Abstract
Symmetries in the p53 response-element (p53RE) encode binding modes for p53 tetramer to recognize DNA. We investigated the molecular mechanisms and biological implications of the possible binding modes. The probabilities evaluated with molecular dynamics simulations and DNA sequence analyses were found to be correlated, indicating that p53 tetramer models studied here are able to read DNA sequence information. The traditionally believed mode with four p53 monomers binding at all four DNA quarter-sites does not cause linear DNA to bend. Alternatively, p53 tetramer can use only two monomers to recognize DNA sequence and induce DNA bending. With an arrangement of dimer of AB dimer observed in p53 trimer–DNA complex crystal, p53 can recognize supercoiled DNA sequence-specifically by binding to quarter-sites one and four (H14 mode) and recognize Holliday junction geometry-specifically. Examining R273H mutation and p53–DNA interactions, we found that at least three R273H monomers are needed to disable the p53 tetramer, consistent with experiments. But just one R273H monomer may greatly shift the binding mode probabilities. Our work suggests that p53 needs balanced binding modes to maintain genome stability. Inverse repeat p53REs favor the H14 mode and direct repeat p53REs may have high possibilities of other modes.
INTRODUCTION
The p53 protein functions as a homo-tetramer and operates as a tumor suppressor, regulating hundreds of genes in response to stress (1). DNA-binding is critical for p53 biological functions, involving sequence-dependent recognition in transcription (2) and geometry-specific binding in transcription-independent pathways (3,4). The p53 response element (p53RE, or p53-binding site) has two half-sites, which may be consecutive or separated by a spacer of 1 to 14 bp. The half-site has two quarter-sites of 10 bp with a consensus sequence of 5′-PuPuPuCWWGPyPyPy-3′, where W can be A or T, and Pu and Py stand for purine and pyrimidine bases, respectively (2). The internal symmetry in the full site is controlled by the spacer (5). When there is no spacer, the full-site palindrome with coupling between quarter-sites one and four (H14 coupling) dominates. With an insertion of a 3 bp spacer in the human genome (8 bp spacer in the mouse genome), two isolated half-sites have a strong palindrome within the half-site. The degree of the H14 coupling distinguishes transcription mechanisms of p53, with the H14 couplings being much stronger for positive regulation than for negatively regulated p53REs. The palindrome sequence couplings may encode potential preferred multiple binding modes of the p53 tetramer to recognize DNA (5).
Besides the sequence-specific DNA recognition, there are geometrical factors in p53–DNA recognition. p53 tetramer binding to linear DNA may cause DNA to bend (6). Consistently, p53 tetramer binds to supercoiled DNA with a higher affinity than to linear DNA (7). p53 protein controls homologous recombination by extremely strong binding to four-way Holliday and other three-way DNA junctions. Visualization using EM (3) and biological evidences (4) indicated that DNA junctions are the predominate p53 binding sites. Clearly, the junctions’ geometric features underlie the strong p53 interactions, in compensation for losing sequence-specific interactions.
Several possibilities for the p53 tetramer–DNA interactions have been discussed (8–11). Still, there is no experimental structure of the p53 tetramer binding to full-site p53RE, or p53 tetramer binding to shape-specific DNA. How does p53 recognize DNA sequence specifically and recognize special geometries without sequence specification? To delineate various aspects of p53–DNA recognition is a difficult task, especially when p53 mutations add more dimensions in p53's structure and functions. Several reports related p53 mutants to geometry-specific DNA binding (12). Furthermore, p53 mutants do not simply lose DNA-binding ability; they also gain functions to activate many genes. But due to the lack of experimental evidence and complex functions of p53, the field is full of black boxes that are often ignored for the sake of simplicity or because they do not fit with the current dominant view (13).
Here we examine possible p53 tetramer–DNA interactions in a systematic way, in order to provide a structural basis to understand p53 tetramer-DNA recognition. We first outline the internal symmetries encoded in the DNA sequences of p53REs and relate the variations of the internal symmetries of p53REs to the variations of binding modes. Accordingly, we constructed several possible models of p53 tetramer–DNA complexes. In Section 2, we report refinement of these models, using molecular dynamics simulations. In Section 3, we examine several models for the p53–Holliday junction recognition, seeking a possible p53 tetramer organization which allows recognition of DNA in a sequence- and geometric-specific way. In Section 4, we correlate the probabilities of four binding modes deduced from DNA sequence analysis with those from energetic calculations. We also check the effects of R273H mutation on p53–DNA interactions.
METHODS
Molecular dynamics simulations
We focus on p53 core domain–DNA interactions. Tetrameric structural models were assembled, based on available X-ray structures of trimeric p53–DNA (PDB code 1tsr) (8) and dimeric p53–DNA (9,10) complexes. The supercoiled DNA was modeled using the X-ray structure of the nucleosome NP147 centered around THY50 (PDB code: 1kx5) (14), with nucleic acids mutated to the corresponding sequence of p53REs. The Holliday junction was modeled using the crystal structure of the Flpe–Holliday junction (PDB code: 1m6x) (15).
The p53 tetramer–DNA complex was solvated with a TIP3P water box with a margin of at least 10 Å from any edge of the water box (typical size of 130 Å × 110 Å × 90 Å) to any protein or DNA atom. The sodium ions were pre-equilibrated in a small water box used in construction of solvation box containing p53 tetramer complex. The initial box had more ions than needed, and the sodium ions too close to solute (5 Å) were removed first, and then sodium ions far from solute were also removed to make overall system neutral. MD simulations were performed using the NAMD package (16) and the Charmm 27 force field (17), with constant pressure ensembles (NPT) at 1 atm and temperature at 300 K°. The time step was 2 fs with a SHAKE constraint on all bonds with hydrogen atoms. Productive MD runs were performed after 5000 steps of minimizations and three 150 ps heating and equilibration runs. In the first 10 ns, electrostatic interactions had a cutoff distance of 10 Å. For the simulations after 10 ns, long-range electrostatic interactions were calculated with the PME method (18).
Three cysteine residues coordinated with Zinc were deprotonated. The bond distances of the three Zn–S (Zn-Cys) and Zn–N (Zn-His) bonds were fixed during the simulations. The charge and vdW parameters of the Zn and deprotonated Cys were taken from Maynard and Covell (19). We selectively protonated five histidine residues, following the suggestion of Wright et al. (20). The only two histidines left in the deprotonated state were His179 (coordinated to zinc) and His214 (close to R174).
DNA bending angles were estimated using angles between normal vector pairs calculated with FREEHELIX98 (21). Protein can cause DNA to bend in different behaviors, including kink, writhe or continuous curvature. A clear definition of bending angle is only valid in the case of DNA kink. The matrix of the angles between all normal vector pairs may characterize DNA bending (21). We use the difference between the average normal vector angles in the first halfsite and the second halfsite of p53 RE as DNA bending angle, based on the average conformation during 13–14 ns MD simulation. Statistical errors are estimated using 8, 9, 10, 11 and 12 bp in each halfsite.
Free energy landscape and Monte Carlo (MC) simulations
MC simulations were used to estimate the binding mode probabilities based on conformations obtained in the last 5 ns of MD simulations. Using the CHARMM 27 force field (17) and Generalized Born using molecular volume (GBMV) (22) implemented in the CHARMM package and, each conformer was first minimized 1000 cycles and then the conformation energy was evaluated by grid-based GBMV. The minimization does not change conformations obtained from MD simulation and only relaxed local geometries due to thermal fluctuation occurred in MD simulations. In the GBMV calculation, no distance cutoff was used, the dielectric constant of water was set to 80 and the Debye-Huckel ionic term was 0.2 to reflect the salt effect.
The total 2000 conformations (500 for each of four binding modes examined) were used to construct the free energy landscape for a p53 tetramer–DNA complex to evaluate binding mode probabilities. Starting from a randomly selected conformation i, another conformation i + 1 was randomly selected from any of the four modes to ensure binding mode change. Then, the Boltzmann factor was used to decide if it is allowed to move from conformation i to i + 1. If e−(Ei+1 – Ei)/KT > Random number, the move is allowed; where Ei and Ei+1 are the conformational energies evaluated using the GBMV calculations for the conformation i and i + 1, respectively; K is the Boltzmann's constant, and T is the absolute temperature (298 K used here). After 1 million steps, the conformations visited for each binding mode were counted. The relative probability of binding mode j was evaluated as:
| 1 |
where Pj is the population, Nj is the total number of conformations visited for the binding mode j, and Ntotal is the total steps.
