Abstract
Genome instability is a fundamentally important component of aging in all eukaryotes. How age-related genome instability occurs remains unclear. The free radical theory of aging posits oxidative damage to DNA and other cellular constituents as a primary determinant of aging. More recent versions of this theory predict that mitochondria are a major source of reactive oxygen species (ROS) that cause oxidative damage. Although substantial support for the free radical theory exists, the results of some tests of this theory have been contradictory or inconclusive. Enhanced growth signaling also has been implicated in aging. Many efforts to understand the effects of growth signaling on aging have focused on inhibition of oxidative stress responses that impact oxidative damage. However, recent experiments in the model organism Saccharomyces cerevisiae (budding yeast) and in higher eukaryotes suggest that growth signaling also impacts aging and/or age-related diseases—including cancer and neurodegeneration—by inducing DNA replication stress, which causes DNA damage. Replication stress, which has not been broadly considered as a factor in aging, may be enhanced by ROS that signal growth. In this article, we review evidence that points to DNA replication stress and replication stress-induced genome instability as important factors in aging.
INTRODUCTION
The importance of genome instability during aging is indicated by the accelerated aging phenotype associated with defects in genome maintenance programs in a variety of organisms, as well as accumulating evidence—most recently in the model organism Saccharomyces cerevisiae (budding yeast) (1,2)—that genome instability and the rate at which this instability occurs increase with age. A number of potential mechanisms, often overlapping, have been proposed to explain age-dependent genome instability. These include the accumulation of oxidative damage to DNA, defects in mitochondrial function that promote oxidative stress and damage to DNA and other cellular constituents, mutations in proteins required for efficient DNA replication, DNA repair and checkpoints, telomere erosion and epigenetic effects on DNA repair and other genome maintenance programs.
Although many of these mechanisms are likely to contribute to aging in some or all eukaryotes, the specific contributions of each and their relative importance remain a matter of debate. For example, tests of one of the more compelling possibilities—that oxidative damage to DNA is an important determinant of life span—have produced equivocal results (3). Although it is clear that oxidative DNA damage accumulates with age in many, if not all, eukaryotic organisms, experimental manipulations that mitigate oxidative stress do not always extend life span. Conversely, although disruption of cellular pathways that mitigate oxidative stress can lead to increases in age-dependent oxidative DNA damage, this does not always coincide with a shorter life span. Also unresolved is the question of whether oxidative DNA damage impacts normal aging in the absence of its contributions to age-related diseases, such as cancer and various neurodegenerative disorders.
How cellular processes that regulate aging impact genome stability also remain unclear. Compelling evidence now exists that in all eukaryotes, aging is regulated by conserved insulin/insulin-like growth factor (I-(IFG-1)) pathways and growth-signaling pathways regulated by the target of rapamycin (TOR) family of kinases (4). In general, experimental manipulations that upregulate these pathways promote aging, and manipulations that downregulate these pathways—including mutational inactivation or caloric restriction—extend life span and mitigate age-related pathologies. Downregulation of these pathways often leads to a reduction in oxidative stress and oxidative damage to DNA and other cellular constituents. For the most part, however, the relationship between aging and changes in oxidative damage downstream of alterations in growth-signaling pathways remains correlative rather than causal.
A number of recent studies have revealed that in mammalian cells, the constitutive activation of highly conserved growth-signaling pathways implicated in aging and in the regulation of oxidative stress responses also causes DNA replication stress (5–8). Replication stress and replication-induced genome instability that occur downstream of oncogenic growth signaling has been detected at the earliest stages of cancer. Replication stress was also recently implicated in cellular senescence, which protects against cancer, but promotes aging (5,7).
Although defects in cellular responses to replication stress mediated by RecQ helicases, DNA repair enzymes and checkpoints have been implicated in accelerated aging by the premature aging phenotypes associated with these defects, replication stress that arises downstream of growth signaling has not been broadly considered as a factor contributing to normal aging. In this article, we review recently published evidence that suggests DNA replication stress is an important contributor to normal aging and age-dependent genome instability in all eukaryotes. We hypothesize that replication stress induced downstream of growth signaling by reactive oxygen produced in mitochondria underlies some of the age-dependent genome instability attributed to oxidative damage to DNA.
DNA REPLICATION STRESS
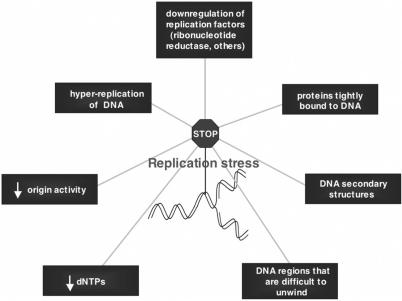
DNA replication stress is defined as inefficient DNA replication that causes DNA replication forks to progress slowly or stall. Factors that cause replication stress and replication stress-induced DNA damage include alterations in pools of dNTP precursors required for DNA synthesis, changes in the expression of proteins required for synthesis of dNTPs or other components of DNA synthesis, decreased frequency with which initiation of DNA replication occurs at origins of replication (producing larger replicons), hyper-DNA replication caused by the activation of origins more than once per S phase, DNA damage lesions that block replication forks, and inhibition of DNA replication by drugs. Replication stress also occurs in regions of DNA that are intrinsically difficult to replicate due to secondary structures or that are difficult to unwind during DNA replication. Proteins bound to DNA can also cause replication forks to pause, and thus causing replication stress [Figure 1; (9)].
Figure 1.
Factors that contribute to replication fork stalling (replication stress). Some of these factors overlap. For example, reduced levels of dNTPs could be a consequence of reduced availability of nutrient precursors or downregulation of proteins required for their synthesis.
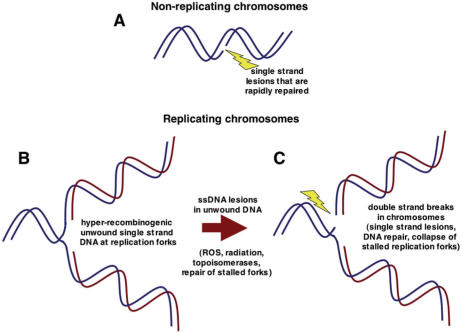
DNA replication stress leads to DNA damage and genome instability in part because of the unique structure of replicating DNA molecules (Figure 2). When single-strand lesions occur in non-replicating molecules of DNA, the overall integrity of chromosomes is maintained by hydrogen bond base pairing on either side of these lesions until they are repaired (Figure 2A). In contrast to non-replicating DNA, replicating DNA at replication forks contains unwound, highly recombinogenic single-stranded template DNA before this DNA is converted to double-strand DNA by template-directed DNA synthesis (Figure 2B). The template strands on each arm of replication forks are no longer base-paired to their original complementary template strands, and instead are base-paired to newly synthesized DNA (indicated by red strands in Figure 2B and C). Consequently, single-strand lesions within unwound DNA at replication forks cause double-strand breaks, which are also highly recombinogenic (Figure 2C). Double-strand breaks are also more difficult to repair compared to single-strand lesions in non-replicating DNA. Double-strand DNA breaks and genome rearrangements triggered at replication forks can occur in association with spontaneously arising single-strand lesions produced by reactive oxygen species (ROS), radiation and other factors. The increased risk for genome instability arising specifically at DNA replication forks posed by all these factors is reflected in the evolution of highly conserved checkpoint pathways that block entry into or progression through S phase in cells that have suffered DNA damage. Checkpoint pathways also stabilize stalled replication forks. Specific mechanisms also exist for repairing stalled or collapsed replication forks (10,11).
Figure 2.
