Abstract
Mammalian stress granules (SGs) harbor untranslated mRNAs that accumulate in cells exposed to environmental stress. Drugs that stabilize polysomes (emetine) inhibit the assembly of SGs, whereas drugs that destabilize polysomes (puromycin) promote the assembly of SGs. Moreover, emetine dissolves preformed SGs as it promotes the assembly of polysomes, suggesting that these mRNP species (i.e., SGs and polysomes) exist in equilibrium. We used green flourescent protein–tagged SG-associated RNA-binding proteins (specifically, TIA-1 and poly[A] binding protein [PABP-I]) to monitor SG assembly, disassembly, and turnover in live cells. Fluorescence recovery after photobleaching shows that both TIA-1 and PABP-I rapidly and continuously shuttle in and out of SGs, indicating that the assembly of SGs is a highly dynamic process. This unexpected result leads us to propose that mammalian SGs are sites at which untranslated mRNAs are sorted and processed for either reinitiation, degradation, or packaging into stable nonpolysomal mRNP complexes. A truncation mutant of TIA-1 (TIA-1ΔRRM), which acts as a transdominant inhibitor of SG assembly, promotes the expression of cotransfected reporter genes in COS transfectants, suggesting that this process of mRNA triage might, directly or indirectly, influence protein expression.
Keywords: TIA-1, stress granules, protein translation, eIF-2α, PABP-1
Introduction
Stress granules (SGs) are phase dense particles that appear in the cytoplasm of plant and animal cells subjected to environmental stress (Nover et al. 1983, Nover et al. 1989; Scharf et al. 1998; Kedersha et al. 1999). In both plant and animal cells, SGs harbor untranslated mRNAs that accumulate as a consequence of stress-induced translational arrest. In mammalian cells, the assembly of SGs is initiated by the phosphorylation of eukaryotic initiation factor (eIF)-2α (Kedersha et al. 1999), an essential component of a heterotrimeric translational regulatory complex that loads the initiator tRNA (Met-tRNAMet) onto the 40S ribosomal subunit (Berlanga et al. 1998; Gray and Wickens 1998). Phosphorylation of eIF-2α inhibits translational initiation (Srivastava et al. 1998), allowing elongating ribosomes to run off their mRNA transcripts, which then accumulate at SGs (Kedersha et al. 1999). In cells allowed to recover from stress, SG-associated mRNAs rapidly move to poly-ribosomes (Nover et al. 1989). If mRNAs sequestered at SGs are unavailable for translational reinitiation, then the assembly and disassembly of these structures could influence the duration of stress-induced translational arrest (Kedersha et al. 1999).
Assembly of SGs in response to the phosphorylation of eIF-2α is dependent on the related RNA-binding proteins TIA-1 and TIAR (Kedersha et al. 1999). These proteins possess three RNA recognition motifs (RRMs) at their NH2 termini and a glutamine-rich, prion-related domain (PrD) at their COOH termini (Tian et al. 1991; Kawakami et al. 1992). These structural domains confer the functions required for the assembly of SGs: RNA binding and self-aggregation. In vitro, RRM1 is devoid of RNA-binding activity, RRM2 binds with high affinity to uridine-rich sequences found in selected mRNAs, and RRM3 binds RNA without known sequence specificity (Dember et al. 1996). These properties allow full-length TIA-1 and TIAR to bind to heterogeneous mRNAs without apparent sequence specificity (Dember et al. 1996). At the same time, both proteins bind to uridine-rich sequences with high affinity and specificity (Dember et al. 1996). Although sequence nonspecific binding could allow these proteins to recruit untranslated mRNAs to SGs, sequence-specific binding allows these proteins to regulate the translation of specific transcripts, including TNF-α mRNA (Piecyk et al. 2000). The PrD is capable of self-aggregation (Kedersha et al. 1999) and is essential for the assembly of SGs (Kedersha, N., and P. Anderson, manuscript in preparation). Overexpression of a truncation mutant encoding the isolated PrD but lacking the RNA-binding domains (TIA-1ΔRRM) prevents arsenite-induced assembly of SGs in COS transfectants (Kedersha et al. 1999). TIA-1ΔRRM forms cytoplasmic aggregates that sequester endogenous TIA-1 and TIAR (Kedersha et al. 1999). In cells transfected with this mutant, mRNA does not accumulate at SGs (Kedersha et al. 1999).
A major unanswered question is whether SGs actively participate in stress-induced translational arrest by sequestering selected mRNAs away from the translational machinery, as has been proposed for plant heat shock granules (Nover et al. 1989). Consistent with this possibility, we show here that TIA-1ΔRRM enhances the expression of cotransfected reporter genes as it prevents the assembly of SGs. Interestingly, the opposing effects of translational inhibitors that stabilize or destabilize polysomes suggest that SGs are in a dynamic equilibrium with polysomes. Surprisingly, photobleaching studies indicate that the major protein components of SGs (TIA-1 and poly[A]+ binding protein I [PABP-1]) are in constant and extremely rapid flux, despite the relatively stable appearance of individual SGs visualized using time lapse photomicroscopy. These observations indicate that mammalian SGs do not passively sequester mRNA from the translational machinery but are highly active cytoplasmic sorting sites for mRNPs. We conclude that SGs are dynamic microdomains that influence the fate of the untranslated mRNAs that accumulate in stressed cells.
Materials and Methods
Cells and Antibodies
DU-145 prostate cancer cells and COS7 cells were obtained from American Type Culture Collection. Goat anti–TIA-1 polyclonal antibody was obtained from Santa Cruz Biotechnology, Inc., and anti-hemagglutinin (HA) was obtained from BAbCo. Antiluciferase polyclonal antibody was obtained from Cortex Biochem, anti–phosphorylated eIF-2α was obtained from Research Genetics, and anti–β-galactosidase was obtained from Promega. Anti–eIF-2α mAb was a kind gift from Dr. Richard Panniers (National Institutes of Health, Bethesda, MD). The anti–PABP antibody 10E10 was a kind gift from Dr. Gideon Dreyfuss (University of Pennsylvania, School of Medicine, Philadelphia, PA). Antibodies against the small ribosomal proteins S3 and S19 were a kind gift from Dr. Joachim Stahl (Max Delbruck Centre for Molecular Medicine, Berlin, Germany). Secondary antibodies (all ML grade, both fluorochrome-tagged and HRP-tagged) were obtained from Jackson ImmunoResearch Laboratories.
