Abstract
Escherichia coli alkA mutants, which are deficient for an inducible DNA glycosylase, 3-methyladenine-DNA glycosylase II, are sensitive to mutagenesis by low doses of the alkylating agent N-methyl-N'-nitro-N-nitrosoguanidine (MNNG). As many as 90% of the alkA-dependent mutations induced by MNNG are also umuC+ dependent and thus are due to DNA lesions that are substrates for the mutagenic functions of the SOS response. A great number of these mutations are base substitutions at A . T sites, particularly A . T transversions. We discuss which DNA lesions may be responsible for these mutations. Our results show that the induction of 3-methyladenine-DNA glycosylase II, which occurs as part of the adaptive response to alkylating agents such as MNNG, significantly reduces the mutagenicity as well as the lethality of alkylation damage.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALLEN M. K., YANOFSKY C. A BIOCHEMICAL AND GENETIC STUDY OF REVERSION WITH THE A-GENE A-PROTEIN SYSTEM OF ESCHERICHIA COLI TRYPTOPHAN SYNTHETASE. Genetics. 1963 Aug;48:1065–1083. doi: 10.1093/genetics/48.8.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev. 1972 Dec;36(4):525–557. doi: 10.1128/br.36.4.525-557.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagg A., Kenyon C. J., Walker G. C. Inducibility of a gene product required for UV and chemical mutagenesis in Escherichia coli. Proc Natl Acad Sci U S A. 1981 Sep;78(9):5749–5753. doi: 10.1073/pnas.78.9.5749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes W. M. Construction of an M13 histidine-transducing phage: a single-stranded cloning vehicle with one EcoRI site. Gene. 1979 Feb;5(2):127–139. doi: 10.1016/0378-1119(79)90098-2. [DOI] [PubMed] [Google Scholar]
- Barnes W. M., Tuley E. DNA sequence changes of mutations in the histidine operon control region that decrease attenuation. J Mol Biol. 1983 Apr 15;165(3):443–459. doi: 10.1016/s0022-2836(83)80212-5. [DOI] [PubMed] [Google Scholar]
- Benzer S. ON THE TOPOGRAPHY OF THE GENETIC FINE STRUCTURE. Proc Natl Acad Sci U S A. 1961 Mar;47(3):403–415. doi: 10.1073/pnas.47.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek D. T., Weis C. C., Swenson D. H. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis. 1980 Jul;1(7):595–606. doi: 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- Boiteux S., Huisman O., Laval J. 3-Methyladenine residues in DNA induce the SOS function sfiA in Escherichia coli. EMBO J. 1984 Nov;3(11):2569–2573. doi: 10.1002/j.1460-2075.1984.tb02175.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetsanga C. J., Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979 Aug 10;6(11):3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J Mol Biol. 1977 Dec 15;117(3):525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H. Genetic studies of the lac repressor. IV. Mutagenic specificity in the lacI gene of Escherichia coli. J Mol Biol. 1977 Dec 15;117(3):577–606. doi: 10.1016/0022-2836(77)90059-6. [DOI] [PubMed] [Google Scholar]