Following the same procedure, we also evaluated the binding probability of p53 R273H mutant; using the exact conformation ensemble obtained from wild-type p53–DNA complex and only relaxing local structural changes due to mutation. Each binding mode had 500 conformations initially. We divided the mutant complexes into four groups. In one mutant monomer group, each of the monomers in the p53 tetramer was mutated in turn, and the other three monomers were unchanged; leading to 8000 combinations used in binding probability evaluation. In two mutant monomer groups, two of the monomers in the p53 tetramer were mutated in turn; and the other two monomers are unchanged. This combination led to 3000 conformations for each binding mode, and 12 000 conformations were used to evaluate the binding probabilities. The binding probabilities in the groups of three mutants and four mutants were similarly evaluated. Even though current approach allows one to test conformational energy changes due to mutation for a given conformation ensemble, it overlooked the mutation effects to change conformational ensemble. However the large combinational possibility prevented us to run additional MD simulations for each possible mutant complex.
The combination of explicit water simulation with subsequent conformational energy analysis using implicit solvation takes the advantage of proper conformation sampling in the molecular dynamics simulation and efficient energy evaluation with MM-GB approach (23). Still the possible shortcomings existed in each of the methods (MD simulations in explicit water, GBMV energy evaluation, MC population estimation and mutation effects) may not be fixed easily. Proper electrostatics calculations and conformation sampling are coupled. Ewald summation introduces an artificial periodicity into non-periodic molecular system in solution. Even though the artifacts maybe not obvious for polyalanine, for charged molecule and ionic solvation the consequences should be addressed (24). The solvation effects in GBMV energy calculations used in the energy landscape calculations are also approximate, and the conformations used in GBMV analysis are taken from explicit water simulations. For systems such as those explored here, simulations may be expected to largely sample conformations reachable from the starting conformations. The energy barriers may be too high to sample conformations away from these states, which prevents a single long MD simulation to sample all relevant binding modes automatically. Thus, the GBMV energy evaluations and subsequence MC simulations do not overcome the shortcoming inherited from early simulations. The advantages of using the MC simulation to estimate binding mode probability rely on that (i) the MC simulation has good numerical stability; and (ii) MC simulations allow transition probabilities among several binding modes to be controlled. Even though all modes are allowed to freely change in the current simulation, one may define transition pathway in future study. Nevertheless, the binding mode probabilities need to be carefully checked. Here we seek to check the correlations of the binding mode probabilities from molecular simulations and DNA sequence analyses. A correlation of two totally different methods would indicate the molecular simulations provided essential structural basis of p53 tetramer to read the information encoded in DNA sequences.
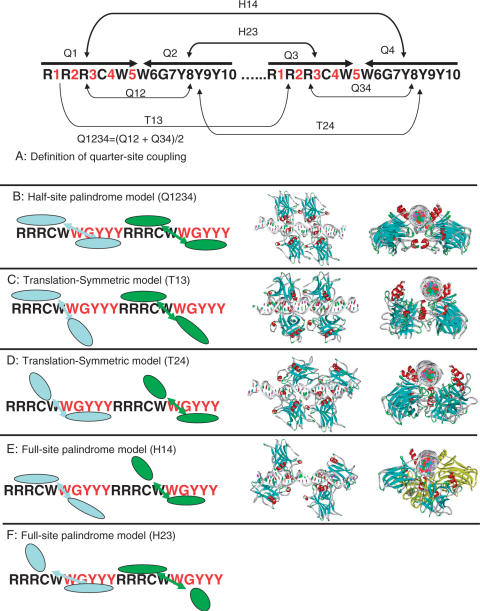
Sequence-dependent quarter-site couplings
A p53RE consists of two half-sites. Each half-site consists of two quarter-sites. We examined the base inversion correlation (A with T, G with C) within the half-site palindrome, as well as in the full-site palindrome and we also examine base repetition between half-sites; based on a sequence analysis of the p53REs in the human genome (5). We define three types of couplings in p53RE (Figure 1A). The first is Q-coupling, between the quarter sites, which is defined by the number of inverted bases between Q1 and Q2, and between Q3 and Q4 (i.e. within the half-sites):
| 2 |
The second is the H-coupling between the two half-sites, to measure the number of inverted bases between the two half-sites:
| 3 |
The third element, T-coupling, describes how many bases are the same in respective positions in the two half-sites:
| 4 |
Figure 1.
Definition of quarter-site coupling (A) and five corresponding binding modes. (B) Q1234 is the mode of fully occupied quarter-sites with half-site palindrome. (C) T13 mode. (D) T24 mode. (E) The H14 mode uses quarter-sites 1 and 4 to tightly bind p53, and (F) H23 where quarter-sites 2 and 3 are fully occupied. Starting models assembled from existing crystal structures for four binding modes (Q1234, T13, T24 and H14) are illustrated. The initial structures are subjected to refinements using molecular dynamics simulations.
For each potential p53RE sequence in the human genome (25), the numbers of bases involved in the Q-coupling, H-coupling and T-coupling are calculated according to the definitions in Equations (2–4), respectively. The overall coupling counts in the dataset are the summation of the individual counts of coupled bases in each position in each p53RE.
The fraction of the bases involved in specific coupling is calculated as follows:
| 5 |
where Fq(i, j) is the fraction of Q-coupled base i at position j; Nq(i, j) is the Number of Q-coupled base i in position j; and N(j) is the overall number of bases in position j in the dataset (total number of sequences). Similarly, the fractions of H-coupled base Fh(i, j) and T-coupled base Ft(i, j) are calculated. For a given sequence, the couplings of two quarter-sites are evaluated by adding the fractions of the coupling of the bases in all positions within the coupled quarter-sites (Figure 1A). The weight of Q1234 is scaled to be comparable with the other four modes where only two quarter sites are evaluated. Finally, the probabilities or the populations of a quarter-site coupling mode are normalized as:
| 6 |
RESULTS
Arrangements of p53 tetramer according to the symmetries encoded in p53 response elements
Figure 1 shows the internal symmetries in the p53RE (5) and possible ways of p53 tetramer–DNA interactions. We used the term Q1234 (Quarter-sites 1, 2, 3, 4) to describe the half-site palindrome mode, where each p53 monomer binds p53RE in the same mode (Figure 1B). p53 may bind DNA at quarter-sites one and three, while allowing variation in quarter-sites two and four (Figure 1C). This mode satisfies the translational symmetry, and we called it T13. Similarly, T24 (Figure 1D) refers to p53 binding with quarter-sites two and four. There are two possibilities to meet the requirement of the full-site palindrome; one is the H14 mode in which quarter-sites one and four tightly bind p53 (Figure 1E); another is H23 where quarter-sites two and three are fully occupied (Figure 1F).
p53 core domains bind the isolated DNA half-site as a symmetric dimer in the dimer-DNA crystal structure (9,10). The construction of the Q1234 mode is straightforward if we assume that all four quarter-sites have the same binding mode with p53, as observed in all three available experimental p53–DNA structures (8–10). In order to examine the Q1234 binding mode, two models were built with slight variations.
Other possible binding modes can be constructed using the crystal structures of the trimeric p53–DNA structure (8). For the T13 and T24 modes, the three p53 chains in the trimer–p53 structure were used. Putting the fourth p53 unit into the T13 and T24 modes required a slight adjustment of the dimer–dimer interface, which was performed without difficulty (Figures 1C and 1D, T13 and T24).
While it was suspected that the AB dimer in the trimeric p53–DNA structure (8) may be caused by crystal packing, it is also possible that the AB dimer has biological significance (11,26,27). Here we make use of this structure in building possible binding modes, to see how its assemblies can fit different sequence and geometry patterns to allow one quarter-site to interact with p53 specifically (chain B) and the second to have a lesser contact (chain A).
It is straightforward to build the H14 mode with quarter-sites one and four specifically binding p53. The starting structure for the H14 mode has no dimer–dimer interaction for linear DNA (Figure 1E). The H14 mode was subjected to refinement using molecular dynamics simulations leading to dimer–dimer interaction.
