Abstract
The intracellular parasite Cryptosporidium parvum develops inside a vacuole at the apex of its epithelial host cell. The developing parasite is separated from the host cell cytoplasm by a zone of attachment that consists of an extensively folded membranous structure known as the feeder organelle. It has been proposed that the feeder organelle is the site of regulation of transport of nutrients and drugs into the parasite. In this report, we localize an ≈200-kDa integral membrane protein, CpABC, from Cryptosporidium parvum to the host–parasite boundary, possibly the feeder organelle. The predicted amino acid sequence of CpABC has significant structural similarity with the cystic fibrosis conductance regulator and the multidrug resistance protein subfamily of ATP-binding cassette proteins. This is an example of a parasite-encoded transport protein localized at the parasite–host interface of an intracellular protozoan.
Cryptosporidium parvum is an intracellular apicomplexan protozoan parasite that causes cryptosporidiosis, an important enteric disease worldwide (1). Cryptosporidiosis is usually self-limiting in immunocompetant individuals, but it is an opportunistic infection in persons with the acquired immunodeficiency syndrome, in whom the disease is profoundly debilitating and life-threatening (2). There is currently no consistently effective treatment for cryptosporidiosis even though a wide spectrum of drugs have been tested, both in vitro and in vivo (3, 4).
Identification of potential drug targets, with a view to develop an effective therapy, is a priority for C. parvum. Transporter proteins that regulate the movement of ions, nutrients, etc. into this intracellular parasite are one possible target. C. parvum invades intestinal epithelial cells of the gastrointestinal tract. In common with other members of the phylum Apicomplexa, it develops inside a vacuole in the host cytoplasm, although it is distinct in that the vacuole and parasite remain at the apex of the host cell (5). The developing parasite is separated from the host cell cytoplasm by a zone of attachment that consists of an extensively folded membrane structure known as the feeder organelle (6). It has been proposed that the feeder organelle regulates access of nutrients or drugs to this parasite (5–7), although there is one report that paromomycin enters the parasitophorous vacuole via the apical host membrane (8).
In an effort to understand the transport pathways that operate between the host cell and the intracellular parasite, we selected to identify transporters that belong to the ATP-binding cassette (ABC) protein superfamily. ABC proteins are found in all major taxa, and most of them are integral membrane proteins (9). They are associated with xenobiotic resistance phenotypes in bacteria, protozoa, fungi, nematodes, and mammals (9). Their transport substrates are extremely diverse (9). Some ABC transport proteins, such as the multidrug resistance P-glycoprotein, transport unmodified substrates (10) whereas other transporters, such as multidrug resistance protein 1 (MRP1) and related proteins, transport a range of substrates that are conjugated to glutathione, glucuronide, or sulfate (11–13). The role of P-glycoprotein and MRP1 in multidrug resistance in cancer cells is well documented (14, 15). Another ABC protein, the cystic fibrosis conductance regulator (CFTR), is a gated chloride channel (16).
In this paper, we report the detailed characterization and localization of a C. parvum ABC protein, CpABC, that shares conserved features of protein structure with MRP1 and CFTR. Significantly, CpABC localizes to the feeder organelle. The localization of CpABC to the host–parasite boundary of an intracellular parasite has important implications for the design of drug therapy for cryptosporidiosis.
MATERIALS AND METHODS
Parasites.
The KSU-1 isolate of C. parvum (genotype 2) was used in this study. Oocysts were a gift from S .J. Upton (Kansas State University) (17). Asexual intracellular stages were cultured in the human colonic carcinoma (Caco-2) cell line on Transwells (Costar) as described (18).
Genomic Libraries and Probes.
