LEU2 | lacZ | ||
| pLexA-ILK136–452 | pB42AD–CH-ILKBP29–372 | + | + |
| pLexA-Lam C | pB42AD–CH-ILKBP29–372 | − | − |
| pLexA-ILK136–452 | pB42AD-PINCH | − | − |
The interactions between the proteins encoded by the pLexA and pB42AD constructs were determined by yeast two-hybrid assays as described in Materials and Methods.
The ILK COOH-terminal Domain but not the NH2-terminal Domain Mediates the Interaction with CH-ILKBP
We next sought to identify regions of ILK involved in the CH-ILKBP binding. To do this, we expressed the full-length and various ILK mutants in yeast cells and analyzed the CH-ILKBP–binding activities (Fig. 1 C). The full-length ILK, like the ILK COOH-terminal fragment (136–452), readily binds to CH-ILKBP (Fig. 1 C). By contrast, the NH2-terminal ANK domain (1–163), which binds PINCH (Fig. 1 C) (Tu et al. 1999), failed to bind to CH-ILKBP. Furthermore, no interaction between an ILK mutant (residues 136–324) containing the pleckstrin homology-like motif but lacking the COOH-terminal most residues was detected (Fig. 1 C), suggesting that the COOH-terminal most residues are required for the binding. However, the COOH-terminal most residues are not sufficient for the binding, since an ILK mutant containing residues 193–452 failed to bind CH-ILKBP (Fig. 1 C). Taken together, these results suggest that the COOH-terminal domain of ILK is both necessary and sufficient for the interaction with CH-ILKBP.
The CH2 Domain Mediates the Interaction with ILK
To confirm the interaction between CH-ILKBP and ILK, we generated GST fusion proteins containing the full-length (Fig. 2 A, lane 1) and the COOH-terminal region (ΔN-ter, residues 29–372) (Fig. 2 A, lane 7) of CH-ILKBP and tested their ability to bind ILK. Both GST–CH-ILKBP (Fig. 2 B, lane 1) and GST–ΔN-ter (29–372) (Fig. 2 B, lane 7), but not GST alone (Fig. 2 B, lane 8), interacted with ILK. These results are consistent with those obtained in the yeast two-hybrid binding assays and confirm that CH-ILKBP forms a complex with ILK in vitro and in yeast cells.
Figure 2.
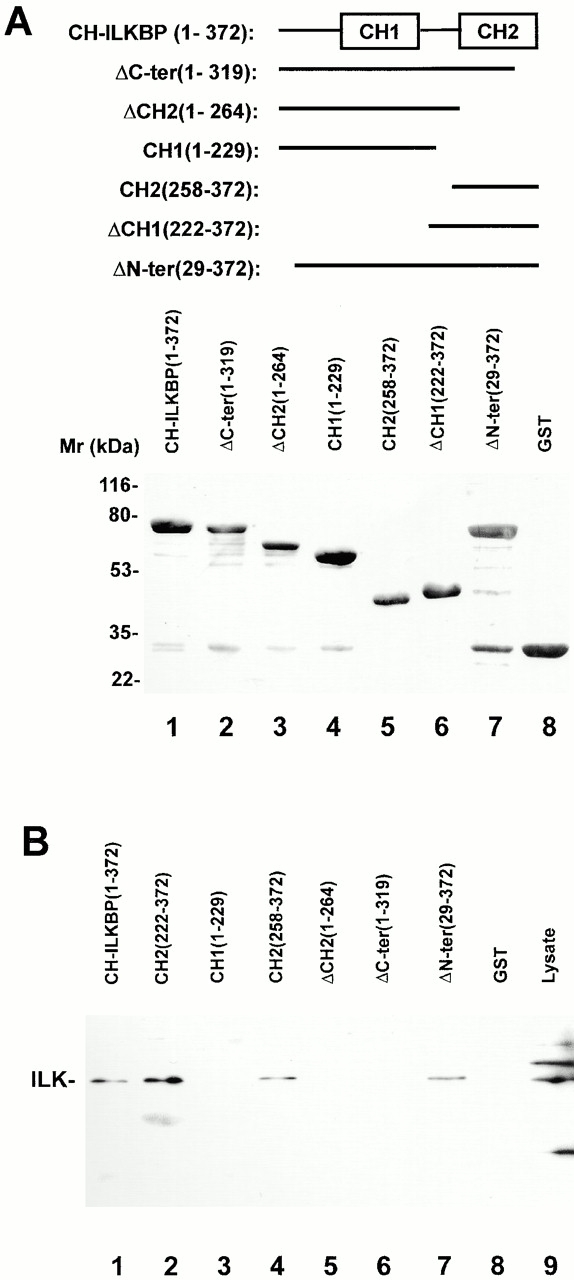
The CH2 domain mediates the interaction with ILK. (A) GST–CH-ILKBP fusion proteins were separated on SDS-PAGE (10 μg/lane) and detected by Coomassie blue R-250 staining. The numbers in parentheses indicate CH-ILKBP residues. (B) ILK binding. C2C12 cell lysates (100 μg) were incubated with equal amount (10 μg) of GST or GST fusion proteins containing CH-ILKBP sequences as indicated in the figure. GST and the GST fusion proteins were precipitated with glutathione-Sepharose 4B beads. ILK was detected by Western blotting with anti-ILK antibody 65.1. (Lane 9) C2C12 cell lysates (10 μg/lane).
CH-ILKBP contains two tandem CH domains (Fig. 1 A). To define the ILK-binding site on CH-ILKBP, we generated a series of GST fusion proteins containing different regions of CH-ILKBP (Fig. 2 A) and analyzed their ILK-binding activities. Deletion of 221 residues from the NH2 terminus did not reduce the ILK-binding activity (Fig. 2 B, lane 2), indicating that the NH2-terminal region including CH1 is not required for the ILK binding. Furthermore, a GST fusion protein containing only the CH2 domain bound to ILK (Fig. 2 B, lane 4). By contrast, deletion of either the entire or part of CH2 abolished the ILK binding (Fig. 2 B, lanes 3, 5, and 6). We conclude from these results that the CH2 domain mediates the interaction with ILK.
CH-ILKBP Associates with ILK in Mammalian Cells
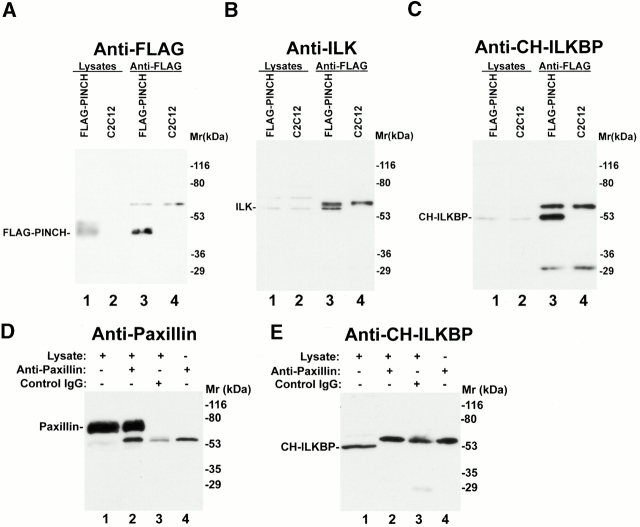
To facilitate studies on CH-ILKBP in mammalian cells, we generated mouse monoclonal anti–CH-ILKBP antibodies. Two antibodies (1D4 and 3B5), which recognize GST–CH-ILKBP (Fig. 3A and Fig. B, lane 2) and MBP–CH-ILKBP29–372 (data not shown) but not GST (Fig. 3A and Fig. B, lane 1), were further characterized. 1D4 recognizes an epitope located within the link region between the two CH domains of CH-ILKBP (Fig. 3 A), whereas 3B5 recognizes an epitope located within the NH2-terminal region (residues 1–229) (Fig. 3 B). To test whether ILK interacts with CH-ILKBP in mammalian cells, we immunoprecipitated CH-ILKBP from C2C12 cell lysates with anti–CH-ILKBP antibody 1D4. Western blotting analyses showed that ILK (Fig. 3 C, lane 2) was coprecipitated with CH-ILKBP (Fig. 3 D, lane 2). Probing the same samples with an antipaxillin antibody failed to detect paxillin in the anti–CH-ILKBP immunoprecipitates (Fig. 3 E, lane 2), despite the presence of abundant paxillin in the cell lysate (Fig. 3 E, lane 1). In additional control experiments, neither CH-ILKBP nor ILK was precipitated in the absence of the anti–CH-ILKBP antibody (Fig. 3C and Fig. D, lane 4). Thus, consistent with the ILK–CH-ILKBP interaction detected in yeast cells and in vitro, ILK and CH-ILKBP form a complex in mammalian cells.
