Abstract
The Arabidopsis CBF1, 2, and 3 genes (also known as DREB1b, c, and a, respectively) encode transcriptional activators that have a central role in cold tolerance. CBF1-3 are rapidly induced upon exposing plants to low temperature, followed by expression of CBF-targeted genes, the CBF regulon, resulting in an increase in plant freezing tolerance. At present, little is known about the cold-sensing mechanism that controls CBF expression. Results presented here indicate that this mechanism does not require a cold shock to bring about the accumulation of CBF transcripts, but instead, absolute temperature is monitored with a greater degree of input, i.e. lower temperature, resulting in a greater output, i.e. higher levels of CBF transcripts. Temperature-shift experiments also indicate that the cold-sensing mechanism becomes desensitized to a given low temperature, such as 4°C, and that resensitization to that temperature requires between 8 and 24 h at warm temperature. Gene fusion experiments identified a 125-bp section of the CBF2 promoter that is sufficient to impart cold-responsive gene expression. Mutational analysis of this cold-responsive region identified two promoter segments that work in concert to impart robust cold-regulated gene expression. These sequences, designated ICEr1 and ICEr2 (induction of CBF expression region 1 or 2), were also shown to stimulate transcription in response to mechanical agitation and the protein synthesis inhibitor, cycloheximide.
Many plants increase in freezing tolerance in response to low nonfreezing temperatures, a phenomenon known as cold acclimation (Guy, 1990; Thomashow, 1999). In Arabidopsis, cold acclimation involves action of the CBF cold-response pathway (Thomashow, 2001). Within 15 min of exposing plants to low temperatures, transcripts accumulate for a family of genes designated CBF1, CBF2, and CBF3 (Gilmour et al., 1998; Jaglo-Ottosen et al., 1998; Medina et al., 1999), or DREB1b, DREB1c, and DREB1a (Liu et al., 1998), respectively, which encode transcriptional activators that are members of the AP2/EREBP family of DNA-binding proteins (Riechmann and Meyerowitz, 1998). These transcription factors bind the cold- and dehydration-responsive DNA regulatory element designated the CRT (C-repeat)/DRE (dehydration response element); (Baker et al., 1994; Yamaguchi-Shinozaki and Shinozaki, 1994; Stockinger et al., 1997) that is present in the promoters of COR and many other cold-responsive genes and stimulate their transcription. Expression of the CBF regulon of target genes then leads to an increase in freezing tolerance (Gilmour et al., 1998; Jaglo-Ottosen et al., 1998; Liu et al., 1998; Kasuga et al., 1999). Multiple mechanisms appear to contribute to the enhancement of freezing tolerance, including the synthesis of cryoprotective polypeptides, such as COR15a (Artus et al., 1996; Steponkus et al., 1998), and the accumulation of compatible solutes that have cryoprotective properties, including Suc, raffinose, and Pro (Nanjo et al., 1999; Gilmour et al., 2000; Taji et al., 2002).
Currently, little is known about how the CBF genes are up-regulated in response to low temperatures, but important insights are beginning to emerge. It has been established that the promoters of the CBF genes are responsive to low temperatures (Shinwari et al., 1998), and a transcription factor, Inducer of CBF Expression 1 (ICE1) that has a role in CBF expression has recently been identified (Chinnusamy et al., 2003). ICE1 encodes a MYC-like basic helix-loophelix (bHLH) protein. A dominant-negative mutation of ICE1, ice1, results in almost complete elimination of CBF3 transcript accumulation in response to low temperatures. Significantly, however, the ice1 mutation has little effect on cold-induced accumulation of CBF2 transcripts, indicating that there are differences in mechanisms of expression within the CBF/DREB1 gene family. Other genes and proteins that affect CBF expression have also been identified. LOS4 encodes a DEAD-box RNA helicase that appears to have a positive role in CBF expression; accumulation of CBF transcripts is reduced or delayed in plants that are homozygous for the recessive los4-1 mutant allele (Gong et al., 2002). In contrast, FRY2 (Xiong et al., 2002) and HOS1 (Lee et al., 2001) appear to down-regulate CBF expression. The FRY2 and HOS1 proteins are, respectively, a novel transcriptional repressor and a novel RING finger protein. It has been suggested that HOS1 may be an E3 ligase that targets CBF regulatory proteins for ubiquitination and protein degradation (Lee et al., 2001).
In this study, we further examine factors that affect the accumulation of CBF transcripts in response to low temperatures. The results indicate that steady-state levels of CBF transcripts increase in response to cold shock and gradual temperature downshifts; that the cold-sensing mechanism becomes desensitized with time at low temperature; that the CBF transcripts have a very short half-life at warm temperature; and that multiple promoter cis-acting regulatory elements function together to stimulate CBF2 transcription in response to low temperature.