Two genomic C. parvum libraries were screened by plaque hybridization: a λZAPII GCH1 genomic DNA (gDNA) library (from the National Institutes of Health AIDS Research and Reference Reagent Program) and a λGEM 11 partial Sau 3A SFGH1 gDNA library (19) (kindly provided by R. Nelson, University of California at San Francisco). The SFGH1 and GCH1 isolates belong to the two recognized C. parvum genotypes (genotypes 1 and 2, respectively) (19, 20). The complete CpABC gene was cloned by screening the SFGH1 gDNA library with the CpABC-p22 probe (see below). The CpABC gene was subcloned from λ clone P4C1 into pBluescript II SK(+) (Stratagene) as a 6.6-kilobase (kb) BamHI restriction fragment and a 4.4-kb PCR product (see below).
The following DNA probes were used in the study. CpABC-ATP, a 405-bp PCR product described previously (21); CpABC-p22, a 966 bp EcoRI restriction fragment from the GCH1 gDNA library identified by screening the library with CpABC-ATP; and CpABC-3.38, a 3.38-kb (SpeI to BamHI) region of the CpABC ORF. Standard protocols were used throughout for the manipulation of phage, plasmids, and DNA (22). DNA probes were prepared from gel-purified fragments of plasmids by using random priming with [α32P]dATP (NEN).
RNA Analysis.
RNA purification and Northern blot analysis were performed as described (21, 23).
PCR.
Standard techniques were used for the PCR. The PCR was used to amplify the 3′ end of the CpABC gene from P4C1 with two primers based on sequences 5′ of the BamHI site in P4C1 (5′ CTCAAGAACCAAACATTCTTACAGG 3′) and the left arm of λ(5′ GCAACGAACAGGTCACTATCAGTC 3′). PCR amplification of purified phage DNA was performed as described in ref. 24 with an annealing temperature of 67°C. The expected ≈4.4-kb PCR product was generated in the presence of P4C1 DNA.
The PCR was used to amplify the 326-bp region spanning the putative start codon of the CpABC ORF from three phage clones. The primers were 5′CACTCACTCAGGTTAAGAGAC3′ and 5′GTATTGAGGAATCCTC3′. Phage stocks were used directly as template as described in ref. 24. The PCR products were cloned into the pCRII plasmid vector (TA cloning kit, Invitrogen).
Sequence Analysis.
Sequences were obtained by the dideoxy-chain termination method through automated sequencing. The sequence of the CpABC gene was determined on both strands. The sequence data have been submitted to the GenBank database under accession number AF110147. Searches of the DNA databases were performed by the blast server service (http://dot.imgen.bcm.tmc.edu:9331/). The sequences were aligned by using clustalw with default parameters and were displayed by using prettybox gcg software (Genetics Computer Group, Madison, WI) (25).
Production of Anti-CpABC Serum.
A region of CpABC that had low amino acid sequence similarity to other members of the ABC superfamily (amino acids 1,115–1218 in Fig. 1) was selected for expression as a glutathione S-transferase (GST) fusion protein. Two primers were designed to amplify this region and to generate EcoRI sites at each end of the PCR product (5′ GGGAATTCAGAAATATATCAGAAAC 3′ and 5′ CCAACTTGAATTCATCCTTG 3′). The template used was the p22 clone (see above). The 104-amino acid peptide (referred to as RNIS) encoded by the p22-derived PCR product differed from that shown in Fig. 1 at seven amino acid residues (1141 T/A; 1144 G/S; 1162 D/N; 1165 A/V; 1193 V/L; 1199 F/L; and 1207 P/A; where the number is the position of the residue and the first and second letters are the residues in genotype 1 and 2, respectively). All of the experiments with the antiserum described in this study used a genotype 2 C. parvum isolate. The PCR product was cloned into the EcoRI site of the pGEX-5X-1 expression vector (Amersham Pharmacia). Production of the fusion protein, GST-RNIS, was according to the manufacturer’s instructions (Amersham Pharmacia). The size of the major protein was 41 kDa, the correct size of a 12-kDa protein fused to GST (29 kDa).