The unique structure of replicating DNA contributes to genome instability at replication forks, which is enhanced by replication fork stalling. (A) When non-replicating DNA molecules suffer single-strand lesions, the integrity of these molecules is maintained by hydrogen bond base pairing on either side of the lesions until they are repaired. (B) Replicating DNA molecules contain single-strand DNA at replication forks after unwinding of double-strand template DNA has occurred in preparation for template-directed DNA synthesis. This single-strand DNA is highly recombinogenic. (C) Single-strand DNA at replication forks is susceptible to single-strand lesions that effectively produce double-strand breaks. Double-strand breaks also occur at stalled replication forks in association with attempts to repair the stalled forks and/or due to replication fork collapse in the absence of checkpoint functions.
BUDDING YEAST AS A MODEL FOR INVESTIGATING AGING IN EUKARYOTES
Studies in budding yeast suggest a role for replication stress in aging. Two complementary and partly overlapping models of aging have been investigated in this model organism—the replicative life span model and the chronological life span model. The replicative life span of budding yeast is determined by counting the number of times cells divide in the presence of nutrients before they senesce and die (12). Replicative life span is regulated by conserved growth-signaling pathways that respond to nutrients. Mutations in nutrient-signaling pathways regulated by protein kinase A (Pka), the Tor1 kinase (Tor1) and Sch9, a budding yeast homolog of AKT/PKB kinases that function in growth signaling downstream of IGF-1 in mammals, extend replicative life span. Caloric restriction induced by reducing the concentration of glucose in growth medium also extends replicative life span (13). The replicative life span-extending effects of caloric restriction may be related to reduced signaling through Pka-, Tor1- and Sch9-dependent pathways. This is suggested by the fact that caloric restriction does not extend replicative life span further when these pathways have been inactivated (14,15).
Chronological life span in budding yeast is defined as the length of time cells survive after they are driven into a non-dividing state by depletion of nutrients from the medium (12,16). Entry into this state coincides with an extensive reorganization of transcriptional regulatory programs leading to changes in the expression of a significant fraction of all budding yeast genes (17,18). These changes include the downregulation of many genes encoding proteins required for DNA replication and cell division when nutrients are in excess and the upregulation of genes required for transcriptional responses to a variety of stresses, including oxidative stress. Budding yeast cells are capable of surviving in this non-dividing state for an extended time, during which growth can be restored by replenishing the medium with nutrients. The non-dividing state of nutrient-depleted budding yeast cells shares features with post-mitotic differentiated cells in higher eukaryotes, including growth arrest of many cells with an apparent G1 content of DNA. Consequently, compared to the replicative aging model, the chronological aging model is considered by some investigators to more accurately reflect aging of post-mitotic, differentiated cells in more complex eukaryotes.
Chronological life span in budding yeast is regulated by the same highly conserved Pka-, Tor1- and Sch9-dependent nutrient-signaling pathways that regulate replicative life span in this organism. Similar to the extension of replicative life span by downregulation of these pathways or by caloric restriction, chronological life span is extended by these experimental manipulations as well (19–22). A mechanistic relationship between chronological and replicative life span is suggested by the observation that chronologically aged cells cultured for an extended time in depleted medium exhibit a shortened replicative life span when nutrients are restored (23).
DNA replication stress and replicative aging of budding yeast
Replication stress is a primary determinant of replicative aging in budding yeast. Although the specific details are complex and involve a diverse array of cellular programs that impact replicative aging—metabolism, energy production, stress responses, transcriptional changes and chromatin modification among others—ultimately, the contributions of these programs to replicative aging converge on effects that cause genome instability in the highly repeated rDNA locus (12). Instability in this locus is promoted by replication fork stalling at a Fob1-dependent replication fork barrier, which prevents replication forks from proceeding through rDNA transcription units in a direction opposite to that of transcription. Recombinational repair mechanisms triggered by replication forks stalled at the Fob1 barrier produce extra-chromosomal rDNA circles (ERCs). During cell division, ERCs are mostly retained in mother cells, where their accumulation eventually triggers age-dependent replicative senescence. Although the mechanisms underlying ERC-dependent senescence remain unknown, one interesting possibility is that in old cells, the large number of ERCs—each of which harbors an origin of DNA replication—titrates proteins required for initiation of DNA replication of cellular chromosomes. This would amplify replication stress.
Although accumulation of ERCs in old budding yeast cells causes replicative senescence, this senescent state also occurs in the absence of ERCs (24–27). Furthermore, the rate at which genome instability occurs outside the rDNA locus in budding yeast dramatically increases with replicative age, and ERC accumulation is not responsible for this increased instability (1). ERC-independent replicative aging is also accelerated by mutations in proteins required for efficient DNA replication and genome stability. These include the budding yeast RecQ helicase Sgs1 (28), the related Dna2 helicase/nuclease and the Fen-1 nuclease (26), all of which are required for maintaining genome instability during DNA replication. In some cases, replicative senescence produced by mutations in these proteins may be related to general toxic effects associated with the loss of function of these proteins rather than normal aging processes. However, replicative senescence of dna2 mutants duplicates morphological and biochemical features of aging observed in replicatively senescent wild-type cells (26), as is the case for some, but not all replicatively senescent sgs1Δ cells (28). These findings—some of which are rarely cited in the aging literature—clearly point to a role for replication stress in replicative aging of budding yeast that occurs independently of ERCs, in addition to the well-established role of replication stress in ERC-dependent replicative aging.
Replication stress and chronological aging of budding yeast
Until recently, replication stress was not considered a factor in chronological aging of budding yeast. This is likely due to the fact that during chronological aging, most cells appear to arrest growth without buds and with a G1 content of DNA, which suggests that they are not in S phase. Many studies of chronological aging have focused instead on changes in cellular responses to oxidative and other stresses (16). As in many other eukaryotes, proteins that mitigate oxidative stress in budding yeast are induced by caloric restriction and by the mutational inactivation of growth-signaling pathways. The induction of oxidative stress responses during nutrient depletion by these experimental manipulations occurs in parallel with chronological life span extension. Although the connections between the induction of oxidative stress responses and chronological life span extension revealed by these studies are largely correlative, it is often assumed they are causally related. This is not always the case. For example, although mutational inactivation of Tor1-dependent growth signaling in budding yeast upregulates the expression of the superoxide dismutase-encoding SOD2 gene and enhances resistance to oxidative stress in stationary phase cells, deletion of SOD2 from tor1Δ cells does not alter their chronological life span (21).
The frequent assumption that budding yeast cells arrest in G1 during nutrient depletion also is not always correct. For example, the constitutive activation of nutrient-signaling pathways during nutrient depletion—which in general is accompanied by shorter chronological life span—can dramatically increase the number of cells that growth arrest in other phases of the cell cycle, including S phase. Mutations that have this effect include the constitutively activating RAS2val19 mutation (29,30) and deletion of RIM15 (30,31) encoding a regulator of oxidative and other stress responses downstream of the function of Ras2 and other nutrient-signaling proteins. Both of these mutations shorten chronological life span.
We recently reported that the converse is true as well—that is, mutational inactivation of nutrient-signaling pathways promotes a tighter G1 arrest during nutrient depletion in concert with a longer chronological life span (30). This includes inactivation of the AKT homolog Sch9, which was previously shown to extend chronological life span in concert with reduced chronological age-dependent genome instability (2). Other experimental manipulations that extend chronological life span, including caloric restriction (30), growth in respiratory medium (ethanol or glycerol; our unpublished data), growth in nutrient-rich medium (YPD) and osmotic stress (30), also promote a tighter G1 arrest in chronological aging experiments. Thus, a strong correlation exists between chronological life span and the efficiency with which nutrient-depleted cells arrest in G1 in association with a variety of experimental manipulations that impact growth signaling (Table 1).