The H23 mode (Figure 1F) does not appear compatible with known p53–DNA interactions, and it is not possible to allow AB dimeric interactions and H23 binding (Supplementary Figure 1). Monomer A in one AB dimer (in quarter-site two) would completely overlap the position of monomer B in the second AB dimer (in quarter-site three). Possible DNA bending does not relieve the conflict. Given the difficulty of constructing an H23 mode with known p53–DNA interactions, we excluded the H23 binding mode for the p53REs without base pair insertions. Therefore, we considered four possible binding modes (Q1234, T13, T24 and H14) for p53REs without spacers. Excluding AB dimers from binding in the H23 mode appears consistent with the features that correlations between quarter-sites two and three are consistently suppressed in p53REs. This may also explain the correlation of the H23 coupling with the binding affinity of p53 tetramer to its response elements: the lower the H23 coupling is, the stronger the p53 binding (5).
Refinement of binding models for sequence-specific p53–DNA interactions with molecular dynamics simulations
We constructed four binding models (Q1234, T13, T24 and H14) for each of five natural p53REs (p21-5′, GADD45, pDINP1, p53AIP1 and Puma BS2, Table 1), which cover a range of half-site and full-site palindrome properties. p21-5′ and GADD45 have more inverted bps (A with T, G with C) in half-sites than in full-site; pDINP1 has the same number of inverted basepairs in the half-sites than in the full-site; and p53AIP1 and Puma-BS2 have more inverted basepairs in the full-site than in the half-sites. There is no spacer between the half-sites. Each binding model was refined by 14 ns molecular dynamics simulations with explicit water. Three key structures (two Q1234 models and one H14 model) were subjected to longer simulation times from 35 to 40 ns.
Table 1.
Five p53REs and possible p53 binding modes examined by sequence analysis, molecular dynamics simulations, and molecular free energy evaluations (GBMV)
| Gene | Sequence | Q1234 | T13 | T24 | H14 | |
|---|---|---|---|---|---|---|
| P21-5′ | CTATGAG GAACATGTCC CAACATGTTG AGCTC | Probability (sequence) % | 21.2 | 25.4 | 29.4 | 24.0 |
| Probability (GBMV) % | 24.9 (25.4) | 25.3 (25.5) | 22.6 (23.1) | 27.2 (26.0) | ||
| Gadd45 | TGTACA GAACATGTCT AAGCATGCTG GGGTC | Probability (sequence) % | 21.5 | 25.7 | 23.8 | 29.0 |
| Probability (GBMV) % | 24.5 (26.2) | 28.0 (25.3) | 19.4 (19.6) | 27.8 (28.9) | ||
| pDINP1 | TTTATA GAACTTGGGG GAACATGTTT ACCAA | Probability (sequence) % | 19.6 | 30.2 | 20.8 | 29.4 |
| Probability (GBMV) % | 23.4 (23.4) | 28.7 (28.5) | 21.5 (20.5) | 27.5 (27.6) | ||
| P53Aip1 | TCCTCC TCTCTTGCCC GGGCTTGTCG AGATG | Probability (sequence) % | 19.9 | 21.5 | 32.7 | 26.0 |
| Probability (GBMV) % | 19.7 (18.9) | 24.3 (24.0) | 27.1 (28.1) | 28.9 (29.1) | ||
| Puma-BS2 | CGCGC CTGCAAGTCC TGACTTGTCC GCGGCA | Probability (sequence) % | 16.6 | 25.8 | 18.3 | 39.3 |
| Probability (GBMV) % | 23.1 (24.2) | 27.1 (27.0) | 20.3 (19.2) | 29.5 (29.5) |
The probabilities (sequence) were evaluated based on quarter-site couplings, and the probabilities (GBMV) were evaluated using 2000 molecular dynamics-refined structures. The numbers in parenthesis were evaluated using 1200 molecular dynamics-refined structures.
Traditional half-site palindrome Q1234 mode
Two symmetric Q1234-mode structures were tested and refined with slightly different dimer–dimer interfaces. Our simulations indicated that half-site palindrome tetramers Q1234 were a possible binding mode for linear DNA binding. It was proposed that crowding of the p53 tetramer within p53RE pushes the DNA to bend (28,29). But in current simulations of nine Q1234 models [two from each p53RE for p21-5′, GADD45, pDINP1 and p53AIP1, and one from Puma-BS2, (Supplementary Figure 2)], we observed linear-like DNA conformations, rather than p53 sitting outside of curved DNA as previously proposed (28,29). Earlier 10 ns MD simulations using duplicated half-sites of two non-native p53REs illustrated a bending of the p53REs (29). However, such bending in the Q1234 mode may be DNA sequence-dependent or due to special simulation setups.
Overall, large-scale DNA bending does not appear in the current simulations of five native p53REs in the Q1234 model. In the prolonged 42 ns simulation of the puma BS2 complex in Q1234 mode, we still can not observe the DNA bending induced by the p53 tetramer.
To further check the Q1234 model, we extended DNA sequences to consider a p53 response element with insertion of a 2 bp spacer between two half-sites. The crystal packing of a p53 dimer–DNA 12mer complex (9) may represent the best opportunity to examine p53 tetramer binding to DNA in the Q1234 mode. Using the crystal dimer-of dimer structure (PDB code 2ahi), we linked two DNA fragments to create a continuous DNA sequence, representing a p53 response element with insertion of a 2 bp spacer (CGGACATGTCCGCGGACATGTCC). During MD simulation, comparing with starting crystal structures, the RMSD of the p53–DNA complex increases quickly to about 3 Å around 5 ns, and stabilized around 4 Å from 14 ns to 35 ns of the simulation and the DNA is essentially linear. (Supplementary Figure 3).
Direct repeats T13 and T24 modes
After 14 ns simulations, there is no consistent trend in the direction of DNA bending in the T13 and T24 modes (Supplementary Figure 4). The p21-5′ DNA bends (10°) with p53 tetramer sitting the outside of DNA loop. The GADD45 DNA in T13 mode shows significant bending (16°) in the flanking region with the p53 tetramer sitting outside of the DNA loop. But for the pDINP1 DNA in T13 mode, we observed that the p53 tetramer sitting inside the DNA bending loop, which again occurred in the flanking region. The DNA conformations for all other T13 and T24 modes are almost linear.
Full-site palindrome H14 mode causes linear DNA to bend
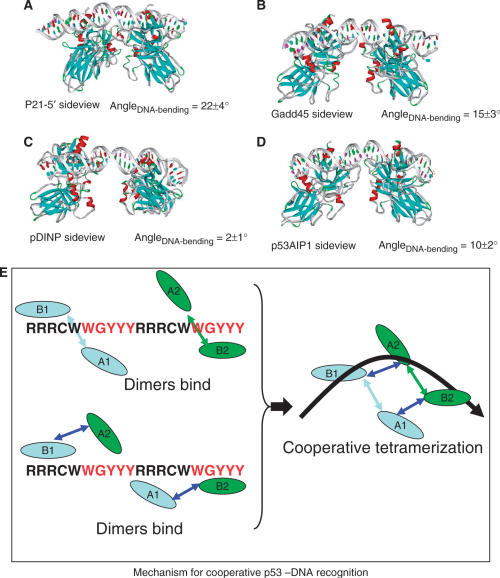
DNA bending accompanying cooperative p53 core domain tetramerization consistently appears in the simulations of full-site palindrome H14 models based on the asymmetric AB dimer from the crystal p53–trimer DNA complex (8) (Figure 3). Here, starting from a linear DNA conformation (Figure 1E), the DNA bends in the same direction during the simulations of four p53REs (p21-5′, GADD45 and p53AIP1, Figure 2) in complex with two separated p53 dimers, and the bending is significant (10° to 23°). pDINP DNA bends less (2°). The DNA surrounds the p53 core domain tetramer. The conformational evolution of the full-site palindromic H14 model during the MD simulations suggests cooperative p53–DNA binding and DNA conformational change. As shown in Figure 2 box E, even though the p53 dimers initially bind at quarter-sites one and four separately, the driving force to form stable protein–protein interface pulls the two dimers closer. The dimer–dimer association, driven by the electrostatic interactions of four pairs of salt-bridges of E180–R181, completes the core domain tetramerization and DNA bending simultaneously. Such a mechanism explains why the DNA sequence in two distant quarter-sites one and four are coupled, a phenomenon dominated by p53REs without spacer.