RESULTS
CBF Transcripts Accumulate in Response to Cold Shock and a Gradual Decrease in Temperature
Transferring Arabidopsis plants abruptly from 20°C to 4°C results in the rapid accumulation of CBF transcripts (Fig. 1A; Gilmour et al., 1998; Liu et al., 1998; Medina et al., 1999). In our experiments, CBF transcript levels reached a maximum at about 3 h and then declined significantly, but remained elevated over those found in warm-grown plants over the course of the 3-week experiment. The magnitude of the cold shock affected the peak levels of the CBF transcripts (Fig. 1B). That is, when plants were transferred from 20°C to 10°C, the CBF levels after 2 h were less than if the plants were transferred from 20°C to 4°C. Similarly, higher levels of CBF transcripts were observed when the cold shock was from 20°C to –5°C compared with the 20°C to 4°C treatment.
Figure 1.
Accumulation of CBF transcripts in response to low temperatures. A, Plants were abruptly transferred from 22°C to 4°C for the indicated periods and CBF transcript levels were determined by RNA-blot hybridization. Gene-specific probes were used to detect, independently, CBF1, 2, and 3 transcripts. B, Plants were abruptly transferred from 22°C to the indicated cold temperatures for 2 h and CBF transcript levels were determined by RNA-blot hybridization. The probe used, a full-length cDNA of CBF1, cross-hybridized with CBF1, 2, and 3 transcripts. C, Plants grown at warm temperatures were subjected to a cold shock or to a gradual temperature decrease, and CBF and COR15a transcript levels were determined by RNA-blot hybridization. The full-length cDNA for CBF1 was used to detect CBF1, 2, and 3 transcripts. In each experiment, rRNA stained with ethidium bromide was used to compare loading.
To determine whether the accumulation of CBF transcripts was dependent upon a rapid cold shock, plants were slowly cooled at a rate of 2°C h–1, and CBF transcript levels were determined at various times (Fig. 1C). The results indicated that a gradual drop in temperature from 20°C to 4°C (over an 8-h time period) resulted in CBF levels that were essentially the same as those obtained with an abrupt 20°C to 4°C cold-shock treatment (Fig. 1C). The threshold temperature at which accumulation of CBF transcripts became detectable was 14°C. As temperatures continued to drop, the levels of CBF transcripts continued to increase as did the transcript levels of the CBF target gene, COR15a. Taken together, the results of the cold shock and gradual temperature downshift experiments indicated that the cold-sensing mechanism is not a “binary” on and off system, but instead, consists of a circuit that can monitor absolute temperature and act like a rheostat to adjust the output— the level of CBF transcript accumulation—to the level of low temperature input.
CBF Induction Involves a Cold-Sensing Mechanism That Becomes Desensitized with Time at Low Temperatures
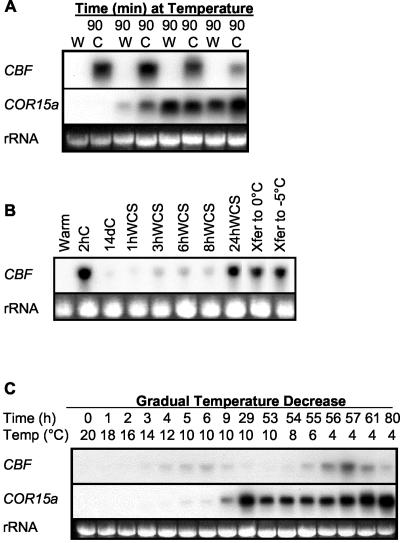
In the cold-shock and gradual temperature down-shift experiments, the levels of CBF transcripts decreased upon continued exposure of the plants to low temperatures. To investigate this phenomenon further, plants were moved back and forth between 20°C and 4°C at 90-min intervals and the levels of CBF transcripts were determined (Fig. 2A). The results indicated that after four rounds of transfer, the levels of CBF transcripts obtained upon cold shock were significantly diminished. Moreover, when plants that had been cold-acclimated at 4°C for 14 d were returned to warm temperatures for 1 h and then abruptly transferred to 4°C, there was no detectable increase in CBF transcript levels (Fig. 2B). If, however, cold-acclimated plants were allowed to adjust to warm temperatures for 24 h and then transferred to 4°C, normal CBF transcript levels were attained. These data suggested that the cold-sensing mechanism became desensitized to 4°C upon extended incubation and that it could become resensitized to 4°C after 24 h at warm temperatures. The resensitization process took between 8 and 24 h, as cold-acclimated plants that had been returned to warm temperature for 3, 6, and 8 h were incapable of mounting a normal cold-shock induction of CBF transcripts (Fig. 2B).
Figure 2.
Desensitization and resensitization of the regulatory circuitry controlling accumulation of CBF transcripts. A, Plants were subjected to repeated transfer between 20°C (W) and 4°C (C), and CBF and COR15a transcript levels were determined by RNA-blot hybridization. B, CBF transcript levels of in plants that were cold acclimated at 4°C (C) for 14 d, returned to warm temperatures for the indicated times, and then subjected to a cold shock (WCS) at 4°C. CBF transcript levels were also determined in cold-acclimated plants (14 d) that were transferred (Xfer) to 0°C or –5°C 2 h. C, CBF and COR15a transcripts levels in plants cooled at 2°C h–1. Plants were held at 10°C for 48 h before cooling to 4°C. The probe used hybridized with CBF1, 2, and 3 transcripts. rRNA stained with ethidium bromide was used to compare loading in each experiment.