Figure 3.
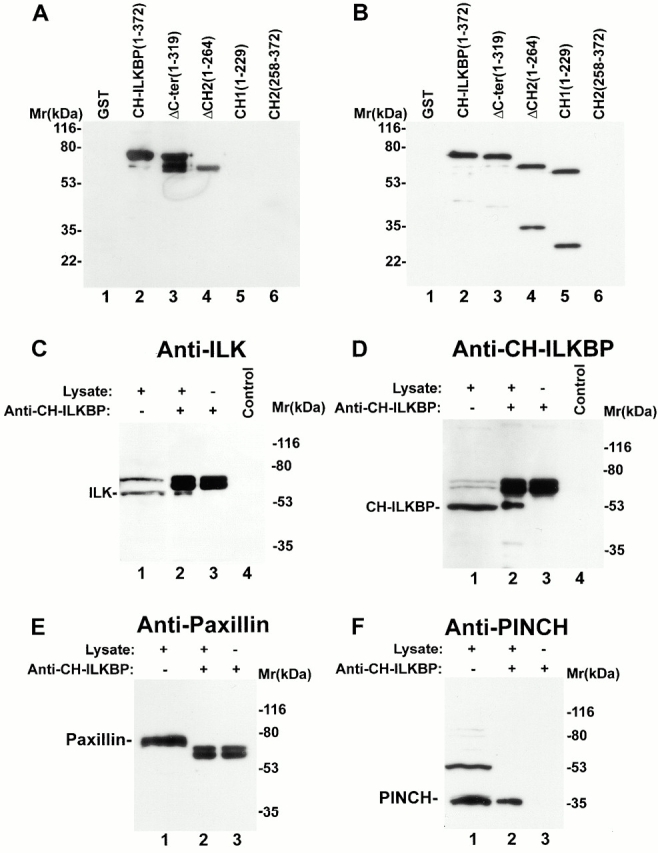
CH-ILKBP forms a complex with ILK and PINCH in mammalian cells. (A and B) GST and GST–CH-ILKBP proteins (0.1 μg protein/lane) were analyzed by Western blotting with anti–CH-ILKBP antibody 1D4 (A) and 3B5 (B). (C and D) Coimmunoprecipitation of ILK with CH-ILKBP. Anti–CH-ILKBP immunoprecipitates (lane 2) and control precipitates (lane 4) were prepared using C2C12 cell lysates as described in Materials and Methods. ILK and CH-ILKBP were detected by Western blotting with anti-ILK antibody 65.1 (C) and anti–CH-ILKBP antibody 3B5 (D), respectively. Lane 1 was loaded with C2C12 cell lysates (20 μg protein/lane). (E and F) Cell lysates (lane 1) and anti–CH-ILKBP immunoprecipitates (lane 2) were analyzed by Western blotting using mouse antipaxillin (E) or rabbit anti-PINCH (F) antibodies. Lane 3 was loaded with samples prepared like those of lane 2 except that the lysates were omitted.
CH-ILKBP, ILK, and PINCH Form a Ternary Complex in Mammalian Cells
We next tested whether the CH-ILKBP–ILK complex associates with PINCH, an FA protein that interacts with the NH2-terminal ANK domain of ILK (Tu et al. 1999). Western blotting analyses of the anti–CH-ILKBP immunoprecipitates with anti-PINCH antibodies showed that PINCH (Fig. 3 F, lane 2) was coprecipitated with ILK (Fig. 3 C, lane 2) and CH-ILKBP (Fig. 3 D, lane 2). These results suggest that CH-ILKBP, ILK, and PINCH form a complex in mammalian cells. To confirm this, we expressed FLAG-PINCH in C2C12 cells (Fig. 4 A, lane 1). FLAG-PINCH was immunoprecipitated from the cell lysates with anti-FLAG antibody (Fig. 4 A, lane 3). Western blotting analyses of the immunoprecipitates showed that, as expected, ILK (Fig. 4 B, lane 3) was coimmunoprecipitated with FLAG-PINCH (Fig. 4 A, lane 3). Consistent with coimmunoprecipitation of PINCH with ILK and CH-ILKBP (Fig. 3C, Fig. D, and Fig. F), CH-ILKBP was coprecipitated with ILK and FLAG-PINCH (Fig. 4 C, lane 3). In control experiments, neither ILK (Fig. 4 B, lane 4) nor CH-ILKBP (Fig. 4 C, lane 4) was precipitated with the anti-FLAG antibody in the absence of FLAG-PINCH (Fig. 4 A, lane 4), confirming the specificity of the assays. In additional control experiments, we immunoprecipitated paxillin and found that CH-ILKBP was not coprecipitated with paxillin (Fig. 4D and Fig. E). Taken together, these results show that CH-ILKBP forms a complex with ILK and PINCH but not with paxillin in mammalian cells.
Figure 4.
Coimmunoprecipitation of CH-ILKBP and ILK with FLAG-PINCH. (A and B) FLAG-PINCH was immunoprecipitated from lysates of C2C12 cells expressing FLAG-PINCH. FLAG-PINCH immunoprecipitates (lane 3) or control precipitates (lane 4) were analyzed by Western blotting with anti-FLAG antibody M2 (A), anti-ILK antibody 65.1 (B), or anti–CH-ILKBP antibody 3B5 (C). Lanes 1 and 2 were loaded with lysates (7.5 μg protein/lane) of the FLAG-PINCH–expressing C2C12 cells and the control C2C12 cells, respectively. (D and E) Antipaxillin immunoprecipitates (lane 2) or control IgG precipitates (lane 3) were blotted with antipaxillin antibody (D) or anti–CH-ILKBP antibody 3B5 (E). (Lane 1) 10 μg lysates; (lane 4) 0.4 μg antipaxillin IgG.
CH-ILKBP Localizes to FAs and Associates with the Cytoskeleton
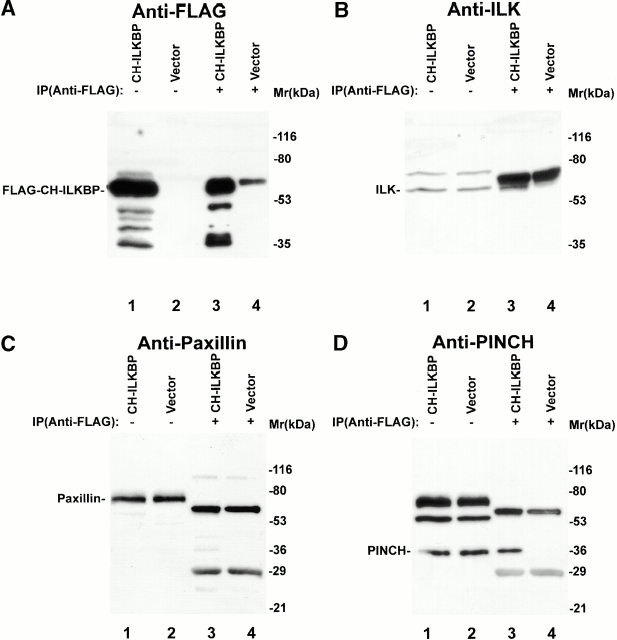
Previous studies have shown that ILK is a component of FAs (Hannigan et al. 1996; Li et al. 1999), sites where actin cytoskeleton connects to integrins and the ECM. The finding that CH-ILKBP binds to ILK prompted us to examine whether CH-ILKBP localizes to FAs. To do this, we expressed FLAG–CH-ILKBP in mammalian cells. We first analyzed the ability of FLAG–CH-ILKBP to interact with ILK by coimmunoprecipitation assays. The results showed that ILK (Fig. 5 B, lane 3) was coimmunoprecipitated with FLAG–CH-ILKBP (Fig. 5 A, lane 3). Analyses of the same immunoprecipitates showed that paxillin was not coimmunoprecipitated with FLAG–CH-ILKBP (Fig. 5 C, lane 3). Reprobing the membrane with anti-PINCH antibodies showed that, as expected, PINCH was coimmunoprecipitated with ILK and FLAG–CH-ILKBP (Fig. 5 D, lane 3). In control experiments, neither ILK (Fig. 5 B, lane 4) nor PINCH (Fig. 5 C, lane 4) was precipitated in the absence of FLAG–CH-ILKBP (Fig. 5 A, lane 4). Thus, FLAG–CH-ILKBP like endogenous CH-ILKBP forms a complex with ILK and PINCH in mammalian cells.