COS Cell Transfections
COS7 cells were transfected using SuperFect (QIAGEN) according to the manufacturer's instructions. Cells plated in 6-well plates (2 × 105 cells/well plated 20 h before transfection) were transfected for 2–4 h, then trypsinized and replated into parallel plates for both immunofluorescence (24-well plates containing 11-mm coverslips) and Western blotting or immunoprecipitation (12-well plates). This allowed us to ascertain by immunofluorescence that similar transfection efficiencies were obtained with different constructs (i.e., <20% variation within individual experiments), in order to confirm that differences in protein expression seen by Western blot were not due to differences in transfection efficiency.
Western Blot Analysis
Cells were lysed in SDS sample buffer, boiled, and sonicated to shear DNA. Proteins were resolved on 4–20% gradient gels (Invitrogen) using SDS-PAGE. Proteins were transferred to nitrocellulose, and the protein loading was assessed by Ponceau red staining. Blots were then blocked in 5% milk or 5% horse serum and probed using standard procedures. Proteins were detected using HRP-conjugated secondary antibodies (Jackson ImmunoResearch Laboratories), chemiluminescence (Far Western; Pierce Chemical Co.), and BioMax MR film (Eastman Kodak Co.).
Inhibition of Protein Synthesis
Cells were washed with HBSS and incubated in cysteine/methionine-free media (DME containing 5% dialyzed FBS) for 30 min. During this time, cells were exposed to the drugs at concentrations indicated in the figure legends. Immediately after the 30-min drug treatment period, one set of cells was metabolically labeled by the addition of 35S Trans-label (NEN Life Science Products) for 30 min, harvested by trypsinization, and lysed in SDS-PAGE sample buffer. After sonication, aliquots of cell lysates were separated using SDS-PAGE and stained to insure equal protein recovery from each sample. Aliquots of each sample were precipitated using 60% acetone, washed, resuspended in 1% SDS, then counted in Hydrofluor (National Diagnostics) using a liquid scintillation counter. A parallel set of cells was preincubated with either emetine, puromycin, or no drugs for 30 min, at which point SGs were induced by the addition of arsenite (0.5 mM) to the media, and the incubation was continued for 30 min. Cells were then processed for immunofluorescence staining using anti–TIA-1 as described below, and the percentage of cells with SGs was determined microscopically. To assess the ability of different drugs to reverse SGs, cells were exposed to arsenite for 30 min, various concentrations of emetine, puromycin, or media alone were added, and the cells were incubated for an additional 1 h before fixation and staining. Results shown are typical of three independent experiments.
Sucrose Gradient Analysis
Cells were plated and used 48 h after plating. Cells were treated with various agents as indicated in the figure legends for the times indicated. Monolayers were washed with cold HBSS, incubated in cold HBSS containing 10 μg/ml cycloheximide for 10 min, and scrape harvested and centrifuged. Cell pellets were lysed in 1.0 ml of ice-cold lysis buffer (140 mM KCl, 1 mM DTT, 20 mM Tris, pH 8.5, mM MgCl2, 0.5% NP-40, 0.5 U/ml RNAsin [Promega], 10 mM cycloheximide, 1 mM PMSF, 5 μM leupeptin, 5 mM benzamidine, 5 μg/ml aprotinin) and mechanically disrupted using 12 strokes of a teflon–glass homogenizer at low speed. Nuclei and debris were removed by microfuge centrifugation at 1,000 g for 10 min, and the postnuclear supernatant was then centrifuged at 15,000 g for 20 min. The final supernatants were layered onto preformed 11-ml 20–47% linear sucrose gradients (containing a 60% sucrose cushion) made up in 140 mM KCl, 1 mM DTT, 20 mM Tris, pH 7.8, 1.5 mM MgCl2. Centrifugation was performed at 40,000 rpm for 2 h 45 min using a Beckman Coulter SW40Ti rotor. Fractions were eluted from the top of the gradient using an ISCO gradient elution system, ∼1-ml fractions were collected, and OD was measured at 260 nm to obtain the polysome profile. Aliquots of individual fractions were acetone-precipitated to remove sucrose and to concentrate the proteins, which were resuspended in reducing SDS sample buffer, and processed for Western blot analysis.
Plasmid Constructs
pMT2-TIA-1 and pMT2-TIA-1ΔRRM have been described previously (Tian et al. 1991). The coding regions of TIA-1 and PABP (a kind gift from R. Pictet, Institut Jacques Monod, University of Paris, Paris, France) were cloned into pSRα–HA-GFP (a kind gift from Michel Streuli, Dana Farber Cancer Institute, Boston, MA), using a PCR strategy to produce the fusion proteins HA-GFP–TIA-1 and HA-GFP–PABP. In brief, TIA-1 was amplified from pMT2–TIA-1 for 25 cycles (94°C for 1 min, 50°C for 1 min, and 74°C for 1 min) using Tli polymerase (Promega) and primers with EcoRI and XbaI cloning sites (CCGGAATTCATGGAGGACGAGATGCCCA and GCTCTAGATTCACTGGGTTTCATACCCTGC, respectively). PABP was similarly amplified from pMA-PABP using primers with XbaI and KpnI cloning sites (CTAGTCTAGAATGAACCCCAGTGCCCCC and CGGGGTACCTTAAACAGTTGGAACACC, respectively). The inserts were cut with EcoRI and XbaI or XbaI and KpnI, respectively, and cloned in-frame with a fusion HA-GFP tag in pSRα–HA-GFP that was similarly cut. The final clones were verified by sequencing. pcDNA3–β-galactosidase was obtained from Invitrogen. Plasmids encoding luciferase followed by the TNF-α 3′ untranslated region, either with or without its AU-rich element (ARE), were a kind gift from Veronique Kruys (Université Libre de Bruxelles, Brussels, Belgium).
Fluorescence Microscopy
Cells were grown, fixed, and stained as described previously (Kedersha et al. 1999), with minor modifications as follows. Normal horse serum was used as a blocking agent in lieu of normal goat serum, and secondary antibodies were raised in donkey (ML grade; Jackson ImmunoResearch Laboratories). Cy3-conjugated antibodies were used instead of Texas red conjugates. In situ hybridization was performed as described previously (Kedersha et al. 1999). Cells were viewed using a Nikon Eclipse 800 microscope, and images were digitally captured using a CCD-SPOT RT digital camera and compiled using Adobe Photoshop® software (v5.5).
Video Fluorescence Microscopy
Epifluorescence video microscopy was used to obtain digitized fluorescence images of live COS cells transfected with plasmids encoding GFP–TIA-1, GFP-PABP, or GFP only. Cells were treated as indicated in the figure legends, placed on a temperature-controlled microscope stage (37 ± 0.5°C), and observed using a ZEISS Axioskop microscope. The illumination source was a 100-W mercury arc lamp. Illuminating light was passed through heat and dichroic filters and focused on the sample through a 25×/0.8 NA oil immersion objective. Fluorescence emission was imaged using a cooled CCD camera (Roper Scientific) and processed for pseudocolor by a Metamorph image processor (Universal Imaging Corp.). Background intensity was subtracted from each image. The final images were compiled using Adobe Photoshop® (v5.5).