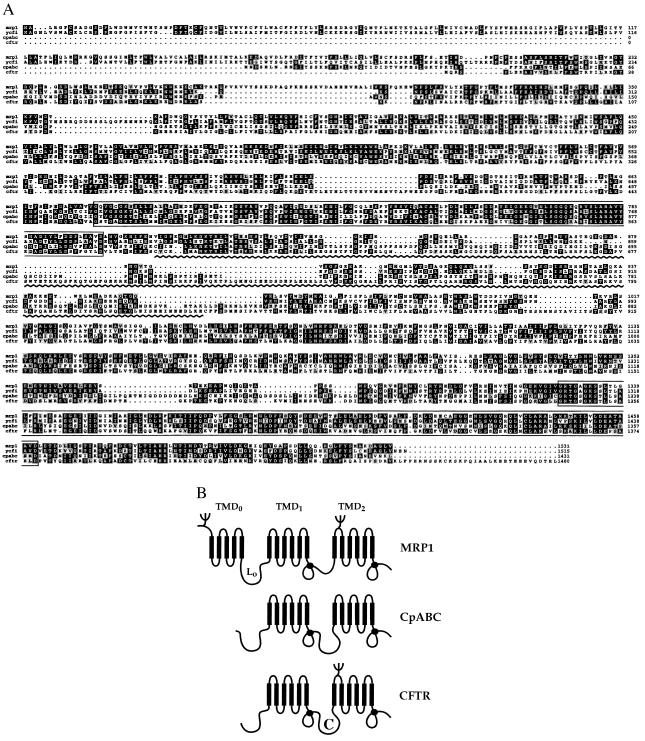
Figure 1.
CpABC resembles CFTR and the MRP subfamily of ABC proteins. (A) Alignment of the predicted amino acid sequence of CpABC with human MRP1 (49), S. cerevisiae YCF1 (50), and human CFTR (29). Database accession numbers are AF110147 (CpABC), P33527 (MRP), P39109 (YCF1), and P13569 (CFTR). Shading indicates agreement with the consensus (two or more identical residues in a column). White on black indicates that the residue is identical to the consensus, black on gray indicates that it is similar or somewhat-similar, and black on white indicates that it is nonsimilar (25). The nucleotide binding folds are boxed. The wavy line indicates the connector domain in CFTR. (B) Schematic representation of proposed transmembrane topology of CpABC based on computer-assisted hydropathy analyses (G. Tusnády, personal communication) and sequence alignments. The transmembrane topology of MRP and CFTR have been established experimentally (35–37, 51, 52). The drawing is modified from ref. 32. L0 is the cytoplasmic linker between TMD0 and TMD1 in MRP. The solid circles indicate the NBFs. C is the connector domain in CFTR.
The GST-RNIS fusion protein was used to immunize a New Zealand white rabbit that was seronegative for Cryptosporidium and GST antibodies (Cocalico Biologicals, Reamstown, PA). The rabbit serum, referred to as RabαGST-CpABC serum, was depleted of GST antibodies by absorption over Sepharose conjugated to GST. Absorbed antiserum was tested by immunoblotting against C. parvum oocysts, GST-RNIS fusion protein, and pMAL-RNIS fusion protein (see below).
The 324 bp RNIS EcoRI fragment also was cloned into the pMAL-c2 expression vector (New England Biolabs) to generate a maltose binding protein (MBP)–RNIS fusion protein. Expression of the MBP–RNIS fusion protein was according to the manufacturer’s instructions. MBP–RNIS coupled to cyanogen bromide-activated Sepharose (Amersham Pharmacia) was mixed with serum for 1 hr and was washed in 0.1 M NaHCO3/0.5 M NaCl (pH 8.3). Antibodies binding to MBP–RNIS were eluted with 0.2 M glycine (pH 2.0) and were dialyzed against PBS. Affinity-purified RabαGST-CpABC antibodies were tested for reactivity against CpABC by immunoblotting C. parvum oocysts.
Immunoblotting.