Figure 5.
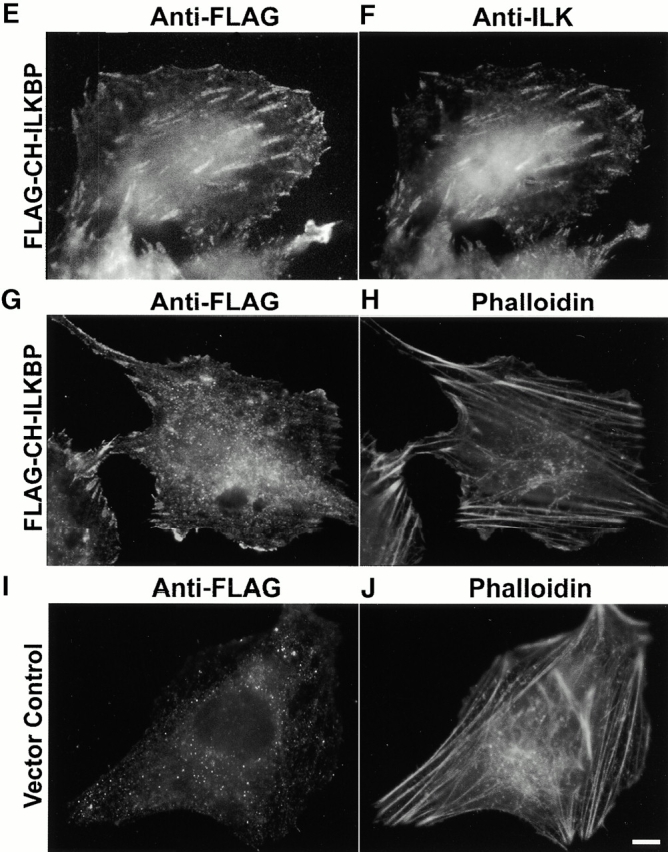
Complex formation and subcellular localization of FLAG–CH-ILKBP. (A–D) Coimmunoprecipitation of ILK and PINCH with FLAG–CH-ILKBP. The FLAG–CH-ILKBP immunoprecipitates (lane 3) and control precipitates (lane 4) were prepared as described in Materials and Methods and analyzed by Western blotting with anti-FLAG antibody M2 (A), anti-ILK antibody 65.1 (B), and antipaxillin antibody (clone 349) (C), respectively. Lanes 1 and 2 were loaded with cell lysates (20 μg protein/lane). In D, the membrane used in C was reprobed (without striping) with anti-PINCH antibodies. (E–J) FA localization of FLAG–CH-ILKBP. Rat mesangial cells were transfected with the FLAG–CH-ILKBP vector (E–H) or a FLAG vector as a control (I and J). The FLAG–CH-ILKBP, FAK, and actin stress fibers were detected by staining the cells with mouse anti-FLAG antibody M2 (E, G, and I), rabbit anti-FAK antibody (F), or rhodamine-labeled phalloidin (H and J). We also analyzed the subcellular localization of FLAG–CH-ILKBP in C2C12 cells and obtained similar results (not shown). Bar, 5 μm.
To test whether FLAG–CH-ILKBP localizes to FAs, we stained C2C12 cells and rat mesangial cells that express FLAG–CH-ILKBP with anti-FLAG antibodies. The results showed that FLAG–CH-ILKBP is targeted to FAs (Fig. 5 E), where clusters of FAK (a marker of FAs) were also detected (Fig. 5 F). Clusters of FLAG–CH-ILKBP were concentrated at the ends of actin stress fibers (Fig. 5G and Fig. H). In control experiments, no specific staining was observed in cells transfected with a control FLAG vector (Fig. 5I and Fig. J), confirming the specificity of the staining. In additional experiments, we expressed GFP–CH-ILKBP in mammalian cells and found that it also localized to FAs (see below).
To analyze the subcellular localization of endogenous CH-ILKBP, we stained mammalian cells with monoclonal anti–CH-ILKBP antibodies. The results showed that endogenous CH-ILKBP (Fig. 6 A) like FLAG–CH-ILKBP (Fig. 5E and Fig. G) was clustered in FAs where FAKs were also detected (Fig. 6 B). Clusters of CH-ILKBP were concentrated at the ends of actin stress fibers (Fig. 6C and Fig. D). In epithelial cells that had formed cell–cell contacts, CH-ILKBP localized to cell matrix FAs but not to cell–cell adhesions where abundant β-catenin was detected (Fig. 6E and Fig. F). In control experiments, incubation of the anti–CH-ILKBP antibody with GST–CH-ILKBP eliminated the staining (data not shown), confirming the specificity of the antibody staining. The observation that CH-ILKBP is concentrated at the ends of actin stress fibers suggested a possibility that CH-ILKBP is physically associated with the cytoskeleton fractions. To test this, we isolated cytoskeleton fractions from C2C12 cells. Under the conditions used, ∼5–10% of CH-ILKBP was found in the Triton X-100–insoluble cytoskeleton fractions (Fig. 6 G). As a comparison, we found that a similar percentage of FAK (Fig. 6 H), an abundant component of FAs, and no detectable amount of extracellular signal–regulated kinase (ERK) (Fig. 6 I), a more dynamic component of FAs (Fincham et al. 2000), was present in the cytoskeleton fractions.
Figure 6.
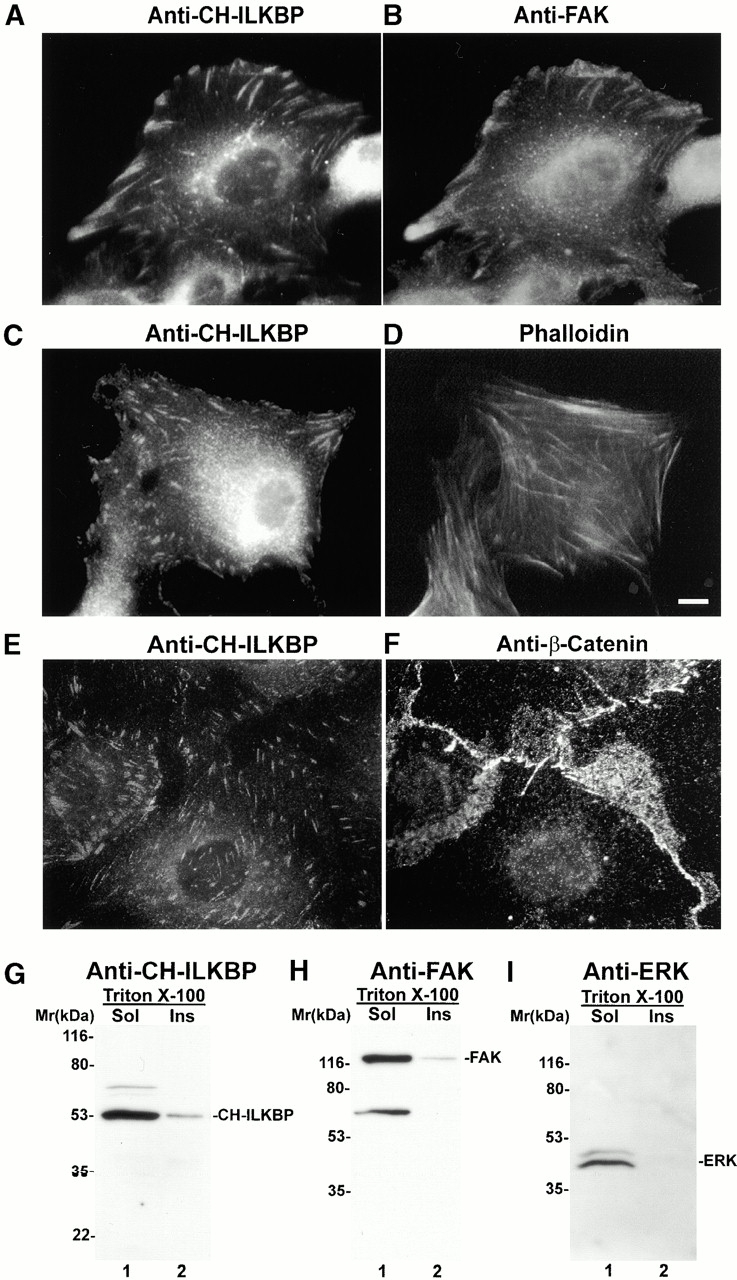
FA localization and cytoskeleton association of CH-ILKBP. (A–F) Subcellular localization of CH-ILKBP. Primary rat mesangial cells were stained with mouse anti–CH-ILKBP antibody 1D4 and rabbit anti-FAK antibodies (A and B) or anti–CH-ILKBP antibody 1D4 and phalloidin (C and D). We have also stained IEC18 rat intestinal epithelial cells, CHO cells, and mouse C2C12 cells with anti–CH-ILKBP antibodies and found that CH-ILKBP localizes to FA in these cells as well. Immunofluorescence images of IEC18 cells stained with mouse anti–CH-ILKBP antibody 1D4 (E) and rabbit anti–β-catenin antibody (F). (G–I) Association of CH-ILKBP with Triton X-100–insoluble cytoskeleton fractions. CH-ILKBP, FAK, and p44/42 ERK in the Triton X-100–soluble and –insoluble fractions were detected by Western blotting with anti–CH-ILKBP antibody 3B5 (G), anti-FAK antibody (H), and anti-p44/42 ERK antibody (New England Biolabs, Inc.) (I), respectively. Bar, 5 μm.
