Abstract
Peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1α (PGC-1α) is a highly regulated transcriptional coactivator that coordinates energy metabolism in mammals. Misregulation of PGC-1α has been implicated in the pathogenesis of several human diseases, including diabetes, obesity, and neurological disorders. We identified SCFCdc4 as an E3 ubiquitin ligase that regulates PGC-1α through ubiquitin-mediated proteolysis. PGC-1α contains two Cdc4 phosphodegrons that bind Cdc4 when phosphorylated by Glycogen Synthase Kinase 3β (GSK3β) and p38 MAPK, leading to SCFCdc4-dependent ubiquitylation and proteasomal degradation of PGC-1α. Furthermore, SCFCdc4 negatively regulates PGC-1α-dependent transcription. We demonstrate that RNAi-mediated reduction of Cdc4 in primary neurons results in an increase of endogenous PGC-1α protein, while ectopic expression of Cdc4 leads to a reduction of endogenous PGC-1α protein. Finally, under conditions of oxidative stress in neurons, Cdc4 levels are decreased, leading to an increase in PGC-1α protein and PGC-1α-dependent transcription. These results suggest that attenuation of SCFCdc4-dependent proteasomal degradation of PGC-1α has a role in mediating the PGC-1α-dependent transcriptional response to oxidative stress.
[Keywords: Cdc4, Fbw7, PGC-1, protein degradation, ubiquitin-mediated proteolysis, oxidative stress]
Members of the peroxisome proliferator-activated receptor γ (PPARγ) coactivator-1 (PGC-1) family of transcriptional coactivators are important regulators of energy metabolism in mammals. The three family members, PGC-1α, PGC-1β (also known as PERC), and PGC-1-related coactivator (PRC), coactivate a diverse set of transcription factors and coordinate the expression of genes that promote mitochondrial biogenesis and oxidative metabolism (Finck and Kelly 2006; Handschin and Spiegelman 2006). PGC-1α, the best studied member of the family, also induces hepatic gluconeogenesis, glucose uptake in muscle (Finck and Kelly 2006; Handschin and Spiegelman 2006), and the mitochondrial antioxidant defense system in neurons and endothelial cells (Valle et al. 2005; St-Pierre et al. 2006). Thus, PGC-1α promotes oxidative metabolism directly, by enhancing cellular respiratory capacity, as well as indirectly, by stimulating substrate uptake and protecting cells from oxidative damage (Valle et al. 2005; Finck and Kelly 2006; Handschin and Spiegelman 2006; St-Pierre et al. 2006).
Mitochondrial dysfunction and alterations in PGC-1 levels have been implicated in the pathogenesis of several diseases, including diabetes, heart disease, and neurological disorders (Lin et al. 2004; Leone et al. 2005; Cui et al. 2006; Finck and Kelly 2006; St-Pierre et al. 2006). The reduced levels of PGC-1α and PGC-1β in the muscle of type 2 diabetic patients and individuals predisposed to diabetes, have been proposed to underlie decreased mitochondrial function and the development of insulin resistance (Mootha et al. 2003; Patti et al. 2003). Loss of PGC-1α activity in the heart correlates with heart failure, while chronic PGC-1α overexpression leads to cardiomyopathy (Finck and Kelly 2006; Handschin and Spiegelman 2006). Mice lacking PGC-1α also display hyperactivity, signs of increased anxiety, and neurological defects in the striatum, a region of the brain that is affected in neurodegenerative movement disorders (Lin et al. 2004; Leone et al. 2005). Notably, decreased PGC-1α expression is likely part of the mechanism by which a mutant huntingtin protein leads to mitochondrial dysfunction, striatal neurodegeneration, and Huntington’s disease (Cui et al. 2006). Finally, loss of PGC-1α expression potentiates chemical-induced neurodegeneration in the substantia nigra and hippocampus of mouse brains, potentially linking PGC-1α dysfunction and the ensuing decrease in the capacity of the oxidative defense system to Parkinson’s disease (St-Pierre et al. 2006).
PGC-1α function is regulated at multiple levels. Several studies have addressed the transcriptional induction of PGC-1α gene expression by cAMP, nitric oxide, calcium-dependent signaling pathways, and AMP-dependent protein kinase (Handschin and Spiegelman 2006). At the post-translational level, PGC-1α is phosphorylated by p38 MAPK on Thr263, Thr299, and Ser266 (Puigserver et al. 2001). Phosphorylation at these sites increases the transcriptional activity of PGC-1α, at least partly via the release of inhibitory factors such as p160 myb-binding protein (Knutti et al. 2001; Puigserver et al. 2001; Fan et al. 2004). PGC-1α activity is also modulated by lysine acetylation, arginine methylation, and likely by phosphorylation of sites that have yet to be identified (Nemoto et al. 2005; Rodgers et al. 2005; Teyssier et al. 2005). Finally, PGC-1α protein is rapidly turned over (Puigserver et al. 2001; Sano et al. 2007). Although phosphorylation has been shown to affect the half-life of PGC-1α protein, the mechanisms that regulate and mediate PGC-1α proteolysis have remained elusive.
Ubiquitin-mediated proteolysis is an important regulatory mechanism involved in a diverse set of cellular processes including development, cell cycle control, and transcription. The recognition of substrates for ubiquitylation is performed by E3 ubiquitin ligases (Pickart and Eddins 2004). One class of E3 ligases collectively known as Skp1/Cullin/F-box (SCF) (Cardozo and Pagano 2004; Willems et al. 2004) contains Skp1, Cul1, Rbx1/Roc1, and one of a number of F-box proteins. Cul1 serves as a scaffold for the complex, Skp1 links Cul1 to the F-box protein, and Rbx1, a ring-finger protein, recruits an E2 ubiquitin conjugating enzyme to the complex. Each F-box protein captures a particular set of substrates to the Skp1–Cullin–Rbx1–E2 complex through a unique protein–protein interaction domain.
Cdc4 (also known as Fbw7) is the F-box component of the SCFCdc4 ubiquitin ligase that targets cyclin E, c-Myc, c-Jun, Notch, and SREBP for ubiquitylation (Hubbard et al. 1997; Gupta-Rossi et al. 2001; Koepp et al. 2001; Moberg et al. 2001, 2004; Oberg et al. 2001; Strohmaier et al. 2001; Nateri et al. 2004; Welcker et al. 2004b; Yada et al. 2004; Sundqvist et al. 2005). Cdc4 recognizes a short, phosphothreonine-containing motif known as the Cdc4 phosphodegron (CPD) present in each of these substrates (Nash et al. 2001). The recognition of CPDs and thus the ubiquitylation of SCFCdc4 substrates is regulated by phosphorylation of the substrate at the threonine residue in position three of the CPD. The phosphorylated CPD associates with three conserved arginine residues in a domain of eight WD40 repeats in the C terminus of Cdc4 (Orlicky et al. 2003). Thus, SCFCdc4 provides regulated protein degradation in response to specific phosphorylation signals.
While yeast and invertebrates express only one Cdc4 isoform, mammals express three splice-variant isoforms termed α, β, and γ (Spruck et al. 2002). Cdc4α resides in the nucleoplasm, Cdc4γ is concentrated in the nucleolus, and Cdc4β is retained in the cytoplasm (Welcker et al. 2004a). Surprisingly, we recently discovered that SCF complexes containing both of the nuclear Cdc4 isoforms are required sequentially for cyclin E degradation (van Drogen et al. 2006). SCFCdc4α acts first to promote Pin1-dependent prolyl-isomerization of the CPD, after which SCFCdc4γ carries out ubiquitylation (van Drogen et al. 2006).
The PGC-1α protein contains two sequences that conform to known CPD motifs. We therefore investigated whether SCFCdc4 targets PGC-1α for proteasomal degradation. Here we demonstrate that the PGC-1α is indeed ubiquitylated by SCFCdc4 in a phosphorylation-dependent manner. Furthermore, we demonstrate that SCFCdc4 antagonizes PGC-1α protein levels and PGC-1α-dependent transcription. Finally, we demonstrate that SCFCdc4 regulates PGC-1α in primary neurons in response to oxidative stress.