Significantly, the desensitization that occurred upon exposure to 4°C did not eliminate the ability of the plants to sense and respond to further drops in temperature. When plants that had been cold-acclimated at 4°C for 14 d were directly transferred to 0°C or –5°C, an increase in CBF levels occurred (Fig. 2B). Also, desensitization was not unique to 4°C. When plants were subjected to a gradual decrease in temperature from 20°C to 10°C, accumulation of CBF transcripts occurred, but the levels declined upon continued exposure to this temperature (Fig. 2C). Upon renewed gradual decrease in temperature, CBF transcripts again increased. As in the cold-shock experiments, the levels of CBF transcripts attained at 4°C were greater than those attained at 10°C (Fig. 2C).
CBF Transcripts Have a Short Half-Life at Warm Temperatures
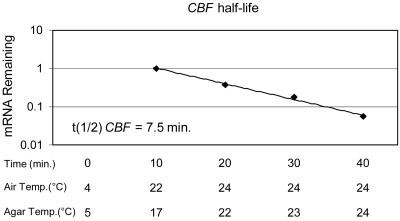
The results of the temperature transfer experiments described above indicated that that the half-life of the CBF transcripts at warm temperatures was very short, as no transcripts were detected 90 min after transferring plants from cold to warm temperatures (Fig. 2A). To estimate the half-life of the CBF transcripts, plants were transferred from cold to warm temperatures and the transcript levels were determined at 10-min intervals (Fig. 3). The results indicate that the CBF transcripts had a half-life of only 7.5 min at warm temperatures, a value that is among the shortest described for plant genes (Gutierrez et al., 2002). This value is a maximum estimate as it assumes that the promoters of the CBF genes become inactive within minutes of transferring plants from low to warm temperatures.
Figure 3.
Estimation of CBF transcript half-life at warm temperatures. Plants that had been exposed to a low temperature (4°C) for 2 h were returned to a warm temperature (24°C) and CBF transcript levels were determined at 10-min intervals by RNA-blot hybridization. Air and agar temperatures in a plate were recorded during the experiment. Transcript levels for eif-4A did not change significantly during the time course and were used as a reference for loading. The probe for CBF hybridized with CBF1, 2, and 3 transcripts.
A 125-bp Region of the CBF2 Promoter Contains Multiple cis-Acting Regulatory Elements Contributing to Cold-Responsive Gene Expression
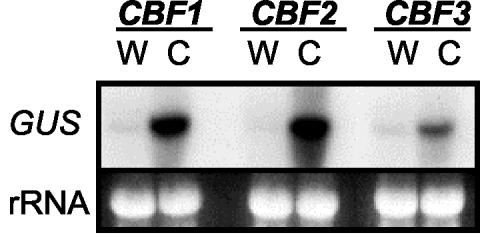
Shinwari et al. (1998) reported that the promoters of CBF1/DREB1b, CBF2/DREB1c, and CBF3/DREB1a are responsive to low temperatures. Our results confirmed this finding (Fig. 4). Approximately 1 kb of each CBF promoter (including the 5′-untranslated region up to the ATG start codon) was fused to the β-glucuronidase (GUS) reporter gene, transformed into Arabidopsis plants, and T2 and T3 populations of plants were analyzed for expression of the promoter::GUS fusions in response to low temperature. RNA-blot analysis indicated that the transcript levels for the reporter genes increased markedly in response to low temperatures (Fig. 4). However, surprisingly, histochemical staining of the plants for GUS activity did not reflect the differences in GUS transcript levels; staining was very similar in intensity regardless of whether the plants were grown at warm or low temperatures for varying lengths of time (up to 3 d; not shown). This suggested that the CBF promoters were active at normal warm temperatures during development of the seedlings. GUS activity resulting from GUS gene activation was detectable in the hypocotyl of seedlings 4 d after germination and could be detected throughout seedlings after 10 and 14 d of growth (not shown). Several different transgenic lines containing any of the three CBF promoters fused to GUS showed similar staining results. Whether the staining at later times is due to continued activation of the CBF promoters or GUS protein stability is uncertain. Regardless, GUS enzyme activity could not be used to faithfully reflect cold induction of the CBF promoters. Therefore, RNA-blot hybridizations were used for all further analysis of promoter activity.
Figure 4.
Activation of the CBF1, 2, and 3 promoters in response to low temperatures. Representative transgenic lines of Arabidopsis carrying the CBF1, 2, or 3 promoter fused to the GUS reporter gene were transferred from 22°C (W) to 4°C (C) for 2 h. The transcript levels of the gene fusions were determine using a probe for the GUS sequences.