The FA Localization of CH-ILKBP Is Mediated by the ILK-binding CH2 Domain
We next sought to determine the domain of CH-ILKBP that mediates the FA localization of CH-ILKBP. To do this, we expressed GFP fusion proteins containing the full-length CH-ILKBP (GFP–CH-ILKBP), the CH2 deletion mutant (GFP-ΔCH2), and the CH2 fragment (GFP-CH2) in mammalian cells. Consistent with the results obtained with anti–CH-ILKBP antibody (Fig. 6) and FLAG–CH-ILKBP (Fig. 5), GFP–CH-ILKBP localized to FAs where ILK (Fig. 7A and Fig. B) and FAK (data not shown) were clustered. Deletion of the ILK-binding CH2 domain eliminated its ability to localize to FAs (Fig. 7C and Fig. D), indicating that the ILK-binding CH2 domain is required for the FA localization. Furthermore, GFP fusion protein containing the CH2 fragment was able to cocluster with ILK in FAs (Fig. 7E and Fig. F), albeit the level of fluorescence was lower than that of GFP–CH-ILKBP (Fig. 7, compare A and E). Taken together, these results suggest that the ILK-binding CH2 domain mediates the FA localization of CH-ILKBP.
Figure 7.
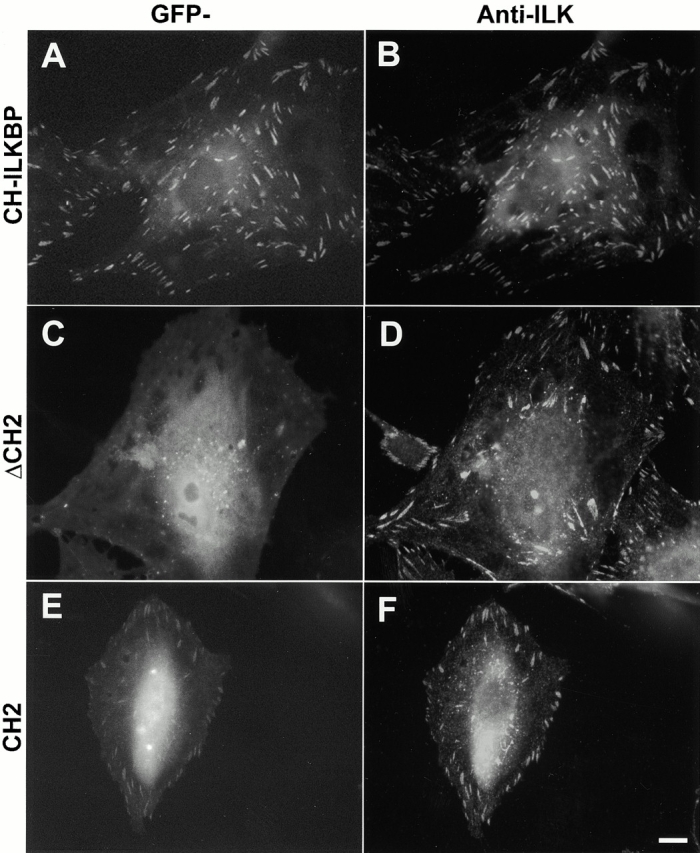
The ILK-binding CH2 domain mediates the localization of CH-ILKBP to FAs. Rat mesangial cells expressing GFP–CH-ILKBP (A and B), GFP-ΔCH2 (residues 1–229) (C and D), or GFP-CH2 (residues 222–372) (E and F) were plated on fibronectin-coated coverslips and stained with mouse anti-ILK antibody 65.1 and Rhodamine redTX-conjugated anti–mouse IgG antibody. GFP fusion proteins (A, C, and E) and ILK (B, D, and F) were visualized under a fluorescence microscope equipped with GFP and rhodamine filters. We have also expressed GFP–CH-ILKBP, GFP-ΔCH2, and GFP-CH2 in C2C12 cells and found that GFP–CH-ILKBP and GFP-CH2 but not GFP-ΔCH2 localized to FAs in these cells as well (not shown). Bar, 5 μm.
A Point Mutation That Disrupts the ILK-binding Impairs the FA Localization of CH-ILKBP
To further analyze the ILK-binding activity and FA localization of CH-ILKBP, we introduced a point mutation (F271D) into the ILK-binding CH2 domain of CH-ILKBP. GST–CH-ILKBP fusion proteins bearing the F271D point mutation were generated (Fig. 8 A), and their ILK-binding activities were determined (Fig. 8 B). Neither the full-length CH-ILKBP containing the F271D point mutation (Fig. 8 B, lane 6) nor the CH2 domain containing the F271D point mutation (Fig. 8 B, lane 2) bound to ILK. In control experiments, both GST–CH-ILKBP (Fig. 8 B, lane 5) and the GST fusion protein containing the CH2 fragment (Fig. 8 B, lane 3), but not GST alone (Fig. 8 B, lane 4), bound to ILK. These results indicate that the F271D point mutation ablated the ILK-binding activity. We next determined the effect of disrupting the ILK-binding on FA localization of CH-ILKBP. To do this, we expressed a GFP–CH-ILKBP protein bearing the F271D point mutation (GFP-F271D) in mammalian cells. In contrast to GFP–CH-ILKBP, which was coclustered with ILK in FAs (Fig. 7A and Fig. B), GFP-F271D was unable to localize to FAs where abundant ILK was detected (Fig. 8C and Fig. D). Thus, ablation of the ILK-binding activity by a single amino acid substitution impairs the FA localization of CH-ILKBP.
Figure 8.
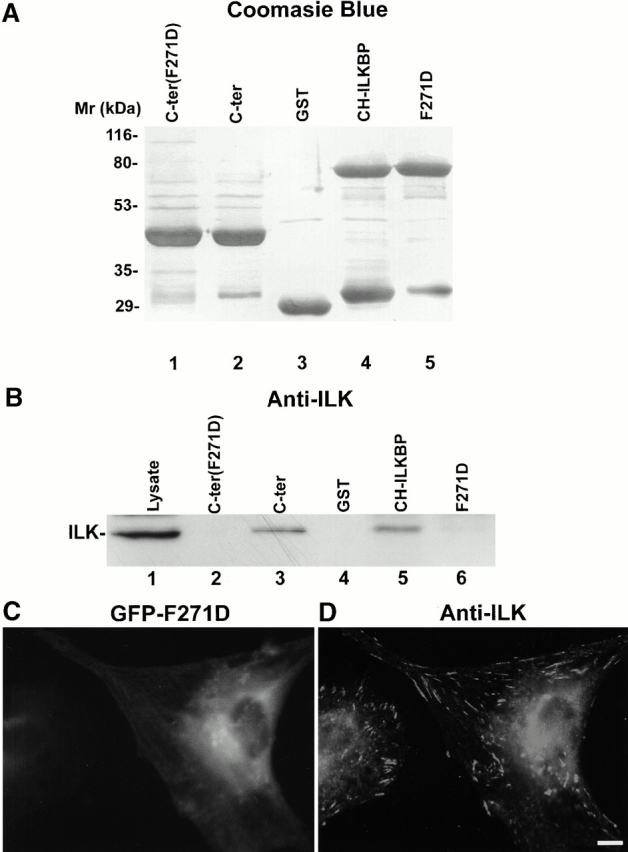
A point mutation that disrupts the ILK binding impairs the FA localization of CH-ILKBP. (A) GST–CH-ILKBP proteins. GST fusion proteins containing the mutant form (F271D) of CH2 domain (residues 258–372) (lane 1), the CH2 domain (residues 258–372) (lane 2), CH-ILKBP (lane 4), and the F271D point mutant (lane 5) and GST (lane 3) were separated on SDS-PAGE (10 μg/lane) and detected by Coomassie blue R-250 staining. (B) ILK binding. C2C12 cell lysates (260 μg) were incubated and precipitated with equal amount (10 μg) of GST or GST–CH-ILKBP proteins as indicated. ILK was detected by Western blotting with anti-ILK antibody 65.1. Lane 1 was loaded with C2C12 cell lysates (9 μg/lane). (C and D) FA localization. Rat mesangial cells expressing GFP fusion protein containing the full-length F271D point mutant were plated on fibronectin-coated coverslips and stained with anti-ILK antibody 65.1 and Rhodamine red™-conjugated anti–mouse IgG antibody. GFP-F271D (C) and ILK (D) were visualized under a fluorescence microscope equipped with GFP and rhodamine filters. Bar, 5 μm.
