Abstract
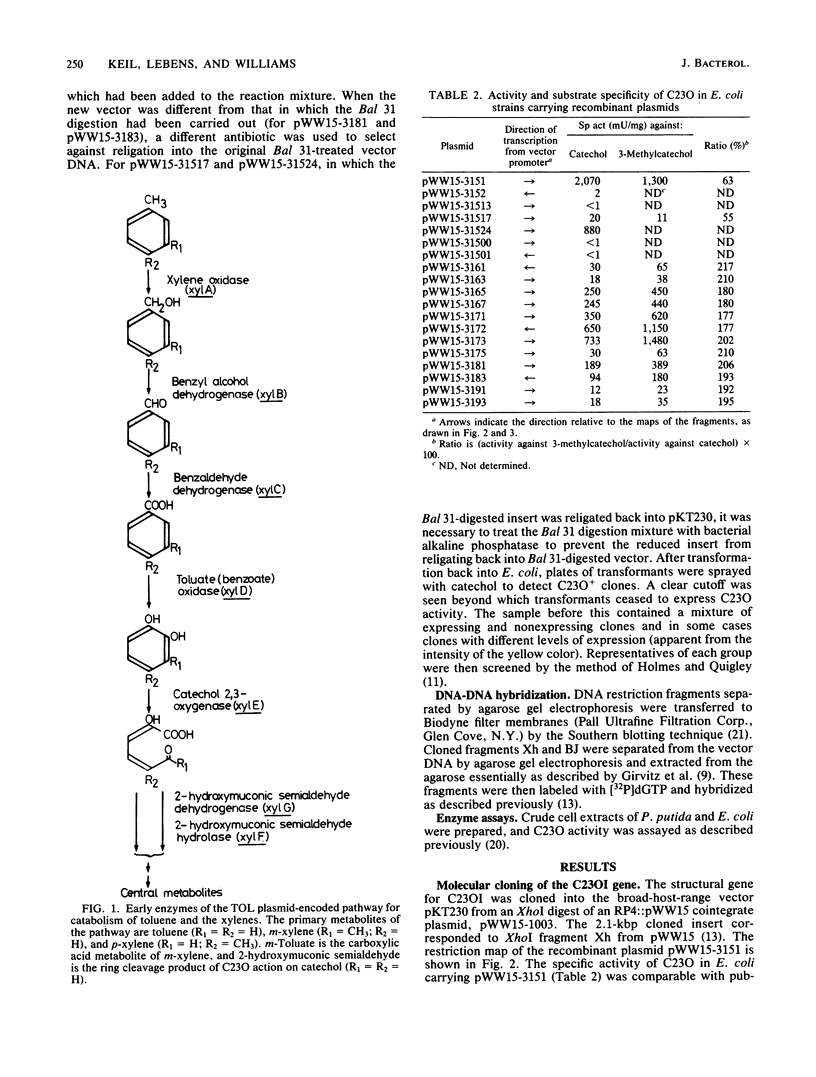
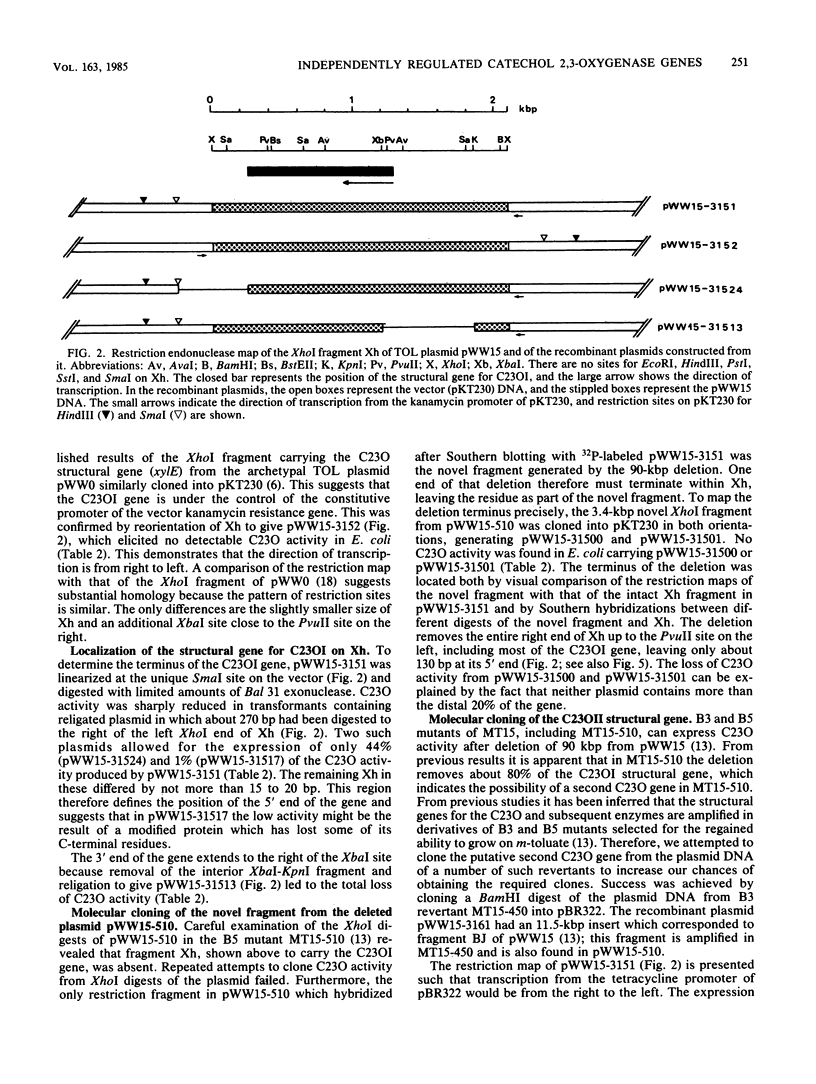
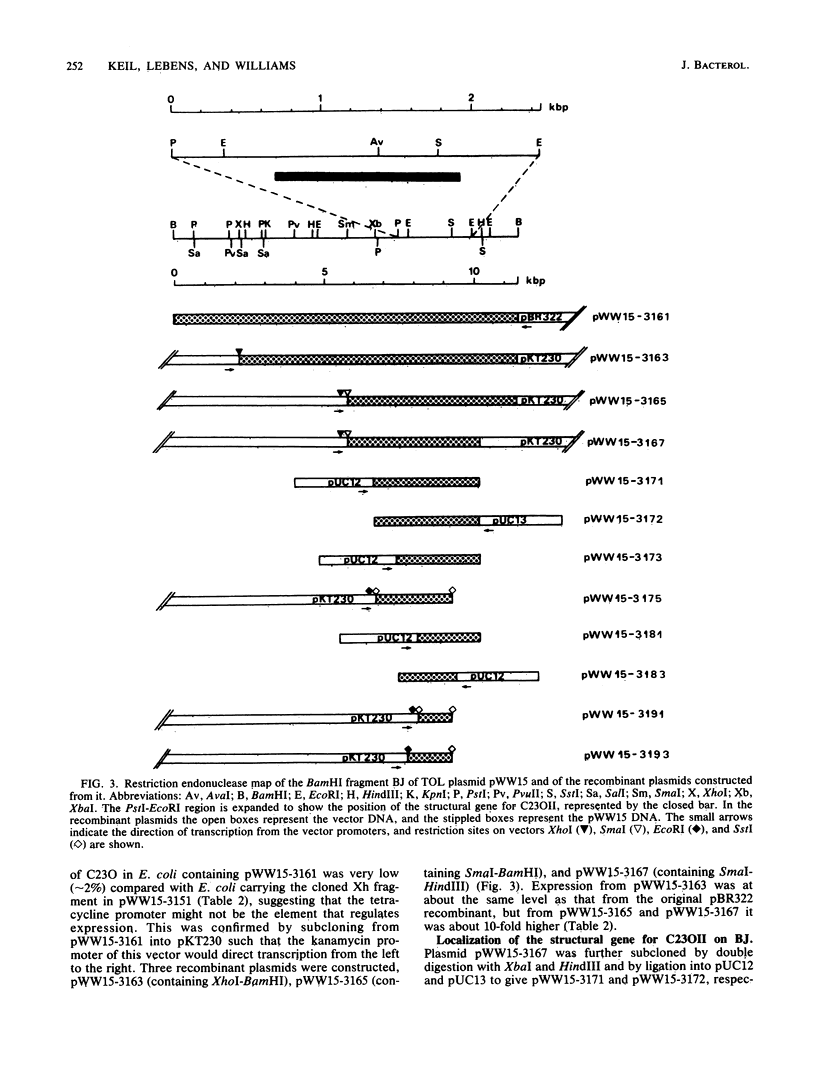
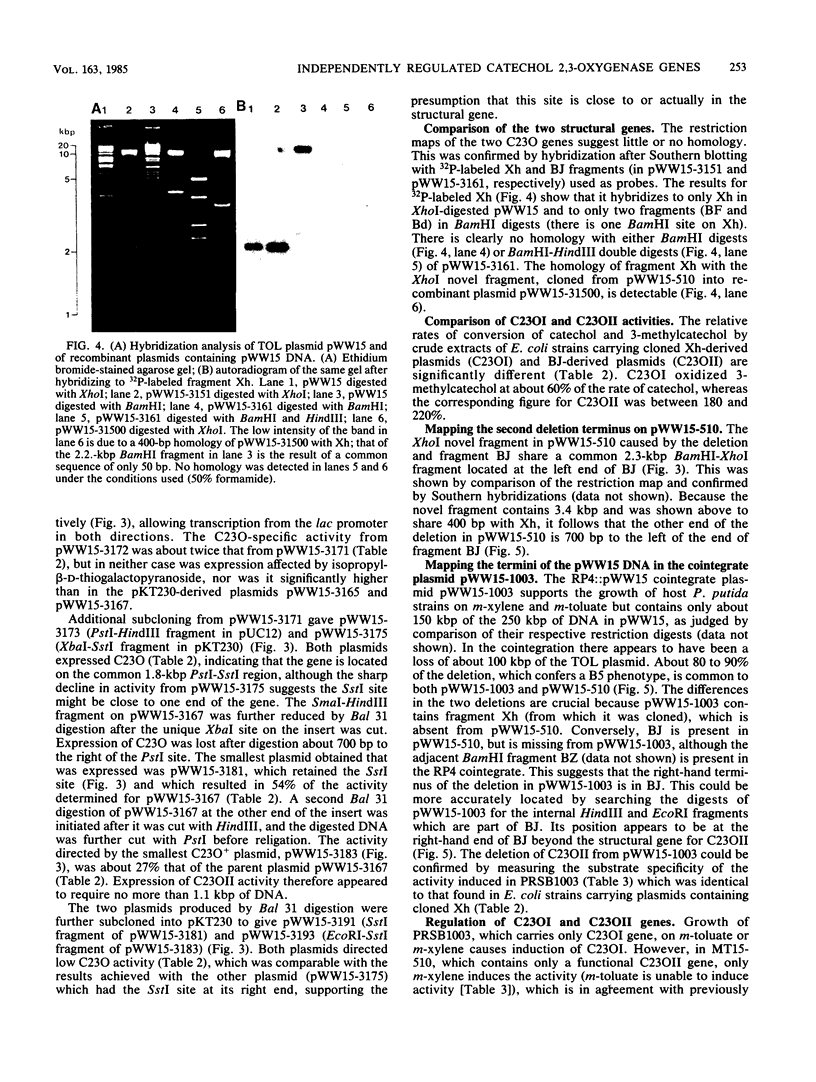
Pseudomonas putida MT15 contains a 250-kilobase-pair (kbp) TOL plasmid pWW15, encoding toluene and xylene catabolism, which undergoes large spontaneous deletions to give two classes of mutants with altered catabolic phenotypes (H. Keil and P. A. Williams, J. Gen. Microbiol, 131:1023-1033, 1985). Two structural genes for catechol 2,3-oxygenase (C23O) were cloned from pWW15. The gene for C23OI was located on the 2.1-kbp XhoI fragment Xh, whereas that for C23OII was found on the 11.5-kbp BamHI fragment BJ. The two restriction fragments and the subcloned regions of them showed no similarity in the pattern of restriction digestion, nor did they hybridize with each other. The substrate specificities of the two enzymes were also substantially different. The two structural genes were separated on pWW15 by about 100 kbp. In plasmid pWW15-510 of a B5 mutant, the 90-kbp deletion in the plasmid removed most of the intervening DNA, but it also deleted 80% of the gene for C23OI from its 3' end. Thus, only C23OII was expressed in the host MT15-510. Conversely, in RP4::pWW15 cointegrate plasmid pWW15-1003, only the C23OI gene was present. The expression of C23O