Results
Cdc4 binds to PGC-1 through two phosphodegron sequences
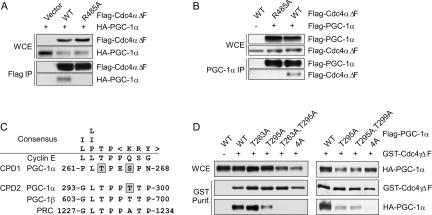
To determine whether Cdc4 interacts with PGC-1α, we transfected HEK293T cells with plasmids expressing epitope-tagged alleles and performed coimmunoprecipitations (co-IP). To prevent degradation and preserve the Cdc4–substrate interactions, we used alleles of Cdc4 deleted of their F-box (ΔF) (Strohmaier et al. 2001). Indeed, PGC-1α copurified with Flag-tagged Cdc4αΔF but not with an allele of Cdc4αΔF mutated at Arg485 (one of the three arginines critical for CPD binding) (Koepp et al. 2001; Nash et al. 2001), nor with the Flag epitope alone (Fig. 1A). When the co-IPs were performed in reverse, Cdc4αΔF copurified with PGC-1α but not with vector alone (Fig. 1B). As expected, Cdc4αΔF-R485A did not copurify with PGC-1α (Fig. 1B). These results demonstrate that PGC-1α associates with Cdc4αΔF, and that this association depends on an intact Cdc4 substrate-binding domain. Importantly, PGC-1α also copurified with wild-type Cdc4α when a proteasomal inhibitor was added prior to cell lysis (data not shown).
Figure 1.
Cdc4 binds PGC-1α through two CPDs. HEK293T cells were transiently transfected with plasmids expressing tagged alleles of PGC-1α and/or Cdc4. Whole-cell extracts (WCE) were made and immunoprecipitated as indicated. (A) Flag-Cdc4αΔF was immumoprecipitated using anti-Flag beads and whole-cell extracts and immunoprecipitations were separated and immunoblotted for the proteins indicated. (B) PGC-1α was immumoprecipitated from whole-cell extracts using anti-PGC-1α polyclonal antibody. Whole-cell extracts and immunoprecipitations were separated by SDS-PAGE and immunoblotted for the proteins indicated. (C) The sequence of a consensus CPD and the genuine CPD of cyclin E are compared with sequences found in the PGC-1 family members. Amino acids in PGC-1α previously demonstrated to be phosphorylated by p38 MAPK are boxed in gray. The numbers surrounding the phosphodegrons correspond to the first and last amino acid shown in the human PGC-1 family members. (D) GST-Cdc4γΔF was affinity-purified using glutathione beads. Whole-cell extracts and purifications were separated and immunoblotted for the tagged proteins indicated. PGC-1α-4A corresponds to the quadruple mutation of T263, S266, T295, and T299 to alanine.
PGC-1α contains two sequences that conform to the consensus CPD (Fig. 1C). One of these CPDs is almost completely conserved in the other two members of the PGC-1 family of transcription factors (PGC-1β and PRC), suggesting an important role in PGC-1 function. Three of the four residues in the two putative CPDs of PGC-1α whose phosphorylation is predicted to be important for Cdc4 binding have been shown previously to be phosphorylated by p38 MAPK (Fig. 1C; Puigserver et al. 2001). To determine if these two CPD-like sequences are indeed the binding site of Cdc4, we mutated the four predicted phosphorylation sites to alanine individually or in different combinations and performed co-IPs from HEK293T cells. Mutation of CPD1 had no effect, while mutation of CPD2 reduced Cdc4γΔF binding. However, mutation of both CPDs simultaneously abolished the interaction between PGC1α and Cdc4γΔF (Fig. 1D). These results demonstrate that these two CPDs represent the sites of interaction between PGC-1α and Cdc4, and suggest that PGC-1α is a genuine substrate of SCFCdc4.
SCFCdc4 targets PGC-1α for ubiquitylation
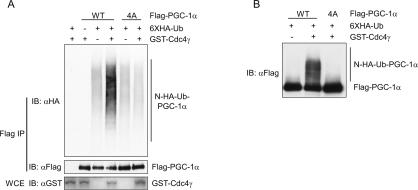
We next asked if the CPDs in PGC-1α promote Cdc4-dependent ubiquitylation of PGC-1α. To test this, we performed an in vivo ubiquitylation assay after transiently transfecting HEK293T cells with plasmids expressing Flag-PGC-1α, HA-ubiquitin, and GST-Cdc4γ or GST alone. To stabilize ubiquitylated proteins, the proteasome inhibitor MG132 was added to the culture medium 4 h prior to lysis. PGC-1α was immunoprecipitated and the immunoprecipitations were immunoblotted for HA-ubiquitin-labeled proteins (Fig. 2A) or Flag-PGC-1 (Fig. 2B, long exposure of separate experiment). The addition of Cdc4γ greatly increased the amount of ubiquitylated PGC-1α but had no effect on the CPD mutant of PGC-1α that lacked all four phosphorylation sites (PGC-1α-4A) (Fig. 2A,B). Similar amounts of nonubiquitylated PGC-1α were immunoprecipitated from each sample in Figure 2A, as demonstrated in the bottom panel. These results demonstrate that the CPDs of PGC-1α direct Cdc4-dependent ubiquitylation. In this assay, PGC-1α was also ubiquitylated in the absence of exogenous Cdc4 (Fig. 2, lane 3). Because PGC-1α and PGC-1α-4A were ubiquitylated equally well in the absence of Cdc4, we believe that this represents CPD-independent ubiquitylation via another ubiquitin ligase present in HEK293T cells. Altogether, these results clearly demonstrate CPD-dependent ubiquitylation of PGC-1α by SCFCdc4 in cultured cells.
Figure 2.
SCFCdc4 ubiquitylates PGC-1α in vivo. An in vivo ubiquitylation assay was performed by coexpressing the proteins indicated with HA-ubiquitin. (A) Whole-cell extracts (WCE; bottom panel) and Flag-PGC-1α immunoprecipitations (top two panels) were immunoblotted for the indicated proteins. (B) Flag-PGC-1α immunoprecipitated with anti-Flag antibody was immunoblotted with anti-Flag antibody.
SCFCdc4 targets PGC-1α for degradation by the proteasome
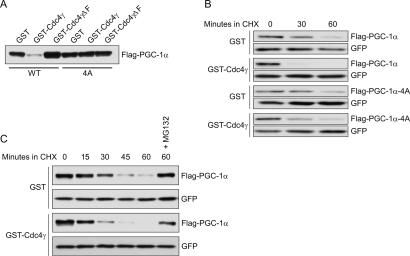
To determine whether Cdc4-dependent ubiquitylation of PGC-1α leads to proteasomal degradation, we coexpressed alleles of PGC-1α and Cdc4 in HEK293T cells and analyzed PGC-1α protein levels and stability. As compared with GST alone, expression of GST-Cdc4γ reduced the levels of PGC-1α but not the CPD mutant PGC-1α-4A (Fig. 3A). Neither mutation of the CPDs of PGC-1α in the absence of exogenous Cdc4 nor deletion of the F-box of Cdc4 significantly affected the levels of PGC1α, consistent with the observation that endogenous Cdc4 is expressed at extremely low levels in HEK293 cells (B.L. Olson and S.I. Reed, unpubl.). To demonstrate that the loss of PGC-1α protein in the presence of exogenous Cdc4γ was due to proteolysis, we carried out PGC-1α protein half-life determinations after inhibition of protein synthesis by treatment with cycloheximide. Compared with the expression of GST alone, the expression of exogenous GST-Cdc4γ led to a significant decrease in the half-life of cotransfected PGC-1α, while having no effect on the CPD mutant PGC-1α-4A (Fig. 3B). The Cdc4γ-mediated decrease in PGC-1α half-life was alleviated by the inclusion of MG132 in the experiment, suggesting that ubiquitylated PGC-1α is degraded by the 26S proteasome (Fig. 3C).
Figure 3.
SCFCdc4 targets phosphorylated PGC-1α for proteolysis. (A) HEK293T cells were cotransfected with the indicated alleles of PGC-1α and Cdc4γ. PGC-1α protein levels were determined by immunoblotting with anti-Flag antibody. (B,C) HEK293T cells were transfected with vector alone or Cdc4γ and different alleles of PGC-1α. Cyclohexamide (CHX) was added to cell cultures 48 h post-transfection, whole-cell extracts were prepared at the indicated times after CHX addition, and PGC-1α protein levels were analyzed by immunoblotting. GFP was cotransfected as a transfection and loading control.