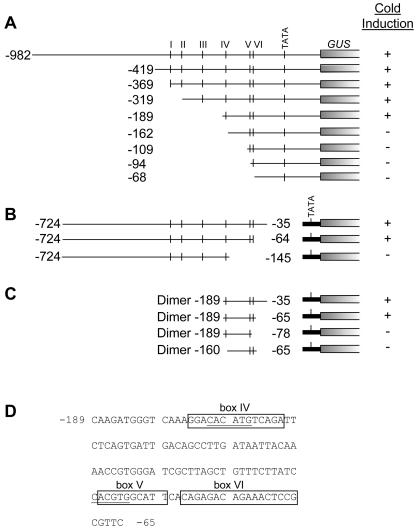
A deletion analysis was conducted to identify sequences within the CBF2 promoter that were involved in cold-regulated gene expression. We chose to work with CBF2 based on the observation that transcripts for CBF2 accumulated to higher levels than CBF1 and CBF3, suggesting that the promoter might be more active and thus potentially easier to analyze than the others. One set of deletions removed sequences from the 5′ end of the promoter and included the 5′-untranslated region up to the ATG start codon (Fig. 5A). Another set of deletions removed sequences from the 3′ end beginning just upstream of the TATA sequence (Fig. 5B). With these later constructs, the CBF2 promoter fragments were placed upstream of a minimal promoter fragment from the CaMV35S gene that provides the TATA sequence. In this vector, –46 CaMV:GUS, the minimal promoter drives expression of the GUS reporter gene. These constructs were designed to test whether any of the sequences that are conserved between the three CBF/DREB1 promoters, designated boxes I to VI by Shinwari et al. (1998), were involved in cold responsiveness.
Figure 5.
CBF2 promoter fragments used to identify cold-regulatory elements. Each fragment was inserted into a binary vector upstream of the GUS reporter gene. Fragments that were deleted from the 3′ end were inserted into a binary vector upstream of the cauliflower mosaic virus (cauliflower mosaic virus) 35S minimal promoter (–46 minimal promoter) fused to GUS. A, Promoter fragments with deletions from the 5′ end. B, Promoter fragments with deletions from the 3′ end. C, Dimer constructs. D, Sequence of the 125-bp region of the CBF2 promoter (positions –189 to –65) that imparts cold-regulated gene expression. The locations of the conserved boxes IV, V, and VI are indicated and the E-box/ABRE-like sequences are underlined. Relative levels of cold induction (data from Figs. 6, 7, 8) are indicated as “+” (strong) and “–” (weak).
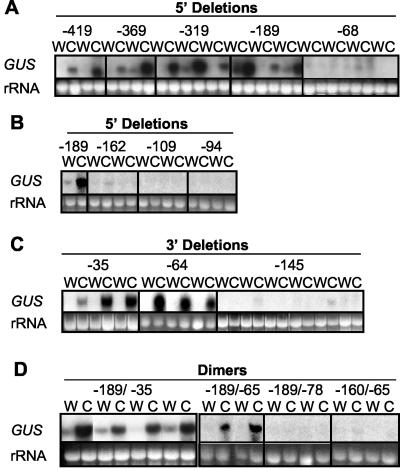
The results indicated that 5′ deletions to –419, –369, –319, and –189 did not greatly impair the cold responsiveness of the CBF2 promoter (Fig. 6A). Thus, boxes I, II, and III were not required for cold induction of the promoter (Fig. 5A). However, deletion of the promoter to –68, which removed boxes IV, V, and VI, resulted in almost complete elimination of cold responsiveness (Fig. 6A). Additional constructs indicated that 5′ deletions to –162, –109, and –94 also severely reduced responsiveness of the promoter (Fig. 6B). Thus, sequences between –189 and –162, which includes box IV (Fig. 5A), appeared to have an important role in cold-regulated expression of the promoter.
Figure 6.
Activities of CBF2::GUS promoter constructs. RNA isolated from plants grown at 22°C (W) or grown at 22°C and transferred to 4°C for 2 h (C) was analyzed for GUS transcript levels by RNA-blot hybridization. A and B, Plants containing 5′ deletion constructs. C, Plants containing 3′ deletion constructs. D, Plants containing dimer constructs. rRNA stained with ethidium bromide was used to compare loading in each experiment.
The sequence from –724 to –35 could impart cold-regulated gene expression when fused to the –46 CaMV:GUS reporter gene (Figs. 5Band 6C). Deletion from the 3′ end to –64 did not have a dramatic effect on the cold responsiveness, but deletion to –145 all but eliminated cold responsiveness of the promoter fragment (Fig. 6C). These results indicated that boxes I, II, III, and IV were insufficient to impart robust cold-regulated gene expression and that box V or VI or both might contain a cold-responsive element(s) (Fig. 5B).
The results of the 5′ and 3′ deletion analysis suggested that sequences between –189 and –65, which contained boxes IV, V, and VI, might be sufficient to impart cold-responsive gene expression (Fig., A and B). Dimers of the –189/–35 or –189/–65 sequences were responsive to low temperatures (Fig. 6D). Dimer fragments consisting of –189/–78 and –146/–64 sequences, which resulted in deletion of box IV and the right one-half of box VI, respectively (Fig. 5C), eliminated strong cold responsiveness (Fig. 6D). These results were consistent with the hypothesis that boxes IV and VI were involved in cold-regulated expression of the CBF2 promoter (Fig. 5C). In addition, the possibility remained that box V was also required for robust cold responsiveness.