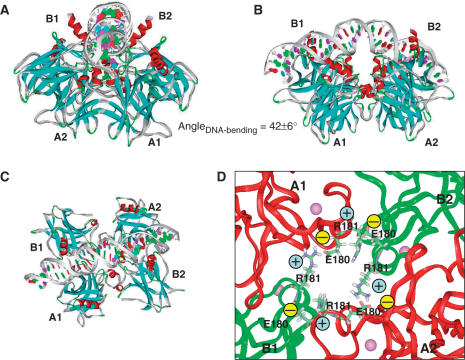
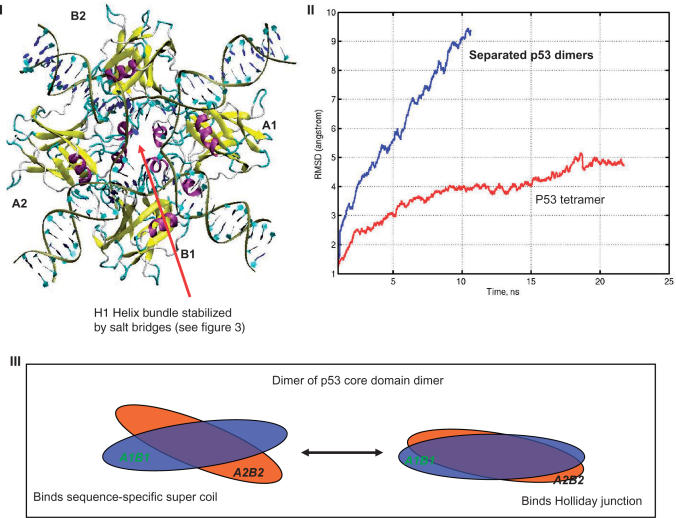
Figure 3.
Structural details of the potential H14 binding mode, in which two swapped copies of p53 dimer bind DNA symmetrically in respect to full-site palindrome. p53-tetramer–Puma BS2 complex is used here. Chains binding DNA specifically at quarter-sites one and four are the B chains in the p53-trimer–DNA complex. B1 and B2 are used to term their position in two dimers, respectively. The A chains assist in the DNA recognition and provide tetramer stability. A1 and A2 are used to name the chain position. (A) A view along the DNA chain. (B) A view perpendicular to the DNA chain, in which the p53 tetramer is inside the DNA loop. (C) A view from the top of the complex illustrating the arrangement of the core domain tetramer. (D) Atomic details of the salt bridges stabilizing the p53 tetramer.
Figure 2.
Change of linearity of the DNA double helix during molecular dynamics simulations of the tetramer bound to the DNA in the H14 mode with the p53 interacting with four p53REs (A), p21-5′ binding site; (B) GADD45; (C) pDINP1; and (D) p53AIP1. A potential mechanism in these simulations for cooperative p53–DNA interaction and DNA bending is illustrated in box E.
Refined H14 binding mode to supercoil DNA
To further examine the H14 binding and p53 tetramer's preference to the DNA supercoil, we reconstructed an H14 mode with DNA in the supercoil structure. We used a DNA fragment in a nucleosome as template and mutated the sequences to the five p53REs to be examined (p21-5′, GADD45, pDINP1, p53AIP1 and PUMA-BS2). Using the crystal structure of AB dimer, we manually docked two p53 dimers into the DNA supercoil. We found that a compacted p53 tetramer (with swapped dimer of AB dimmer) fits perfectly to the DNA supercoil structure and satisfies the sequence-specific DNA recognition (Figure 3). Five additional MD simulations of the H14 mode were carried out for the five p53 complexes in the compact H14 mode to refine the p53 tetramer–DNA complex. The simulation time for the p21-5′, GADD45, pDINP1 and p53AIP1 complexes is 14 ns, and that for PUMA–BS2 complex lasts 42 ns. The simulation results provided a possible structural rationale for p53–DNA recognition (Figure 3 and Supplementary Figure 5).
The refined H14 model keeps the essential features observed in the p53 trimer–DNA complex and satisfies chemical and structural complementarities for the protein–protein and protein–DNA recognition, exactly as observed in the p53–DNA crystal structures (8–10). Figure 3 indicates how the p53 tetramer recognizes DNA supercoil geometry in three views. For the protein–protein interactions, the p53 core domain tetramer arranges as swapped dimer of AB dimer (8). The newly formed dimer–dimer interface is similar to that of the monomer–monomer interface in the asymmetric AB dimer from the crystal p53–trimer DNA complex (8). Therefore, by utilizing one protein–protein interaction interface, p53 proteins form dimer and then a dimer of dimer, helping with DNA binding.
In the center of the tetramer complex, the H1 helix from each monomer may form a helix bundle (Supplementary Figure 5I), which is stabilized by salt bridges among the four monomers [R181(B1)-E180(A1), R181(A1)-E180(B2), R181(B2)-E180(A2), R181(A2)-E180(B1); Figure 3D]. The E180–R181 salt bridge has been shown to be important for p53 DNA interaction and function (30). Note that in both crystal structures of p53 dimer–DNA complexes (9,10), the salt bridges between E180 and R181 are not obvious. In the Ho's structure (10), two R181 side chains are closer than the distance between E180 and R181. In the Kitayner's structure (9), two R181 side chains are even closer and the salt bridges are intramolecular rather than between two p53 monomers. Thus, the experimentally observed E180-R181 salt bridge (30) occurred only in the p53 trimer–DNA complex(8), which we used in the H14 model. The circular salt bridges in the H14 mode explain the key role of the E180 and R181 interaction to stabilize the p53 tetramer and hence to make p53 functional.
In addition to the salt bridges, there are several interactions involving most residues observed to have large changes in NMR chemical shifts (31) upon p53 tetramerization, including Glu171, Val172, Arg174, Asp184, Gln192 and Phe212. Gln192 forms hydrogen bonds with the side chain of ser183 and the backbone of Arg181 (Supplementary Figure 5II). Several other interactions (with His178, Gly244, Glu171 and Arg174) formed in the helix bundle and can reinforce the salt-bridge interactions. His178 hydrogen bonds with the Gly244 backbone (Supplementary Figure 5III) and with the Glu171 side chain (Supplementary Figure 5IV). There is a hydrogen bond between the backbone of Gly244 and Arg174 (Supplementary Figure 5IV). Val172 is between monomers A and B (Supplementary Figure 5V), as in the p53 trimer–DNA crystal structure (8). Another contact in the p53 trimer–DNA complex is the Asp184 and Phe212 (data not shown), where the side-chain of Phe212 sits right on the backbone of Asp184. The Asp184–Phe212 contact also exists in the mouse p53 trimer crystal structure (32). Previous simulations indicated that the Asp184–Phe212 contact plays an important role in stabilizing the monomer–monomer interaction in the AB dimer (11). The Gly244 backbone hydrogen bonds may explain the mutation of Gly244Ala, which weakens p53 binding to DNA 6–8-fold (31). Overall, the p53 core domain tetramer and its interactions with DNA supercoils are very stable during MD simulations.