Glycogen Synthase Kinase (GSK3) may regulate PGC-1α turnover by phosphorylating T295
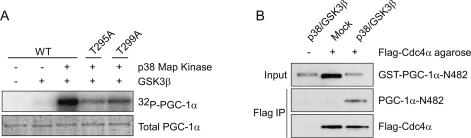
Full CPD recognition by Cdc4 requires phosphorylation at position 3 and position 6 or 7 (Ye et al. 2004). Since mutation of T263, S266, and T299 almost completely ablates p38 MAPK-dependent phosphorylation of PGC-1α (Puigserver et al. 2001), we expected that another kinase was responsible for phosphorylation of T295. The phosphothreonine that forms the core of the CPD consensus sequence has been shown to be phosphorylated by GSK3β in a number of SCFCdc4 substrates (Gregory et al. 2003; Welcker et al. 2003; Sundqvist et al. 2005; Wei et al. 2005). In addition, GSK3β often requires a priming phosphorylation at position +4, another residue frequently phosphorylated in CPDs of known SCFCdc4 targets and phosphorylated by p38 MAPK in PGC-1α (Puigserver et al. 2001). Therefore, we tested whether GSK3β participates in phosphorylating T295 by performing in vitro kinase assays with activated forms of recombinant p38α and GSK3β. Kinase reactions were carried out in two steps, using a purified truncated derivative of PGC-1α that includes the two CPDs and expresses well in Escherichia coli (GST-PGC-1α-N482, amino acids 1–482). First, a priming phosphorylation with p38 MAPK and unlabeled ATP was carried out; this was followed by incubation with GSK3β, γ-[32P]-ATP, and the p38 MAPK inhibitor SB202190. Incubation with p38 MAPK led to a reduction in PGC-1α-N482 mobility based on SDS-PAGE, consistent with previously demonstrated phosphorylation of PGC-1α by p38 MAPK (Fig. 4A, bottom panel; Puigserver et al. 2001). GSK3β phosphorylated PGC-1α-N482 only after a p38 priming phosphorylation (Fig. 4A). Importantly, phosphorylation of PGC-1α-N482-T295A was significantly reduced relative to wild-type, demonstrating that the major site targeted by GSK3β phosphorylation is T295 (Fig. 4A). Furthermore, mutation of the p38 MAPK-targeted priming site, T299, also significantly reduced phosphorylation by GSK3β (Fig. 4A). These data demonstrate that following phosphorylation of T299 by p38 MAPK, GSK3β can phosphorylate T295.
Figure 4.
T295 of PGC-1α is phosphorylated by GSK3β. (A) Recombinant GST-PGC-1α N482 was purified from E. coli and was phosphorylated in vitro with p38 MAPK in the presence of unlabeled ATP. Following the priming phosphorylation, GSK3β was added in the presence of γ-radiolabeled ATP and the p38 inhibitor SB202190. (Bottom) Proteins were resolved by SDS PAGE, stained with colloidal blue for total protein levels, and analyzed by PhosphorImager for 32P incorporation. (B) Recombinant PGC-1α was purified from E. coli, phosphorylated by the indicated kinases, and allowed to bind to Cdc4α-coated beads. After washing, input and bound fractions were immunoblotted for the indicated proteins.
We next determined whether phosphorylation of PGC-1α by these two kinases is required for Cdc4 binding. Flag epitope or Flag-tagged Cdc4α was retrovirally expressed in HEK293 cells and bound to anti-Flag beads. These beads were incubated with recombinant GST-PGC-1α-N482 that had been phosphorylated by p38 and GSK3β or mock-phosphorylated. Phosphorylated PGC-1α-N482 was retained on the Cdc4 beads but not on control beads, while mock-phosphorylated PGC-1α was not retained (Fig. 4B). These results demonstrate that the binding of PGC-1α to Cdc4 is phosphorylation-dependent, and suggest that the interaction is direct. They also confirm that p38 MAPK and GSK3β indeed phosphorylate a PGC-1α CPD recognized by Cdc4.
SCFCdc4 negatively regulates PGC-1α-dependent transcription
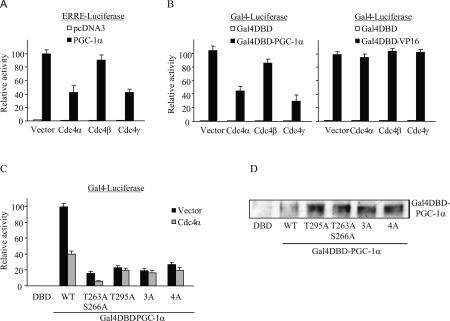
To determine how SCFCdc4 affects PGC-1α function, we evaluated the ability of PGC-1α to induce transcription from two simple reporter constructs in the presence and absence of exogenous Cdc4. In the first construct, luciferase expression was driven by the response element of the ESRRA promoter, a physiologically relevant target of PGC-1α (Mootha et al. 2004; Schreiber et al. 2004) where PGC-1α is recruited via interactions with endogenous transcription factors. In the second construct, PGC-1α transcriptional activity was measured directly, independent of interactions with transcription factors, using a fusion of PGC-1α to the Gal4-DNA-binding domain (Gal4-DBD) and a Gal4-responsive luciferase reporter. In both of these assays, coexpression of the nuclear isoforms of Cdc4 led to a significant decrease in PGC-1α-dependent activation of the reporters, while coexpression of the cytoplasmic form of Cdc4 caused only a minor reduction (Fig. 5A,B, left panel). The Cdc4 effect was specific for PGC-1α, as exogenous Cdc4 had no effect on reporter constructs in the absence of PGC-1α (Fig. 5A,B, left panel), nor on the ability of another transcriptional activator, Gal4-VP16, to stimulate transcription from the same reporter (Fig. 5B, right panel). These results suggest that overexpression of Cdc4 targets PGC-1α for degradation, thereby restricting its availability for transcriptional activation. We next tested whether prevention of Cdc4 binding to PGC-1α by mutation of the CPDs would alter PGC-1α transcriptional activity in the direct transactivation assay. All of the CPD mutants tested were expressed at levels equal to, or greater than, the levels of wild-type PGC-1α (Fig. 5D). Consistent with published studies (Knutti et al. 2001; Puigserver et al. 2001), mutation of previously described p38 MAPK-dependent phosphorylation sites decreased PGC-1α activity in HEK293T (data not shown) and COS7 cells (Fig. 5C). Similarly, mutation of the GSK3β site T295 significantly reduced the transcriptional activity of PGC-1α. These results suggest that phosphorylation of the PGC-1α CPDs is important for the transcriptional function of PGC-1α. Importantly, mutation of phosphorylation sites in CPD2 alone (T295A) or both CPDs together (3A and 4A) rendered PGC-1α activity insensitive to Cdc4, demonstrating that the Cdc4 effect on PGC-1α-dependent transcription is mediated through PGC-1α CPDs, and in particular, CPD2 (Fig. 5C).
Figure 5.
SCFCdc4 inhibits PGC-1α activity. COS7 cells were transiently transfected with the indicated contructs and luciferase activity was measured and normalized to cotransfected βgal activity. Relative activity represents the average normalized luciferase activity of at least three independent experiments. Error bars represent the standard error of the mean. (A) ERRα coactivation assay: PGC1α (or vector control pcDNA3) and GST-Cdc4 (or vector control) were transiently transfected into COS7 cells with an ERRα-responsive luciferase reporter construct. (B) COS7 cells were transfected with plasmids expressing Gal4-DBD, Gal4DBD-VP16, or Gal4DBD-PGC1α, and the indicated isoforms of GST-Cdc4 (or vector control) with a Gal4-responsive luciferase reporter. (C) The indicated CPD mutants of Gal4-PGC1α were transiently cotransfected into COS7 cells together with a Gal4-responsive luciferase reporter construct and GST or GST-Cdc4α. 3A and 4A correspond to mutations in which alanines are substituted for the three p38 phosphorylation sites (T263, S266, T299) or all four phosphorylation sites (T263, S266, T295, and T299), respectively. (D) COS7 cells were cotransfected with Gal4 fusion constructs (Gal4-DBD, Gal4DBD-PGC-1α wild type, or the indicated mutants) and βgal. Cytoplasmic and nuclear extracts were harvested 48 h later. The amount of nuclear extract loaded on the gel was normalized for transfection efficiency based on βgal activity in the cytoplasmic extracts. Expression levels of Gal4 fusion proteins were determined by anti-PGC-1α Western blot.