Table 1.
Experimental manipulations during nutrient depletion of budding yeast that impact chronological life span and G1 arrest
| Experimental manipulation | GI arrest | Chronological life span |
|---|---|---|
| sch9Δ | Increased (30) | Increased (20,30) |
| ras2Δ | Increased (30) | Increased (30) |
| RAS2val19 | Decreased (29,30) | Decreased (29,30) |
| rim15Δ | Decreased (30,31) | Decreased (30,31) |
| tor1Δ | Increaseda | Increased (21) |
| Caloric restriction (reduced glucose) | Increased (30) | Increased (22,30) |
| Osmotic stress | Increased (30) | Increased (22,30) |
| Culture in YPD | Increased (30) | Increased (30) |
| Culture in ethanol | Increaseda | Increased (22) |
| Culture in glycerol | Increaseda | Increased (22) |
| mec1-21 | Decreased (30) | Decreased (30) |
| rad53-21 | Decreased (30) | Decreased (30) |
| cln3Δ | Increased (87) | Increased (87) |
| CLN3 ox | Decreased (30,32) | Decreased (30,32) |
Numbers in parentheses indicate references to articles containing data.
aOur unpublished data.
Growth arrest in S phase is expected to produce replication stress. Consistent with this possibility, during nutrient depletion, cells harboring mutations in the DNA damage and replication stress response proteins Mec1 and Rad53 loose viability faster than wild-type cells or cells that fail to express the DNA damage response protein Rad9, which responds to DNA damage, but not replication stress (30). Ectopic expression during nutrient depletion of the G1 cyclin Cln3—which promotes cyclin-dependent kinase activity required for entry into and progression through S phase—abrogates the G1 arrest induced by nutrient depletion and causes many cells to growth arrest in S phase instead. These cells also rapidly loose viability (30,32) in concert with a dramatic increase in chronological age-dependent genome instability in surviving cells (30). Ectopic expression of Cln3 also abrogates the G1 arrest induced by rapamycin, an inhibitor of Tor1 signaling, and also leads to growth arrest in S phase and rapid cell death (32). These findings establish that during nutrient depletion, growth arrest in S phase leads to genome instability and shortens chronological life span due to the accumulation of replication stress. Importantly, the effects of ectopic Cln3 expression on genome instability and chronological life span occur in the absence of upstream alterations in growth signaling that would impact oxidative and other stress responses.
The tighter G1 arrest promoted by caloric restriction and mutations that downregulate growth-signaling pathways—which were previously shown to induce oxidative stress responses—likely extends chronological life span in part by protecting against replication stress. A role for replication stress in chronological aging of budding yeast is consistent with the recent finding that a distinct subpopulation of nutrient-depleted cells—which includes all the cells that remain budded after growth arrest has occurred—expresses a number of genes encoding proteins required for the resolution of stalled replication forks (33). It could also explain the shorter replicative life span of chronologically aged cells when nutrients are subsequently restored (23). The accumulation of replication stress and replication stress-induced DNA damage during chronological aging would be expected to shorten the subsequent replicative life span of these cells, which (as discussed above) is also impacted by replication stress.
Replication stress and hormesis effects on aging in budding yeast
Similar to caloric restriction, chronic low-level osmotic stress extends both the replicative (34) and chronological (22,30) life span of budding yeast. Also similar to caloric restriction, the extended chronological life span of osmotically stressed cells is associated with a tighter arrest in G1 induced during nutrient depletion (30). This tighter G1 arrest is likely related to the transient G1 arrest induced by osmotic stress in cycling populations of cells (35,36), which protects against genome instability by blocking entry into S phase (35). The genome-protecting effect of this transient G1 arrest suggests that osmotic stress also induces replication stress. A transient G1 arrest is also detected in cycling cells exposed to other stresses, including oxidative stress (37,38), heat shock (39) and DNA damage (40). This likely reflects the existence of replication stress-inducing effects of these other stresses as well.
The ‘hormesis’ hypothesis of aging is based on the observation that caloric restriction or chronic low-level exposure to any of these stresses induces cross-resistance to other stresses at the same time that it extends life span (41). Hormesis effects on aging are observed in many eukaryotes in addition to budding yeast. Although the mechanistic details of these effects remain unclear, we have argued that they include a general response to environmental stresses that blocks entry into S phase under environmentally stressful conditions that are suboptimal for replicating DNA, thus protecting cells from replication stress (30).
REPLICATION STRESS AND AGING IN HIGHER EUKARYOTES
RecQ helicases, replication stress and aging
A universal role for replication stress in aging is suggested by the premature aging phenotypes observed in many eukaryotes harboring mutations in highly conserved RecQ helicases (including Sgs1 in budding yeast). Although RecQ helicases are not essential for viability, they contribute to genome stability at stalled replication forks by activating checkpoints, stabilizing stalled replication complexes, preventing the formation of aberrant recombination intermediates and facilitating the resolution of these intermediates when they form (42). In humans, mutations in the human WRN RecQ helicase cause genome instability and the premature aging syndrome Werner syndrome. In addition to shortened life span, Werner syndrome is characterized by premature graying and thinning of hair, osteoporosis, Type II (‘late onset’) diabetes, cataracts and an increased incidence of cancer (43). Defects in two other human RecQ helicases that are also required for cellular responses to replication stress—BLM and RECQL4—also lead to genome instability and cancer-predisposing, premature aging syndromes [Bloom syndrome and Rothmund–Thomson syndromes, respectively (44)]. Not all the pathologies associated with these premature aging syndromes are related to normal aging. However, based on the overlap between features of these syndromes with various phenotypes of aged individuals, there is little doubt that they mimic many aspects of normal aging, perhaps by accelerating the consequences of replication stress.
Growth signaling, replication stress and aging in higher eukaryotes
The possibility that replication stress arising downstream of growth signaling in higher eukaryotes might contribute to normal aging in the absence of defective responses to stalled replication forks (such as responses mediated by RecQ helicases or other proteins) has not been widely considered. Consistent with this possibility, however, several recent studies have established previously unknown connections between replication stress and growth signaling implicated in cancer and aging in mammals (5–8). Initially, this included the detection of DNA damage and activated checkpoints (6,8) as well as apoptosis (8) in a variety of pre-neoplastic lesions in humans and mice. In these studies, DNA damage was detected specifically at chromosomal fragile sites that are induced in cultured cells by inhibiting DNA replication with drugs. These findings strongly implicate replication stress-induced DNA damage in the etiology of cancer at its earliest stages. Similar to the effects of RAS2val19 expression or ectopic expression of CLN3 during nutrient depletion in budding yeast (30), replication stress in pre-neoplastic lesions is induced by enhanced growth signaling. For example, increased replication stress and DNA damage were detected in hyperplastic lesions induced in mice by the ectopic expression of genes encoding oncogenic growth factors, as well as cyclins that (similar to Cln3 in budding yeast) stimulate cyclin-dependent kinase activity required for entry into and progression through S phase (6,8).
Even more recently, replication stress associated with enhanced growth signaling was implicated in the phenomenon of cellular senescence, which is more directly linked to normal aging. Senescence is a non-dividing state that cannot be reversed by stimulating growth. It is triggered by a variety of stresses, including DNA damage, telomere erosion that mimics DNA damage and induces DNA damage signals and enhanced growth signaling that leads to DNA damage (45). Senescence induced by oncogene-enhanced growth signaling is referred to as ‘oncogene-induced senescence’ (OIS) and requires the induction of DNA damage responses, including those mediated by p53. Inhibition of cyclin-dependent kinases required for progression into and through S phase by the cyclin-dependent kinase inhibitors p21—which is induced by p53—and p16INK4a underlies the cell cycle arrest that characterizes the senescent state. This cell cycle arrest protects against the formation of tumors downstream of oncogenic growth signaling, because abrogation of this arrest (e.g. by mutations in p53- or p16INK4a-dependent pathways) can lead to tumorigenesis.