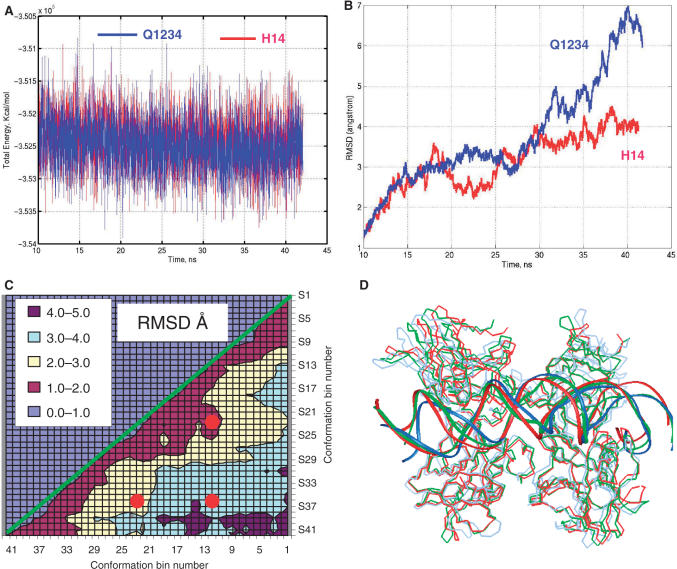
To further validate the novel H14 model, we extended the MD simulations to 42 ns for the PUMA BS2 complex in Q1234 and H14 binding modes. As can be seen in Figure 4A, the total energies during our initial simulations (14 ns) and extended simulations (up to 42 ns) are stable, and both Q1234 and H14 binding modes are similar. In terms of structural fluctuation, we found that the H14 binding mode has a smaller RMSD (root-mean-square-derivations) than the Q1234 mode (Figure 4B), indicating that the p53 tetramer–DNA complex should be structurally more stable in H14 mode than in Q1234 mode for Puma BS2. To further check the structural stability of the H14 mode, we cross-examined the 2-dimensional RMSD map of the H14 binding mode trajectory (Figure 4C). For a flexible DNA molecule, a map of RMSD differences between each couple of MD snapshots may have ‘islands’ of low RMSD (33). In the case of the large complex we studied here, we found small RMSD area dominants (63% <3 Å, Figure 4C). We superimposed three structures (Figure 4D), averaged from 11–12 ns, 21–22 ns and 36–38 ns trajectory, respectively. Their RMSD relationships are indicated as red dots on the 2D-RMSD map in the low 1–2 Å, 2–3 Å and 3–4 Å regions (Figure 4C). Even with large RMSD differences, the conformations are very close (Figure 4D). For example, the RMSD between the conformers at 12 ns and 37 ns was 3.6 Å for overall structure; and it decreased to 2.9 Å if we only measure the core region around the salt-bridge stabilized helix bundle (residues 153 to 255). Therefore, for the H14 binding mode trajectory, the RMSD are mostly contributed by loop fluctuation in each monomer, rather than large change of quaternary structure.
Figure 4.
Trajectories of prolonged simulations of PUMA BS2 binds p53 in Q1234 and H14 modes. (A) Total energies of simulated systems. (B) RMSDs of backbone atoms of p53 tetramer in Q1234 and H14 modes, respective to the structures averaged from first 10 ns simulations. (C) Two-dimensional RMSD map for the H14 mode. 42 conformations are obtained by average conformations during 1 ns windows in the 42 ns trajectory. Then the RMSD between all pair of conformers are calculated. The red dots indicated the RMSD positions among three conformations (12 ns, 22 ns and 37 ns). (D) The superimposed structures for the three conformations are illustrated: 12 ns, redline; 22 ns greenline; 37 ns, blueline.
The results indicate that the MD-refined structures of the models can provide sequence-specific DNA recognition and allow DNA structural flexibility. In the Supplementary Data table, we report the atomic coordinates used in MD simulations for PUMA BS2 in the H14-binding mode. We further noticed that the compact p53 tetramer that recognizes the p53 supercoil in the H14 mode can also recognize the Holliday junction.
p53 tetramer–Holliday junction interactions: crossroad for sequence-specific and geometry-specific p53–DNA interactions
To investigate geometry-specific p53–DNA interactions, we model a p53–Holliday junction complex by fitting the p53 core domains to a Holliday junction from a Flpe-Holliday junction crystal structure (15). The symmetric nature of the Flpe-Holliday junction sequence simplifies the modeling of the interaction of the p53 with a different sequence (TAAGTTCCTATTCTTTTAAAAGAATAGGAACTTC). If we assume that each p53 core domain monomer interacts with one double-helical DNA stem of the Holliday junction, it appears impossible for the p53 core domain to bind the Holliday junction by a symmetrically positioned p53 dimer observed in p53 dimer–DNA crystal structure (9,10), where the p53 dimer binds one double-stranded DNA using both dimeric DNA-binding surfaces. The asymmetric AB dimer in the p53 trimer–DNA complex allows simultaneous binding of two DNA double strands with one p53 dimer, illustrating more biological significance of the AB dimmer.
We consider three possible AB-based arrangements. In the first (Supplementary Figure 6A), an isolated p53 core domain binds DNA at the major grove away from the junction. The second arrangement allows each p53 core domain dimer to interact with two DNA arms simultaneously, but there is no interaction between two p53 dimers (Supplementary Figure 6B). The third arrangement allows the tetramer of four p53 core domains to bind DNA in the center of the junction (Figure 5) and maintain the protein–protein interface of the p53-trimer DNA structure. In the third arrangement, the p53 core domains bind again as two swaped copies of the AB dimers. In the center of the tetramer complex, a helix bundle also can be formed from the H1 helix, stabilized by salt bridges. The protein–protein interfaces between the A1-B1 and A2-B2 are the same as in the crystal structure, while the new interfaces, A1-B2 and A2-B1, are also similar to the crystal form.
Figure 5.
Models of the p53–Holliday junction interactions. The model presents tetrameric arrangements similar to the DNA sequence-specific p53 core domain (Figures 3 and 4). (I) shows p53 core domain tetramer interacting with the Holliday junction; (II) trajectories of MD simulation of Holliday junctions binding with two separated p53 dimers (Supplementary Figure 5B) and p53 tetramer (Figure 5I). (III) Illustrates the p53 tetramer binding the DNA supercoil and the Holliday junction using the same arrangement.
Subsequent MD simulations revealed that neither four single p53 core domains nor the two core domain dimers stabilize the p53-Holliday junction complex. As shown in Figure 5II, the RMSD of the separated core domain dimer–DNA complex quickly rises to about 8 Å, mainly due to bending of DNA away from planer conformation. However, the core domain tetramer appears to provide a stable framework for p53 to bind the Holliday junction during the 20 ns structural refinement (Figure 5).
The geometric match between the Holliday junction topology and the p53 core domain tetramer (a dimer of AB dimer) is similar to the arrangement of the sequence-specific H14 binding mode described in earlier sections. The H14 binding mode appears to provide both sequence-specific and conformation-specific p53–DNA interactions. The twist angle between the two p53 dimers is close to planar in the p53 Holliday junction complex, while the dimers twist more in the sequence-specific p53–DNA binding (Figure 5III). In both arrangements, the H1 helix from each monomer contributes to a helix bundle in the center.
Recombination intermediates can have several conformations. The extended open-X form of the junction exists in all known protein–Holliday junction complexes, but the more compact stacked-X structures have been observed in free DNA (34). While our simulations have shown that the p53 can recognize the open-X form of the Holliday junction, it is interesting to ask if the p53 tetramer can bind the Holliday junction in the stacked-X conformation, as proposed in the model of the junction-resolving enzyme Hje (35). In principle, the rotation of the p53 dimer of dimer is flexible enough to fit the stacked-X Holliday junction, however with less dimer–dimer interactions. Thus, while the p53 dimer of dimer can interact with the Holliday junction at various intermediate states, the most stable form of the p53 tetramer–Holliday junction complex should be the open-X conformation, as observed in all other protein–Holliday junction complexes (34).
Probability of binding modes for sequence-specific p53–DNA interactions and effects of p53 mutation
Evaluation of the free energy landscape and the probability of binding modes
The quarter-site coupling probabilities (H14 and H23) obtained from statistics of potential p53REs in the human genome correlate with the p53–DNA-binding affinity (5). If we assume that the possibility of a specific binding mode is sequence-dependent and equal to its corresponding quarter-site coupling probability (i.e. Q1234, T13, T24, H14, H23 binding mode probability equals the Q1234, T13, T24, H14 and H23 quarter-site coupling probabilities, respectively), the binding mode probabilities (H14 and H23) should correlate with the measured p53–DNA-binding affinity. Since we have excluded the possibility of the H23 binding mode for the p53REs without insertion, we re-evaluate the binding mode probability based on the four binding modes (Q1234, T13, T24 and H14) for the p53REs without insertion and all five modes for the rest of the p53REs. The revised correlations are essentially the same as those reported in our sequence analysis work (5), now with the correlation between the H14 mode with experiments being R2=0.74. To further analyze the structural models of p53 tetramer–DNA complexes with respect to their binding probability, we examined the correlations of the binding probabilities obtained from the p53RE sequence analysis with the binding probabilities derived from our molecular simulations of the structural models (Figure 6A).
Figure 6.