SCFCdc4 regulates PGC-1α levels and transcriptional activity in primary neurons
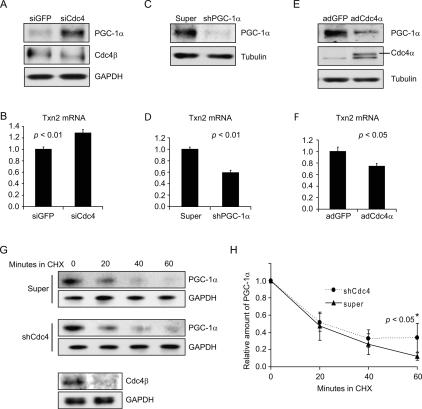
To determine if SCFCdc4 plays a role in the regulation of endogenous PGC-1α protein levels and PGC-1α-dependent transcriptional coactivation, we examined the effect of manipulating Cdc4 levels on endogenous PGC-1α levels and on PGC-1α target gene expression in primary embryonic mouse brain neurons. When Cdc4 levels were reduced by siRNA-mediated silencing, endogenous PGC-1α levels increased (Fig. 6A). At the same time, the transcript encoding an antioxidant defense protein, thioredoxin 2 (Txn2), previously shown to be under PGC-1α control (Valle et al. 2005; Rangwala et al. 2007), was increased (Fig. 6B). Consistent with Txn2 transcription being under PGC-1α control in primary neurons, shRNA-mediated silencing of endogenous PGC-1α led to a decrease in Txn2 transcript levels (Fig. 6C,D). When Cdc4 levels were increased by transduction with a recombinant Cdc4α-expressing adenovirus, endogenous PGC-1α levels (Fig. 6E) and Txn2 transcript levels (Fig. 6F) were reduced similarly to the effect of silencing PGC-1α (Fig. 6C,D). Taken together, these results suggest that SCFCdc4 regulates the level of endogenous PGC-1α in primary neurons by controlling its ubiquitin-mediated proteolysis, and that this, in turn, has an impact on the expression of PGC-1α targets such as Txn2. To confirm that SCFCdc4 regulates endogenous PGC-1α at the level of turnover, we carried out a cycloheximide chase experiment in conjunction with Cdc4 shRNA-mediated gene silencing to determine the effect on PGC-1α half-life (Fig. 6G,H). Although the initial rates of PGC-1α turnover in the Cdc4-silenced and control neurons were similar, the rate of turnover at later times was decreased in the Cdc4-silenced neurons. A trend was observed throughout the time course, but the difference in turnover became statistically significant at 60 min. These data suggest the existence of two populations of PGC-1α in primary neurons: one that is highly unstable and not targeted by SCFCdc4, and one that is less unstable and targeted by SCFCdc4. The existence of a Cdc4-independent pathway for PGC-1α turnover is consistent with data in Figure 3, where PGC-1α phosphodegron mutants are unstable when transfected into HEK293 cells even though they are no longer targeted for turnover by SCFCdc4, as well as data in Figure 2 showing SCFCdc4-independent ubiquitylation of PGC-1α.
Figure 6.
PGC-1α levels are regulated by SCFCdc4-dependent ubiquitin-mediated proteolysis in primary embryonic mouse brain neurons. (A,B) siRNA-mediated silencing of Cdc4 in primary neurons leads to an increase in PGC-1α levels and an increase in Txn2 transcription. Primary neurons were transfected twice with control (GFP-specific) or Cdc4-specific siRNAs at 24-h intervals and then neurons were harvested 24 h later. (A) PGC-1α and Cdc4 protein levels determined by Western blotting. (B) Txn2 transcript levels were determined by real-time PCR. (C,D) shRNA-mediated silencing of PGC-1α in primary neurons leads to a decrease in transcription of the antioxidant defense gene TXN2. Either control or recombinant adenovirus expressing PGC-1α-specific shRNA was transduced into primary embryonic mouse brain neurons, which were then harvested 24 h later. (C) PGC-1α protein levels were determined by Western blotting. (D) Txn2 transcript levels were determined by real-time PCR. (E,F) Increased expression of Cdc4α in primary neurons leads to a reduction in PGC-1α levels and Txn2 transcription. Primary neurons were transduced with adenoviruses expressing GFP or Cdc4α and then harvested 24 h later. (E) PGC-1α and Cdc4 protein levels were determined by Western blotting. (F) Txn2 transcript levels were determined by real-time PCR. (G,H) Reduction of Cdc4 levels by shRNA-mediated gene silencing decreases the rate of PGC-1α turnover. Primary neurons were transduced with control (super) or Cdc4 shRNA adenovirus and then 24 h later a cycloheximide chase experiment was performed. Time 0 corresponds to the time of cycloheximde addition. Neurons were harvested at the indicated times after cycloheximide addition. (G) PGC-1α and GAPDH (loading control) levels were determined by Western blotting. Cdc4β levels were determined for time 0 to confirm effective Cdc4 silencing. (H) Quantitation of PGC-1α levels normalized to GAPDH in three cycloheximide chase experiments, including the one shown in G. Error bars correspond to one standard deviation. The difference between the decay curves achieves statistical significance (P < 0.05, Student’s t-test) at the 60-min time point, although a trend is observable at all time points. (B,D,F) For Txn2 mRNA level determinations, values given are normalized to 36B4 mRNA levels. Error bars represent the standard error from three experiments. P values given are based on Student’s t-test.
SCFCdc4 regulates PGC-1α levels in primary neurons in response to oxidative stress
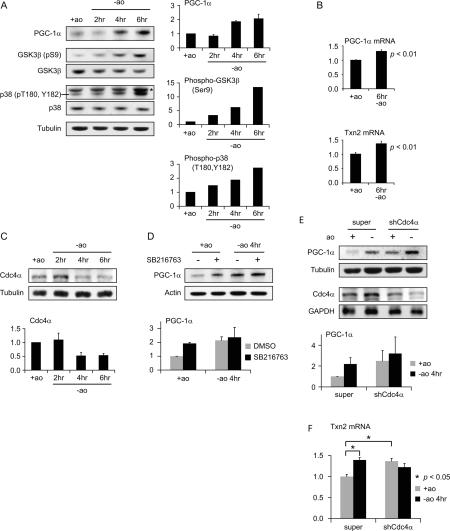
It has been proposed that PGC-1α contributes to the protective response to oxidative stress by transactivating antioxidant genes. Embryonic mouse brain primary neurons are cultured in medium containing a cocktail of antioxidants because they are extremely sensitive to oxidative stress. We found that removing this antioxidant cocktail is sufficient to trigger a defensive response as seen by increased PGC-1α levels and increased transcription of the PGC-1α-responsive antioxidant gene, Txn2 (Fig. 7A,B). During this same time course, we also observed a modest increase in PGC-1α transcript levels consistent with reports that PGC-1α stimulates its own transcription (Handschin et al. 2003), and at least partially explaining the increase in PGC-1α protein levels (Fig. 7B). In order to determine whether SCFCdc4-mediated ubiquitylation and proteolysis might contribute to determining the levels of PGC-1α protein under these circumstances, we first investigated the activation status of protein kinases required for phosphorylating the PGC-1α phosphodegrons, thereby priming it for turnover. After removal of antioxidants from the medium, GSK3β, which phosphorylates T295 on CPD2 (Fig. 4), the primary phosphodegron (Figs. 2, 3), became increasingly phosphorylated on S9, indicating progressive inactivation after induction of oxidative stress (Fig. 7A). Therefore, inactivation of GSK3β could stabilize PGC-1α under conditions of oxidative stress, contributing to its accumulation. p38 MAPK, another kinase that phosphorylates the PGC-1α phosphodegrons (Fig. 4; Knutti et al. 2001; Puigserver et al. 2001), also becomes progressively phosphorylated during the time course (Fig. 7A). In this case, phosphorylation of p38 MAPK (on T180 and Y182) is stimulatory, consistent with previous reports that p38 MAPK is activated by oxidative stress (Mielke and Herdegen 2000; Chang and Karin 2001). Since activation of p38 MAPK could promote degradation of PGC-1α via CPD1, this oxidative stress-mediated change in p38 MAPK status cannot explain the increase in PGC-1α levels in response to oxidative stress. However, it may help to explain the induction of PGC-1α-dependent transcription of Txn2 after oxidative stress. We also determined the levels of Cdc4α, the principal nuclear Cdc4 isoform, during the oxidative stress time course (Fig. 7C). Cdc4α levels consistently decreased by a factor of 2 at the 4- and 6-h time points, consistent with a role in PGC-1α stabilization. Indeed, the kinetics of Cdc4α decrease correlated well with the kinetics of PGC-1α increase. In order to determine whether inhibition of GSK3β could contribute to PGC-1α level increases during the oxidative stress response, we treated unstressed cells and cells subjected to oxidative stress for 4 h with the GSK3β inhibitor SB216763. In unstressed cells, PGC-1α levels increased approximately twofold when treated with the inhibitor (Fig. 7D) without a change in the PGC-1α mRNA levels (data not shown), consistent with GSK3β-dependent phosphorylation playing a role in PGC-1α turnover in the absence of oxidative stress. Under conditions of oxidative stress, inhibition of GSK3β had a much less pronounced effect on PGC-1α stabilization. These results suggest that either GSK3β is already largely inhibited and/or that the SCFCdc4-dependent degradation pathway for PGC-1α is already inactive, possibilities that are not mutually exclusive. In order to assess the role of Cdc4 in the oxidative stress-dependent accumulation of PGC-1α, we subjected neurons to oxidative stress after reducing Cdc4 levels by shRNA-mediated gene silencing. Cdc4α levels were reduced to ∼50% of controls based on Western blotting (Fig. 7D, bottom panels) and real-time PCR (data not shown). In control cells, PGC-1α levels were increased 4 h after antioxidant removal, as expected and shown in Figure 7A. When Cdc4 was silenced, PGC-1α levels were already elevated under the nonstressed conditions and exhibited only a marginal increase in response to removal of antioxidants (Fig. 7E). Consistent with these changes in PGC-1α protein levels, Txn2 mRNA levels increased in response to oxidative stress in control neurons but were already elevated and did not increase further in neurons in which Cdc4 was silenced. These data are consistent with oxidative stress-mediated reduction of Cdc4 levels being central to increasing PGC-1α levels and PGC-1α-activated transcription.