activity from these two derivative plasmids and from the wild-type pWW15 showed that only C23OI was induced by growth in the presence of m-toluate, whereas both activities were induced in the only C23OI was induced by growth in the presence of m-toluate, whereas both activities were induced in the presence of m-xylene. These findings cast doubt on the earlier hypothesis that the deletions in B3 and B5 mutants remove a regulatory gene by which m-toluate induces the enzymes necessary for its own catabolism.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bayley S. A., Duggleby C. J., Worsey M. J., Williams P. A., Hardy K. G., Broda P. Two modes of loss of the Tol function from Pseudomonas putida mt-2. Mol Gen Genet. 1977 Jul 20;154(2):203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Williams P. A. Construction of a partial diploid for the degradative pathway encoded by the TOL plasmid (pWWO) from Pseudomonas putida mt-2: evidence for the positive nature of the regulation by the xyIR gene. Mol Gen Genet. 1980 Jan;177(2):321–328. doi: 10.1007/BF00267445. [DOI] [PubMed] [Google Scholar]
- Girvitz S. C., Bacchetti S., Rainbow A. J., Graham F. L. A rapid and efficient procedure for the purification of DNA from agarose gels. Anal Biochem. 1980 Aug;106(2):492–496. doi: 10.1016/0003-2697(80)90553-9. [DOI] [PubMed] [Google Scholar]