Proteins isolated from both oocysts and intracellular stages were immunoblotted with RabαGST-CpABC and preimmune sera. Before solubilization, oocysts were preincubated in 5 mM EDTA and 2 mM PMSF for 15 min on ice. Treated oocysts (107) were mixed with PBS containing 1% Triton X-100 and were centrifuged. The supernatant was solubilized in sample buffer (0.1M Tris⋅HCl/2% SDS/10% glycerol/0.1M DTT/bromophenol blue). The pellet was boiled with sample buffer and was loaded immediately on a 7.5% SDS/PAGE gel. For immunoblotting of asexual intracellular stages, an enriched fraction of parasites was released from the infected monolayer by the following procedure. Monolayers of infected cells cultured for 40 hr were washed twice with PBS and then twice with cold water (4°C) containing EDTA and PMSF. The water washes were combined and centrifuged, and the pellet was mixed with PBS containing 1% Triton X-100. The mixture was centrifuged, and the pellet was mixed with PBS containing 1% SDS and was centrifuged. The pellet was boiled with sample buffer. The supernatants from the Triton X-100 and SDS extracts also were boiled with sample buffer and were loaded on a 7.5% SDS/PAGE gel. After electrophoresis, the gel was transferred to nitrocellulose, was blocked in 1% BSA in PBS, and was blotted with GST-absorbed RabαGST-CpABC serum (1:50 dilution), RabαGST-CpABC serum (1:75 dilution), or MBP–RNIS affinity-purified RabαGST-CpABC antiserum. After washing in Tris-saline, 0.1% Tween buffer, the paper was incubated with a second antibody, alkaline phosphatase-conjugated goat anti-rabbit (Bio-Rad), and was color developed by using nitroblue tetrazolium and 5-bromo-4-chloro-3-indoyl phosphate.
Immunofluorescence.
For intracellular stages, it was necessary to release infected cells from the monolayer to visualize CpABC. Release of schizonts and Caco-2 infected cells by water lysis from the monolayer was as described above. Smears of cells and parasites released from the monolayer were air-dried, were fixed in EtOH, and were permeabilized by treatment with −20°C acetone for 5 min. Smears were overlaid with RabαGST-CpABC (diluted 1:75) for 1 hr, were washed in PBS, and incubated with FITC goat anti-rabbit antibody containing 5 mM 4′,6-diamidino-2-phenylindole. The slides were viewed with a Nikon epifluorescent microscope. Preimmune serum was included as a negative control.
RESULTS
Cloning, Sequencing, and RNA Analysis of CpABC.
A PCR product corresponding to a putative ABC transporter, CpABC, in C. parvum has been described (21). To isolate the complete ORF, we cloned and sequenced the CpABC gene from the partial Sau 3A SFGH1 gDNA λGEM11 library (19). There was one large ORF within a 5,206-bp sequence that encoded a putative 1431-amino acid protein (Fig. 1) with a predicted molecular weight of 162 kDa. The CpABC ORF begins with an ATG that is immediately preceded by an in-frame stop codon. The same sequence also was seen in three independent phage clones, and no introns were identified in the 892-bp sequence upstream of the ATG start site (data not shown). Therefore, although we have not identified the translation start site unequivocally, the rarity of introns in C. parvum genes [only 1 of 20 genes analyzed thus far (26)] supports the interpretation that the ATG at the beginning of the ORF is indeed the start site. Northern analysis with the CpABC-3.38 probe reproducibly identified two transcripts in C. parvum sporozoites: a major band of 5.65 kb and a minor band of 7.7 kb (data not shown). The significance of the existence of two transcripts is not known.
CpABC Resembles CFTR and the MRP Subfamily of ABC Proteins.