Figure 7.
SCFCdc4 regulates PGC-1α in response to oxidative stress in primary embryonic mouse brain neurons. (A–C) Response of primary embryonic mouse brain neurons to oxidative stress. Neurons were harvested in the presence of antioxidant (ao) cocktail and at the indicated times after removal of antioxidants. (A) PGC-1α, GSK3β (pS9), total GSK3β, p38 MAPK (pT180, pY182), total p38 MAPK, and tubulin levels were determined by Western blotting. All data shown are from the same blot that was stripped and reprobed. The band denoted with an asterisk in the phospho-p38 MAPK panel corresponds to a background band. Quantification of PGC-1α, GSK3β (pS9), and p38 MAPK (pT180, pY182) normalized to tubulin is shown to the right. PGC-1α data represent the average of three experiments. Error bars correspond to one standard deviation. (B) Oxidative stress leads to induction of PGC-1α and Txn2 mRNAs. Values given are normalized to the 36B4 mRNA levels. Error bars correspond to the standard error from six experiments. P values given are based on Student’s t-test. (C) Cdc4α levels in primary neurons decrease in response to oxidative stress. Cdc4α levels were determined by Western blotting. Quantification of Cdc4α levels normalized to tubulin is shown to the right. Data shown represent the average of three experiments. Error bars correspond to one standard deviation. (D) Inhibition of GSK3β leads to stabilization of PGC-1α in nonoxidatively stressed primary neurons but not in oxidatively stressed neurons. Primary embryonic mouse brain neurons in the presence of antioxidants or upon removal of antioxidants were treated with the GSK3β inhibitor SB216763 for 4 h prior to harvest. Western blot showing PGC-1α protein level is shown on the left. Quantification of PGC-1α relative to β-actin is shown on the right. Data shown represent the average of two experiments. Error bars correspond to the variance of the means. (E,F) Reduction of Cdc4 levels and oxidative stress lead to similar increases in PGC-1α protein levels and in Txn2 transcript levels. (E) Primary mouse brain neurons were transduced with control (super) or recombinant Cdc4 shRNA adenovirus and then 24 h later neurons were harvested in the presence of antioxidants or 4 h after removal of antioxidants. (Left) PGC-1α and Cdc4α levels were determined by Western blotting. Quantification of PGC-1α relative to tubulin is shown on the right. Data shown represent the average of two experiments. Error bars correspond to the variance of the means. (F) Txn2 transcript levels determined by real-time PCR in the same experiment described in E. Normalization is to the cyclophilin transcript. Error bars correspond to the standard error from three experiments. P values given are based on Student’s t-test.
Discussion
PGC-1α is an important regulator of diverse energy metabolism programs in mammals, and decreases in PGC-1α expression have been linked to diabetes, heart disease, and neurodegenerative disorders (Finck and Kelly 2006; Handschin and Spiegelman 2006). While several studies have elucidated mechanisms that regulate PGC-1α mRNA expression (Handschin and Spiegelman 2006), the mechanisms that determine PGC-1α protein levels and ultimately PGC-1α function are not known. Here we demonstrate that one mechanism that regulates PGC-1α protein levels is ubiquitin-mediated proteolysis initiated by the E3 ubiquitin ligase SCFCdc4. Our results clearly demonstrate that the nuclear isoforms of Cdc4 function as negative regulators of PGC-1α. We also demonstrate that SCFCdc4 targets an active form of PGC-1α that has been phosphorylated by p38 MAPK and GSK3β. Cdc4 recognizes PGC-1α protein motifs that are highly conserved in the other two PGC-1 family members, PGC-1β and PRC. Thus, our findings are likely to have wider implications for a regulatory role of Cdc4 in PGC-1-dependent pathways beyond PGC-1α.
Recently, numerous observations have led to the realization that ubiquitylation and ubiquitin-dependent protein degradation are important components of transcriptional regulation (Perissi and Rosenfeld 2005). Many transcriptional activation domains also serve as potent degradation motifs, and the two activities are often inseparable (Molinari et al. 1999; Salghetti et al. 2001). Frequently, activation is coupled to degradation such that one signal both activates a transcription factor and targets it for degradation (Lonard et al. 2000). In some cases, ubiquitylation is in fact stimulatory for transcription (Lipford et al. 2005). The most attractive explanation for this seemingly unproductive relationship is that the coupling of degradation to transcription ensures that transcription can be exquisitely responsive to regulatory signals. We demonstrated that PGC-1α proteolysis is similarly linked to transcriptional activation: Phosphorylation of CPD sequences both activates PGC-1α (phosphorylation site mutants exhibit reduced transcriptional activity) and provides the signal for ubiquitin-mediated degradation by SCFCdc4. Ubiquitylation of another transcription factor, SREBP, by SCFCdc4 has been demonstrated to occur specifically at relevant promoters (Punga et al. 2006). It is possible that this may, in fact, be the case for SCFCdc4-mediated ubiquitylation of PGC-1α as well. Indeed, the kinetics of PGC-1α degradation in neurons suggests that two pools exist, only one of which is targeted by SCFCdc4. We speculate that the SCFCdc4-sensitive pool corresponds to transciptionally active PGC-1α, since the same phosphorylation events that render PGC-1α transcriptionally active also mediate its ubiquitylation and degradation (Figs. 1D, 5C). For such transcriptional regulators, a single molecule that is sequentially activated and degraded by phosphorylation and ubiquitylation in situ will be limited to activating only one round of transcription, thereby ensuring that the activation signal does not persist. It is also possible that degradation of PGC-1α at target promoters facilitates sequential recruitment of factors in ordered assemblies, allowing rapid conversion to activating complexes or potentiating transition from the initiation phase to the elongation phase of transcription (Conaway et al. 2002; Muratani and Tansey 2003).