- Drapeau G. R., Brammar W. J., Yanofsky C. Amino acid replacements of the glutamic acid residue at position 48 in the tryptophan synthetase A protein of Escherichia coli. J Mol Biol. 1968 Jul 28;35(2):357–367. doi: 10.1016/s0022-2836(68)80030-0. [DOI] [PubMed] [Google Scholar]
- Elledge S. J., Walker G. C. Proteins required for ultraviolet light and chemical mutagenesis. Identification of the products of the umuC locus of Escherichia coli. J Mol Biol. 1983 Feb 25;164(2):175–192. doi: 10.1016/0022-2836(83)90074-8. [DOI] [PubMed] [Google Scholar]
- Evensen G., Seeberg E. Adaptation to alkylation resistance involves the induction of a DNA glycosylase. Nature. 1982 Apr 22;296(5859):773–775. doi: 10.1038/296773a0. [DOI] [PubMed] [Google Scholar]
- Foster P. L., Eisenstadt E., Cairns J. Random components in mutagenesis. Nature. 1982 Sep 23;299(5881):365–367. doi: 10.1038/299365a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler R. G., Degnen G. E., Cox E. C. Mutational specificity of a conditional Escherichia coli mutator, mutD5. Mol Gen Genet. 1974;133(3):179–191. doi: 10.1007/BF00267667. [DOI] [PubMed] [Google Scholar]
- Fowler R. G., McGinty L., Mortelmans K. E. Mutational specificity of ultraviolet light in Escherichia coli with and without the R plasmid pKM101. Genetics. 1981 Sep;99(1):25–40. doi: 10.1093/genetics/99.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe I., Johnston H. M., Biek D., Roth J. R. A refined map of the hisG gene of Salmonella typhimurium. Genetics. 1979 May;92(1):17–26. doi: 10.1093/genetics/92.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson L., Rossi J., Landy A. Dual function transcripts specifying tRNA and mRNA. Nature. 1981 Dec 3;294(5840):422–427. doi: 10.1038/294422a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P., Defais M., Samson L., Schendel P. The adaptive response of E.coli to low levels of alkylating agent: the role of polA in killing adaptation. Mol Gen Genet. 1978 Jul 4;162(3):299–305. doi: 10.1007/BF00268855. [DOI] [PubMed] [Google Scholar]
- Jeggo P. Isolation and characterization of Escherichia coli K-12 mutants unable to induce the adaptive response to simple alkylating agents. J Bacteriol. 1979 Sep;139(3):783–791. doi: 10.1128/jb.139.3.783-791.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Hjelmgren T., Lindahl T. Induction of a DNA glycosylase for N-methylated purines is part of the adaptive response to alkylating agents. Nature. 1982 Apr 22;296(5859):770–773. doi: 10.1038/296770a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Griffin B. Adaptive response to alkylating agents involves alteration in situ of O6-methylguanine residues in DNA. Nature. 1979 Jul 5;280(5717):76–77. doi: 10.1038/280076a0. [DOI] [PubMed] [Google Scholar]