- Guerry P., LeBlanc D. J., Falkow S. General method for the isolation of plasmid deoxyribonucleic acid. J Bacteriol. 1973 Nov;116(2):1064–1066. doi: 10.1128/jb.116.2.1064-1066.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Jeenes D. J., Williams P. A. Excision and integration of degradative pathway genes from TOL plasmid pWW0. J Bacteriol. 1982 Apr;150(1):188–194. doi: 10.1128/jb.150.1.188-194.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz D. A., Chapman P. J. Catabolism of pseudocumene and 3-ethyltoluene by Pseudomonas putida (arvilla) mt-2: evidence for new functions of the TOL (pWWO) plasmid. J Bacteriol. 1981 Apr;146(1):179–191. doi: 10.1128/jb.146.1.179-191.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz D. A., Chapman P. J. Isolation and characterization of spontaneously occurring TOL plasmid mutants of Pseudomonas putida HS1. J Bacteriol. 1981 Jun;146(3):952–964. doi: 10.1128/jb.146.3.952-964.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nakai C., Kagamiyama H., Nozaki M., Nakazawa T., Inouye S., Ebina Y., Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983 Mar 10;258(5):2923–2928. [PubMed] [Google Scholar]
- Pickup R. W., Williams P. A. Spontaneous deletions in the TOL plasmid pWW20 which give rise to the B3 regulatory mutants of Pseudomonas putida MT20. J Gen Microbiol. 1982 Jul;128(7):1385–1390. doi: 10.1099/00221287-128-7-1385. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Wheatcroft R., Williams P. A. Rapid methods for the study of both stable and unstable plasmids in Pseudomonas. J Gen Microbiol. 1981 Jun;124(2):433–437. doi: 10.1099/00221287-124-2-433. [DOI] [PubMed] [Google Scholar]
- Williams P. A., Murray K. Metabolism of benzoate and the methylbenzoates by Pseudomonas putida (arvilla) mt-2: evidence for the existence of a TOL plasmid. J Bacteriol. 1974 Oct;120(1):416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams P. A., Worsey M. J. Ubiquity of plasmids in coding for toluene and xylene metabolism in soil bacteria: evidence for the existence of new TOL plasmids. J Bacteriol. 1976 Mar;125(3):818–828. doi: 10.1128/jb.125.3.818-828.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Characterization of a spontaneously occurring mutant of the TOL20 plasmid in Pseudomonas putida MT20: possible regulatory implications. J Bacteriol. 1977 Jun;130(3):1149–1158. doi: 10.1128/jb.130.3.1149-1158.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K. M., Gunsalus I. C. Plasmid gene organization: naphthalene/salicylate oxidation. Proc Natl Acad Sci U S A. 1982 Feb;79(3):874–878. doi: 10.1073/pnas.79.3.874. [DOI] [PMC free article] [PubMed] [Google Scholar]