A recently described function of PGC-1α is the induction of genes involved in the antioxidant defense system (Valle et al. 2005). These findings suggest that PGC-1α does not only enhance cellular oxidative metabolism but also endows cells with an increased ability to neutralize toxic byproducts of metabolism, such as reactive oxygen species. Negative regulation of PGC-1α by SCFCdc4 suggests a potential mechanism for allowing PGC-1α to accumulate in response to oxidative stress. We found that in neurons responding to oxidative stress, PGC-1α levels and transcriptional target gene induction increase with similar kinetics to Cdc4α reduction (Fig. 7). This reduction in Cdc4 levels can, in principle, account for the observed concomitant increase in PGC-1α levels, since a similar reduction of Cdc4 mediated by shRNA gene silencing leads to an equivalent increase in PGC-1α levels and PGC-1α-dependent induction of the antioxidant gene Txn2. Indeed, since the phosphorylation events that target PGC-1α to SCFCdc4 are those that render PGC-1α transcriptionally active, reducing Cdc4 levels would be expected to have a positive impact on PGC-1α-dependent transcription. The mechanism for Cdc4 protein reduction is under investigation, but appears to be post-transcriptional, since there is no apparent change in Cdc4 mRNA levels (data not shown). Another contributing factor to PGC-1α accumulation may be inhibition of GSK3β. Phosphorylation of GSK3β on Ser9 by AKT/PKB has been shown to be inhibitory (Cross et al. 1995; Pap and Cooper 1998; Datta et al. 1999). We show in Figure 7 that GSK3β Ser9 phosphorylation increases significantly in neurons in response to oxidative stress. Since the primary phosphophodegron of PGC-1α, CPD2, depends on GSK3β phosphorylation at T295 (Figs. 3, 4), inhibition of GSK3β would be expected to stabilize PGC-1α. These data, taken together, suggest that under conditions of oxidative stress, PGC-1α stability in neurons may be regulated both at the level of substrate availability and ubiquitin ligase concentration. Although it has been presumed that SCF ubiquitin ligases such as SCFCdc4 are regulated primarily at the level of substrate phosphorylation, there is increasing evidence in mammalian cells of regulation of F-box protein expression as well. For example, targeting of p27Kip1 by the ubiquitin ligase SCFSkp2 has been shown to be regulated both at the level of p27 phosphodegron phosphorylation and Skp2 abundance (Carrano et al. 1999; Sutterluty et al. 1999; Tsvetkov et al. 1999).
The GSK3β phosphorylation data presented here suggest that AKT/PKB is activated in response to oxidative stress in primary embryonic mouse brain neurons, as has been determined for other neuronal systems (Crossthwaite et al. 2002; Halvorsen et al. 2002). It has been recently reported that in hepatocytes, activation of AKT/PKB by insulin directly inhibits PGC-1α by phosphorylation on Ser570 (Li et al. 2007). It is unclear whether similar negative regulation of PGC-1α by AKT/PKB occurs in neurons in response to oxidative stress, although the transcriptional data reported here would suggest not. Certainly, the role of AKT/PKB activation in repressing gluconeogenesis and fatty acid oxidation in response to insulin in hepatocytes is quite tissue-specific and distinct from its role in neurons, where different transcription factors and target genes are coactivated. Therefore, alternative regulatory consequences for PGC-1α are likely with respect to AKT/PKB activation in neurons responding to oxidative stress.
Overexpression of Cdc4 in neurons resulted in reduced expression of the PGC-1α-dependent antioxidant defense gene Txn2 (Fig. 6). The finding that elevated Cdc4 levels reduce PGC-1α levels and target gene transcription may be significant for human diseases, and in particular neurodegenerative diseases associated with oxidative damage (Lin and Beal 2006). For example, Parkinson’s disease is thought to result from cumulative oxidative damage to the dopaminergic neurons of the substantia nigra region of the brain (Abou-Sleiman et al. 2006; Farrer 2006). It will be interesting to determine if Cdc4 levels are also increased in Parkinson’s-affected brains leading to reduced PGC-1α levels and possibly accounting for increased oxidative damage.
Materials and methods
Cell culture, media, and reagents
HEK293A and HEK293T cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen) with 25 mM glucose and supplemented with 10% newborn calf serum, 2 mM L-glutamine, penicillin, and streptomycin. Primary neurons were isolated from E16 C57/B6 mouse embryos using established protocols. Briefly, whole brains were harvested into Hank’s buffered saline (HBSS; Invitrogen, catalog no. 14,175) and incubated with 0.125% trypsin for 15 min at 37°C. Trypsinized brains were washed five times with HBSS, mechanically dissociated into single-cell suspension, and plated at a density of 4 × 104 cells per milliliter in six-well culture dishes (final density ∼106 cells per well) in DMEM + GlutaMAX-1 (Invitrogen, catalog no. 10566) with 4.5 mM glucose and supplemented with 10% fetal bovine serum, penicillin, and streptomycin. After 24 h, the medium was replaced with Neurobasal medium (Invitrogen, catalog no. 21,103), supplemented with B-27 (Invitrogen, catalog no. 17504), penicillin, and streptomycin. The medium was changed every day after day in vitro 3 (DIV3).
Oxidative stress in primary embryonic mouse neurons
Neuronal cultures from E16 mice were subjected to oxidative stress by transferring cells from the regular growth medium: Neurobasal, 2% B-27, 0.5 mM GlutaMAX, pen/strep, to medium lacking antioxidants: Neurobasal, 2% B-27 minus AO (Invitrogen, catalog no. 10,889), 0.5 mM GlutaMAX and pen/strep. The antioxidants contained in B-27 are vitamin E, vitamin E acetate, superoxide dismutase, catalase, and glutathione. For protein analysis, cells were scraped off the tissue culture plates, spun down, and washed in HBSS before lysing in 0.5% NP-40, 10 mM Tris-HCl (pH, 7.5), 5 mM EDTA, and 150 mM NaCl, 10 μg/mL aprotinin, 5 μg/mL leupeptin, 5 μg/mL pepstatin A, and 100 μM PMSF. Lysates were sonicated for 10 sec and boiled for 10 min in 2× Lammeli buffer. For RNA analysis, the medium was removed and cells were lysed in Trizol and stored at −80°C until analysis.
Transient transfections and immunoprecipitations
HEK293T cells were transiently transfected using the calcium phosphate precipitation method. Cells were lysed 48 h later in lysis buffer 1 (100 mM Tris-HCl at pH 8.0, 100 mM NaCl, 5 mM EDTA, 5% glycerol, 0.1% NP-40) for purification of Flag-tagged proteins, or lysis buffer 2 (50 mM HEPES at pH 7.5, 500 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.5% Tween) for purification of GST-tagged proteins. Lysis buffers were supplemented with 1 mM DTT, phosphatase inhibitors (50 mM NaF, 1 mM Na3VO4), and protease inhibitors (10 μg/mL aprotinin, 5 μg/mL leupeptin, 5 μg/mL pepstatin A, 100 μM PMSF). Lysates were clarified by centrifugation and bound overnight to 10 μL of M2 agarose (Sigma #A2220) or glutathione sepharose 4B (Amersham Biosciences). For PGC-1α immunoprecipitations, 1 μL of rabbit polyclonal anti-PGC-1α antibody was incubated with the above lysates overnight and 10 μL of GammaBind G sepharose (Amersham Biosciences) were added for 2 h prior to washing. Lysates and immonoprecipitates were separated on 8% SDS-PAGE gels, transferred to Immobilon-P PVDF membranes (Millipore #IPVH00010), and incubated with primary antibodies overnight.
In vivo ubiquitylation assay
HEK293T cells were transfected as above. Forty hours post-transfection, cells were transferred to medium containing 25 μM MG-132 (Biomol #PI102) for 4 h prior to harvesting. Cells were then lysed in RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) containing the phosphatase and protease inhibitors described above, 5 μM MG-132, and 10 mM N-ethylmaleimide (Calbiochem #34115). Lysates were precleared with GammaBind G sepharose, and Flag-tagged PGC-1α was immunoprecipitated overnight using 3 μL of M2 Agarose. The immunoprecipitates were washed extensively with RIPA buffer adjusted to 500 mM NaCl, separated on 3%–8% NuPAGE Tris-Acetate gradient gels (Invitrogen #EA0375), and blotted as described above.
Cycloheximide half-life experiments
HEK293T cells were transfected as described above in 10-cm dishes. Twenty-four hours later, the transfected cells were split into six-well plates. After an additional 24 h, cells were transferred to medium containing 500 μM cycloheximide (Sigma #C7698). Cells were lysed in RIPA buffer containing protease and phosphatase inhibitors as described above at incremental times after cycloheximide addition. Neuronal cultures from E16 mice were transduced with a recombinant adenovirus expressing shRNA against Cdc4 or with a control adenovirus, on DIV8. Twenty-four hours later, 500 μM cycloheximide was added to the cultures and cells were harvested at indicated time points after cycloheximide addition. For extraction and detection of PGC-1α protein by Western blotting, neurons were harvested as described above.
In vitro kinase assay
Alleles of PGC-1α-N-482 were expressed and purified from E. coli as described previously. GSK3β and activated p38 MAPK were purchased from Calbiochem (catalog nos. 361524 and 559324, respectively). The priming phosphorylation was performed at 30°C in 10 mM Tris-HCl (pH 8.0), 1 mM DTT, and 100 μM ATP with activated p38 MAPK. After 30 min, samples were diluted 1–4 into buffer containing 10 μM SB202190, γ-[32P]-ATP, and GSK3β and allowed to incubate for 30 additional minutes. Radiolabel incorporation was determined using a PhosphorImager screen.