One of the more important recent findings in the cancer research arena is the discovery that OIS is triggered by replication stress in pre-neoplastic cells (5,7). In addition to chromosome instability at fragile sites detected in both pre-neoplastic and senescent cells, a role for replication stress in OIS is indicated by the detection in these cells of aberrant DNA replication and partly replicated chromosomes (5,7). Furthermore, oncogene-enhanced growth signaling does not lead to OIS in cells that are first arrested outside of S phase by contact inhibition or serum deprivation.
Similar to the effects of replication stress in nutrient-depleted budding yeast cells that are assumed (often incorrectly) to arrest in G1, the role of replication stress in OIS may have escaped detection earlier in part because of the frequent, but perhaps erroneous assumption that senescent cells growth arrest in G1. Although cells that have undergone OIS often growth arrest with an apparent G1 content of DNA measured by flow cytometry (45), this technique cannot detect small increases in DNA content that would occur in cells that arrest growth shortly after entering S phase. In fact, the detection of partly replicated chromosomes in OIS cells using FISH technology (7) clearly suggests growth arrest of these cells occurs within S phase. The importance of these findings for understanding cancer is emphasized by recent paradigm-shifting studies suggesting that the tumor-suppressing properties of p53 are related to its induction of OIS in response to replication stress, and not the acute responses to DNA damage previously believed to be responsible for p53-dependent tumor suppression (46).
The discoveries of oncogene-induced replication stress and replication stress-induced senescence have important implications for understanding aging, in addition to understanding the etiology of cancer. First, these findings establish that, as in budding yeast, growth signaling implicated in aging can cause replication stress-induced DNA damage and genome instability in higher eukaryotes, in addition to inhibitory effects on oxidative and other stress responses. Second, the induction of DNA damage, cellular senescence and apoptosis by replication stress points to a specific mechanism by which replication stress associated with growth signaling might impact aging in mammals and other higher eukaryotes. Senescent cells increase in number with age, and increased senescence mediated by p53 and/or p16ink4a has been implicated in a number of age-related pathologies (45,47). In fact, another remarkable (and unexpected) recent discovery in the cancer research arena is the premature aging phenotype of mice expressing a hyperactive form of p53, despite its inhibitory effects on tumorigenesis (47). Most likely, senescence and/or apoptosis mediated by p53 and p16ink4a promote aging by reducing the proliferative capacity or number of stem cells or their proliferating progeny, leading to the decline in capacity for tissue renewal that underlies many aging phenotypes (47,48). Senescent cells may also contribute to aging by secreting substances capable of altering the tissue microenvironment (49).
A role for replication stress-related genome instability in the age-dependent decline of stem cells and their proliferating progeny is consistent with the recent detection of age-dependent chromosome instability specifically in proliferating cells in various tissues of mice, including brain and intestines (50). In fact, a compelling argument for replication stress as a trigger for age-dependent decline in stem cells populations can be made based on the findings of a recent study of mice expressing a hypomorphic allele of the DNA replication protein Mcm2 (51). Reduced expression of Mcm2 in mice homozygous for this mutation does not impact normal development. However, beginning at 9 weeks of age, these mice exhibit a constellation of aging phenotypes, including the development of tumors, a reduction in the number of stem cells in brain, skeletal muscle and intestinal crypts, increased DNA damage and the expression of markers of premature aging. To our knowledge, this is the first report of a progeroid syndrome directly caused by replication stress rather than by defects in cellular ‘responses’ to replication stress associated with mutations in RecQ helicases and other proteins.
Life span extension by inhibition of growth signaling leading to reduced replication stress
An important corollary to this model is that, as in budding yeast, reduced growth signaling promotes longevity in higher eukaryotes by ‘reducing’ replication stress. Several observations support this notion. First, decades ago it was reported that caloric restriction—which inhibits tumorigenesis and prolongs life span in all eukaryotes—also inhibits DNA replication and reduces the number of proliferating cells in a variety of mammalian tissues (52,53). Similar to the tighter G1 arrest that protects against replication stress induced by caloric restriction in chronologically aged budding yeast (30), the anti-proliferative effects of caloric restriction in mammals may protect against replication stress by prolonging residence of cells in G1.
Second, it was recently discovered that mice (54) and Drosophila (55) harbor ‘metabolic checkpoints’ that depend on AMP-activating protein kinase (AMPK) and p53 to arrest cells in G1 under conditions of energy deprivation, such as caloric restriction. In mice (and presumably in other organisms as well), this checkpoint responds to low glucose levels, and abrogation of the checkpoint causes lethality associated with an increased fraction of cells in S phase (54). Presumably, this checkpoint underlies some of the anti-proliferative effects of caloric restriction reported earlier. Importantly, unlike the irreversible growth arrest with partially replicated chromosomes associated with senescence, the metabolic checkpoint is reversible, at least after transient exposures to low glucose—when glucose levels are restored, mouse cells resume dividing. However, prolonged exposures to low glucose induces senescence (54).
In fact, AMPK exhibits p53-dependent and mTOR-dependent anti-proliferative effects in many types of normal and tumor cells (56). Inhibition of proliferation by AMPK has been implicated in the neuroprotective effects of both caloric restriction and resveratrol, a plant polyphenol that mimics caloric restriction and inhibits aging in yeast and mice (57). AMPK is required for longevity in Caenorhabditis elegans (58), and it has been suggested that an AMPK- and p53-dependent metabolic checkpoint regulated by mitochondrial events contributes to longevity in this organism (59).
Numerous other recent studies have reported unexpected roles for p53 in responses to (and regulation of) glucose metabolism in mammalian cells, in addition to its well-established roles in DNA damage responses.
These studies shed new light on long-standing connections between metabolism and cancer, as well as aging (60). They also point to an anti-aging role for p53. Consistent with this notion is a recent report that p53 negatively regulates IGF-1-AKT and mTOR pathways implicated in aging (61). An anti-aging role for p53 does not contradict the pro-aging role of p53 in cellular senescence described above. In contrast to premature aging of mice associated with high level of expression of a mutant form of p53, mice expressing increased, but normally regulated p53—which may mimic the modest levels of p53 expression transiently induced by nutrient-limiting conditions—exhibit an extended life span (62).
In budding yeast, the AMPK homolog Snf1 is also activated by low glucose and promotes chronological longevity [(63) and references therein]. Snf1 null cells also fail to arrest in G1 during nutrient depletion (our unpublished data) and thus are exposed to increased replication stress during chronological aging experiments. Compared to wild-type cells, Snf1 null cells are also more sensitive to the replication stress-inducing compounds hydroxyurea and methymethane sulfonate (63). Together, these findings suggest that energy deprivation is a source of replication stress in all eukaryotes and triggers a conserved metabolic checkpoint that protects against replication stress by inducing growth arrest in G1. We propose that this G1 arrest and the protection from replication stress it affords contribute to the beneficial effects of caloric restriction.