- Karran P., Lindahl T., Ofsteng I., Evensen G. B., Seeberg E. Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase. J Mol Biol. 1980 Jun 15;140(1):101–127. doi: 10.1016/0022-2836(80)90358-7. [DOI] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980 May;77(5):2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Ichikawa H., Iwo K., Kato T. Base-change mutagenesis and prophage induction in strains of Escherichia coli with different DNA repair capacities. Genetics. 1970 Oct;66(2):187–217. doi: 10.1093/genetics/66.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Mutational specificity of depurination. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1494–1498. doi: 10.1073/pnas.81.5.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Hollstein M., Christman M. F., Schwiers E. A., Ames B. N. A new Salmonella tester strain (TA102) with A X T base pairs at the site of mutation detects oxidative mutagens. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7445–7449. doi: 10.1073/pnas.79.23.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., Marnett L. J., Ames B. N. Spontaneous and mutagen-induced deletions: mechanistic studies in Salmonella tester strain TA102. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4457–4461. doi: 10.1073/pnas.81.14.4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
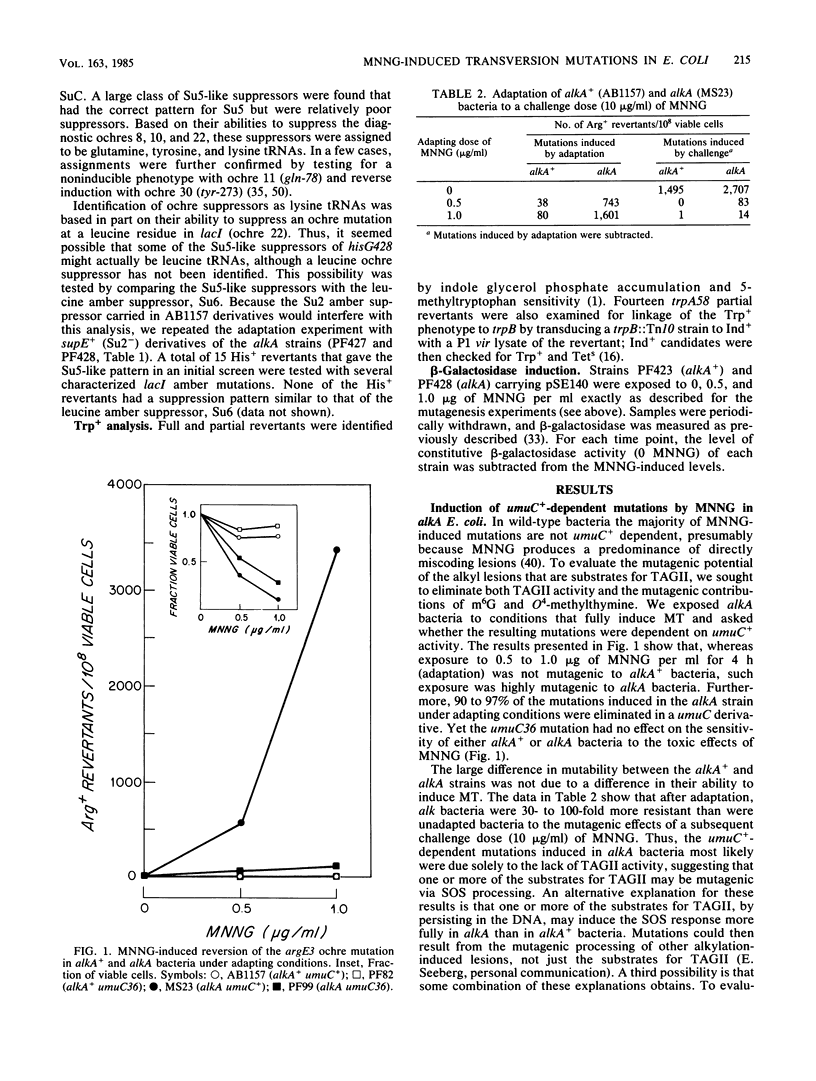
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- McCarthy T. V., Karran P., Lindahl T. Inducible repair of O-alkylated DNA pyrimidines in Escherichia coli. EMBO J. 1984 Mar;3(3):545–550. doi: 10.1002/j.1460-2075.1984.tb01844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H. Carcinogens induce targeted mutations in Escherichia coli. Cell. 1982 Nov;31(1):5–7. doi: 10.1016/0092-8674(82)90398-1. [DOI] [PubMed] [Google Scholar]
- Miller J. H., Coulondre C., Hofer M., Schmeissner U., Sommer H., Schmitz A., Lu P. Genetic studies of the lac repressor. IX. Generation of altered proteins by the suppression of nonsence mutations. J Mol Biol. 1979 Jun 25;131(2):191–222. doi: 10.1016/0022-2836(79)90073-1. [DOI] [PubMed] [Google Scholar]
- Riazuddin S., Lindahl T. Properties of 3-methyladenine-DNA glycosylase from Escherichia coli. Biochemistry. 1978 May 30;17(11):2110–2118. doi: 10.1021/bi00604a014. [DOI] [PubMed] [Google Scholar]
- Samson L., Cairns J. A new pathway for DNA repair in Escherichia coli. Nature. 1977 May 19;267(5608):281–283. doi: 10.1038/267281a0. [DOI] [PubMed] [Google Scholar]
- Schaaper R. M., Kunkel T. A., Loeb L. A. Infidelity of DNA synthesis associated with bypass of apurinic sites. Proc Natl Acad Sci U S A. 1983 Jan;80(2):487–491. doi: 10.1073/pnas.80.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schendel P. F., Defais M. The role of umuC gene product in mutagenesis by simple alkylating agents. Mol Gen Genet. 1980;177(4):661–665. doi: 10.1007/BF00272677. [DOI] [PubMed] [Google Scholar]