Transient transfection and transcriptional activity reporter assays
COS7 cells were grown in DMEM containing 25 mM glucose and supplemented with 10% FBS, 2 mM L-glutamine, penicillin, and streptomycin. Cells were transfected by calcium phosphate precipitation. For direct transactivation experiments, COS7 cells plated in 12-well plates received 50 ng of primary effector (pcDNA3-Gal4DBD, pcDNA3-Gal4DBD-VP16, pcDNA-Gal4DBD-hPGC1α, or the indicated mutants of pcDNA-Gal4DBD-hPGC1α), 100 ng of secondary effector (pcDNA3-GST, GST-hCdc4α, GST-hCdc4β, or GST-hCdc4γ), 50 ng of CMV-βGal, and 250 ng of the Gal4-responsive luciferase reporter pGK-1. For PGC-1α coactivation experiments, COS7 cells were transfected with 50 ng of primary effector (pcDNA3 or pcDNA3-HA-hPGC1α), 100 ng of secondary effector (pcDNA3-GST, GST-hCdc4α, GST-hCdc4β, or GST-hCdc4γ), 50 ng of CMV-βGal, and 100 ng of the estrogen-related receptor (ERR)-responsive promoter ERRE-luciferase (derived from a fragment of the ERRα promoter) (Teyssier et al. 2005). Cell lysates from both direct transactivation and coactivation experiments were harvested 48 h post-transfection and assayed for both β-galactosidase and luciferase activities. Luciferase activity was first normalized against β-galactosidase activity (to control for transfection efficiency), and is expressed relative to the activity of wild-type Gal4-VP16, Gal4-PGC1α, or PGC1α. Data from two or three experiments (each performed in triplicate) are expressed as the average ± the standard error of the mean.
Quantitative analysis of mRNA abundance
Total RNA was isolated from primary murine neuronal cultures Trizol (Invitrogen). Four-hundred nanograms of total RNA and oligo dT primers were used to synthesize cDNA with SuperScript II RT (Invitrogen). cDNA levels were determined by real-time PCR incorporating SYBR green on a Chromo4 real-time PCR machine (Bio-Rad). Transcript levels were normalized against the abundance of ribosomal subunit protein Arbp (36B4) or cyclophilin mRNAs, and are expressed relative to the message levels in control samples. Primer sequences will be provided on request. Values represent the average ± the standard error of the mean of three to six independent samples.
siRNA transfection and adenoviral transduction
Cultures were transfected 2 d in a row, on DIV7 and DIV8, with siRNA oligos using Transmessenger transfection reagent (Qiagen), or transduced with recombinant adenovirus on DIV8. Adenoviral transductions were performed by adding 2 × 1010 adenovirus particles per well of a six-well plate, directly to the medium (2 mL), with no medium change. Twenty-four hours after the second transfection, or after adenoviral transduction, cells were harvested for either protein or RNA analysis as described above. An adenovirus expressing shRNA targeting expression of Cdc4 was generated using previously described vectors and adenovirus construction techniques (Schreiber et al. 2003). The Cdc4 target sequence is 5′-GAGTGGATCTCTT GATACA-3′, which resides in Exon 10.
GSK3β inhibition experiment
Neuronal cultures from E16 mice were incubated with the specific GSK3β inhibitor SB216763 (Sigma) for 4 h on DIV8. The inhibitor was used at a final concentration of 50 μM. Control cells were incubated with an equal volume of DMSO. The experiment was performed in the presence or absence of antioxidants. Cells were harvested for protein and RNA analysis as described above.
Antibodies
Rabbit polyclonal antibodies against Cdc4 were prepared as described (Strohmaier et al. 2001). Rabbit polyclonal PGC-1α antibodies have been described previously (Schreiber et al. 2003). Mouse monoclonal antibody against α-tubulin (DMA1) and against Flag epitope (M2, #A2220) were purchased from Sigma. GSK3β rabbit monoclonal antibody (#9315), phospho-GSK3β (Ser9) rabbit polyclonal antibody (#9336), p38 MAPK mouse monoclonal and antibody (#9228), and phospho-p38 MAPK rabbit monoclonal antibody (#9215) were purchased from Cell Signaling Technology. GST mouse monoclonal antibody (#sc-138) and GAPDH rabbit polyclonal antibody (#sc-25778) were purchased from Santa Cruz Biotechnology, Inc. HA rabbit polyclonal antibody (#18848-01) was purchased from QED Bioscience, Inc. β-Actin rabbit polyclonal (#ab-8227) was purchased from Abcam, Inc.
Acknowledgments
We thank Donato Tedesco for constructing the Cdc4α-expressing adenovirus and Marc Montminy for the PGC-1α shRNA adenovirus. We also thank Frank van Drogen for help with experiments and Erin Olson for critical reading of the manuscript. B.L.O. and S.E.R were supported by fellowships from the American Cancer Society. This work was supported in part by grant SPF-1574 from Novartis and NIH grant CA78343 to S.I.R. and NIH Grant DK64951 to A.K.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1624208
References
- Abou-Sleiman P.M., Muqit M.M., Wood N.W. Expanding insights of mitochondrial dysfunction in Parkinson’s disease. Nat. Rev. Neurosci. 2006;7:207–219. doi: 10.1038/nrn1868. [DOI] [PubMed] [Google Scholar]
- Cardozo T., Pagano M. The SCF ubiquitin ligase: Insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Carrano A.C., Eytan E., Hershko A., Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Chang L., Karin M. Mammalian MAP kinase signalling cascades. Nature. 2001;410:37–40. doi: 10.1038/35065000. [DOI] [PubMed] [Google Scholar]
- Conaway R.C., Brower C.S., Conaway J.W. Emerging roles of ubiquitin in transcription regulation. Science. 2002;296:1254–1258. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- Cross D.A., Alessi D.R., Cohen P., Andjelkovich M., Hemmings B.A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Crossthwaite A.J., Hasan S., Williams R.J. Hydrogen peroxide-mediated phosphorylation of ERK1/2, Akt/PKB and JNK in cortical neurones: Dependence on Ca2+ and PI3-kinase. J. Neurochem. 2002;80:24–35. doi: 10.1046/j.0022-3042.2001.00637.x. [DOI] [PubMed] [Google Scholar]
- Cui L., Jeong H., Borovecki F., Parkhurst C.N., Tanese N., Krainc D. Transcriptional repression of PGC-1α by mutant huntingtin leads to mitochondrial dysfunction and neurodegeneration. Cell. 2006;127:59–69. doi: 10.1016/j.cell.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Datta S.R., Brunet A., Greenberg M.E. Cellular survival: A play in three Akts. Genes & Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- Fan M., Rhee J., St-Pierre J., Handschin C., Puigserver P., Lin J., Jaeger S., Erdjument-Bromage H., Tempst P., Spiegelman B.M. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: Modulation by p38 MAPK. Genes & Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer M.J. Genetics of Parkinson disease: Paradigm shifts and future prospects. Nat. Rev. Genet. 2006;7:306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- Finck B.N., Kelly D.P. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J. Clin. Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory M.A., Qi Y., Hann S.R. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J. Biol. Chem. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israel A. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Halvorsen E.M., Dennis J., Keeney P., Sturgill T.W., Tuttle J.B., Bennett J.B. Methylpyridinium (MPP+)- and nerve growth factor-induced changes in pro- and anti-apoptotic signaling pathways in SH-SY5Y neuroblastoma cells. Brain Res. 2002;952:98–110. doi: 10.1016/s0006-8993(02)03216-x. [DOI] [PubMed] [Google Scholar]
- Handschin C., Spiegelman B.M.2006. Peroxisome proliferator-activated receptor γ coactivator 1 coactivators, energy homeostasis, and metabolism Endocr. Rev. 27728–735. [DOI] [PubMed] [Google Scholar]
- Handschin C., Rhee J., Lin J., Tarr P.T., Spiegelman B.M. An autoregulatory loop controls peroxisome proliferator-activated receptor γ coactivator 1α expression in muscle. Proc. Natl. Acad. Sci. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard E.J., Wu G., Kitajewski J., Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes & Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutti D., Kressler D., Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc. Natl. Acad. Sci. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepp D.M., Schaefer L.K., Ye X., Keyomarsi K., Chu C., Harper J.W., Elledge S.J. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294:173–177. doi: 10.1126/science.1065203. [DOI] [PubMed] [Google Scholar]