Analysis of the putative 1,431-amino acid protein revealed CpABC to be a member of the ABC transport protein family (27). Four domains were identified based on protein primary structure. There are two nucleotide binding folds (NBF) with Walker A and B motifs and the characteristic ABC transporter signature sequence (27) (Fig. 1A). Computer-assisted hydropathy analyses identified two membrane-spanning domains [transmembrane domains (TMDs)] with six individual spans per domain (Fig. 1B and data not shown) (28). These domains are arranged as follows: TMD1, NBF1, TMD2, NBF2. The N terminus of CpABC is predicted to be cytoplasmic because no signal sequence was seen. The region between NBF1 and TMD2, called the “regulatory” or “connector” region (C) in CFTR (29), is comprised of 23% charged amino acids and contained two consensus sites for cAMP- and cGMP-dependent protein kinase phosphorylation. Twenty-three potential N-glycosylation sites are present in CpABC.
Sequence comparisons of CpABC with other ABC proteins showed that it grouped with the MRP subfamily and CFTR (12, 30). An alignment of CpABC with MRP1, the yeast cadmium transporter (YCF1), and CFTR is shown in Fig. 1A. As expected, the NBFs show the highest sequence identity. Indeed, in each of these proteins, NBF1 and NBF2 have 122 and 135 residues, respectively. Significantly, the identity and similarity extended beyond the NBFs (Fig. 1A). The percentage similarity and identity of CpABC to YCF1, MRP1, and CFTR are 50 and 33, 48 and 33, and 44 and 28, respectively. CpABC holds some features in common with CFTR; that is, the connector domain is larger than the truncated region found in MRP1 and a MRP1 N-terminal transmembrane domain (TMD0) is absent (31). However, the N-terminal stretch of 104 amino acids preceding TMD1 in CpABC is similar to the cytoplasmic linker (L0) in MRP and YCF1 (32) (Fig. 1). A proposed transmembrane topology for CpABC is shown in Fig. 1B.
CpABC Is an ≈200-kDa Protein Present in Sporozoites and Intracellular Stages.
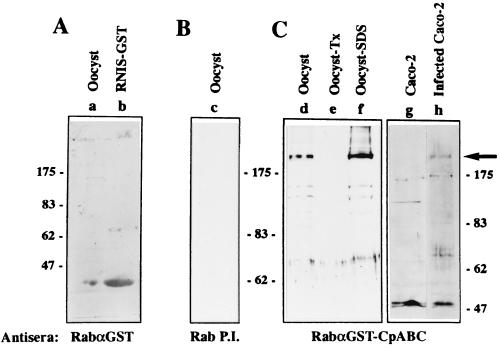
The RabαGST-CpABC serum recognized an ≈200-kDa protein in C. parvum oocysts and in intracellular stages (Fig. 2, lanes d, f, and h). The antiserum also recognized a smaller protein of 70 kDa in intracellular stages. The detection of a 70-kDa protein in intracellular stages was a consistent observation in numerous immunoblots and suggests that, despite the presence of protease inhibitors, there is a rapid breakdown of this protein when isolated from cells. A less likely interpretation is that the endogenous protein is present in two forms, i.e., as 200- and 70-kDa proteins, in intracellular stages. Preimmune serum did not recognize an ≈200-kDa protein (Fig. 2, lane c). Several proteins were recognized in uninfected Caco-2 cells (Fig. 2, lane g). An anti-GST serum did not recognize a 200-kDa protein in oocysts, thereby demonstrating that the ≈200-kDa protein was not a C. parvum GST (Fig. 2, lane a). Furthermore, affinity-purified RabαGST-CpABC antibody recognized a 200-kDa protein and did not react with GST (data not shown). The specificity of the RabαGST-CpABC serum allows us to conclude that the protein is CpABC. CpABC in both oocysts and intracellular stages was only solubilized by boiling in 2% SDS, although the smaller fragments could be solubilized by less severe detergent treatment [i.e., solubilization in 1% Triton X-100 (Fig. 2, lane e)]. This is consistent with it being an integral membrane protein.
Figure 2.