Third, unlike the premature aging phenotype of mice expressing a hypomorphic allele of the replication initiation protein Mcm2 described above (51), in adult C. elegans, RNAi ablation of the mcm-2 gene extends life span (64). Furthermore, mcm-2 and other mcm proteins that form replication initiation complexes are downregulated in long-lived C. elegans daf-2 insulin receptor mutants, and this downregulation may contribute to their extended life span (65). Initiation proteins are required for entering S phase and establishing replication forks, and Mcm2 is not expressed in quiescent mammalian cells. In fact, the absence of Mcm2 and other (but not all) replication initiation proteins may be a defining characteristic of the quiescent state (66). In contrast to the partial inhibitory effects on Mcm2 expression likely produced by the hypomorphic Mcm2 allele in mice—which would permit entry into S phase, but with a reduced number of replication forks—the life span-extending effects of RNAi knockdown of mcm-2 or its downregulation in daf-2 mutants may be related to more robust downregulation of mcm-2 expression that mimics metabolic checkpoints by blocking entry into S phase.
A caveat to this hypothesis is that except for the germline, cells in adult C. elegans are post-mitotic, and therefore do not enter S phase. It is possible that the anti-aging effects of inhibiting mcm-2 expression in adult C. elegans are indirectly related to replication stress-induced genotoxicity in dividing germline cells, the ablation of which enhances longevity (67). An alternative possibility is that on rare occasions, post-mitotic cells undergo unscheduled DNA replication leading to partial replication of chromosomes, which would require Mcm2. Relevant here are the many reports that unscheduled DNA replication in post-mitotic neurons is a feature of numerous age-related neurodegenerative disorders (68,69). Even more relevant is a recent report that in a Drosophila model for age-related neurodegeneration, growth signaling by TOR pathways leads to unscheduled DNA replication in post-mitotic neurons, which is a causal factor in neurodegeneration (70).
Replication stress, mitochondria and growth signaling
Increased oxidative damage to DNA and other cellular constituents by ROS produced in dysfunctional mitochondria is an important component of modern versions of the ‘free radical theory’ of aging (3,71). It is often assumed that the production of ROS in mitochondria is directly proportional to the rate of mitochondrial respiration, and that increased respiration promotes aging. A number of recent studies in budding yeast and mammals argue that these long-held assumptions are incorrect (72). For example, caloric restriction and other experimental manipulations that enhance respiration in budding yeast reduce, rather than increase levels of ROS at the same time that they enhance life span (73). Similarly, budding yeast cells cultured in medium containing glycerol or ethanol, which are metabolized via respiratory pathways, exhibit a longer chronological life span (22). Furthermore, deletion of TOR1 extends chronological life span of budding yeast by enhancing respiration, but reducing ROS (21). As might be expected based on these reports, experimental manipulations that increase the production of ROS in mitochondria shorten the chronological life span of this organism (73,74).
The counterintuitive findings that increased respiration can lead to reduced ROS and longer life span extends to higher eukaryotes as well. For example, the life span-extending effects of caloric restriction in rats and cultured human cells (75), mice (76) and humans (77) is accompanied by increased respiration, and in the case of rats and humans, reduced levels of ROS (ROS were not examined in the mouse study). In all three studies, caloric restriction also stimulated mitochondrial biogenesis. In budding yeast, the tighter G1 arrest induced by caloric restriction in nutrient-depleted cells in concert with chronological life span extension is also accompanied by increased mitochondrial biogenesis (30). The chronological life span-extending effects of deleting TOR1 that increase respiration and reduce ROS referred to above are also accompanied by evidence for mitochondrial biogenesis (21). Thus, mitochondrial biogenesis that increases respiration, but reduces ROS and is inhibited by growth signaling may be a general feature of the life span-extending effects of caloric restriction in all eukaryotes.
Interestingly, mammalian mitochondrial biogenesis is stimulated by AMPK (78–80), and the neuroprotective effects of resveratrol associated with its stimulation of AMPK-dependent inhibitory effects on proliferation discussed above are accompanied by mitochondrial biogenesis (57). Thus, AMPK appears to play an important role in mitochondrial biogenesis that connects this process to growth signaling. These findings suggest a revised model for the role of mitochondria in aging. In this model, mitochondria lie at the nexus of growth-signaling pathways that promote aging by inhibiting mitochondrial biogenesis, reducing respiration and increasing ROS. Conversely, inhibition of growth-signaling pathways extends life span in part by stimulating mitochondrial biogenesis, increasing respiration and reducing ROS.
This model is consistent with the free radical theory that posits oxidative damage as a primary determinant of aging, although it conflicts with the long-held assumption that higher rates of aerobic metabolism shorten life span by increasing ROS. However, accumulating evidence indicates that in mammals, superoxide anions and other species of reactive oxygen produced in mitochondria signal growth (81). In the context of this information as well as the emerging evidence connecting growth signaling to replication stress and the evidence for a role for replication stress in aging, another possibility must be considered—that inhibition of mitochondrial biogenesis by growth signaling produces ROS that contribute to aging by transducing replication stress-enhancing growth signals downstream of mitochondrial events.
Although a role for ROS in growth signaling in budding yeast has not been directly addressed, budding yeast exhibit a metabolic redox cycle that is coordinated with the cell cycle (82), which suggests that ROS signal growth in this organism, as in mammals and other eukaryotes. Although the evidence is strictly correlative, the reduced ROS reported to occur in calorie-restricted budding yeast cells during nutrient depletion (73) might contribute to the tighter G1 arrest accompanied by evidence for mitochondrial biogenesis we recently observed in calorie-restricted cells during chronological aging (30). Similarly, since ROS contribute to growth signaling in mammals, AMPK-dependent mitochondrial biogenesis in response to low glucose in mammalian cells might contribute to the anti-proliferative effects of the metabolic checkpoint by reducing ROS.
In fact, a role for ROS-dependent growth signaling in aging of eukaryotes is clearly evident in a recent study employing the Drosophila model of TOR-dependent, age-related neurodegeneration described above. In this study, unscheduled DNA replication and neurodegeneration occurring in post-mitotic neurons downstream of TOR signaling were blocked by genetic manipulations or pharmacological treatments that reduce ROS (83). We think it is likely that some of the well-documented pro-aging effects of growth signaling and increased oxidative stress in all eukaryotes could be related to enhanced growth signaling by ROS that promotes replication stress-induced DNA damage, in addition to oxidative DNA damage, and is inhibited by caloric restriction.
SUMMARY AND FUTURE PERSPECTIVES
Despite many tests of the free radical theory during the several decades that have elapsed since it was first proposed, in many organisms (including humans), the nature of age-dependent genome instability and whether oxidative damage to DNA limits life span remains unclear. The emerging connections between growth signaling and DNA replication stress in budding yeast and in higher eukaryotes outlined in this article suggest a revised model for understanding aging and age-dependent genome instability: in addition to oxidative damage, age-dependent DNA damage and genome instability are caused by DNA replication stress arising downstream of deregulated growth signaling.
According to this model, replication stress that impacts aging is likely caused by the downregulation of some, but not all, growth-signaling pathways during entry into a non-dividing state. This leads to growth arrest within S phase in the absence of a full complement of factors required for efficient DNA replication. This can occur during nutrient depletion of budding yeast cells expressing a constitutively active Ras2 protein, for example, which leads to the transcriptional induction of CLN3 encoding a protein required for the G1-S transition, but not genes required for DNA replication [(84), including Supplementary data]. In higher eukaryotes, replication stress that impacts aging likely occurs in stem cells or their proliferating progeny when the constitutive activation of some growth-signaling pathways by mutations or other factors coincides with the downregulation of other growth-signaling pathways (in quiescent stem cells or during differentiation, for example) required for efficient DNA replication. This leads to apoptosis or the irreversible growth arrest in S phase with partially replicated chromosomes that characterizes OIS. Both outcomes would cause the reduced capacity for tissue renewal that underlies many age-related pathologies. Age-related replication stress also likely arises in post-mitotic neurons. As discussed above, inappropriate activation of growth signaling and ectopic entry into S phase after neuronal differentiation has occurred have been implicated in a variety of age-related neurodegenerative disorders.