FRAP
COS cells were transiently transfected as described above, stimulated with arsenite for 30 min at 37°C, and viewed using a 40×/0.85 NA oil objective. FRAP experiments (Axelrod et al. 1976) were performed at room temperature using an Ultima interactive laser cytometer (Meridian Instruments). The Gaussian beam radius was 1 μm and excitation and emission wavelengths were 488 nm and 510 ± 5 nm, respectively. Photobleaching power at the sample was ∼0.5 mW, and the bleaching time was 1 s. The photobleaching beam was positioned directly over each SG. The fractional recovery (i.e., the fraction of GFP-conjugated protein that was capable of diffusion into the bleached SG) was obtained by using nonlinear least squares analysis of fluorescence recovery curves (Golan et al. 1986).
Online Supplemental Material
Digitized images were captured every 2 min as described above (Video Fluorescence Microscopy), and pseudocolored using Metamorph software. Images were overlaid using Adobe Photoshop® (v5.5) and animated using Adobe ImageReady™ (v2.0) software, using a 0.1-s delay between frames taken 2 min apart. Intensity scale is the same as shown in Fig. 6. Videos containing animated versions of the data also shown in Fig. 5 and Fig. 6 are available at http://www.jcb.org/cgi/content/full/151/6/1257/DC1.
Figure 6.
Disassembly of GFP–TIA-1+ SGs. COS cells were transfected with GFP–TIA-1 and cultured for 24 h before the addition of arsenite (0.5 mM, 30 min). Cells were then washed and cultured in the absence of arsenite. At the indicated times, fluorescence images from a field including live transfectants were recorded, pseudocolored, and compiled as described in the legend to Fig. 5. The relative fluorescence intensity is indicated by the scale at the left in which green is the lowest and blue is the highest intensity. A time lapse video of this process is available at http://www.jcb.org/cgi/content/full/151/6/1257/DC1.
Figure 5.
Assembly of GFP–TIA-1+ SGs. COS cells were transfected with GFP–TIA-1 and cultured for 24 h; arsenite (0.5 mM) was then added to initiate the stress response. Cellular fluorescence was viewed and photographed using digital capture, pseudocolored using Meridian software, and compiled using Adobe Photoshop®. The relative fluorescence intensity is indicated by the scale at the left in which green is the lowest and blue is the highest intensity. Times after addition of arsenite are indicated in each panel. A time lapse video of this process is available at http://www.jcb.org/cgi/content/full/151/6/1257/DC1.
Results
Components of Polysomes and SGs Are in Equilibrium
In tomato cells, mRNA moves from polysomes to SGs in cells exposed to heat shock and from SGs to polysomes in cells allowed to recover from heat shock (Nover et al. 1989). In mammalian cells subjected to graded doses of arsenite-induced oxidative stress, translation is inhibited at concentrations of arsenite that are insufficient to induce the assembly of SGs (Fig. 1 A; black bars monitor protein synthesis, gray bars monitor SG assembly of parallel samples). This suggests that mRNA must be liberated from polysomes before it can be recruited to SGs. Consistent with this hypothesis, inhibitors of protein synthesis that stabilize polysomes by freezing ribosomes on translating mRNAs (e.g., emetine and cycloheximide [Ballestra 1991]) inhibit the arsenite-induced assembly of SGs (Fig. 1 B). Similar results were obtained using cycloheximide (data not shown). In contrast, puromycin, an inhibitor of protein synthesis that destabilizes polysomes by promoting premature termination (Ballestra 1991), fails to inhibit and, at some concentrations, enhances the arsenite-induced assembly of SGs (Fig. 1 C). These results suggest that nonpolysomal mRNA could be a rate-limiting determinant of SG assembly.
Figure 1.
Differential effects of drugs that stabilize or destabilize polysomes on the arsenite-induced assembly of SGs. (A) Arsenite inhibits protein synthesis and induces the assembly of SGs in a dose-dependent manner. DU-145 cells were treated with the indicated concentration of arsenite for 30 min before measuring incorporation of [35S]methionine during an additional 30-min labeling period, expressed as a percentage of control incorporation (black bars), or processing for immunofluorescence microscopy to quantitate the percentage of cells containing TIA-1+ SGs (gray bars). (B and C) DU-145 cells were subjected to arsenite-induced stress (0.5 mM, 1 h) in the absence or presence of the indicated concentrations of emetine (B) or puromycin (C). Samples were processed for immunofluorescence microscopy to quantitate the percentage of cells with TIA-1+ SGs. Parallel cultures were pulsed with [35S]methionine for 30 min immediately after the drug treatment to quantitate incorporation into acetone-precipitable proteins as described in Materials and Methods. Gray bars indicate the percentage of cells containing SGs, black bars indicate 35S incorporation as a percentage of control.
The accumulation of untranslated mRNA at SGs could be static or dynamic. If it is static, mRNA (and associated proteins) will move into SGs during stress and out of SGs during recovery. In this case, addition of emetine to stressed cells should prevent further growth of preformed SGs. If it is dynamic, mRNA and associated proteins will continuously shuttle in and out of SGs during both stress and recovery. In this case, addition of emetine to stressed cells might disassemble preformed SGs by trapping mRNA at polysomes. As shown in Fig. 2, emetine dissolves preformed TIA-1+ SGs in the continued presence of stress. Although TIA-1 and TIAR are concentrated in the nucleus of DU-145 cells under control conditions (Fig. 2 A), these proteins move to cytoplasmic SGs in response to arsenite (Fig. 2 B; 0.5 mM, 30 min). In contrast, treatment with emetine at 10 μg/ml, which completely abrogates protein synthesis (see Fig. 1), increases the amount of cytoplasmic TIA-1/R but does not induce its aggregation into SGs (Fig. 2 C). The addition of emetine to arsenite-stressed cells (0.5 mM arsenite for 30 min, followed by the addition of 10 μg/ml emetine for an additional 60 min) dissolves preformed SGs in the continued presence of arsenite (Fig. 2 D; similar results were obtained using cycloheximide, data not shown). In contrast, addition of puromycin (20 μg/ml, a concentration that inhibits protein synthesis to a similar extent as 10 μg/ml emetine, see Fig. 1) to arsenite-stressed cells causes SGs to increase in size (Fig. 2 F). Puromycin alone induces the assembly of SGs in rare cells (Fig. 2 E, arrow), the percentage of which increases with time (data not shown).
Figure 2.