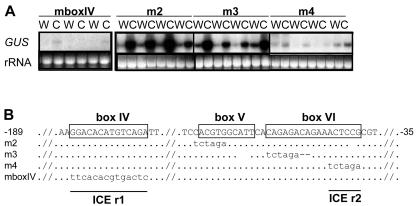
To confirm the importance of the boxes IV and VI sequences in cold-responsive gene expression, and to explore the importance of box V, mutations were introduced into these sequences in the context of the –189 5′ deletion construct and were tested for cold induction (Fig. 7). Whereas the wild-type –189 5′ deletion construct was strongly cold responsive (Fig. 6A), the mutant versions having the entire box IV (mboxIV) or right-hand portion of box VI (m4) substituted with alternative nucleotides displayed very low induction in response to low temperatures (Fig. 7). In contrast, mutagenesis of the left-hand portion of box VI (m3) or the E-box (CANNTG; Massari and Murre, 2000)/ABRE-like (CACGTG; Busk and Pages, 1998) sequences present at the left-side of box V (m2) had no obvious effects on cold-regulated gene expression.
Figure 7.
Mutational analysis identifying regions of the CBF2 promoter involved in cold-regulated gene expression. A, CBF2::GUS promoter constructs with mutations in boxes IV (mboxIV), V (m2), and VI (m3 and m4) were tested for cold responsiveness by transferring plants from 22°C (W) to 4°C (C) for 2 h and determining GUS transcript levels by RNA-blot hybridization. rRNA stained with ethidium bromide was used to compare loading. B, Sequence corresponding to wild-type and mutated (m2, m3, m4, and mboxIV) CBF2::GUS promoter constructs. Regions identified as required for cold induction are underlined and designated ICEr1 and ICEr2.
In summary, the promoter analysis described above indicated that sequences within box IV and the right-hand side of box VI are involved in cold induction of the CBF2 promoter. Therefore, these sequences were designated “Induction of CBF Expression region 1 and 2” (ICEr1 and ICEr2), respectively.
ICEr1 and ICEr2 are Involved in Responsiveness of the CBF2 Promoter to Mechanical Agitation and Cycloheximide Treatment
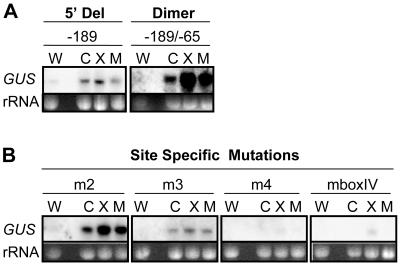
Earlier work established that CBF transcripts accumulate rapidly in response to mechanical agitation (Gilmour et al., 1998) and the protein synthesis inhibitor cycloheximide (Qin, 2001). To determine whether this regulation involved the same sequences involved in cold induction of the CBF2 promoter, we compared cold, mechanical, and cycloheximide induction of specific promoter constructs described above. The results indicated that as with cold, the ICEr1 and ICEr2 sequences were involved in mechanical and cycloheximide regulation of the promoter. Whereas the native promoter deleted 5′ to –189 and the 125-bp –189/–65 dimer fragment conferred responsiveness to treatments with cold, mechanical agitation, and cycloheximide, the site specific mutations m4 and mboxIV virtually eliminated responsiveness to these factors (Fig. 8).
Figure 8.
Sequences of the CBF2 promoter involved in cold, cycloheximide, and mechanical responsiveness. Plants containing CBF2 promoter constructs were grown at 22°C (W) and then exposed to 4°C (C) for 2 h or treated with cycloheximide (X) or mechanical agitation (M) as described in “Materials and Methods.” RNA was isolated and GUS transcript levels were determined by RNA-blot hybridization. A, Plants containing the 5′ deletion to –189 (5′Del) or the –189/–65 dimer (Dimer) construct. B, Plants containing the m2, m3, m4, and mboxIV mutations. rRNA stained with ethidium bromide was used to compare loading in each experiment.
DISCUSSION
The results reported here provide further insights into the regulation of CBF genes by low temperatures. One is that the cold-sensing mechanism that controls CBF expression does not require a cold shock to bring about the accumulation of CBF transcripts (Fig. 1). Instead, the thermosensing circuitry appears to monitor absolute temperature and act like a rheostat increasing output, i.e. the levels of CBF transcripts, with greater degrees of input, i.e. lower temperatures. However, the output from the circuit (which could involve changes in rates of CBF transcription and rates of CBF transcript turnover) does not remain constant with time at a given temperature. Within a few hours of exposure to a temperature, the thermosensing circuitry becomes desensitized to that temperature and produces less output— lower levels of CBF transcripts (Fig. 1). Significantly, this apparent desensitization does not preclude a robust response to a further decrease in temperature. Plants that had been adapted to 4°C for 14 d and had low levels of CBF transcripts produced high levels of CBF transcripts when transferred to 0°C or –5°C (Fig. 2). In contrast, when plants that had been adapted to 4°C for 14 d were returned to warm temperatures for 1 to 8 h, and then abruptly returned to 4°C, CBF transcripts showed little increase in accumulation; resensitizing 4°C-adapted plants to 4°C took between 8 and 24 h of exposure to 22°C (Fig. 2). These results indicate that the thermosensing circuitry controlling CBF expression involves a low temperature “memory” component affecting desensitization.