CH-ILKBP Is Involved in the Regulation of Cell Adhesion and Spreading
The findings that CH-ILKBP interacts with ILK and localizes to FAs prompted us to investigate whether CH-ILKBP plays a role in cell adhesion. To do this, we transfected CHO cells with FLAG vectors encoding the full-length CH-ILKBP, the ILK-binding defective F271D point mutant, the ILK-binding CH2 fragment, the CH2 deletion mutant, or a FLAG vector lacking CH-ILKBP sequence as a control (Fig. 9 A). As expected, the FLAG-tagged CH-ILKBP (Fig. 9 A, lane 2) and CH2 domain (lane 4), but neither FLAG-F271D (lane 3) nor FLAG-ΔCH2 (lane 5), formed a complex with ILK (Fig. 9 B) and PINCH (Fig. 9 D) in CHO cells. Consistent with the results obtained with C2C12 cells (Fig. 3 E and Fig. 5 C), paxillin was not associated with the FLAG-tagged wild-type or mutant forms of CH-ILKBP in CHO cells (Fig. 9 C, lanes 2–5), despite the presence of abundant paxillin in these cells (Fig. 9 C, lanes 6–9). To analyze the effect of overexpression of the different forms of CH-ILKBP on cell adhesion, we plated the CHO cells on collagen IV–coated 96-well plates and quantified the number of adhered cells. The results showed that overexpression of FLAG–CH-ILKBP did not significantly alter cell adhesion (Fig. 9 E). However, overexpression of the ILK-binding defective CH-ILKBP point mutant F271D or the ILK-binding CH2 fragment, but not that of the NH2-terminal fragment lacking CH2, significantly reduced cell adhesion (Fig. 9 E), suggesting that CH-ILKBP, through interactions with ILK and other proteins, plays an important role in the regulation of cell adhesion.
Figure 9.
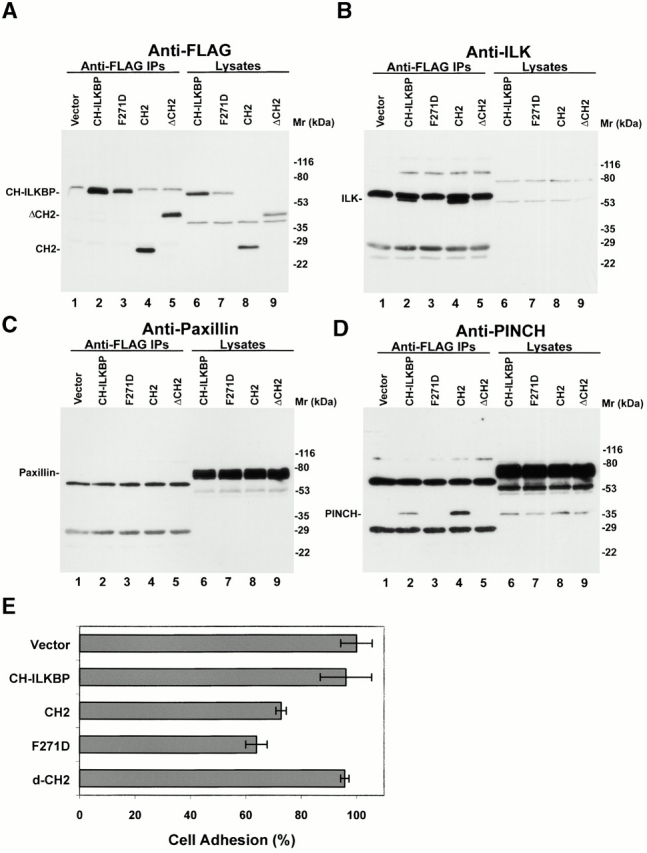
Effects of CH-ILKBP on CHO cell adhesion. (A–D) FLAG-tagged CH-ILKBP (lane 2), F271D (lane 3), CH2 (residues 222–372) (lane 4), and ΔCH2 (residues 1–229) (lane 5) were immunoprecipitated from CHO cells transfected with the corresponding FLAG expression vectors. (Lane 1) Anti-FLAG immunoprecipitates obtained with the vector control cells. The immunoprecipitates were analyzed by Western blotting with anti-FLAG (A), anti-ILK (B), or antipaxillin (C) antibodies. In D, the membrane used in C was reprobed (without striping) with anti-PINCH antibodies. Positions of FLAG–CH-ILKBP (or F271D), FLAG-ΔCH2, FLAG–CH2, ILK, paxillin, and PINCH were indicated. (Lanes 6–9) Lysates (15 μg/lane) of the transfectants as indicated. (E) Cell adhesion. Cells (as indicated in the figure) were allowed to adhere to collage IV–coated 96-well plates for 90 min, and the adhered cells were quantified as described in Materials and Methods. Under the condition used, ∼70% of the vector control cells adhered. The adhesion efficiency of CHO cells expressing different forms of CH-ILKBP was compared with that of the vector control cells and presented as percentages of that of the control cells. Data represent the mean ± SD of two separate experiments.
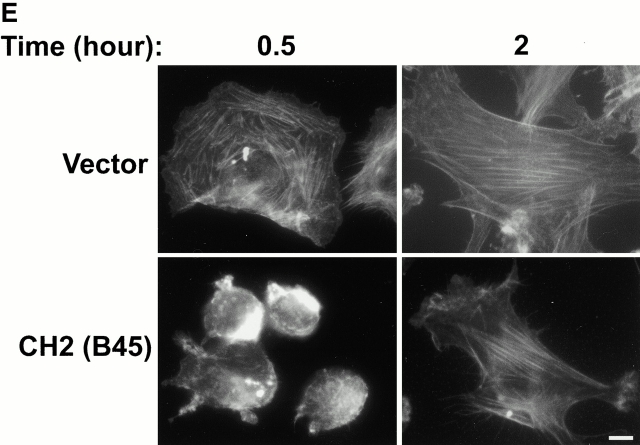
To confirm that CH-ILKBP is involved in cell adhesion, we expressed FLAG-tagged CH-ILKBP, the ILK-binding CH2 fragment, and the CH2 deletion mutant, respectively, in a different cell type (C2C12 cells). Because of the relative low efficiency of transient transfection in C2C12 cells, we isolated C2C12 clones stably expressing different forms of CH-ILKBP. Two independent clones were isolated from each transfection, and the expression of FLAG–CH-ILKBP, FLAG-CH2, or FLAG-ΔCH2 in these cells was confirmed by Western blotting (Fig. 10 A). Analyses of the cell adhesion showed that although C2C12 cells overexpressing CH-ILKBP adhered to collagen I as efficiently as the vector control cells, C2C12 cells overexpressing the ILK-binding CH2 fragment adhered much less efficiently (Fig. 10 B). By contrast, overexpression of the CH2 deletion NH2-terminal fragment did not significantly alter the efficiency of cell adhesion (Fig. 10 B). Noticeably, the spreading of the C2C12 cells overexpressing the ILK-binding CH2 fragment was much slower than that of the vector control cells or those overexpressing FLAG–CH-ILKBP or FLAG-ΔCH2 (Fig. 10C and Fig. d). Staining of the CH2-overexpressing cells with phalloidin reveled that the actin stress fiber formation was retarded (Fig. 10 E). Thus, consistent with the ILK-binding activity and FA localization of CH-ILKBP, CH-ILKBP is critically involved in the regulation of cell adhesion, actin cytoskeleton organization, and cell shape change.