This model also predicts that caloric restriction exerts some of its anti-aging (and anti-cancer) effects by protecting cells from replication stress. Protection from replication stress by caloric restriction is likely provided by the induction of energy-sensing ‘metabolic checkpoints’ that coordinately downregulate all growth-signaling pathways, leading to an efficient growth arrest in G1, rather than S phase. This might include growth-signaling pathways that have been constitutively activated by mutations upstream of cyclin-dependent kinase activity required for progression into S phase, which is likely inhibited by these checkpoints. Mutations in RecQ helicases and other proteins that respond to replication stress accelerate aging by amplifying the consequences of replication stress that develops during normal aging. Although this model can account for some of the failures of the free radical theory of aging to explain various aging phenotypes, it is not inconsistent with this theory. Most likely, the roles of oxidative stress-induced and replication stress-induced DNA damage in aging are inextricably linked by their common origin in deregulated growth signaling, perhaps including growth signaling by ROS produced in mitochondria (Figure 3).
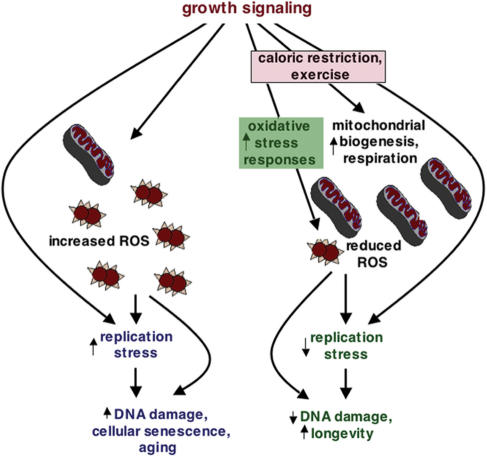
Figure 3.
Replication stress model of aging. Growth signaling inhibits mitochondrial biogenesis and respiration and increases ROS, leading to replication stress, genome instability, cellular senescence and aging. Replication stress is likely enhanced by ROS-dependent growth signaling and by growth signaling that occurs independently of ROS. Caloric restriction and mutational inactivation of growth-signaling pathways stimulate mitochondrial biogenesis, increase respiration and reduce ROS. Reduced ROS-dependent and -independent growth signaling reduces replication stress and genome instability and promotes life span. In mammals, exercise also extends life span extension and promotes mitochondrial biogenesis and increased respiration (77,89). The effects of replication stress on aging likely occur in parallel with oxidative damage to DNA and other cellular constituents.
A more complete understanding of how replication stress potentially impacts aging will require a better understanding of the non-dividing state induced by energy deprivation and during differentiation. Pathways that regulate the G1 to S phase transition in cycling populations of cells and their downregulation in response to DNA damage and other stresses have been extensively investigated. However, much less is known about how these pathways are downregulated during differentiation or in response to nutrient-limiting conditions that lead to quiescence, or how quiescence is maintained. The paucity of details concerning downregulation of these pathways in mammalian cells is reflected by the following fact: only recently was it determined that p53—one of the most thoroughly characterized mammalian proteins—induces a G1 arrest in response to low glucose, in addition to DNA damage.
Even less is known about how cells arrest in G1 in response to nutrient-limiting conditions in budding yeast, the organism that (together with fission yeast) was employed in numerous studies of growth in the presence of excess nutrients that provided the framework for understanding cell cycle regulation in all eukaryotes. For example, although the PUBMED search engine identifies (as of late October 2007) 3145 publications associated with the terms ‘Cln’ (G1 cyclins), Cdc28 (the cyclin-dependent kinase required for entry into S phase regulated by Clns) or ‘Sic1’ (an inhibitor of Cdc28), it identifies just 20 publications associated with these terms and the term ‘stationary phase’ (the growth-arrested state induced by nutrient depletion), most of which are not relevant to stationary phase G1 arrest.
This paucity of information exists despite the likelihood that in budding yeast, events associated with the downregulation of growth during nutrient depletion—such as those detected in chronological aging experiments—more accurately model how replication stress arises and contributes to cancer and aging compared to models based on studies of cells dividing in the presence of excess nutrients. This view runs somewhat counter to the conventional wisdom that the chronological aging model of budding yeast is mostly relevant to aging of post-mitotic cells in higher eukaryotes. However, the chronological life span of this organism is clearly impacted by events during early stages of nutrient depletion that impact a ‘post-mitotic’ growth arrest before this arrest is established. For example, the effects of caloric restriction by reducing glucose concentrations must be limited to the first few days of nutrient depletion when cells have not yet arrested growth. This is because after this time period, the concentration of glucose approaches zero, and thus the reduced glucose levels present at the beginning of these experiments can no longer impact life span. Similarly, the life span-extending effects in budding yeast cells associated with increased respiration when Tor1 is inactivated are limited to the first few days when cells are arresting growth, and are absent after this growth arrest has been established (21). In short, at least some of the molecular events that impact chronological life span in budding yeast likely mimic events in dividing stem cell populations or their proliferating progeny at early stages of differentiation in higher eukaryotes, before the permanent ‘post-mitotic’ growth arrest associated with the differentiation of many cells has occurred.
However, chronological aging experiments can also mimic events in post-mitotic cells. For example, caloric restriction imposed during chronological aging experiments by replacing depleted medium with water after growth arrest is established dramatically extends chronological life span (16). Furthermore, stimulation of growth by adding glucose, but not other nutrients, to growth-arrested stationary phase cultures rapidly induces apoptosis in concert with evidence that cells are attempting to re-enter the cell cycle (85). Apoptosis may occur because the addition of glucose in the absence of other nutrients stimulates some, but not all, growth regulatory pathways required for efficient DNA replication, leading to replication stress-induced DNA damage. These observations may be relevant, for example, to age-dependent neurodegeneration that occurs when unscheduled DNA replication and apoptosis are triggered in post-mitotic neurons downstream of the inappropriate activation of growth-signaling pathways (86).
The role of replication stress in aging may reflect a broader role for replication stress in evolution than previously considered. In addition to its potential impact on the evolution of budding yeast after an ancient whole genome duplication [(87); see Ref. (30) for discussion], replication stress may have been an important factor in the evolution of many multicellular organisms. High concentrations of salt induce mutations in budding yeast and DNA strand breaks in many eukaryotes (88). The increased mutation frequency associated with entry into S phase in osmotically stressed budding yeast cells that fail to arrest in G1 (35) suggests strand breaks produced by increased salinity might be caused by replication stress. It has been hypothesized that DNA strand breaks and mutations produced when the salinity of seawater increased during the Precambrian–Cambrian period enhanced genetic diversity that drove the explosive adaptive radiation of species during the Cambrian period (88). Thus, replication stress may have contributed to this adaptive radiation. As may have been the case for the evolution of budding yeast, this adaptive radiation may have required the evolution of mechanisms for protecting against replication stress that impact aging.