Emetine (but not puromycin) dissolves TIA-1+ SGs in the continued presence of stress. DU-145 cells were untreated (A), treated with 0.5 mM arsenite for 30 min (B), treated with emetine (10 μg/ml) for 1 h (C), treated with arsenite for 90 min with addition of 10 μg/ml emetine after 30 min (D), treated with puromycin (20 μg/ml) for 1 h (E), or treated with arsenite for 90 min with addition of 10 μg/ml puromycin after 30 min (F). Cells were then processed for immunofluorescence using antibodies specific for TIA-1.
To confirm that poly(A)+ RNA is also recruited to SGs, we used in situ hybridization to monitor the subcellular localization of poly(A)+ RNA under these same conditions. As shown in Fig. 3, emetine dissolves preformed poly(A) RNA+ SGs despite the continued presence of stress (compare Fig. 3B with D). In contrast, puromycin causes arsenite-induced poly(A) RNA+ SGs to increase in size (Fig. 3 F). Although emetine alone does not affect the subcellular localization of poly(A)+ RNA (Fig. 3 C), puromycin alone induces the formation of poly(A) RNA+ SGs in rare cells (Fig. 3 E, arrow). The differential effects of drugs that stabilize or destabilize polysomes on the accumulation of poly(A)+RNA at SGs suggest that mRNA may be in a dynamic equilibrium between SGs and polysomes.
Figure 3.

Emetine (but not puromycin) dissolves poly(A) RNA+ SGs in the continued presence of stress. DU-145 cells were subjected to the same conditions described in the legend to Fig. 2 before processing for in situ hybridization using an oligo-dT probe to monitor the subcellular localization of poly(A)+ RNA.
Phosphorylation of eIF-2α is necessary and sufficient to induce the assembly of SGs (Kedersha et al. 1999). Thus, the ability of emetine to dissolve SGs could result from dephosphorylation of eIF-2α. Therefore, we measured the phosphorylation of eIF-2α in DU-145 cells subjected to arsenite-induced oxidative stress in the absence or presence of emetine. As shown in Fig. 4 A, immunoblots of total cell extracts probed with antibodies against phosphorylated eIF-2α or total eIF-2α reveal that eIF-2α is not significantly phosphorylated in untreated cells (Fig. 4 A, lane C). Arsenite (0.5 mM) induces a time-dependent phosphorylation of eIF-2α (A30, 30-min treatment; A90, 90-min treatment). Addition of emetine to cells pretreated with arsenite for 30 min does not reduce the phosphorylation of eIF-2α (Fig. 4 A, lane A+E) and, instead, appears to increase it. Neither emetine (Fig. 4 A, lane E) nor puromycin (Fig. 4 A, lane P) significantly induces the phosphorylation of eIF-2α in the absence of arsenite. These data indicate that emetine does not dissolve preformed SGs by promoting eIF-2α dephosphorylation, nor does puromycin enhance the assembly of SGs by promoting eIF-2α phosphorylation.
Figure 4.
SG dissolution by emetine is due to polysome stabilization and occurs downstream of eIF-2α phosphorylation. (A) DU-145 cells were cultured in media alone (lane C), 0.5 mM arsenite for 30 min (A30), 0.5 mM arsenite for 90 min (A90), 0.5 mM arsenite for 90 min with addition of 10 μg/ml emetine after 30 min (A+E), 10 μg/ml emetine for 60 min (E), or 20 μg/ml puromycin for 60 min (P). Whole cell lysates were then separated by SDS-PAGE, transferred to nitrocellulose, and probed using antibodies specific for eIF-2α (bottom) or phosphorylated eIF-2α (top, p-eIF-2α). (B) Polysome profiles of control and treated cells. DU-145 cells were untreated (black open squares), or treated as described in A and subjected to sucrose gradient analysis as described in Results. A30, blue diamonds; A90 treatment, red squares; A+E treatment, green squares; E alone, blue squares. Polysome profiles were obtained by measuring the absorbance at 260 nm of individual fractions. Monosomes are found in fractions 2–5, and polysomes are found in fractions 7–12 (the top of the gradient is fraction 1). (C) Western blots of individual fractions. Fractions obtained from the gradients were subjected to PAGE in SDS and immunoblotting for PABP-I, TIA-1, and ribosomal subunit (Rib) proteins S3 and S19 as indicated. The lightest fractions are at the left, and the 80S and polysomal regions are indicated. 1, control; 2, arsenite treatment for 30 min; 3, arsenite treatment for 90 min; and 4, arsenite treatment for 90 min with emetine added during the last 60 min.
Further evidence for a dynamic equilibrium between SGs and polysomes was obtained by comparing polysome profiles in arsenite-stressed cells in the absence or presence of emetine. DU-145 cells were treated as described above before separating cytosolic extracts on sucrose gradients (Fig. 4 B). The absorbance of individual fractions clearly distinguishes 80S monosomes (fractions 2–5) from polysomes (fractions 7–11) in untreated DU-145 cells (Fig. 4 B, black open squares). The location of monosomes and polysomes was confirmed by blotting individual fractions for ribosomal proteins S3 and S19 (Fig. 4 C). Emetine treatment alone (10 μg/ml, 1 h) does not significantly change the polysome profile (Fig. 4 B, blue squares). As expected, 30 min of arsenite treatment (0.5 mM) results in the disassembly of polysomes (Fig. 4 B, blue diamonds), whereas 90 min of arsenite treatment (Fig. 4 B, red squares) also disassembles polysomes and appears to shift the monosome peak even further to the top of the gradient, perhaps indicating the accumulation of ribosomal subunits. The addition of emetine to arsenite-stressed cells (0.5 mM arsenite for 30 min followed by 10 μg/ml emetine for an additional 1 h) promotes the assembly of low density polysomes (fractions 8–10, green diamonds), even though arsenite is present throughout the entire 90-min incubation (compare green diamonds with red squares). These results indicate that SG-associated mRNA can move to polysomes during stress, despite the continued phosphorylation of eIF-2α.
Individual fractions from the sucrose gradients were subjected to immunoblotting analysis to localize TIA-1 and PABP-I, the major protein components of SGs (Kedersha et al. 1999) in these same gradients. As shown in Fig. 4 C, panel 1), PABP-I is found in both monosome-containing (fractions 2–5) and polysome-containing (fractions 7–12) fractions in unstressed control cells, but is excluded from fraction 1 of the gradient, indicating that all of the PABP-I in these cells associates with sedimentable complexes. In response to arsenite-induced translational arrest, polysome disassembly shifts PABP-I away from large polysomes into the monosome region of the gradient (Fig. 4 C, panels 2 and 3). The addition of emetine to arsenite-stressed cells causes PABP-I to shift to higher density fractions that contain polysomes (Fig. 4 C, panel 4), suggesting that some translational initiation is still occurring in the arsenite-treated cells and that emetine is trapping the newly assembled polysomes. The distribution of ribosomal proteins S2 and S19 parallels that of PABP-I (Fig. 4 C, panels 1–4), which verifies that the OD260 profiles accurately reflect the distribution of monosomes and polysomes within the gradient.