Figure 10.
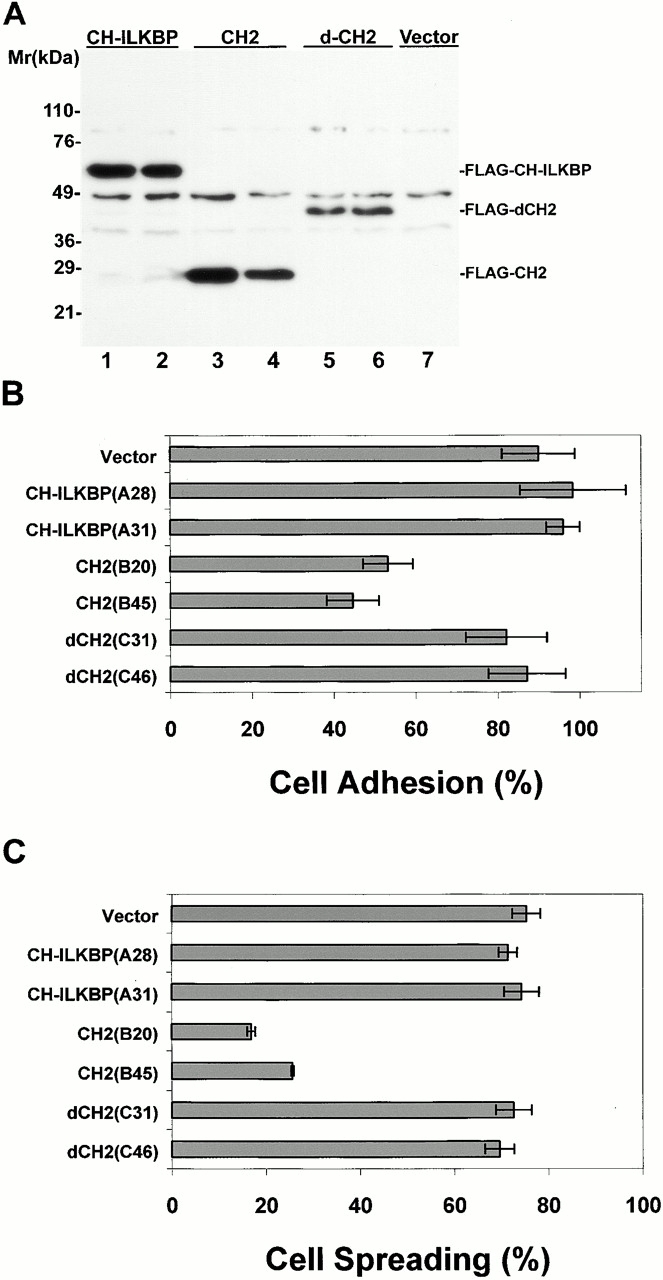
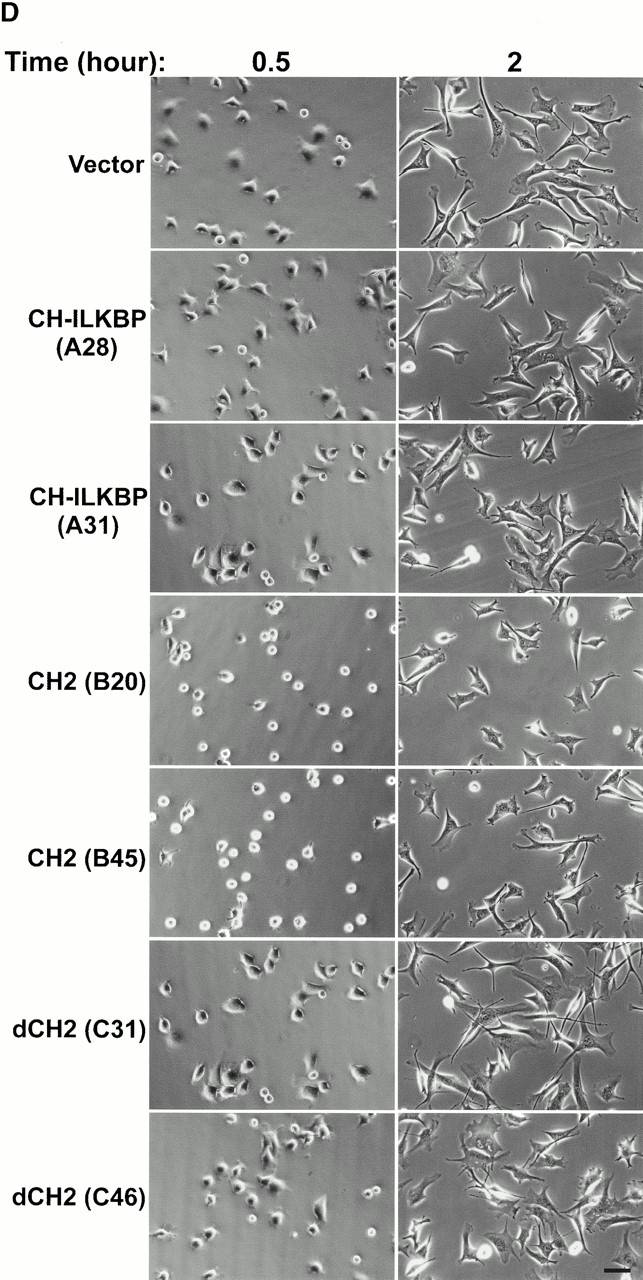
Effects of CH-ILKBP on C2C12 cell adhesion and spreading. (A) Lysates (15 μg/lane) of independent C2C12 clones expressing the FLAG-tagged proteins were probed with anti–FLAG-antibody M2. (Lane 1) FLAG–CH-ILKBP clone A28; (lane 2) FLAG–CH-ILKBP clone A31; (lane 3) FLAG-CH2 (residues 222–372) clone B20; (lane 4) FLAG-CH2 clone B45; (lane 5) FLAG-ΔCH2 (residues 1–229) clone C31; (lane 6) FLAG-ΔCH2 clone C46; (lane 7) lysates (15 μg) from the vector control cells. (B) Cell adhesion. Cells (as indicated in the figure) were allowed to adhere to collage I–coated 12-well plates for 30 min and were then washed. Cell adhesion is presented as the number of adhered cells (after wash) divided by the number of total cells (before wash). Data represent the mean ± SD of four randomly selected fields. (C and D) Cell spreading. C2C12 cells expressing different forms of CH-ILKBP were seeded in collage I–coated 12-well plates. The cell morphology was recorded 0.5 and 2 h after seeding (D). The percentage of cells adopting spread morphology 0.5 h after seeding was quantified by analyzing >300 cells from three randomly selected fields (C). Data represent mean ± SD. (E) C2C12 cells overexpressing FLAG-CH2 (clone B45) and the vector control cells were plated on collage I–coated slides for 0.5 or 2 h and stained with rhodamine-labeled phalloidin. Identical results were obtained with clone B20. Bars: (D) 100 μm; (E) 5 μm.