CpABC is an ≈200-kDa integral membrane protein in oocysts and intracellular stages. RabαGST-CpABC serum from a rabbit immunized with the GST-RNIS fusion protein was absorbed against GST and was used to immunoblot extracts of oocysts and intracellular stages of C. parvum. In addition, GST reactivity of RabαGST-CpABC serum was analyzed. Antisera used were rabbit anti-GST antibodies (RabαGST) (A), rabbit pre-immune serum (Rab P.I.) (B) and absorbed RabαGST-CpABC serum (C). Lanes: a, C. parvum oocysts, solubilized in sample buffer by boiling as described; b, bacterial lysate expressing pGEX-GST-RNIS (note spillover into lane a); c. oocysts boiled in sample buffer (2% SDS); d, oocysts boiled in sample buffer; e, 1% Triton X-100 lysate of oocysts; f, pellet remaining after 1% Triton X-100 extract, solubilized by boiling in sample buffer (2% SDS); g, lysate of uninfected Caco-2 cells; h, lysate of Caco-2 cells infected with C. parvum. The arrow indicates the position of the ≈200 kDa CpABC protein.
According to the CpABC ORF, the predicted molecular weight of CpABC is 162 kDa whereas the actual protein migrates at ≈200 kDa on SDS/PAGE. By analogy with other ABC proteins, it is likely that CpABC is glycosylated, and this would explain at least part of the size disparity. The propensity of integral membrane proteins to migrate anomalously in SDS/PAGE is also a factor. However, the possible existence of an additional exon cannot be ruled out.
CpABC Is Located at the Parasite–Host Boundary in Intracellular Stages.
In sporozoites, CpABC is localized internally in an area posterior to the nucleus that is electron dense when visualized by phase contrast microscopy (data not shown). Permeabilization with 1% Triton X-100 did not disrupt the localization, suggesting that CpABC is localized in an organelle or membrane.
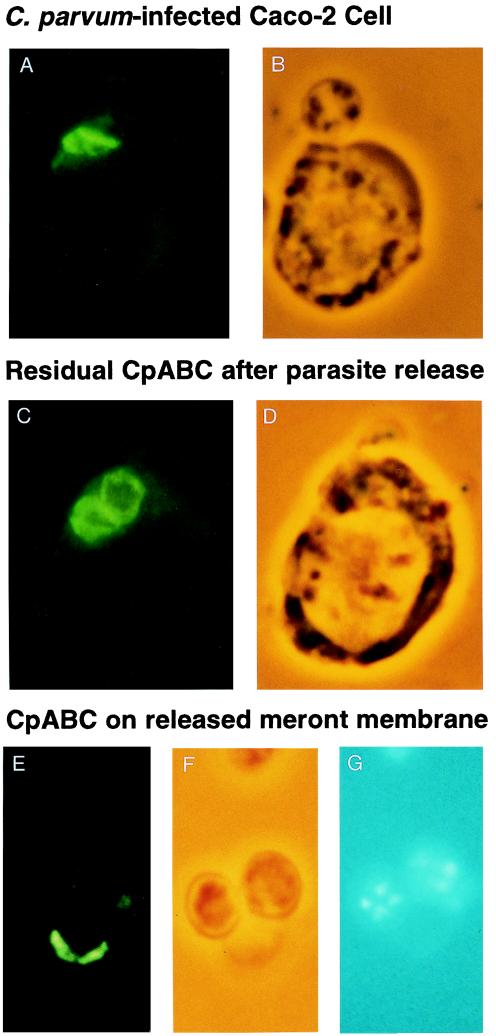
The localization of CpABC in intracellular stages of C. parvum is shown in Fig. 3. CpABC was exclusively localized at the boundary between the parasite and host cell in the region of the feeder organelle (Fig. 3 A and B). The distribution of CpABC between the host cell and the parasite was broad, wider than would be expected for a unit bilayer membrane. This, together with its highly asymmetrical localization, strongly suggests a location at or near the feeder organelle. In infected cells from which the parasite had been released, CpABC remained with the host cell and assumed a donut-like shape (Fig. 3 C and D). In most released parasites (meronts), there was little or no CpABC present; note that two of the three meronts in Fig. 3 E–G were devoid of CpABC. In the few meronts on which CpABC was present, it was localized on one side (Fig. 3 E and F), consistent with it being the attachment zone of the parasite to the host cell. Electron microscopy of infected Caco-2 cells that had been treated with ice-cold water showed that the feeder organelle was present and largely intact and that released meronts were enclosed in an intact parasite membrane (data not shown). This reinforces our interpretation of localization of CpABC and suggests that the feeder organelle is not a modification of the parasite membrane.