In contrast to PABP-I, the bulk of TIA-1 (Fig. 4 C, all panels) and TIAR (data not shown) is concentrated at the top of the gradient (i.e., fractions 1–4) under all conditions, indicating that most TIA-1 protein is present in soluble and low density complexes. The distribution of TIA-1 is not altered in cells treated with arsenite (Fig. 4 C, panels 2 and 3) nor in cells treated with arsenite and emetine (Fig. 4 D). Significant amounts of TIA-1 migrate with the 80S peak, with trace amounts barely detectable in the polysome region. These results suggest that TIA-1 associates with translationally inactive mRNA rather than polysomal mRNA, consistent with data from our lab (Piecyk et al. 2000) and others (Gueydan et al. 1999), indicating that TIA-1 functions as a translational silencer. It is not clear from these data whether SGs migrate as discrete particles in sucrose gradients. Equilibrium banding of tomato heat shock granules required prior fixation (Nover et al. 1989), suggesting that these cytoplasmic microdomains may not remain intact in sucrose gradients.
Real Time Imaging of TIA-1+/PABP-I+ SGs
We monitored the assembly and disassembly of SGs in real time in live cells, using cDNA constructs encoding GFP alone, GFP–TIA-1, and GFP–PABP-I. COS cells were transiently transfected with GFP–TIA-1 and cultured for 24 h. The subcellular localization of GFP–TIA-1 was then monitored using fluorescence microscopy. Like the endogenous protein, GFP–TIA-1 is concentrated in the nuclei of transfected COS cells (Fig. 5, time 0). In cells subjected to arsenite-induced oxidative stress, GFP–TIA-1 moves from the nucleus to the cytoplasm in a time-dependent manner (Fig. 5). At early times (i.e., 6 min), GFP–TIA-1 is diffusely distributed throughout the cytoplasm. At later times (≥14 min), cytoplasmic GFP–TIA-1 accumulates at discrete foci that enlarge and coalesce in the continued presence of stress (time lapse video available at http://www. jcb.org/cgi/content/full/151/6/1257/DC1). Endogenous TIA-1 accumulates at SGs in COS cells with similar kinetics (Kedersha et al. 1999), whereas GFP alone does not associate with SGs upon arsenite treatment. This result confirms that TIA-1 is a constituent of the SG and defines the kinetics of SG assembly in real time.
We also monitored the disassembly of GFP–TIA-1+ SGs that had formed in response to a sublethal dose of arsenite (0.5 mM for 30 min). In cells allowed to recover in the absence of arsenite, GFP–TIA-1+ SGs are disassembled within 60–90 min (Fig. 6). Although small SGs are dissolved faster than large SGs, the rate of dissolution appears to be similar regardless of the initial size. During the disassembly process, GFP–TIA-1+ SGs slowly dissolve while maintaining their position in the cytoplasm. GFP–TIA-1+ SGs that are adjacent to the nucleus appear to move towards the nuclear envelope as they slowly disperse. A time lapse video of this process is available at http://www.jcb.org/cgi/content/full/151/6/1257/DC1. As expected, cycloheximide enhances the disassembly rate of GFP–TIA-1+ SGs in COS transfectants allowed to recover from a sublethal dose of arsenite (data not shown). Similar results were obtained using GFP–PABP-I (data not shown), another invariant marker of SGs (Kedersha et al. 1999).
FRAP
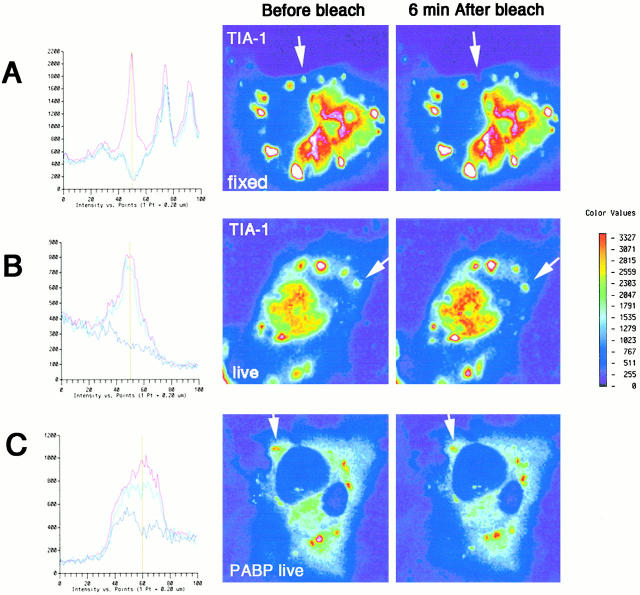
We used FRAP analysis to determine whether GFP–TIA-1 and GFP–PABP-I shuttle in and out of SGs. COS cells expressing either GFP–TIA-1 or GFP–PABP-I were subjected to arsenite-induced oxidative stress to induce the assembly of SGs. Confocal microscopy was used to select SGs for FRAP analysis (Fig. 7, right, arrows point to targeted SGs). Graphical representations of fluorescence intensity were obtained by scanning an argon beam in a linear pattern across the selected field (Fig. 7, left). COS (GFP–TIA-1) transfectants were arsenite-treated and then fixed with paraformaldehyde and postfixed with methanol (Fig. 7 A). These cells exhibit diffuse cytoplasmic fluorescence of 400–600 arbitrary units with peaks that correspond to SGs (Fig. 7 A, the scanned field includes the targeted SG and two adjacent SGs). Before photobleaching, fluorescence from the targeted SG peaks at 2,200 arbitrary units (Fig. 7 A, red). Photobleaching (achieved by irradiating the targeted peak for 1 s using a 1-μm-diameter argon laser beam at 488 nm) eliminates both diffuse cytoplasmic and SG-associated fluorescence (i.e., the fluorescence intensity dips below the background), as revealed by a scan taken 0.65 s after photobleaching (Fig. 7 A, purple). A second scan taken 22 s after photobleaching is similar to the initial postphotobleaching scan (Fig. 7 A, blue), as is the scanned image (Fig. 7 A, right) taken 6 min later, indicating that unirradiated GFP–TIA-1 does not reconstitute the SG in fixed cells. In live COS (GFP–TIA-1) transfectants treated with arsenite, fluorescence from the targeted SG peaks at 800 arbitrary units (Fig. 7 B, red). Although the photobleaching treatment eliminates most (72%) of the SG-associated fluorescence as revealed by a scan taken 0.65 s after irradiation (Fig. 7 B, purple), a second scan taken 22 s after photobleaching reveals that the fluorescence intensity has recovered to that of the prephotobleached SG (Fig. 7 B, blue). This recovery of fluorescence must be due to the recruitment of unbleached GFP–TIA-1 from outside of the photobleached field. Similar results are observed using GFP–PABP-I (Fig. 7 C) (before bleaching, red; 0.65 s after bleaching, purple; 22 s after bleaching, blue). The kinetics of fluorescence recovery in fixed and unfixed cells are compared in Fig. 8. The recovery of GFP–TIA-1 and GFP–PABP-I is biphasic, with a rapid early phase (t 1/2 ∼ 1 s) and a slower late phase (t 1/2 ∼ 5 s). During the early phase, GFP–TIA-1 returns to the SG faster than GFP–PABP-I. During the late phase, the rates at which GFP–TIA-1 and GFP–PABP-I recover are similar. Overall recovery of GFP–TIA-1 is greater than that of GFP–PABP-I (99 vs. 61%). These results are representative of data obtained from photobleaching of 10 GFP–TIA-1+ SGs and 8 GFP–PABP-I+ SGs.