At present, we know nothing of the nature of the putative thermosensing memory component. However, a “cold memory” has been described for cold-induced calcium flux in Arabidopsis (Knight et al., 1996). Transfer of Arabidopsis plants to low temperatures results in a rapid, transient increase in free calcium in the cytoplasm, which comes from extracellular and intracellular calcium stores (Knight et al., 1996; Knight and Knight, 2000). Knight et al. (1996) have shown that the calcium signature—the peak levels and duration of the calcium influx—is altered upon cold acclimation, becoming diminished in magnitude and prolonged in length. The significance here is that the expression of certain cold-regulated genes, including COR and other CBF-targeted genes, appears to involve the action of calcium as second messenger. When cold-induced calcium influx was inhibited using chemicals or pharmacological agents, expression of the COR genes was impaired; when chemical and pharmacological agents were used to raise intracellular calcium levels at warm temperatures, COR gene expression was induced (Knight et al., 1996; Tahtiharju et al., 1997). Thus, an interesting possibility raised is that the desensitization “memory” that we describe here may be related to the calcium cold memory described by Knight et al. (1996).
What are the molecular components of the thermosensing circuitry that regulate accumulation of CBF transcripts? At present, our understanding in this area is limited, but proteins that are potential components of the thermosensing mechanism have been identified by Zhu and colleagues in screens for mutants altered in low-temperature expression of RD29A, a CBF-targeted gene (Ishitani et al., 1997). HOS1 (Lee et al., 2001) and FRY2 (Xiong et al., 2002), a novel RING finger protein and novel transcriptional repressor, respectively, appear to be negative regulators of CBF expression whereas LOS4, a DEAD-box RNA helicase, appears to be a positive regulator (Gong et al., 2002). In addition, mutant analysis indicates that the thermosensing circuitry may include a negative feedback loop that includes the CBF proteins themselves or their downstream targets (Guo et al., 2002).
Key components of the regulatory mechanism that control CBF expression include the transcription factors that bind to the promoter and control transcription. One such factor, ICE1, was recently reported (Chinnusamy et al., 2003). ICE1 encodes a MYC-like bHLH protein that appears to have a fundamental role in the regulation of CBF3. A dominant-negative allele of ICE1, ice1, results in almost complete elimination of CBF3 transcript accumulation in response to low temperatures. Significantly, however, accumulation of CBF2 transcripts in response to low temperatures is little affected by the ice1 mutation (and may, in fact, be somewhat greater in the ice1 mutant). Thus, ICE1 would not appear to have a critical role in CBF2 expression at low temperatures.
As a step toward identifying transcription factors involved in CBF2 expression, we identified a 125-bp promoter segment that is sufficient to impart cold-responsive gene expression (Figs. 5, 6, 7). Within this 125-bp segment are two regions, designated ICEr1 and ICEr2, that by themselves are only weakly responsive to low temperatures, but in combination, impart a robust cold response. There are no obvious known transcription factor-binding sites within the ICEr2 sequence, but ICEr1 contains the sequence CACATG, which includes a consensus recognition site for bHLH proteins, CANNTG (Massari and Murre, 2000). Therefore, ICEr1 is a potential binding site for the ICE1 protein. However, as alluded to above, the ice1 mutation has little effect on CBF2 transcript accumulation at low temperatures. Thus, it is unclear whether ICE1 binds to ICEr1 in vivo and contributes to CBF2 expression. However, Arabidopsis encodes an estimated 139 bHLH proteins (Riechmann and Ratcliffe, 2000), some of which have DNA-binding domains that are similar to that present in ICE1 (Chinnusamy et al., 2003). These must be considered candidates for ICEr1-binding proteins. Another potential bHLH protein-binding site, CACGTG, which is also a potential abscisic acid-responsive element and bZIP protein-binding site (Busk and Pages, 1998), overlaps the left-hand end of box V. However, mutations in this sequence, in the context of the –189 5′ deletion, had no apparent effect on cold responsiveness and thus, a role for this sequence in CBF2 expression is not evident.
The ICEr1 and ICEr2 sequences are not only involved in cold responsiveness of the CBF2 promoter, but also in induction of the promoter by mechanical agitation and cycloheximide. Whether cold, mechanical agitation, or cycloheximide mediate their action through the same regulatory system remains to be determined. However, it is of interest that mechanical agitation, like low temperatures, causes a transient increase in cytoplasmic calcium levels (Knight et al., 1992). Thus, a plausible hypothesis is that cold and mechanical stress cause a calcium flux that stimulates CBF transcription. Whether the increases in calcium brought on by cold and mechanical stress are mediated through the same mechanism is unknown. However, in this regard, it is noteworthy that the activity of an onion-mechanosensitive calcium-selective channel has been shown to increase as temperature is lowered to about 6°C (Ding and Pickard, 1993).
A relationship between calcium flux and cycloheximide-induced gene expression also exists. Berberich and Kusano (1997) have found that transcripts for mlip15, a cold-inducible maize (Zea mays) gene encoding a putative bZIP transcription factor, accumulate in response to cycloheximide and that chelators of calcium reduce mlip15 transcript accumulation in response to cold and cycloheximide treatments. Thus, cycloheximide, like mechanical agitation, may induce CBF2 expression by causing an increase in intracellular calcium levels. The fact that cycloheximide inhibits translation raises the possibility that the mechanism controlling cold-induced calcium flux might involve action of a short-lived repressor protein that keeps a calcium channel “closed” in the absence of a low temperature or mechanical stimulus.