Figure 3.
CpABC localizes at the host–parasite interface in intracellular stages. Shown is immunofluorescent localization of CpABC on C. parvum-infected Caco-2 cells removed from the monolayer by treatment with cold water and smeared on slides, air dried, and incubated with RabαGST-CpABC serum (1:50) and FITC goat anti-rabbit serum. (A) C. parvum-infected Caco-2 cell with parasite in situ. (B) Corresponding phase-contrast image. (C) Caco-2 cell after meront release. Remnants of two parasites are evident. (D) Phase-contrast image of same field. (E) Released meronts. (F) Phase-contrast. (G) 4′,6-diamidino-2-phenylindole stain image of same field.
DISCUSSION
In this report, we have demonstrated that CpABC is a member of a subfamily of integral membrane ABC proteins that includes the MRPs and CFTR and that it is located at the parasite–host boundary in intracellular stages (Fig. 4). The location of CpABC at the major parasite–host interface suggests that it plays an important role in the regulation of transport in the developing parasite.
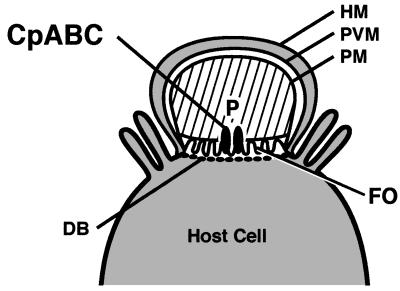
Figure 4.
Schematic of an epithelial cell infected with C. parvum. CpABC is located at the feeder organelle (FO), the major host–parasite boundary. DB, dense band; HM, host cell membrane; P, parasite; PM, parasite membrane; PVM, parasitophorous vacuole membrane.
The structural similarity of CpABC to MRP1, YCF1, and CFTR has important functional implications: It suggests that CpABC is involved in ion transport. Superficially, the structural similarity to CFTR is more compelling than to the MRPs. However, two human MRP subfamily members (MRP4 and MRP5) similarly lack a TMD0 (33, 34). Furthermore, recent work has shown that the TMD0 domain of MRP1 is not required for transport whereas the cytoplasmic linker (L0) between TMD0 and TMD1 is essential (32). This suggests that, despite the absence of a TMD0 domain, CpABC may be capable of transporting large organic anions and may function as a transporter of endogenous or xenobiotic conjugates (11, 30). The size and sequence characteristics of the region of CpABC that corresponds to the connector domain of CFTR is suggestive of some functional similarity (16). Future studies to identify substrate(s) and transport properties of CpABC should clarify this.
The topology of MRP1 and CFTR is well established (16, 35–37), and it is therefore reasonable to propose the transmembrane topology for CpABC shown in Fig. 1B. The orientation of CpABC in the membrane at the parasite–host boundary will therefore determine whether it is responsible for export of substrates out of the parasite or import of substrates into the parasite.
CpABC is present in sporozoites, and its localization is suggestive of an organellar location. Although it is possible that CpABC plays an active role in oocysts and sporozoites—for example, as a vacuolar pump like YCF1 (38, 39)—its major role appears to be in the intracellular stages. Indeed, its distribution in sporozoites is consistent with being part of the dense granules that are exocytosed at invasion (data not shown). It is not yet clear whether the protein is synthesized de novo once the parasite begins to develop intracellularly. Attempts to immunoprecipitate a metabolically labeled protein with RabαGST-CpABC serum so far have been unsuccessful.