Figure 7.
Fluorescence recovery after photobleaching. Fluorescence microscopy of arsenite-treated (0.5 mM, 30 min) COS cells expressing GFP–TIA-1 (A and B) or GFP–PABP-I (C). Formaldehyde-fixed cells are viewed in A. Live cells are viewed in B and C. (left) Fluorescence intensity across a linear scan centered on the individual SG identified in the fluorescence micrograph shown in the right panel. Red, intensity before photobleach treatment; violet, 0.65 s after photobleach; blue, 22 s after photobleach. (right) Pseudocolor photomicrographs of COS transfectants before and 6 min after photobleaching. Arrows indicate the targeted SGs.
Figure 8.
Kinetics of fluorescence recovery. After photobleaching, fluorescence measurements were taken at the peak of the targeted SG (identified by the y axis) at the indicated times. The percent recovery relative to the initial prebleach values is shown as a function of time. The earliest point measured is 0.65 s after photobleaching, at which time some recovery has already occurred in the live cells.
TIA-1ΔRRM Promotes the Expression of Cotransfected Reporter Genes
A TIA-1 truncation mutant lacking its RNA-binding domains (TIA-1ΔRRM) prevents the arsenite-induced assembly of SGs and forms cytoplasmic microaggregates that sequester endogenous TIA-1 and TIAR (Kedersha et al. 1999). A map indicating the coding sequences of the full-length and the TIA-1ΔRRM truncation is shown in Fig. 9 A. COS cells were transfected with plasmids encoding full-length recombinant TIA-1, TIA-1ΔRRM, or empty vector, and expression of the recombinant proteins was confirmed by immunoblotting analysis (Fig. 9 B, panel d). The TIA–1ΔRRM cDNA construct produces two peptides, a consequence of translational initiation at methionines 219 and 230 (Fig. 9 A). The effect of these recombinant proteins on the expression of co-transfected β-galactosidase (Fig. 9 B, panel a) or luciferase (Fig. 9 B, panels b and c) was determined by blotting the same filters with antibodies specific for the reporter proteins. The relative expression of the reporter proteins, determined by densitometric scanning, revealed that TIA-1ΔRRM increases the expression of β-galactosidase (4.2 ± 4–fold, n = 5) compared with the vector control. In contrast, full-length TIA-1 slightly reduces the expression of β-galactosidase (0.9 ± 0.5–fold, n = 5) compared with the vector control. Metabolic labeling and immunoprecipitation experiments (data not shown) give similar results. Because TIA-1 binds to AREs in the 3′ untranslated region of TNF-α transcripts (Piecyk et al. 2000), we compared the expression of luciferase reporters linked to the 3′ untranslated region of TNF-α with or without the ARE (Fig. 9 B, compare panels b and c). The presence of the ARE does not significantly affect luciferase expression in COS (TIA-1ΔRRM) transfectants. In contrast, the ARE may enhance the repressive effect of full-length TIA-1 (Fig. 9 B, compare panel b with c). These results suggest that TIA-1 normally functions to dampen protein expression, consistent with previous results showing that TIA-1 represses TNF-α expression in macrophages (Piecyk et al. 2000). It remains to be determined whether SG assembly is required for this functional effect.
Figure 9.

TIA-1ΔRRM enhances the expression of cotransfected reporter genes. (A) Map represents the sequence of full-length TIA-1 and of two related protein products encoded by the TIA-1ΔRRM construct. The TIA-1ΔRRM DNA begins at the site marked T4T8 on the map of TIA-1. The truncated construct expressed in COS cells results in two protein products initiated at methiones 219 and 230, which are resolved as a doublet (B, lane 3). (B) COS cells were transiently transfected with empty pMT-2 vector control, pMT2 full-length TIA-1, or pMT2-TIA-1ΔRRM. In each case, cells were cotransfected with plasmids encoding β-galactosidase (a), luciferase linked to the TNF-α ARE (b), or luciferase lacking the TNF-α ARE (c). After 48 h, cells were solubilized in SDS sample buffer and subjected to SDS-PAGE. After transfer to nitrocellulose, blots were probed with antibodies specific for β-galactosidase (a), luciferase (b and c), or TIA-1/R (d).
Discussion
Stress-induced translational arrest is initiated by the phosphorylation of eIF-2α, a subunit of the eIF2:GTP:Met-tRNAi ternary complex that is required at the first step of protein synthesis (Gray and Wickens 1998). Phosphorylated eIF-2α (eIF2[αP]) forms an active ternary complex that can initiate a single round of translation (Trachsel and Staehelin 1978). During this event, GTP is hydrolyzed to produce eIF2(αP):GDP, a competitive inhibitor of the guanine nucleotide exchange factor (eIF2B) that normally regenerates eIF2:GTP (Kimball et al. 1998). Inactivation of eIF2B reduces the availability of eIF2:GTP:Met-tRNAi, reducing the rate of translational initiation. Residual eIF2:GTP:Met-tRNAi is selectively available to newly synthesized mRNAs encoding heat shock proteins, which are actively translated during stress. The selective expression of heat shock proteins has been proposed to result from the eIF2(αP)-induced assembly of SGs (Kedersha et al. 1999), which are sites at which “housekeeping” mRNAs but not heat shock mRNAs accumulate in heat shocked plant cells (Nover et al. 1989). By sequestering selected mRNAs from the translational apparatus, SGs could determine which mRNAs (e.g., heat shock versus non-heat shock) are available to the limited pool of initiation factors that are present in stressed cells.