A final point regards a possible relationship between the regulation of CBF genes and the Arabidopsis TCH4 gene, which encodes a xyloglucan endotransglycosylase (Purugganan et al., 1997). Like the CBF genes, TCH4 is induced in response to cold temperature and touch (mechanical agitation; Polisensky and Braam, 1996). Iliev et al. (2002) defined a 102-bp region of the TCH4 promoter that is sufficient to impart cold and touch responsiveness. Within this region is a 24-bp segment of the TCH4 promoter (positions –138 to –114) that is 66% identical in sequence to the CBF2 promoter between positions –189 and –165. Significantly, this region of the CBF2 promoter contains the ICEr1 sequence that is involved in cold and mechanical responsiveness. These observations suggest that TCH4 may be regulated, at least in part, by the same cold- and mechanical-regulatory system that controls CBF2 gene expression.
MATERIALS AND METHODS
Plant Material, Growth, and Treatment Conditions
Arabidopsis ecotype Wassilewskija-2 and transgenic plants in the Wassilewskija-2 background were grown in controlled environment chambers at 22°C under constant illumination from cool-white fluorescent lights (100–125 μmol m–2 s–1) in Baccto planting mix (Michigan Peat, Houston). Pots were subirrigated with deionized water as needed. Plants were also grown on solid agar medium that contained Gamborg's B5 nutrients (Invitrogen, Carlsbad, CA) and 1% (w/v) phytoagar (Invitrogen) for various treatments. Experiments were performed on seedlings that were 10- to 12-d-old. Cold-shock treatments involved transfer of plants from a 22°C chamber to a 4°C chamber with dim light (approximately 25 μmol m–2 s–1). Gradual temperature decreases were achieved by decreasing the temperature of the growth chamber 0.5°C every 15 min until a desired temperature was reached. Treatments with cycloheximide were performed by growing seedlings on filter papers that had been placed on top of the agar so that the seedlings could be lifted off the plate with minimal damage or mechanical stress. The seedlings were then floated on a solution of cycloheximide (10 μg mL–1) in covered dishes for 2 h. Mechanical treatment involved tapping plates on a bench top for 15 min before harvesting tissue.
Constructs and Plant Transformation
Plasmids were constructed using PCR and standard molecular biological techniques (Sambrook et al., 1989). Primers used to amplify fragments of the CBF promoters contained sites for unique restriction enzymes so that after amplification, the PCR product could be restricted and then cloned directly into the final vector in one step.
Site-directed mutations were created in regions of the specific promoter fragments by Gene Editor (Promega, Madison, WI) or Quick Change (Stratagene, La Jolla, CA) mutagenesis kits according to the product guide. Promoter fragments and mutations were sequenced on an Automated Fluorescent Sequencer (Applied Biosystems, Foster City, CA) at the Michigan State University-Department of Energy Plant Research Laboratory Sequencing Facility.
The DNAs were transferred to Arabidopsis via a whole-plant dipping method similar to that described by Clough and Bent (1998). Plants containing transferred DNA were identified by germinating seeds on medium containing kanamycin (50 mg L–1). These kanamycin-resistant seedlings were grown to maturity and the next generation seeds (T2) or T3 seeds were used for experiments.
Staining for GUS Activity
To stain plant tissue to determine location of GUS activity, experimentally treated plants were immersed in a GUS-staining solution as described by Jefferson et al. (1987) and were incubated overnight at 37°C. Tissue was cleared with several rinses of 70% (v/v) ethanol to aid in the localization of the staining.
RNA Isolation and Hybridization
Total RNA was extracted from plant material with the use of RNeasy Plant Mini kits (Qiagen, Valencia, CA) with modifications. To obtain adequate and consistent yields with the kit, the amount of starting plant tissue was doubled. Subsequently, the amount of extraction buffer (RLT) was also doubled. The remaining procedure was performed as described in the Qiagen manual.
Northern transfers were prepared and hybridized as described (Hajela et al., 1990) using normal high-stringency wash conditions (Stockinger et al., 1997).
Probes for specific CBF genes and GUS were prepared as previously described by Baker et al. (1994) and Gilmour et al. (1998). Probes were labeled with 32P by random priming (Feinberg and Vogelstein, 1983).
Acknowledgments
We thank Steve Triezenberg and members of the Michael F. Thomashow laboratory for critical reading of the manuscript.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.027169.
This research was supported in part by the U.S. Department of Agriculture National Research Initiative program (grant no. 98–35100–6999) and by the Department of Energy and the Michigan Agricultural Experiment Station. J.T.V. was a recipient of a U.S. Department of Education Graduate Assistantship in Areas of National Need (GAANN) fellowship.