Discussion
Cell matrix adhesion is a fundamental process that regulates gene expression, differentiation, proliferation, and morphological changes. FAs are important cell adhesion sites through which the ECM is physically linked to the actin cytoskeleton (for reviews see Jockusch et al. 1995; Burridge and Chrzanowska-Wodnicka 1996; Calderwood et al. 2000). In this study, we have identified a novel FA protein (CH-ILKBP) that specifically interacts with ILK, an important regulator of integrin-mediated processes (Hannigan et al. 1996; Wu et al. 1998; Dedhar et al. 1999; Wu 1999). The interaction of ILK with CH-ILKBP is mediated by the COOH-terminal domain of ILK. In previous studies, we have shown that ILK binds to PINCH through ILK NH2-terminal ANK domain (Tu et al. 1999). Using multiple experimental approaches, we show in this study that PINCH, ILK, and CH-ILKBP form a ternary complex in cells. CH-ILKBP localizes to FAs in response to cell matrix but not cell–cell adhesions, a property shared by ILK (Li et al. 1999). ILK likely plays a major role in recruiting CH-ILKBP to FAs, as ILK is the only FA protein that is known to bind to CH-ILKBP. Furthermore, the ILK-binding CH2 domain, which does not bind to paxillin in cells (Fig. 9), is both necessary and sufficient for the FA targeting. Finally, a point mutation that disrupts the ILK binding abolished the FA localization of CH-ILKBP. These data suggest a model by which ILK functions in cell adhesions. In this model, ILK, through its COOH- and NH2-terminal domains, binds to CH-ILKBP and PINCH simultaneously, resulting in the formation of a relatively stable ternary complex. The CH-ILKBP–ILK–PINCH complex together with other CH-ILKBP-, ILK-, and PINCH-binding proteins provides an important physical connection at FAs, linking the ECM to the actin cytoskeleton. Consistent with the notion that ILK recruits CH-ILKBP to FAs where PINCH–ILK–CH-ILKBP provides an anchor for the actin filaments, the association of the ILK-binding defective mutants (ΔCH2 or F271D) with the Triton X-100–insoluble cytoskeleton was significantly reduced (our unpublished preliminary data). Using cells that were either transiently or stably overexpressing mutant forms of CH-ILKBP, we have obtained important functional evidence supporting this model. Cells overexpressing the ILK-binding CH2 fragment exhibited significant reductions in cell matrix adhesion, actin stress fiber formation, and spreading. However, it is interesting to note that the adhesion and spreading of the cells overexpressing the ILK-binding CH2 fragment were only partially impaired. Besides the technical considerations (for example, the transfection efficiency or the expression level of the CH2 fragment), this likely reflects the fact that there are multiple protein complexes, including those containing talin, filamin, or α-actinin, that can physically link the ECM and transmembrane receptors to the intracellular actin cytoskeleton (Jockusch et al. 1995; Burridge and Chrzanowska-Wodnicka 1996; Calderwood et al. 2000).
Cell adhesion is an essential cellular function of all multicellular organisms. Recent studies suggest that cells in different organisms ranging from nematodes to human use strikingly similar strategies to mediate this fundamental process (Hynes and Zhao 2000). In genetic model system C. elegans, mutations in genes encoding integrin α (pat-2) or β (pat-3) cause a PAT phenotype characterized by defects in muscle dense body and M-lines, which resemble FAs in mammalian cells (Williams and Waterston 1994; Gettner et al. 1995). Recent genetic studies have shown that lack of PAT-4–ILK [Mackinnon, A.C., and B.D. Williams. 2000. 40th American Society for Cell Biology Annual Meeting. 2664. (Abstr.)]or UNC-97–PINCH (Hobert et al. 1999) in C. elegans results in a phenotype that is identical to that of the integrins. These studies provide strong genetic evidence for a crucial role of the ILK and its binding partner PINCH in the integrin-mediated cell adhesion. Like ILK and PINCH, CH-ILKBP is an ancient protein with homologues found in different organisms including C. elegans. Human CH-ILKBP shares significant sequence similarity with a predicted C. elegans protein (T21D12.4 protein) (Fig. 1 A). Recently, Lin and Williams [Lin, X., and B.D. Williams. 2000. 40th American Society for Cell Biology Annual Meeting. 2666. (Abstr.)] presented genetic evidence showing that C. elegans gene T21D12.4 corresponds to another PAT gene, pat-6. Thus, the CH-ILKBP–ILK–PINCH complex described in this paper likely represents an evolutionally well-conserved link between cell adhesion receptors and the actin cytoskeleton. Zervas et al. 2001 reported recently that mutations in Drosophila ILK cause defects similar to loss of integrin adhesion, and the actin filaments were detached from the membrane at the muscle attachment sites in these mutants. Given the fact that homologues of ILK, CH-ILKBP, and PINCH are present in Drosophila, a CH-ILKBP–ILK–PINCH complex that is analogous to the complex in mammalian cells likely exists in Drosophila and provides an essential connection linking the actin cytoskeleton to the adhesion sites in Drosophila and in other organisms.
The data obtained in this and other studies suggest that ILK is a protein with dual functions. The first function is to provide a molecular scaffold for the assembly of the PINCH–ILK–CH-ILKBP complex as shown in this study, which together with other interactive proteins at FAs links ECM to the actin cytoskeleton. The second function is to serve as a protein kinase in the regulation of gene expression, survival, differentiation, and proliferation (Hannigan et al. 1996; Radeva et al. 1997; Delcommenne et al. 1998; Novak et al. 1998; Wu et al. 1998; Huang et al. 2000; Persad et al. 2000). Given the importance of compartmentalization in kinase action during signal transduction, it is likely that the protein–protein interactions mediated by ILK, including that between ILK and CH-ILKBP, are involved not only in cell adhesion, actin cytoskeleton organization, and spreading as shown in this study but also in other ILK-regulated processes.
Northern blotting analyses indicate that CH-ILKBP is a member of a family of structurally related proteins (Fig. 1 B). When this manuscript was in preparation, Nikolopoulos and Turner 2000 described a new paxillin- and actin-binding protein termed actopaxin. Actopaxin is structurally related to CH-ILKBP (human actopaxin is 98% identical to human CH-ILKBP at the amino acid level and 90% identical at the cDNA level). Because the difference between actopaxin and CH-ILKBP cDNA sequences is significant and occurs throughout the sequences, they are likely encoded by two different genes. However, the striking similarity in protein sequences suggests that they may share certain common functions. Actopaxin contains a potential ILK-binding sequence that is identical to that in CH-ILKBP, suggesting that actopaxin may also function as an ILK-binding protein. However, it is important to note that although CH-ILKBP contains a potential paxillin-binding sequence as defined by Nikolopoulos and Turner 2000, we could not detect an association between paxillin and CH-ILKBP in vivo, despite using several different cell types, including mouse C2C12 cells (Fig. 3 and Fig. 5), CHO cells (Fig. 9), human kidney 293 cells, and rat embryo fibroblasts (REF-52) (our unpublished results), and multiple experimental approaches (immunoprecipitation of endogenous or epitope-tagged CH-ILKBP, PINCH, or paxillin). On the other hand, in all of the cell types analyzed, we have readily detected a stable CH-ILKBP–ILK–PINCH complex. Thus, although our results do not rule out the possibility that paxillin could bind to CH-ILKBP in vitro or under certain other conditions, they do suggest that CH-ILKBP functions primarily as an ILK-binding protein in cells.
In summary, we have identified a new ILK-binding protein and have shown that it plays an important role in cell adhesion and spreading. The data presented here provide new insights into the molecular mechanism by which ILK functions at adhesion sites. The CH-ILKBP–ILK–PINCH complex described in this report likely functions as an important scaffold in the assembly and signaling through cell matrix adhesion sites.
Supplemental Material
Acknowledgments
This work was supported by National Institutes of Health grant DK54639 and American Cancer Society research project grant 98-220-01-CSM (to C. Wu).
Footnotes
The online version of this article contains supplemental material.
Y. Tu and Y. Huang contributed equally to this work.
Abbreviations used in this paper: ANK, ankyrin; CH, calponin homology; CH-ILKBP, CH domain-containing ILK-binding protein; ECM, extracellular matrix; ERK, extracellular signal–regulated kinase; FA, focal adhesion; FAK, FA kinase; GFP, green fluorescent protein; GST, glutathione S-transferase; ILK, integrin-linked kinase; MBP, maltose-binding protein.