The sequestration model of translational control implies that SGs are stable depots of untranslated mRNPs; however, the different effects of emetine and puromycin on the assembly of SGs are not consistent with this simple model. Emetine is an elongation inhibitor that freezes ribosomes on their translating mRNA (Ballestra 1991). In arsenite-stressed cells, emetine dissolves SGs and concurrently promotes the assembly of polysomes, despite the persistent phosphorylation of eIF-2α. These data indicate that mRNA can shuttle between SGs and polysomes during stress. Emetine reveals this dynamic equilibrium by acting as a polysome trap and verifies that arsenite-induced phosphorylation of eIF-2α (which is not dephosphorylated in response to emetine) reduces (Fig. 1 A and 4 B) but does not eliminate translational initiation. Unlike emetine, puromycin is an aminoacyl tRNA analogue that destabilizes polysomes by promoting premature termination (Ballestra 1991). By disassembling polysomes and liberating untranslated mRNAs, puromycin promotes the assembly of SGs. Thus, the effects of these drugs on polysome stability may be sufficient to explain their ability to promote and inhibit the assembly of SGs. It should be noted, however, that emetine and puromycin also differentially affect translational termination. Emetine prevents ribosomes from reaching the termination codon, whereas puromycin actively promotes translational termination. If the termination machinery participates in the assembly of SGs, the different effects of emetine and puromycin could be related to their effects on translational termination. The relative contribution of these two mechanisms to the assembly of SGs remains to be determined. Regardless of the mechanism, the different effects of emetine and puromycin suggest that SGs are not simply depots of mRNPs. The ability of mRNA to move between SGs and polysomes suggests that SGs may be dynamic microdomains at which mRNA and associated proteins transiently accumulate as they move between functional domains within the cell.
To address this issue, we used time lapse microscopy and FRAP analysis to monitor the recruitment of GFP-tagged TIA-1 (and PABP-I) to SGs. These experiments reveal that GFP–TIA-1 and GFP–PABP-I slowly accumulate at SGs in response to arsenite. The kinetics of SG assembly must be determined by the rate at which proteins enter (the “in-rate”) and leave (the “out-rate”) individual SGs. If SGs are static (i.e., the out-rate is zero in stressed cells), the growth of SGs will be determined by the in-rate. If SGs are dynamic (i.e., components continuously shuttle in and out), the growth of SGs will be determined by the difference between the in-rate and the out-rate. FRAP analysis shows that photobleached GFP–TIA-1+ SGs are >70% reconstituted very rapidly—within 4 s. Thus the in-rate (t 1/2 ∼ 2 s) is two to three orders of magnitude faster than the rate of SG assembly (t 1/2 ∼ 15–30 min). This result indicates that the accumulation of GFP–TIA-1 at SGs is not a simple function of the in-rate. Rather, the slow accumulation of GFP–TIA-1 at SGs indicates that the rapid in-rate is balanced by a slightly less rapid out-rate. Thus, although TIA-1 is necessary for SG assembly, as indicated by the ability of TIA-1ΔRRM to block SG formation, TIA-1 is not a stable component of SGs themselves. Similarly, PABP-I is not a stable component of SGs, although its turnover within the SG appears less rapid and less complete than that of TIA-1.
Recovery of GFP–TIA-1 and GFP–PABP-I fluorescence in photobleached SGs is biphasic. During the early phase (<2 s), the net accumulation of GFP–TIA-1 is significantly greater than that of GFP–PABP-I. During the late phase, the rates at which GFP–TIA-1 and GFP–PABP-I accumulate in SGs are similar. These results suggest that TIA-1 and PABP-I are not components of the same RNP complex during the early phase of reconstitution. PABP-I associates with mRNA in translated (i.e., polysomes) and untranslated (i.e., SGs) mRNP complexes. In contrast, sucrose gradient analysis reveals that TIA-1 is largely excluded from polysomes in unstressed cells, although a very small fraction of cytoplasmic TIA-1/R can be found in high density, polysome-containing fractions in sucrose gradients. It is therefore possible that TIA-1/R participate in stress-induced translational termination, polysome disassembly and/or delivery of nonpolysomal mRNA to SGs. In the latter case, the later phase of reconstitution of both GFP–TIA-1 and GFP–PABP-I could occur in association with mRNA.
Our data reveal that SGs are highly dynamic structures despite their apparent stability in time lapse videos. In this respect, SGs resemble several nuclear RNA–containing structures (e.g., speckles, coiled bodies, Gemini of coiled bodies, and nucleoli) that are sites of active RNA metabolism (Zeng et al. 1997; Gall et al. 1999; Mattern et al. 1999; Mintz and Spector 2000). At the microscopic level, these metabolic domains appear to be stable structures that maintain a distinct morphology. Despite their stable appearance, many of the molecular components of these nuclear substructures are in constant flux (Sleeman and Lamond 1999). SGs form when the rate of translational termination exceeds the rate of translational initiation, which results in the release of ribosomes from translating mRNAs. Thus, SGs may be sites of mRNA triage at which untranslated mRNAs accumulate during stress before being directed to sites of degradation, reinitiation, or repackaging as mRNPs. When the accumulation of nonpolysomal mRNA exceeds the capacity for triage, untranslated mRNA transiently accumulates at the SG. Under these conditions, mRNA (and associated proteins) will rapidly move in and out of SGs, consistent with the results obtained using FRAP analysis. The RNA-stabilizing protein HuR has been shown to accumulate at SGs in response to heat shock (Gallouzi et al. 2000), suggesting that the proteins that regulate transcript stability might determine whether transcripts that arrive at the SG are translated or degraded. Although mRNA moves in and out of SGs in arsenite-treated cells, the rate of mRNA shuttling may be significantly slower than the rate of protein shuttling. If this is true, sequestration of mRNA at SGs could participate in translational regulation, as proposed previously (Nover et al. 1989). In this case, the observed shuttling of TIA-1 and PABP-I might reflect the recruitment of multiple mRNA transcripts to the SG.
Previously, we described a truncation mutant of TIA-1 (TIA-1ΔRRM) that lacks the ability to bind RNA (Dember et al. 1996), forms cytoplasmic microaggregates that sequester endogenous TIA-1 and TIAR (Kedersha et al. 1999), and prevents the accumulation of mRNA at SGs. We now report that overexpression of this dominant negative mutant also promotes the expression of cotransfected reporter genes in COS transfectants. These results suggest that TIA-1 normally functions to dampen protein expression, which is consistent with the ability of TIA-1 to repress the expression of TNF-α in LPS-stimulated macrophages (Piecyk et al. 2000). Although COS transfectants were cultured in the absence of exogenous stress, transfection with foreign DNA can activate double-stranded RNA–dependent kinase (PKR), which phosphorylates eIF-2α and induces the assembly of SGs (Kaufman 1997; Kedersha et al. 1999). Therefore, it remains to be determined whether TIA-1 (and/or the assembly of TIA-1+ SGs) can dampen protein expression in the absence of stress. In either case, the inverse correlation between SG assembly and protein expression suggests that the proposed mRNA triage mechanism might influence protein expression.