- Schendel P. F., Robins P. E. Repair of O6-methylguanine in adapted Escherichia coli. Proc Natl Acad Sci U S A. 1978 Dec;75(12):6017–6020. doi: 10.1073/pnas.75.12.6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
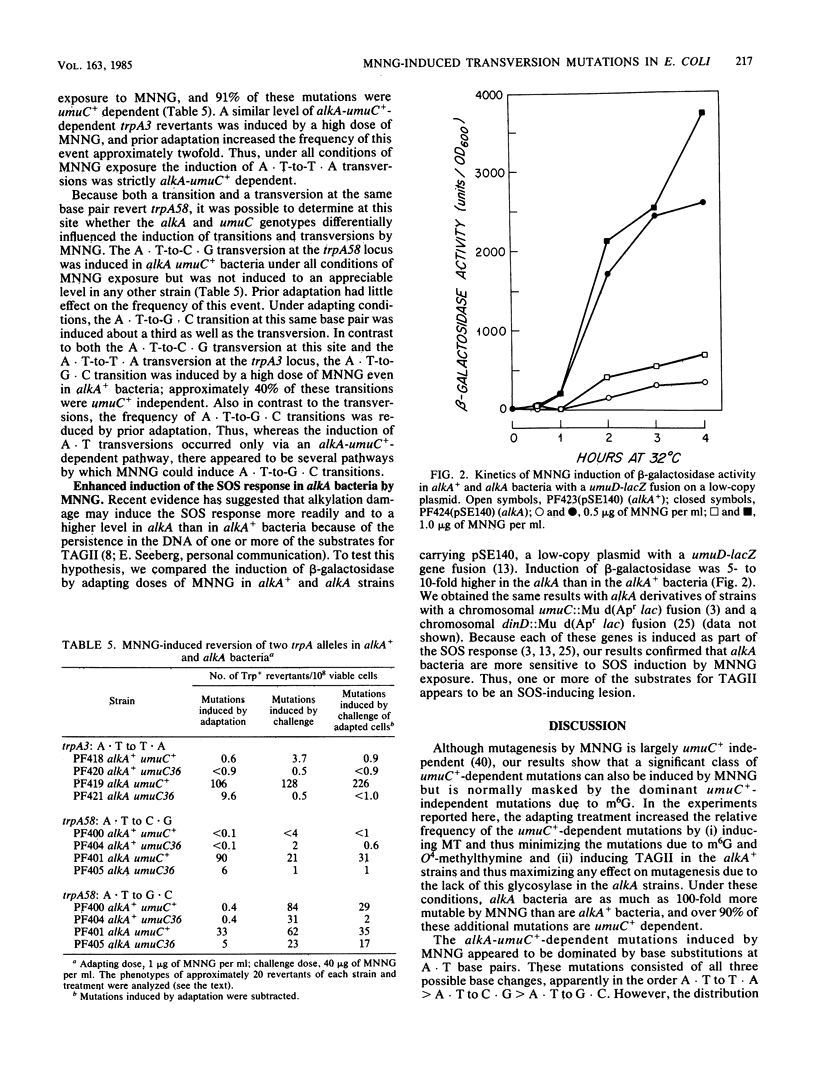
- Sedgwick B. Molecular cloning of a gene which regulates the adaptive response to alkylating agents in Escherichia coli. Mol Gen Genet. 1983;191(3):466–472. doi: 10.1007/BF00425764. [DOI] [PubMed] [Google Scholar]
- Singer B., Sági J., Kuśmierek J. T. Escherichia coli polymerase I can use O2-methyldeoxythymidine or O4-methyldeoxythymidine in place of deoxythymidine in primed poly(dA-dT).poly(dA-dT) synthesis. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4884–4888. doi: 10.1073/pnas.80.16.4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow E. T., Foote R. S., Mitra S. Base-pairing properties of O6-methylguanine in template DNA during in vitro DNA replication. J Biol Chem. 1984 Jul 10;259(13):8095–8100. [PubMed] [Google Scholar]
- Strauss B., Rabkin S., Sagher D., Moore P. The role of DNA polymerase in base substitution mutagenesis on non-instructional templates. Biochimie. 1982 Aug-Sep;64(8-9):829–838. doi: 10.1016/s0300-9084(82)80138-7. [DOI] [PubMed] [Google Scholar]
- Teo I., Sedgwick B., Demple B., Li B., Lindahl T. Induction of resistance to alkylating agents in E. coli: the ada+ gene product serves both as a regulatory protein and as an enzyme for repair of mutagenic damage. EMBO J. 1984 Sep;3(9):2151–2157. doi: 10.1002/j.1460-2075.1984.tb02105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston F., Botstein D., Miller J. H. Characterization of amber and ochre suppressors in Salmonella typhimurium. J Bacteriol. 1979 Jan;137(1):433–439. doi: 10.1128/jb.137.1.433-439.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkin E. M. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev. 1976 Dec;40(4):869–907. doi: 10.1128/br.40.4.869-907.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Katsuki M., Sekiguchi M., Otsuji N. Escherichia coli gene that controls sensitivity to alkylating agents. J Bacteriol. 1978 Jul;135(1):144–152. doi: 10.1128/jb.135.1.144-152.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Sekiguchi M. Pathways for repair of DNA damaged by alkylating agent in Escherichia coli. Mol Gen Genet. 1979 Mar 27;171(3):251–256. doi: 10.1007/BF00267579. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Ito J., Horn V. Amino acid replacements and the genetic code. Cold Spring Harb Symp Quant Biol. 1966;31:151–162. doi: 10.1101/sqb.1966.031.01.023. [DOI] [PubMed] [Google Scholar]