- Leone T.C., Lehman J.J., Finck B.N., Schaeffer P.J., Wende A.R., Boudina S., Courtois M., Wozniak D.F., Sambandam N., Bernal-Mizrachi C., et al. PGC-1α deficiency causes multi-system energy metabolic derangements: Muscle dysfunction, abnormal weight control and hepatic steatosis. PLoS Biol. 2005;3:e101. doi: 10.1371/journal.pbio.0030101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Monks B., Ge Q., Birnbaum M.J. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1α transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- Lin M.T., Beal M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- Lin J., Wu P.H., Tarr P.T., Lindenberg K.S., St-Pierre J., Zhang C.Y., Mootha V.K., Jager S., Vianna C.R., Reznick R.M., et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell. 2004;119:121–135. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lipford J.R., Smith G.T., Chi Y., Deshaies R.J. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- Lonard D.M., Nawaz Z., Smith C.L., O’Malley B.W. The 26S proteasome is required for estrogen receptor-α and coactivator turnover and for efficient estrogen receptor-α transactivation. Mol. Cell. 2000;5:939–948. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- Mielke K., Herdegen T. JNK and p38 stresskinases—Degenerative effectors of signal-transduction-cascades in the nervous system. Prog. Neurobiol. 2000;61:45–60. doi: 10.1016/s0301-0082(99)00042-8. [DOI] [PubMed] [Google Scholar]
- Moberg K.H., Bell D.W., Wahrer D.C., Haber D.A., Hariharan I.K. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature. 2001;413:311–316. doi: 10.1038/35095068. [DOI] [PubMed] [Google Scholar]
- Moberg K.H., Mukherjee A., Veraksa A., Artavanis-Tsakonas S., Hariharan I.K. The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr. Biol. 2004;14:965–974. doi: 10.1016/j.cub.2004.04.040. [DOI] [PubMed] [Google Scholar]
- Molinari E., Gilman M., Natesan S. Proteasome-mediated degradation of transcriptional activators correlates with activation domain potency in vivo. EMBO J. 1999;18:6439–6447. doi: 10.1093/emboj/18.22.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Mootha V.K., Handschin C., Arlow D., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., et al. Errα and Gabpa/b specify PGC-1α-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. 2004;101:6570–6575. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M., Tansey W.P. How the ubiquitin–proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Nash P., Tang X., Orlicky S., Chen Q., Gertler F.B., Mendenhall M.D., Sicheri F., Pawson T., Tyers M. Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature. 2001;414:514–521. doi: 10.1038/35107009. [DOI] [PubMed] [Google Scholar]
- Nateri A.S., Riera-Sans L., Da Costa C., Behrens A. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science. 2004;303:1374–1378. doi: 10.1126/science.1092880. [DOI] [PubMed] [Google Scholar]
- Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- Oberg C., Li J., Pauley A., Wolf E., Gurney M., Lendahl U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 2001;276:35847–35853. doi: 10.1074/jbc.M103992200. [DOI] [PubMed] [Google Scholar]
- Orlicky S., Tang X., Willems A., Tyers M., Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Pap M., Cooper G.M. Role of glycogen synthase kinase-3 in the phosphatidylinositol 3-Kinase/Akt cell survival pathway. J. Biol. Chem. 1998;273:19929–19932. doi: 10.1074/jbc.273.32.19929. [DOI] [PubMed] [Google Scholar]
- Patti M.E., Butte A.J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V., Rosenfeld M.G. Controlling nuclear receptors: The circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Pickart C.M., Eddins M.J. Ubiquitin: Structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Puigserver P., Rhee J., Lin J., Wu Z., Yoon J.C., Zhang C.Y., Krauss S., Mootha V.K., Lowell B.B., Spiegelman B.M. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell. 2001;8:971–982. doi: 10.1016/s1097-2765(01)00390-2. [DOI] [PubMed] [Google Scholar]
- Punga T., Bengoechea-Alonso M.T., Ericsson J. Phosphorylation and ubiquitination of the transcription factor sterol regulatory element-binding protein-1 in response to DNA binding. J. Biol. Chem. 2006;281:25278–25286. doi: 10.1074/jbc.M604983200. [DOI] [PubMed] [Google Scholar]
- Rangwala S.M., Li X., Lindsley L., Wang X., Shaughnessy S., Daniels T.G., Szustakowski J., Nirmala N.R., Wu Z., Stevenson S.C. Estrogen-related receptor α is essential for the expression of antioxidant protection genes and mitochondrial function. Biochem. Biophys. Res. Commun. 2007;357:231–236. doi: 10.1016/j.bbrc.2007.03.126. [DOI] [PubMed] [Google Scholar]
- Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Salghetti S.E., Caudy A.A., Chenoweth J.G., Tansey W.P. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- Sano M., Tokudome S., Shimizu N., Yoshikawa N., Ogawa C., Shirakawa K., Endo J., Katayama T., Yuasa S., Ieda M., et al. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor γ coactivator 1α. J. Biol. Chem. 2007;282:25970–25980. doi: 10.1074/jbc.M703634200. [DOI] [PubMed] [Google Scholar]
- Schreiber S.N., Knutti D., Brogli K., Uhlmann T., Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor α (ERRα) J. Biol. Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- Schreiber S.N., Emter R., Hock M.B., Knutti D., Cardenas J., Podvinec M., Oakeley E.J., Kralli A. The estrogen-related receptor α (ERRα) functions in PPARγ coactivator 1α (PGC-1α)-induced mitochondrial biogenesis. Proc. Natl. Acad. Sci. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck C.H., Strohmaier H., Sangfelt O., Muller H.M., Hubalek M., Muller-Holzner E., Marth C., Widschwendter M., Reed S.I. hCDC4 gene mutations in endometrial cancer. Cancer Res. 2002;62:4535–4539. [PubMed] [Google Scholar]
- St-Pierre J., Drori S., Uldry M., Silvaggi J.M., Rhee J., Jager S., Handschin C., Zheng K., Lin J., Yang W., et al. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- Strohmaier H., Spruck C.H., Kaiser P., Won K.A., Sangfelt O., Reed S.I. Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature. 2001;413:316–322. doi: 10.1038/35095076. [DOI] [PubMed] [Google Scholar]
- Sundqvist A., Bengoechea-Alonso M.T., Ye X., Lukiyanchuk V., Jin J., Harper J.W., Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7) Cell Metab. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Sutterluty H., Chatelain E., Marti A., Wirbelauer C., Senften M., Muller U., Krek W. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1999;1:207–214. doi: 10.1038/12027. [DOI] [PubMed] [Google Scholar]
- Teyssier C., Ma H., Emter R., Kralli A., Stallcup M.R. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes & Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov L.M., Yeh K.H., Lee S.J., Sun H., Zhang H. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 1999;9:661–664. doi: 10.1016/s0960-9822(99)80290-5. [DOI] [PubMed] [Google Scholar]
- Valle I., Alvarez-Barrientos A., Arza E., Lamas S., Monsalve M. PGC-1α regulates the mitochondrial antioxidant defense system in vascular endothelial cells. Cardiovasc. Res. 2005;66:562–573. doi: 10.1016/j.cardiores.2005.01.026. [DOI] [PubMed] [Google Scholar]
- van Drogen F., Sangfelt O., Malyukova A., Matskova L., Yeh E., Means A.R., Reed S.I. Ubiquitylation of cyclin E requires the sequential function of SCF complexes containing distinct hCdc4 isoforms. Mol. Cell. 2006;23:37–48. doi: 10.1016/j.molcel.2006.05.020. [DOI] [PubMed] [Google Scholar]
- Wei W., Jin J., Schlisio S., Harper J.W., Kaelin W.G. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Welcker M., Singer J., Loeb K.R., Grim J., Bloecher A., Gurien-West M., Clurman B.E., Roberts J.M. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- Welcker M., Orian A., Grim J.E., Eisenman R.N., Clurman B.E. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr. Biol. 2004a;14:1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- Welcker M., Orian A., Jin J., Grim J.E., Harper J.W., Eisenman R.N., Clurman B.E. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. 2004b;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems A.R., Schwab M., Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochim. Biophys. Acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K.I. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23:2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X., Nalepa G., Welcker M., Kessler B.M., Spooner E., Qin J., Elledge S.J., Clurman B.E., Harper J.W. Recognition of phosphodegron motifs in human cyclin E by the SCF(Fbw7) ubiquitin ligase. J. Biol. Chem. 2004;279:50110–50119. doi: 10.1074/jbc.M409226200. [DOI] [PubMed] [Google Scholar]