References
- Burridge K., Chrzanowska-Wodnicka M. Focal adhesions, contractility, and signaling. Annu. Rev. Cell Dev. Biol. 1996;12:463–518. doi: 10.1146/annurev.cellbio.12.1.463. [DOI] [PubMed] [Google Scholar]
- Calderwood D.A., Shattil S.J., Ginsberg M.H. Integrins and actin filamentsreciprocal regulation of cell adhesion and signaling. J. Biol. Chem. 2000;275:22607–22610. doi: 10.1074/jbc.R900037199. [DOI] [PubMed] [Google Scholar]
- Clark E.A., Brugge J.S. Integrins and signal transduction pathwaysthe road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- Dedhar S., Hannigan G.E. Integrin cytoplasmic interactions and bidirectional transmembrane signalling. Curr. Opin. Cell Biol. 1996;8:657–669. doi: 10.1016/s0955-0674(96)80107-4. [DOI] [PubMed] [Google Scholar]
- Dedhar S., Williams B., Hannigan G. Integrin linked kinase (ILK)a regulator of integrin and growth-factor signaling. Trends Cell Biol. 1999;9:319–323. doi: 10.1016/s0962-8924(99)01612-8. [DOI] [PubMed] [Google Scholar]
- Delcommenne M., Tan C., Gray V., Rue L., Woodgett J., Dedhar S. Phosphoinositide-3-OH kinase-dependent regulation of glycogen synthase kinase 3 and protein kinase B/AKT by the integrin-linked kinase. Proc. Natl. Acad. Sci. USA. 1998;95:11211–11216. doi: 10.1073/pnas.95.19.11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham V.J., James M., Frame M.C., Winder S.J. Active ERK/MAP kinase is targeted to newly forming cell-matrix adhesions by integrin engagement and v-Src. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:2911–2923. doi: 10.1093/emboj/19.12.2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gettner S.N., Kenyon C., Reichardt L.F. Characterization of beta pat-3 heterodimers, a family of essential integrin receptors in C. elegans . J. Cell Biol. 1995;129:1127–1141. doi: 10.1083/jcb.129.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannigan G.E., Leung-Hagesteijn C., Fitz-Gibbon L., Coppolino M.G., Radeva G., Filmus J., Bell J.C., Dedhar S. Regulation of cell adhesion and anchorage-dependent growth by a new beta 1-integrin-linked protein kinase. Nature. 1996;379:91–96. doi: 10.1038/379091a0. [DOI] [PubMed] [Google Scholar]
- Hobert O., Moerman D.G., Clark K.A., Beckerle M.C., Ruvkun G. A conserved LIM protein that affects muscular adherens junction integrity and mechanosensory function in Caenorhabditis elegans . J. Cell Biol. 1999;144:45–57. doi: 10.1083/jcb.144.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Li J., Zhang Z., Wu C. The roles of integrin-linked kinase in the regulation of myogenic differentiation. J. Cell Biol. 2000;150:861–871. doi: 10.1083/jcb.150.4.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes R.O. Integrinsversatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- Hynes R., Zhao Q. The evolution of cell adhesion. J. Cell Biol. 2000;150:F89–F96. doi: 10.1083/jcb.150.2.f89. [DOI] [PubMed] [Google Scholar]
- Ilic D., Damsky C.H., Yamamoto T. Focal adhesion kinaseat the crossroads of signal transduction. J. Cell Sci. 1997;110:401–407. doi: 10.1242/jcs.110.4.401. [DOI] [PubMed] [Google Scholar]
- Jockusch B.M., Bubeck P., Giehl K., Kroemker M., Moschner J., Rothkegel M., Rudiger M., Schluter K., Stanke G., Winkler J. The molecular architecture of focal adhesions. Annu. Rev. Cell Dev. Biol. 1995;11:379–416. doi: 10.1146/annurev.cb.11.110195.002115. [DOI] [PubMed] [Google Scholar]
- Komoriya A., Green L.J., Mervic M., Yamada S.S., Yamada K.M., Humphries M.J. The minimal essential sequence for a major cell type-specific adhesion site (CS1) within the alternatively spliced type III connecting segment domain of fibronectin is leucine-aspartic acid-valine. J. Biol. Chem. 1991;266:15075–15079. [PubMed] [Google Scholar]
- Landegren U. Measurement of cell numbers by means of the endogenous enzyme hexosaminidase. Applications to detection of lymphokines and cell surface antigens. J. Immunol. Methods. 1984;67:379–388. doi: 10.1016/0022-1759(84)90477-0. [DOI] [PubMed] [Google Scholar]
- Li F., Zhang Y., Wu C. Integrin-linked kinase is localized to cell-matrix focal adhesions but not cell-cell adhesion sites and the focal adhesion localization of integrin-linked kinase is regulated by the PINCH-binding ANK repeats. J. Cell Sci. 1999;112:4589–4599. doi: 10.1242/jcs.112.24.4589. [DOI] [PubMed] [Google Scholar]
- Nikolopoulos S.N., Turner C.E. Actopaxin, a new focal adhesion protein that binds paxillin LD motifs and actin and regulates cell adhesion. J. Cell Biol. 2000;151:1435–1448. doi: 10.1083/jcb.151.7.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak A., Hsu S.C., Leung-Hagesteijn C., Radeva G., Papkoff J., Montesano R., Roskelley C., Grosschedl R., Dedhar S. Cell adhesion and the integrin-linked kinase regulate the LEF-1 and beta-catenin signaling pathways. Proc. Natl. Acad. Sci. USA. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.T., Schaller M.D., Hildebrand J., Leu T.H., Richardson A., Otey C. Focal adhesion kinasestructure and signalling. J. Cell Sci. Suppl. 1994;18:109–113. doi: 10.1242/jcs.1994.supplement_18.16. [DOI] [PubMed] [Google Scholar]
- Persad S., Attwell S., Gray V., Delcommenne M., Troussard A., Sanghera J., Dedhar S. Inhibition of integrin-linked kinase (ILK) suppresses activation of protein kinase B/Akt and induces cell cycle arrest and apoptosis of PTEN-mutant prostate cancer cells. Proc. Natl. Acad. Sci. USA. 2000;97:3207–3212. doi: 10.1073/pnas.060579697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radeva G., Petrocelli T., Behrend E., Leung-Hagesteijn C., Filmus J., Slingerland J., Dedhar S. Overexpression of the integrin-linked kinase promotes anchorage-independent cell cycle progression. J. Biol. Chem. 1997;272:13937–13944. doi: 10.1074/jbc.272.21.13937. [DOI] [PubMed] [Google Scholar]
- Richardson A., Malik R.K., Hildebrand J.D., Parsons J.T. Inhibition of cell spreading by expression of the C-terminal domain of focal adhesion kinase (FAK) is rescued by coexpression of Src or catalytically inactive FAKa role for paxillin tyrosine phosphorylation. Mol. Cell. Biol. 1997;17:6906–6914. doi: 10.1128/mcb.17.12.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry S.K., Horwitz A.F. Adhesion-growth factor interactions during differentiationan integrated biological response. Dev. Biol. 1996;180:455–467. doi: 10.1006/dbio.1996.0319. [DOI] [PubMed] [Google Scholar]
- Schwartz M.A., Schaller M.D., Ginsberg M.H. Integrinsemerging paradigms of signal transduction. Annu. Rev. Cell Dev. Biol. 1995;11:549–599. doi: 10.1146/annurev.cb.11.110195.003001. [DOI] [PubMed] [Google Scholar]
- Tu Y., Li F., Wu C. Nck-2, a novel Src homology2/3-containing adaptor protein that interacts with the LIM-only protein PINCH and components of growth factor receptor kinase signaling pathways. Mol. Biol. Cell. 1998;9:3367–3382. doi: 10.1091/mbc.9.12.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y., Li F., Goicoechea S., Wu C. The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol. Cell. Biol. 1999;19:2425–2434. doi: 10.1128/mcb.19.3.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner C.E. Paxillin. Int. J. Biochem. Cell Biol. 1998;30:955–959. doi: 10.1016/s1357-2725(98)00062-4. [DOI] [PubMed] [Google Scholar]
- Williams B.D., Waterston R.H. Genes critical for muscle development and function in Caenorhabditis elegans identified through lethal mutations. J. Cell Biol. 1994;124:475–490. doi: 10.1083/jcb.124.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. Integrin-linked kinase and PINCHpartners in regulation of cell-extracellular matrix interaction and signal transduction. J. Cell Sci. 1999;112:4485–4489. doi: 10.1242/jcs.112.24.4485. [DOI] [PubMed] [Google Scholar]
- Wu C., Fields A.J., Kapteijn B.A., McDonald J.A. The role of alpha 4 beta 1 integrin in cell motility and fibronectin matrix assembly. J. Cell Sci. 1995;108:821–829. doi: 10.1242/jcs.108.2.821. [DOI] [PubMed] [Google Scholar]
- Wu C., Keightley S.Y., Leung-Hagesteijn C., Radeva G., Coppolino M., Goicoechea S., McDonald J.A., Dedhar S. Integrin-linked protein kinase regulates fibronectin matrix assembly, E-cadherin expression, and tumorigenicity. J. Biol. Chem. 1998;273:528–536. doi: 10.1074/jbc.273.1.528. [DOI] [PubMed] [Google Scholar]
- Yang L.J., Rhee S.G., Williamson J.R. Epidermal grwoth factor-induced activation and translocation of phospholipase C-γ1 to the cytoskeleton in rat hepatocytes. J. Biol. Chem. 1994;269:7156–7162. [PubMed] [Google Scholar]
- Zervas C.G., Gregory S.L., Brown N.H. Drosophila integrin-linked kinase is required at sites of integrin adhesion to link the cytoskeleton to the plasma membrane. J. Cell Biol. 2001;152:1007–1018. doi: 10.1083/jcb.152.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.