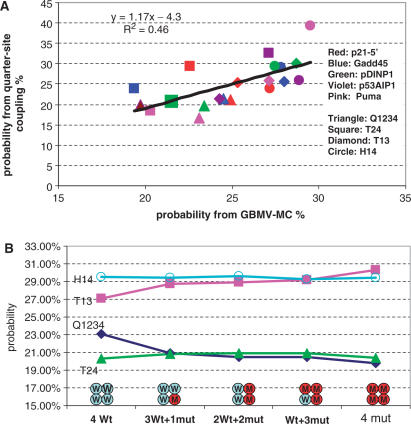
(A) The correlation of the binding mode probability derived from statistics of DNA sequences of p53REs (Y-axis) with the probability derived from molecular simulations (X-axis). The detailed values are provided in Table 1. The points are colored according to DNA sequences (Red: p21-5′, Blue: Gadd45, Green: pDINP1, Violet: p53AIP1, Pink: Puma BS2 and shaped according to binding modes (Triangle: Q1234, Square: T24, Diamond: T13, Circle: H14). (B). Change of binding mode probability with increasing of R273H mutant in p53-tetramer–Puma BS2 complex. p53 is able to have attractive interaction with DNA when two monomers in the tetramer are wild-type p53 (Table 2).
The relative probabilities of the models are evaluated using a MC approach based on the free energy landscape (Methods section). The energy landscape for Puma-BS2 is illustrated in Supplementary Figure 7. Table 1 reports the probabilities evaluated from the two (sequence and simulation) approaches. Overall, for the 20 binding modes (four for each of the five p53REs), the two methods correlated with R2 = 0.46. (Figure 6, Table 1), sufficiently to indicate that the quarter-site couplings may correspond with the binding models of p53 tetramer–DNA interaction. Five hundred conformations were used for each binding mode. We also checked the binding mode probability based on 300 conformations from 12 to 14 ns simulations; and results only differ by 0.7% and have the same correlation with sequence analysis.
Figure 7.
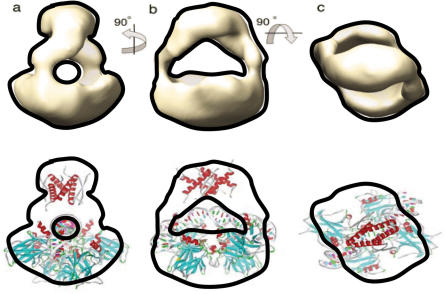
Upper panel: EM image of p53 tetramer in p53–DNA complex in solution, reference (48). (Reproduced with permission). The see-through channel in the EM map indicates the possible DNA position. Lower panel: fit of p53–DNA complex in H14 binding mode to the EM image in the upper panel. The tetramerization domain was added to the Puma BS2–p53 core domain tetramer in Figure 3.
Table 1 shows some discrepancies as to which model may be the best for a given p53RE. Both methods rank the half-site palindrome mode Q1234 as having minor contributions for the tested models. In the GBMV energy and MC population calculations, the H14-binding models are the best for p21-5′, p53AIP1 and PUMA-BS2. For GADD45 and pDINP1, the H14 model is very close to the T13 model.
Effects of R273H mutation: three mutant monomers are needed to inactivate a p53 tetramer
DNA binding-defective p53 mutants (R249S and R273H) were found to be ineffective in impairing the transcriptional activity of p53: at least three p53 mutant molecules are required to inactivate a tetramer (36). Here we tested the effects of R273H mutation on the probabilities of four binding modes for PUMA BS2 complex. R273 binds DNA backbone. The crystal structure of R273H indicates that the mutation only removes DNA contact without inducing p53 structural perturbations (37).
Starting from the minimized structures of wild-type p53 tetramer–DNA complex for PUMA BS2 (Section 4A), we mutated Arg273 to protonated histidine. The distribution of the binding mode probability changes with the numbers of mutant in the tetramer (Figure 6B). The H14 mode still dominates with introducing two mutants into the tetramer. With three monomers mutated, the T13 mode overlaps with H14 to become an equally possible binding mode. When all monomers in the p53 tetramer mutated, the T13 mode dominates. Both the Q1234 and the T24 mode have fewer possibilities, no matter how many monomers are mutated.
The probability analysis measures the ‘relative’ distribution of possible binding modes if p53 tetramer is able to bind DNA corresponding to p53RE symmetries in Q1234, T13, T24 and H24 modes. However, it still possible that p53 tetramer is not able to bind in certain modes due to DNA sequence variation or p53 mutation. Interaction energy calculation can provide additional metric to the binding mode possibility. Therefore, we also evaluated the interaction energies (Table 2) between the p53 tetramer and DNA by the following equation:
| 7 |
where E(p53 tetramer -DNA) is average energy for p53 tetramer–DNA complex evaluated with the GBMV method, and the E(DNA), E(p53-a1), E(p53-a2), E(p53-b1) and E(p53-b2) are the average energies for isolated DNA and p53 monomers, respectively. The conformations of the isolated DNA and p53 monomers have been re-sampled in their isolated states.
Table 2.
Change of p53-DNA interaction energies (kcal/mol) with R273H mutations in Puma BS2 complex
| Mutated Monomersa | H14 mode | T13 mode |
|---|---|---|
| Double mutants | ||
| a1b1 | 48 | 52 |
| a1a2 | −132 | −119 |
| a1b2 | 12 | 23 |
| b1a2 | 44 | 54 |
| b1b2 | 177 | 187 |
| a20b2 | 9 | 49 |
| Triple mutants | ||
| b1a2b2 | 204 | 226 |
| a1a2b2 | 46 | 75 |
| a1b1b2 | 233 | 232 |
| a1b1a2 | 72 | 91 |
aFour p53 monomers are labeled with b1 and b2 binding DNA sequence specifically and a1 and a2 binding DNA non-specifically. Positive interaction energy means repulsion and negative number means attraction. For T24 and Q1234 modes, the interaction energies are always repulsive.
The p53–DNA interaction energies for the two leading binding modes, H14 and T13, are reported in Table 2. In Table 2, four p53 monomers are labeled with the b1 and b2 binding the DNA sequence specifically, and a1 and a2 binding DNA non-specifically. As can be seen in Table 2, with two monomers in the p53 tetramer mutated (a1a2), the H14 and T13 modes still can use wild-type p53 to bind DNA sequence specifically. As a result, there is still an attraction between the p53 tetramer and DNA. However, when three monomers in the tetramer are mutated, neither combination can have attractions between p53 and DNA anymore. The loss of attractive interactions between p53 and DNA with three monomers mutated may explain the experimental finding that at least three p53 R273H mutant molecules are required to inactivate a tetramer (36).
DISCUSSION
Previously, we found that symmetries in p53 REs encode several possible binding modes (5). Nevertheless, the ways that p53 recognizes and accommodates DNA instructions and what happens in the cases of p53 mutations remain unknown. Here we move a step further to study the molecular mechanisms of various binding modes, focusing on native p53REs without insertion and considering DNA structures and p53 mutation. We were encouraged by the converged computational results, which are consistent with experimental findings and provide new insight into p53–DNA interactions.
H14 mode explains many experimental observations
The picture emerging from this work corroborates and provides a molecular mechanism of the highly preferred H14 mode found in DNA analysis for p53REs without spacer (5). (I) In the H14 mode, p53 may use only one type of protein–protein interface existing in the p53 trimer–DNA complex. (II) The core domain tetramerization in the H14 mode induces DNA bending. Subsequently, the H14 mode fits the DNA supercoil structure with sequence-specific recognition. (III) With adjustment of angles between two p53 dimers, the same p53 tetramer can recognize the p53RE sequence and the Holliday junction geometry. (IV) The highly stable salt-bridges in the H14 mode verify the experimental finding that the E180–R181 interaction is crucial for p53's function. (V) The specific interactions of p53 with only two quarter-sites (one and four) in the H14 mode are consistent, in that at least three p53 mutant molecules are required to inactivate a tetramer (36). Our computational mutagenesis study confirmed that the p53 tetramer can bind puma BS2 with two R273H mutant monomers and two wild-type p53 monomers, while three R273H mutant monomers disable p53 tetramer–DNA recognition.
In the H14 model, the p53 DNA-binding site surface is partially exposed in the DNA–p53 tetramer complex. Such a feature allows other proteins to regulate p53–DNA interaction. The binding site overlapping with the core domain DNA-binding site on the p53 protein was observed to be promiscuous (38) since it interacted with different proteins which regulate its function, for example, the 53BP1 protein (39). In another hand, the quarter-sites two and four are still possible to binding other parts (like C-terminal domain) of p53. For example, variation in the placement of the C-terminal domain can modulate the affinity to DNA by adopting two distinct binding modes: a high-affinity mode at the nanomolar range, and a low-affinity mode at the micromolar range (40).