ACKNOWLEDGEMENT
Research in the author's laboratory was funded in part by a NCI Cancer Center Support Grant (PO CA016056). The authors wish to thank Joel Huberman, Keshav Singh and Joris Winderickx for very helpful discussions and support and Molly Burhans for assistance with graphics. Funding to pay the Open Access publication charges for this article was provided by Department of Cell Stress Biology, Roswell Park Cancer Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.McMurray MA, Gottschling DE. An age-induced switch to a hyper-recombinational state. Science. 2003;301:1908–1911. doi: 10.1126/science.1087706. [DOI] [PubMed] [Google Scholar]
- 2.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–667. doi: 10.1016/j.cell.2005.08.042. [DOI] [PubMed] [Google Scholar]
- 3.Muller FL, Lustgarten MS, Jang Y, Richardson A, Van Remmen H. Trends in oxidative aging theories. Free Radic. Biol. Med. 2007;43:477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 4.Longo VD, Finch CE. Evolutionary medicine: from dwarf model systems to healthy centenarians? Science. 2003;299:1342–1346. doi: 10.1126/science.1077991. [DOI] [PubMed] [Google Scholar]
- 5.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 6.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 7.Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 8.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio R.A., Jr, Kastrinakis NG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 9.Hyrien O. Mechanisms and consequences of replication fork arrest. Biochimie. 2000;82:5–17. doi: 10.1016/s0300-9084(00)00344-8. [DOI] [PubMed] [Google Scholar]
- 10.Branzei D, Foiani M. The DNA damage response during DNA replication. Curr. Opin. Cell Biol. 2005;17:568–575. doi: 10.1016/j.ceb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Branzei D, Foiani M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair. 2007;6:994–1003. doi: 10.1016/j.dnarep.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 12.Bitterman KJ, Medvedik O, Sinclair DA. Longevity regulation in Saccharomyces cerevisiae: linking metabolism, genome stability, and heterochromatin. Microbiol. Mol. Biol. Rev. 2003;67:376–399. doi: 10.1128/MMBR.67.3.376-399.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- 14.Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 15.Kaeberlein M, Powers R.W., III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, et al. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–1196. doi: 10.1126/science.1115535. [DOI] [PubMed] [Google Scholar]
- 16.Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- 17.Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 19.Longo VD. The Ras and Sch9 pathways regulate stress resistance and longevity. Exp. Gerontol. 2003;38:807–811. doi: 10.1016/s0531-5565(03)00113-x. [DOI] [PubMed] [Google Scholar]
- 20.Fabrizio P, Pozza F, Pletcher SD, Gendron CM, Longo VD. Regulation of longevity and stress resistance by Sch9 in yeast. Science. 2001;292:288–290. doi: 10.1126/science.1059497. [DOI] [PubMed] [Google Scholar]
- 21.Bonawitz ND, Chatenay-Lapointe M, Pan Y, Shadel GS. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith DL, McClure JM, Matecic M, Smith JS. Calorie restriction extends the chronological life span of Saccharomyces cerevisiae independently of the sirtuins. Ageing Cell. 2007;6:649–662. doi: 10.1111/j.1474-9726.2007.00326.x. [DOI] [PubMed] [Google Scholar]
- 23.Ashrafi K, Sinclair D, Gordon JI, Guarente L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1999;96:9100–9105. doi: 10.1073/pnas.96.16.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Benguria A, Lai CY, Jazwinski SM. Modulation of life-span by histone deacetylase genes in Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:3125–3136. doi: 10.1091/mbc.10.10.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heo SJ, Tatebayashi K, Ohsugi I, Shimamoto A, Furuichi Y, Ikeda H. Bloom's syndrome gene suppresses premature ageing caused by Sgs1 deficiency in yeast. Genes Cells. 1999;4:619–625. doi: 10.1046/j.1365-2443.1999.00288.x. [DOI] [PubMed] [Google Scholar]
- 26.Hoopes LL, Budd M, Choe W, Weitao T, Campbell JL. Mutations in DNA replication genes reduce yeast life span. Mol. Cell. Biol. 2002;22:4136–4146. doi: 10.1128/MCB.22.12.4136-4146.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weitao T, Budd M, Hoopes LL, Campbell JL. Dna2 helicase/nuclease causes replicative fork stalling and double-strand breaks in the ribosomal DNA of Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:22513–22522. doi: 10.1074/jbc.M301610200. [DOI] [PubMed] [Google Scholar]
- 28.McVey M, Kaeberlein M, Tissenbaum HA, Guarente L. The short life span of Saccharomyces cerevisiae sgs1 and srs2 mutants is a composite of normal aging processes and mitotic arrest due to defective recombination. Genetics. 2001;157:1531–1542. doi: 10.1093/genetics/157.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- 30.Weinberger M, Feng L, Paul A, Smith DL, Hontz RD, Smith JS, Vujcic M, Singh KK, Huberman J, et al. DNA replication stress is a determinant of chronological lifespan in budding yeast. PLOS One. 2007 doi: 10.1371/journal.pone.0000748. 2, e748. doi:10.1371/journal.pone.0000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedruzzi I, Dubouloz F, Cameroni E, Wanke V, Roosen J, Winderickx J, De Virgilio C. TOR and PKA signaling pathways converge on the protein kinase Rim15 to control entry into G0. Mol. Cell. 2003;12:1607–1613. doi: 10.1016/s1097-2765(03)00485-4. [DOI] [PubMed] [Google Scholar]
- 32.Zinzalla V, Graziola M, Mastriani A, Vanoni M, Alberghina L. Rapamycin-mediated G1 arrest involves regulation of the Cdk inhibitor Sic1 in Saccharomyces cerevisiae. Mol. Microbiol. 2007;63:1482–1494. doi: 10.1111/j.1365-2958.2007.05599.x. [DOI] [PubMed] [Google Scholar]
- 33.Allen C, Buttner S, Aragon AD, Thomas JA, Meirelles O, Jaetao JE, Benn D, Ruby SW, Veenhuis M, et al. Isolation of quiescent and nonquiescent cells from yeast stationary-phase cultures. J. Cell Biol. 2006;174:89–100. doi: 10.1083/jcb.200604072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaeberlein M, Andalis AA, Fink GR, Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol. Cell. Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escote X, Zapater M, Clotet J, Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat. Cell. Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- 36.Alexander MR, Tyers M, Perret M, Craig BM, Fang KS, Gustin MC. Regulation of cell cycle progression by Swe1p and Hog1p following hypertonic stress. Mol. Biol. Cell. 2001;12:53–62. doi: 10.1091/mbc.12.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flattery-O'Brien JA, Dawes IW. Hydrogen peroxide causes RAD9-dependent cell cycle arrest in G2 in Saccharomyces cerevisiae whereas menadione causes G1 arrest independent of RAD9 function. J. Biol. Chem. 1998;273:8564–8571. doi: 10.1074/jbc.273.15.8564. [DOI] [PubMed] [Google Scholar]
- 38.Lee J, Romeo A, Kosman DJ. Transcriptional remodeling and G1 arrest in dioxygen stress in Saccharomyces cerevisiae. J. Biol. Chem. 1996;271:24885–24893. doi: 10.1074/jbc.271.40.24885. [DOI] [PubMed] [Google Scholar]
- 39.Johnston GC, Singer RA. Ribosomal precursor RNA metabolism and cell division in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 1980;178:357–360. doi: 10.1007/BF00270484. [DOI] [PubMed] [Google Scholar]
- 40.Sidorova JM, Breeden LL. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinclair DA. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 2005;126:987–1002. doi: 10.1016/j.mad.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 42.Bennett RJ, Keck JL. Structure and function of RecQ DNA helicases. Crit. Rev. Biochem. Mol. Biol. 2004;39:79–97. doi: 10.1080/10409230490460756. [DOI] [PubMed] [Google Scholar]
- 43.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larizza L, Magnani I, Roversi G. Rothmund-Thomson syndrome and RECQL4 defect: splitting and lumping. Cancer Lett. 2006;232:107–120. doi: 10.1016/j.canlet.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 45.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 46.Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle. 2007;6:1006–1010. doi: 10.4161/cc.6.9.4211. [DOI] [PubMed] [Google Scholar]