In conclusion, the data presented indicate that SGs are dynamic microdomains containing translationally inactive mRNA into which TIA-1 and PABP-I rapidly shuttle and which are in equilibrium with polysomes. As TIA-1 is a translational silencer and PABP-I a translational enhancer, we favor the view that SGs constitute mRNA triage domains where mRNAs are converted to silenced mRNPs by the addition of TIA-1. Although the molecular details of this process await further investigation, our data establish that SGs are highly dynamic domains within which mRNA processing, sorting, and/or remodeling events are likely to regulate the expression of specific mRNA transcripts.
Supplemental Material
Acknowledgments
We thank members of the Anderson laboratory for helpful discussions.
P. Anderson was supported by National Institutes of Health grant AI33600, a Biomedical Science Grant from the Arthritis Foundation, and a Leukemia Scholar Award. D.E. Golan was supported by grants from the National Institutes of Health (HL32854 and HL15157). M.R. Cho was supported by a Whitaker Biomedical Engineering Research grant.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: ARE, AU-rich element; eIF, eukaryotic initiation factor; HA, hemagglutinin; PABP-I, poly(A)+ binding protein I; PrD, prion-related domain; RRM, RNA recognition motif; SG, stress granule.
References
- Axelrod D., Koppel D., Schlessinger J., Elson E., Webb W. Mobility measurements by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestra, J. 1991. Inhibitors of Eukaryotic Protein Synthesis. H. Trachseh, editor. CRC Press, London.
- Berlanga J., Herrero S., DeHaro C. Characterization of the hemin-sensitive eukaryotic initiation factor 2a kinase from mouse nonerythroid cells. J. Cell Biol. 1998;273:32340–32346. doi: 10.1074/jbc.273.48.32340. [DOI] [PubMed] [Google Scholar]
- Dember L.M., Kim N.D., Liu K.Q., Anderson P. Individual RNA recognition motifs of TIA-1 and TIAR have different RNA binding specificities. J. Biol. Chem. 1996;271:2783–2788. doi: 10.1074/jbc.271.5.2783. [DOI] [PubMed] [Google Scholar]
- Gall J., Bellini M., Wu Z., Murphy C. Assembly of the nuclear transcription and processing machinerycajal bodies (coiled bodies) and transcriptosomes. Mol. Biol. Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallouzi I.E., Brennan C.M., Stenberg M.G., Swanson M.S., Eversole A., Maizels N., Steitz J.A. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan D., Brown C., Cianci C., Furlong S., Caulfield J. Schistosomula of Schistosoma mansoni use lysophosphatidylcholine to lyse adherent human red blood cells and immobilize red cell membrane components. J. Cell Biol. 1986;103:819–828. doi: 10.1083/jcb.103.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray N., Wickens M. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998;14:399–458. doi: 10.1146/annurev.cellbio.14.1.399. [DOI] [PubMed] [Google Scholar]
- Gueydan C., Droogmans L., Chalon P., Huez G., Caput D., Kruys V. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor α mRNA. J. Biol. Chem. 1999;274:2322–2326. doi: 10.1074/jbc.274.4.2322. [DOI] [PubMed] [Google Scholar]
- Kaufman R. DNA transfection to study translational control in mammalian cells. Methods Enzymol. 1997;11:361–370. doi: 10.1006/meth.1996.0434. [DOI] [PubMed] [Google Scholar]
- Kawakami A., Tian Q., Duan X., Streuli M., Schlossman S.F., Anderson P. Identification and functional characterization of a TIA-1-related nucleolysin. Proc. Natl. Acad. Sci. USA. 1992;89:8681–8685. doi: 10.1073/pnas.89.18.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., Miller I., Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 1999;147:1431–1441. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimball S.R., Fabian J.R., Pavitt G.D., Hinnebusch A.G., Jefferson L.S. Regulation of guanine nucleotide exchange through phosphorylation of eukaryotic initiation factor eIF2alpha. Role of the alpha- and delta-subunits of eiF2b. J. Biol. Chem. 1998;273:12841–12845. doi: 10.1074/jbc.273.21.12841. [DOI] [PubMed] [Google Scholar]
- Mattern K., van der Kraan I., Schu W., de Jong L., van Driel R. Spatial organization of four hnRNP proteins in relation to sites of transcription, to nuclear speckles, and to each other in interphase nuclei and nuclear matrices of HeLa cells. Exp. Cell Res. 1999;246:461–470. doi: 10.1006/excr.1998.4267. [DOI] [PubMed] [Google Scholar]
- Mintz P., Spector D. Compartmentalization of RNA processing factors within nuclear speckles. J. Struct. Biol. 2000;129:241–251. doi: 10.1006/jsbi.2000.4213. [DOI] [PubMed] [Google Scholar]
- Nover L., Scharf K.D., Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol. Cell. Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nover L., Scharf K., Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol. Cell. Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piecyk M., Wax S., Beck A., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., Anderson P. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:4154–4163. doi: 10.1093/emboj/19.15.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K.D., Heider H., Hohfeld I., Lyck R., Schmidt E., Nover L. The tomato Hsf systemHsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol. Cell. Biol. 1998;18:2240–2251. doi: 10.1128/mcb.18.4.2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J., Lamond A. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 1999;9:1065–1074. doi: 10.1016/s0960-9822(99)80475-8. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Kumar K., Kaufman R. Phosphorylation of eukaryotic initiation factor 2 mediates apoptosis in response to activation of the double-stranded RNA-dependent protein kinase. J. Biol. Chem. 1998;273:2416–2423. doi: 10.1074/jbc.273.4.2416. [DOI] [PubMed] [Google Scholar]
- Tian Q., Streuli M., Saito H., Schlossman S.F., Anderson P. A polyadenylate binding protein localized to the granules of cytolytic lymphocytes induces DNA fragmentation in target cells. Cell. 1991;67:629–639. doi: 10.1016/0092-8674(91)90536-8. [DOI] [PubMed] [Google Scholar]
- Trachsel H., Staehelin T. Binding and release of eukaryotic initiation factor eIF-2 and GTP during protein synthesis initiation. Proc. Natl. Acad. Sci. USA. 1978;75:204–208. doi: 10.1073/pnas.75.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C., Kim E., Warren S., Berget S. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.