Q1234 mode may be stabilized with spacer between half-sites
The specific binding modes and the details of the sequence-specific interactions may depend on the spacers between two half-sites (9), on gene-specific p53 functions (41) and may be affected by p53 mutations (42). The half-site palindrome dominates the p53REs with insertions of 3 bp spacers for the human genome and 8 bp spacers for the mouse genome (5). The corresponding binding mode Q1234 may be expected to bind p53REs with insertions of 3 bp spacers for the human genome and 8 bp spacers for the mouse genome. In fact, two available structures of p53 dimer–DNA half-site complexes may already corroborate with the role of insertion to stabilize the half-site palindrome mode. While one p53 dimer–DNA structure is helped by chemically cross-linking of p53 to DNA (10), another is stabilized by the p53 dimer–dimer interaction, which is made possible by insertions between two isolated DNA half-sites (9).
For the five native p53 REs without spacer studied here, we found that the Q1234 mode is a linear DNA-binding mode. The Q1234 binding probabilities are lower and are structurally less stable than other modes. The Q1234 mode may have better probabilities for the p53REs with highly symmetric half-sites. The p21-5′ site is one of the native p53RE with the highest half-site palindrome. Consistently, the Q1234 mode in the p21-5′ complex has the best energetic (Table 1). The half-sites in the two dimer DNA complexes(9,10) are 100% symmetric, while half-site in the p53-trimer DNA complex is less symmetric. Nevertheless, insertion of spacer increases the probability of the Q1234 mode greatly. In the future, it needs to systematically examine the effects of insertion of 3 bp and 8 bp spacer on the binding mode probability, which are beyond the scope of the current work.
Variation of binding modes depends on biological functions: T13 and T24 modes
P53 regulates hundreds genes with verities of biological functions. We have shown that the degree of the H14 coupling distinguishes transcription mechanisms of p53, with the H14 couplings being much stronger for positive regulation than for negatively regulated p53REs (5). Accordingly, p53REs in different functional group might have different binding modes. Inverse repeat p53REs may favor the H14 mode and direct repeat p53REs may have high possibilities of T13 and T24 modes. As may be seen in Table 1, both DNA sequence analysis and molecular simulations indicated that T13 is favorable for pDINP1. For p53Aip1, DNA sequence analysis indicated that the T24 is the most favorable binding mode, while molecular simulations shows the T24 is only slightly less favorable than the H14 mode.
Similarly to p53REs, T-box genes also have palindromic response elements with two 10 bp half-sites. In the T-box proteins, the binding modes of transcriptional factors may vary with the arrangement of the DNA-binding site and the insertion of spacers (43). Xenopus T-box protein Xbra, VegT and Eomesodermin bind two half-site T-box response elements with different symmetrical arrangements: The Xbra binds DNA sites with inverted palindrome without insertion; Vegt selects mirror palindrome sites with insertion of 4 bp spacer; and Emoes binds translational repeats with spacers of 3-, 4-, 5- and 12 bp (43). The Xbra binds DNA as a dimer to two half-sites (44) in a way similar to the H14 mode of p53–DNA interaction (Supplementary Figure 8).
Balance of p53-binding modes and genome stability
p53–DNA recognition necessitates flexibility and specificity (45). The possibility of multiple binding modes is consistent with biological experiments suggesting that p53–DNA binding can be modulated by factors that can alter the p53 conformational equilibrium (46). c-Abl can selectively stabilize the p53 tetramer conformation to enhance p53 binding and transcription for p21 but not Bax (47). mAb 421 differentially affects the binding of p53 to two p53REs, suggesting that optimal binding of p53 may require different conformations of the p53 tetramer (41).
The existing and balance of several p53-binding modes may be needed to maintain genome stability. The p53 mutation will shift the balance maintained by native p53. Even though both experiments and our simulations confirm that at least three R273H mutant molecules are needed to completely disable the p53 tetramer, the existence of merely one R273H mutant monomer greatly shifts the possibility of a binding mode (Figure 6B). The finding has many implications to understand the gain of function of p53 mutation and accumulations of p53 mutants in the cancer cell. Changing of binding modes balance might selectively disable certain pathways while maintaining some of wild-type functions, switch wild-type functions to different functions and activate response elements which are not wild-type p53REs.
Comparison with latest electron microscopy of p53–DNA complex
After submitting this work, the quaternary structure of p53–DNA complex in solution was solved by a combination of small-angle X-ray scattering (SAXS), NMR and electron microscopy (EM) (48). Even though the low resolution (25–30 Å) cannot provide atomic details, their experiments provided new information to compare with our computational predictions. Here we highlight three features described in the quaternary structure work (48). (i) Multiple binding modes have been observed. Two leading conformations were identified by the SAXS. Even though it was suggested that the two conformations differ only by the position of the tetramerization domain with respect to the core domains–DNA complex, it is very likely that variation of core domain–DNA-binding modes leads to the two conformations observed experimentally. (ii) EM images have shown that tetramerization domain connects core domains in the p53–DNA complex via only ‘two’ well-defined linkers which correspond to quarter sites one and four (upper panel, Figure 7). This EM feature strongly suggests that it may take only two monomers in p53 tetramer to tightly bind DNA, confirming H14 interaction pattern. (iii) p53 dimer–dimer interface has to be highly twisted to fit EM image. With large twist angle, p53 tetramer cannot use its four monomers to bind p53RE without insertion in the Q1234 mode. Instead, we found our p53–DNA supercoil complex in H14 mode fits EM image nicely (Figure 7). The EM experiments did not provide DNA position in the p53–DNA complex. Therefore, the see-through channel in the EM map imply the possible DNA position (48). As can be seen in the Figure 8, our predicted DNA position and highly bended shape match the see-through channel in the EM map. Combined all these features together, our computational results agree with the observed quaternary structure of p53–DNA complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Drs R. Nussinov, Y. Pan, J. Zheng, C. Tsai, K. Gunasekaran and B. Venkataraghavan for discussions. We thank Maritta Perry Grau for reading the manuscript. This research was supported (in part) by the U. S. Army Medical Research Acquisition Activity under grant W81XWH-05-1-0002. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO-12400. A.-J. Levine thanks grants from NCI and the Verto Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Funding to pay the Open Access publication charges for this article was provided by NCI and the Verto Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat. Genet. 1992;1:45–49. doi: 10.1038/ng0492-45. [DOI] [PubMed] [Google Scholar]
- 3.Lee S, Cavallo L, Griffith J. Human p53 binds Holliday junctions strongly and facilitates their cleavage. J. Biol. Chem. 1997;272:7532–7539. doi: 10.1074/jbc.272.11.7532. [DOI] [PubMed] [Google Scholar]
- 4.Janz C, Susse S, Wiesmuller L. p53 and recombination intermediates: role of tetramerization at DNA junctions in complex formation and exonucleolytic degradation. Oncogene. 2002;21:2130–2140. doi: 10.1038/sj.onc.1205292. [DOI] [PubMed] [Google Scholar]
- 5.Ma B, Pan Y, Zheng J, Levine AJ, Nussinov R. Sequence analysis of p53 response-elements suggests multiple binding modes of the p53 tetramer to DNA targets. Nucleic Acids Res. 2007;35:2986–3001. doi: 10.1093/nar/gkm192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cherny DI, Striker G, Subramaniam V, Jett SD, Palecek E, Jovin TM. DNA bending due to specific p53 and p53 core domain-DNA interactions visualized by electron microscopy. J. Mol. Biol. 1999;294:1015–1026. doi: 10.1006/jmbi.1999.3299. [DOI] [PubMed] [Google Scholar]