References
- Artus NN, Uemura M, Steponkus PL, Gilmour SJ, Lin C, Thomashow MF (1996) Constitutive expression of the cold-regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc Natl Acad Sci USA 93: 13404–13409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SS, Wilhelm KS, Thomashow MF (1994) The 5′ region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol Biol 24: 701–713 [DOI] [PubMed] [Google Scholar]
- Berberich T, Kusano T (1997) Cycloheximide induces a subset of low temperature-inducible genes in maize. Mol Gen Genet 254: 275–283 [DOI] [PubMed] [Google Scholar]
- Busk PK, Pages M (1998) Regulation of abscisic acid-induced transcription. Plant Mol Biol 37: 425–435 [DOI] [PubMed] [Google Scholar]
- Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK (2003) ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Ding JP, Pickard BG (1993) Modulation of mechanosensitive calcium-selective cation channels by temperature. Plant J 3: 713–720 [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132: 6–13 [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF (1998) Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16: 433–442 [DOI] [PubMed] [Google Scholar]
- Gong Z, Lee H, Xiong L, Jagendorf A, Stevenson B, Zhu JK (2002) RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc Natl Acad Sci USA 99: 11507–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xiong L, Ishitani M, Zhu JK (2002) An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc Natl Acad Sci USA 99: 7786–7791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez RA, Ewing RM, Cherry JM, Green PJ (2002) Identification of unstable transcripts in Arabidopsis by cDNA microarray analysis: rapid decay is associated with a group of touch- and specific clock-controlled genes. Proc Natl Acad Sci USA 99: 11513–11518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL (1990) Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol 41: 187–223 [Google Scholar]
- Hajela RK, Horvath DP, Gilmour SJ, Thomashow MF (1990) Molecular cloning and expression of COR (cold-regulated) genes in Arabidopsis thaliana. Plant Physiol 93: 1246–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliev EA, Xu W, Polisensky DH, Oh MH, Torisky RS, Clouse SD, Braam J (2002) Transcriptional and posttranscriptional regulation of Arabidopsis TCH4 expression by diverse stimuli: roles of cis regions and brassinosteroids. Plant Physiol 130: 770–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Stevenson B, Zhu JK (1997) Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9: 1935–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17: 287–291 [DOI] [PubMed] [Google Scholar]
- Knight H, Knight MR (2000) Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J Exp Bot 51: 1679–1686 [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR (1996) Cold calcium signaling in Arabidopsis involves two cellular pools and a change in calcium signature after acclimation. Plant Cell 8: 489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight MR, Smith SM, Trewavas AJ (1992) Wind-induced plant motion immediately increases cytosolic calcium. Proc Natl Acad Sci USA 89: 4967–4971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HJ, Xiong LM, Gong ZZ, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleocytoplasmic partitioning. Genes Dev 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10: 1391–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massari ME, Murre C (2000) Helix-loop-helix proteins: regulators of transcription in eukaryotic organisms. Mol Cell Biol 20: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J (1999) The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119: 463–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjo T, Kobayashi M, Yoshiba Y, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (1999) Antisense suppression of proline degradation improves tolerance to freezing and salinity in Arabidopsis thaliana. FEBS Lett 461: 205–210 [DOI] [PubMed] [Google Scholar]
- Polisensky DH, Braam J (1996) Cold-shock regulation of the Arabidopsis TCH genes and the effects of modulating intracellular calcium levels. Plant Physiol 111: 1271–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purugganan MM, Braam J, Fry SC (1997) The Arabidopsis TCH4 xyloglucan endotransglycosylase: substrate specificity, pH optimum, and cold tolerance. Plant Physiol 115: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H (2001) Genetic analysis of a CBF-mediated signaling pathway in Arabidopsis thaliana. MS thesis. Michigan State University, East Lansing
- Riechmann JL, Meyerowitz EM (1998) The AP2/EREBP family of plant transcription factors. Biol Chem 379: 633–646 [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ (2000) A genomic perspective on plant transcription factors. Curr Opin Plant Biol 3: 423–434 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K (1998) An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem Biophys Res Commun 250: 161–170 [DOI] [PubMed] [Google Scholar]
- Steponkus PL, Uemura M, Joseph RA, Gilmour SJ, Thomashow MF (1998) Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc Natl Acad Sci USA 95: 14570–14575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahtiharju S, Sangwan V, Monroy AF, Dhindsa RS, Borg M (1997) The induction of kin genes in cold-acclimating Arabidopsis thaliana: evidence of a role for calcium. Planta 203: 442–447 [DOI] [PubMed] [Google Scholar]
- Taji T, Ohsumi C, Iuchi S, Seki M, Kasuga M, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K (2002) Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. Plant J 29: 417–426 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50: 571–599 [DOI] [PubMed] [Google Scholar]
- Thomashow MF (2001) So what's new in the field of plant cold acclimation? Lots! Plant Physiol 125: 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee H, Ishitani M, Tanaka Y, Stevenson B, Koiwa H, Bressan RA, Hasegawa PM, Zhu JK (2002) Repression of stress-responsive genes by FIERY2, a novel transcriptional regulator in Arabidopsis. Proc Natl Acad Sci USA 99: 10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K (1994) A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6: 251–264 [DOI] [PMC free article] [PubMed] [Google Scholar]