- 47.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell Biol. 2007;8:703–713. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 48.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 49.Campisi J. Senescent cells, tumor suppression, and organismal aging: good citizens, bad neighbors. Cell. 2005;120:513–522. doi: 10.1016/j.cell.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 50.Busuttil RA, Garcia AM, Reddick RL, Dolle ME, Calder RB, Nelson JF, Vijg J. Intra-organ variation in age-related mutation accumulation in the mouse. PLoS ONE. 2007;2:e876. doi: 10.1371/journal.pone.0000876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pruitt SC, Bailey KJ, Freeland A. Reduced Mcm2 expression results in severe stem/progenitor cell deficiency and cancer. Stem Cells. 2007 doi: 10.1634/stemcells.2007-0483. in press. [DOI] [PubMed] [Google Scholar]
- 52.Ogura M, Ogura H, Ikehara S, Dao ML, Good RA. Decrease by chronic energy intake restriction of cellular proliferation in the intestinal epithelium and lymphoid organs in autoimmunity-prone mice. Proc. Natl Acad. Sci. USA. 1989;86:5918–5922. doi: 10.1073/pnas.86.15.5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hursting SD, Lavigne JA, Berrigan D, Perkins SN, Barrett JC. Calorie restriction, aging, and cancer prevention: mechanisms of action and applicability to humans. Annu. Rev. Med. 2003;54:131–152. doi: 10.1146/annurev.med.54.101601.152156. [DOI] [PubMed] [Google Scholar]
- 54.Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- 55.Mandal S, Guptan P, Owusu-Ansah E, Banerjee U. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell. 2005;9:843–854. doi: 10.1016/j.devcel.2005.11.006. [DOI] [PubMed] [Google Scholar]
- 56.Motoshima H, Goldstein BJ, Igata M, Araki E. AMPK and cell proliferation—AMPK as a therapeutic target for atherosclerosis and cancer. J. Physiol. 2006;574:63–71. doi: 10.1113/jphysiol.2006.108324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dasgupta B, Milbrandt J. Resveratrol stimulates AMP kinase activity in neurons. Proc. Natl Acad. Sci. USA. 2007;104:7217–7222. doi: 10.1073/pnas.0610068104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Apfeld J, O'Connor G, McDonagh T, DiStefano PS, Curtis R. The AMP-activated protein kinase AAK-2 links energy levels and insulin-like signals to lifespan in C. elegans. Genes Dev. 2004;18:3004–3009. doi: 10.1101/gad.1255404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rea SL, Ventura N, Johnson TE. Relationship between mitochondrial electron transport chain dysfunction, development, and life extension in Caenorhabditis elegans. PLoS Biol. 2007;5:e259. doi: 10.1371/journal.pbio.0050259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bensaad K, Vousden KH. p53: new roles in metabolism. Trends Cell Biol. 2007;17:286–291. doi: 10.1016/j.tcb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 61.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 62.Matheu A, Maraver A, Klatt P, Flores I, Garcia-Cao I, Borras C, Flores JM, Vina J, Blasco MA, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–379. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 63.Harkness TA, Shea KA, Legrand C, Brahmania M, Davies GF. A functional analysis reveals dependence on the anaphase-promoting complex for prolonged life span in yeast. Genetics. 2004;168:759–774. doi: 10.1534/genetics.104.027771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3:e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Halaschek-Wiener J, Khattra JS, McKay S, Pouzyrev A, Stott JM, Yang GS, Holt RA, Jones SJ, Marra MA, et al. Analysis of long-lived C. elegans daf-2 mutants using serial analysis of gene expression. Genome Res. 2005;15:603–615. doi: 10.1101/gr.3274805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coller HA. What's taking so long? S-phase entry from quiescence versus proliferation. Nat. Rev. Mol. Cell Biol. 2007;8:667–670. doi: 10.1038/nrm2223. [DOI] [PubMed] [Google Scholar]
- 67.Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 68.Copani A, Caraci F, Hoozemans JJ, Calafiore M, Sortino MA, Nicoletti F. The nature of the cell cycle in neurons: focus on a "non-canonical" pathway of DNA replication causally related to death. Biochim. Biophys. Acta. 2007;1772:409–412. doi: 10.1016/j.bbadis.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 69.Khurana V, Feany MB. Connecting cell-cycle activation to neurodegeneration in Drosophila. Biochim. Biophys. Acta. 2007;1772:446–456. doi: 10.1016/j.bbadis.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Khurana V, Lu Y, Steinhilb ML, Oldham S, Shulman JM, Feany MB. TOR-mediated cell-cycle activation causes neurodegeneration in a Drosophila tauopathy model. Curr. Biol. 2006;16:230–241. doi: 10.1016/j.cub.2005.12.042. [DOI] [PubMed] [Google Scholar]
- 71.Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–495. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 72.Barja G. Mitochondrial oxygen consumption and reactive oxygen species production are independently modulated: implications for aging studies. Rejuvenation Res. 2007;10:215–224. doi: 10.1089/rej.2006.0516. [DOI] [PubMed] [Google Scholar]
- 73.Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:49883–49888. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- 74.Bonawitz ND, Rodeheffer MS, Shadel GS. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol. Cell. Biol. 2006;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc. Natl Acad. Sci. USA. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, et al. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 77.Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, Shulman GI. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am. J. Physiol. Endocrinol. Metab. 2001;281:E1340–E1346. doi: 10.1152/ajpendo.2001.281.6.E1340. [DOI] [PubMed] [Google Scholar]
- 79.Kukidome D, Nishikawa T, Sonoda K, Imoto K, Fujisawa K, Yano M, Motoshima H, Taguchi T, Matsumura T, et al. Activation of AMP-activated protein kinase reduces hyperglycemia-induced mitochondrial reactive oxygen species production and promotes mitochondrial biogenesis in human umbilical vein endothelial cells. Diabetes. 2006;55:120–127. [PubMed] [Google Scholar]
- 80.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc. Natl Acad. Sci. USA. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Menon SG, Goswami PC. A redox cycle within the cell cycle: ring in the old with the new. Oncogene. 2006;26:1101–1109. doi: 10.1038/sj.onc.1209895. [DOI] [PubMed] [Google Scholar]
- 82.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 83.Dias-Santagata D, Fulga TA, Duttaroy A, Feany MB. Oxidative stress mediates tau-induced neurodegeneration in Drosophila. J. Clin. Invest. 2007;117:236–245. doi: 10.1172/JCI28769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang Y, Pierce M, Schneper L, Guldal CG, Zhang X, Tavazoie S, Broach JR. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004;2:E128. doi: 10.1371/journal.pbio.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Granot D, Levine A, Dor-Hefetz E. Sugar-induced apoptosis in yeast cells. FEMS Yeast Res. 2003;4:7–13. doi: 10.1016/S1567-1356(03)00154-5. [DOI] [PubMed] [Google Scholar]
- 86.Herrup K, Neve R, Ackerman SL, Copani A. Divide and die: cell cycle events as triggers of nerve cell death. J. Neurosci. 2004;24:9232–9239. doi: 10.1523/JNEUROSCI.3347-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Andalis AA, Storchova Z, Styles C, Galitski T, Pellman D, Fink GR. Defects arising from whole-genome duplications in Saccharomyces cerevisiae. Genetics. 2004;167:1109–1121. doi: 10.1534/genetics.104.029256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dmitrieva NI, Ferraris JD, Norenburg JL, Burg MB. The saltiness of the sea breaks DNA in marine invertebrates: possible implications for animal evolution. Cell Cycle. 2006;5:1320–1323. doi: 10.4161/cc.5.12.2867. [DOI] [PubMed] [Google Scholar]
- 89.Hood DA. Invited review: contractile activity-induced mitochondrial biogenesis in skeletal muscle. J. Appl. Physiol. 2001;90:1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]