- 7.Palecek E, Brazdova M, Brazda V, Palecek J, Billova S, Subramaniam V, Jovin TM. Binding of p53 and its core domain to supercoiled DNA. Eur. J. Biochem. 2001;268:573–581. doi: 10.1046/j.1432-1327.2001.01898.x. [DOI] [PubMed] [Google Scholar]
- 8.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor–DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 9.Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Ho WC, Fitzgerald MX, Marmorstein R. Structure of the p53 core domain dimer bound to DNA. J. Biol. Chem. 2006;281:20494–20502. doi: 10.1074/jbc.M603634200. [DOI] [PubMed] [Google Scholar]
- 11.Ma B, Pan Y, Gunasekaran K, Venkataraghavan RB, Levine AJ, Nussinov R. Comparison of the protein-protein interfaces in the p53–DNA crystal structures: towards elucidation of the biological interface. Proc. Natl Acad. Sci. USA. 2005;102:3988–3993. doi: 10.1073/pnas.0500215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gohler T, Jager S, Warnecke G, Yasuda H, Kim E, Deppert W. Mutant p53 proteins bind DNA in a DNA structure-selective mode. Nucleic Acids Res. 2005;33:1087–1100. doi: 10.1093/nar/gki252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soussi T. p53 alterations in human cancer: more questions than answers. Oncogene. 2007;26:2145–2156. doi: 10.1038/sj.onc.1210280. [DOI] [PubMed] [Google Scholar]
- 14.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 15.Conway AB, Chen Y, Rice PA. Structural plasticity of the Flp-Holliday junction complex. J. Mol. Biol. 2003;326:425–434. doi: 10.1016/s0022-2836(02)01370-0. [DOI] [PubMed] [Google Scholar]
- 16.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 18.Darden T, York D, Pedersen L. Particle Mesh Ewald - an N.Log(N) Method for Ewald Sums in Large Systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 19.Maynard AT, Covell DG. Reactivity of zinc finger cores: analysis of protein packing and electrostatic screening. J. Am. Chem. Soc. 2001;123:1047–1058. doi: 10.1021/ja0011616. [DOI] [PubMed] [Google Scholar]
- 20.Wright JD, Noskov SY, Lim C. Factors governing loss and rescue of DNA binding upon single and double mutations in the p53 core domain. Nucleic Acids Res. 2002;30:1563–1574. doi: 10.1093/nar/30.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickerson RE. DNA bending: the prevalence of kinkiness and the virtues of normality. Nucleic Acids Res. 1998;26:1906–1926. doi: 10.1093/nar/26.8.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee MS, Salsbury FR, Brooks CL. Novel generalized Born methods. J. Chem. Phys. 2002;116:10606–10614. [Google Scholar]
- 23.Kormos BL, Benitex Y, Baranger AM, Beveridge DL. Affinity and specificity of protein U1A-RNA complex formation based on an additive component free energy model. J. Mol. Biol. 2007;371:1405–1419. doi: 10.1016/j.jmb.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunenberger PH, McCammon JA. Effect of artificial periodicity in simulations of biomolecules under Ewald boundary conditions: a continuum electrostatics study. Biophys. Chem. 1999;78:69–88. doi: 10.1016/s0301-4622(99)00007-1. [DOI] [PubMed] [Google Scholar]
- 25.Hoh J, Jin S, Parrado T, Edington J, Levine AJ, Ott J. The p53MH algorithm and its application in detecting p53-responsive genes. Proc. Natl Acad. Sci. USA. 2002;99:8467–8472. doi: 10.1073/pnas.132268899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantarci N, Doruker P, Haliloglu T. Cooperative fluctuations point to the dimerization interface of p53 core domain. Biophys. J. 2006;91:421–432. doi: 10.1529/biophysj.106.077800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 2006;20:2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagaich AK, Zhurkin VB, Durell SR, Jernigan RL, Appella E, Harrington RE. p53-induced DNA bending and twisting: p53 tetramer binds on the outer side of a DNA loop and increases DNA twisting. Proc. Natl Acad. Sci. USA. 1999;96:1875–1880. doi: 10.1073/pnas.96.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan Y, Nussinov R. Structural basis for p53 binding-induced DNA bending. J. Biol. Chem. 2007;282:691–699. doi: 10.1074/jbc.M605908200. [DOI] [PubMed] [Google Scholar]
- 30.Dehner A, Klein C, Hansen S, Muller L, Buchner J, Schwaiger M, Kessler H. Cooperative binding of p53 to DNA: regulation by protein–protein interactions through a double salt bridge. Angew. Chem. Int. Ed. Engl. 2005;44:5247–5251. doi: 10.1002/anie.200501887. [DOI] [PubMed] [Google Scholar]
- 31.Veprintsev DB, Freund SM, Andreeva A, Rutledge SE, Tidow H, Canadillas JM, Blair CM, Fersht AR. Core domain interactions in full-length p53 in solution. Proc. Natl Acad. Sci. USA. 2006;103:2115–2119. doi: 10.1073/pnas.0511130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao K, Chai X, Johnston K, Clements A, Marmorstein R. Crystal structure of the mouse p53 core DNA-binding domain at 2.7 Å resolution. J. Biol. Chem. 2001;276:12120–12127. doi: 10.1074/jbc.M011644200. [DOI] [PubMed] [Google Scholar]
- 33.Beveridge DL, Ravishanker G. Molecular dynamics studies of DNA. Curr. Opin. Struct. Biol. 1994;4:246–255. [Google Scholar]
- 34.Khuu PA, Voth AR, Hays FA, Ho PS. The stacked-X DNA Holliday junction and protein recognition. J. Mol. Recognit. 2006;19:234–242. doi: 10.1002/jmr.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Middleton CL, Parker JL, Richard DJ, White MF, Bond CS. Substrate recognition and catalysis by the Holliday junction resolving enzyme Hje. Nucleic Acids Res. 2004;32:5442–5451. doi: 10.1093/nar/gkh869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan WM, Siu WY, Lau A, Poon RY. How many mutant p53 molecules are needed to inactivate a tetramer? Mol. Cell. Biol. 2004;24:3536–3551. doi: 10.1128/MCB.24.8.3536-3551.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J. Biol. Chem. 2005;280:16030–16037. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- 38.Friedler A, Veprintsev DB, Rutherford T, von Glos KI, Fersht AR. Binding of Rad51 and other peptide sequences to a promiscuous, highly electrostatic, binding site in p53. J. Biol. Chem. 2005;280:8051–8059. doi: 10.1074/jbc.M411176200. [DOI] [PubMed] [Google Scholar]
- 39.Glover JN, Williams RS, Lee MS. Interactions between BRCT repeats and phosphoproteins: tangled up in two. Trends Biochem. Sci. 2004;29:579–585. doi: 10.1016/j.tibs.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 40.Shakked Z, Yavnilovitch M, Kalb Gilboa AJ, Kessler N, Wolkowicz R, Rotter V, Haran TE. DNA binding and 3′-5′ exonuclease activity in the murine alternatively-spliced p53 protein. Oncogene. 2002;21:5117–5126. doi: 10.1038/sj.onc.1205667. [DOI] [PubMed] [Google Scholar]
- 41.Resnick-Silverman L, Manfredi JJ. Gene-specific mechanisms of p53 transcriptional control and prospects for cancer therapy. J. Cell Biochem. 2006;99:679–689. doi: 10.1002/jcb.20925. [DOI] [PubMed] [Google Scholar]
- 42.Menendez D, Inga A, Resnick MA. The biological impact of the human master regulator p53 can be altered by mutations that change the spectrum and expression of its target genes. Mol. Cell. Biol. 2006;26:2297–2308. doi: 10.1128/MCB.26.6.2297-2308.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conlon FL, Fairclough L, Price BM, Casey ES, Smith JC. Determinants of T box protein specificity. Development. 2001;128:3749–3758. doi: 10.1242/dev.128.19.3749. [DOI] [PubMed] [Google Scholar]
- 44.Muller CW, Herrmann BG. Crystallographic structure of the T domain–DNA complex of the Brachyury transcription factor. Nature. 1997;389:884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- 45.Kim E, Deppert W. The versatile interactions of p53 with DNA: when flexibility serves specificity. Cell Death Differ. 2006;13:885–889. doi: 10.1038/sj.cdd.4401909. [DOI] [PubMed] [Google Scholar]
- 46.McLure KG, Lee PW. p53 DNA binding can be modulated by factors that alter the conformational equilibrium. EMBO J. 1999;18:763–770. doi: 10.1093/emboj/18.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinberg RL, Veprintsev DB, Bycroft M, Fersht AR. Comparative binding of p53 to its promoter and DNA recognition elements. J. Mol. Biol. 2005;348:589–596. doi: 10.1016/j.jmb.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 48.Tidow H, Melero R, Mylonas E, Freund SM, Grossmann JG, Carazo JM, Svergun DI, Valle M, Fersht AR. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc. Natl Acad. Sci. USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.