The structure of the feeder organelle that serves as the primary interface between C. parvum and its host cell is unusual for an intracellular parasite in that it is not a unit bilayer membrane but a highly folded membrane structure (5, 6). The origin of this membrane is of considerable interest, and the localization of a parasite-encoded protein, CpABC, to this region will now allow us to examine this question in more detail. It is difficult to determine from ultrastructural studies whether the feeder organelle is a modification of the parasite membrane, as has been suggested by Tzipori and Griffiths (7), of the vacuole membrane, or neither. The fact that CpABC remains with the host cell after meront release suggests that it may not be part of the meront plasma membrane (Fig. 3). Electron microscopy showed that the membrane of released meronts, devoid of the feeder organelle, is intact (data not shown).
Protein components of the intracellular parasite’s membrane and the vacuole membrane may be assembled from both secretory organelles of the invasive stage and de novo synthesis (40–42). Although it has been postulated that some of these proteins may be transporters, to date, none have been identified directly in Apicomplexans. For most other intracellular microorganisms, the origin and constituents of the vacuole membrane are unknown. Recently, three putative transporter proteins were localized to the inclusion (vacuole) membrane in Chlamydia psittaci (43). Secretion of transporters into the host cell or vacuole membrane at invasion or shortly thereafter would have the advantage that the intracellular parasite would be able to regulate the movement of material immediately, before parasite protein synthesis begins.
Finally, CpABC may play a role in the intrinsic resistance to drug therapies exhibited by C. parvum (3). Overexpression of P-glycoprotein and MRP1 confers resistance to a range of cytotoxic xenobiotics. The presence of a transporter with similar capabilities at the boundary between the parasite and host could contribute to C. parvum’s drug refractory phenotype. Drugs that are effective reversers of the multidrug resistance phenotype (44), such as cyclosporin, have been shown to bind to P-glycoprotein (45). Significantly, it has been shown recently that SDZ-PSC833 and other cyclosporin analogs are potent inhibitors of intracellular growth of C. parvum (18). In addition, cyclosporin analogs also have been reported to be potent inhibitors of Plasmodium (46, 47) and Toxoplasma (48).
Acknowledgments
We thank Steve Upton, Rick Nelson, and the National Institutes of Health AIDS Research and Reference Reagent Program for reagents, Richard Friedman for sequence alignments, András Váradi and Alexandra Fairfield for useful discussions, and Robert Krauss for critical reading of the manuscript. This work was supported by National Institutes of Health Grant AI 41351 and the Center for Environmental Research and Conservation.
ABBREVIATIONS
- ABC
ATP-binding cassette
- CFTR
cystic fibrosis conductance regulator
- MRP
multidrug resistance protein
- gDNA
genomic DNA
- kb
kilobase
- GST
glutathione S-transferase
- NBF
nucleotide binding fold
- TMD
transmembrane domain
- MBP
maltose binding protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF110147).
References
- 1.Guerrant R L. Emerg Infect Dis. 1997;3:51–57. doi: 10.3201/eid0301.970106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colford J M J, Tager I B, Hirozawa A M, Lemp G F, Aragon T, Petersen C. Am J Epidemiol. 1996;144:807–816. doi: 10.1093/oxfordjournals.aje.a009015. [DOI] [PubMed] [Google Scholar]
- 3.Woods K M, Nesterenko M V, Upton S J. Ann Trop Med Parasitol. 1996;90:603–615. doi: 10.1080/00034983.1996.11813090. [DOI] [PubMed] [Google Scholar]
- 4.Blagburn B L, Soave R. In: Cryptosporidium and Cryptosporidiosis. Fayer R, editor. Boca Raton, FL: CRC; 1997. [Google Scholar]
- 5.Marcial M A, Madara J L. Gastroenterology. 1986;90:583–594. doi: 10.1016/0016-5085(86)91112-1. [DOI] [PubMed] [Google Scholar]
- 6.Current W L, Reese N C. J Protozool. 1986;33:98–108. doi: 10.1111/j.1550-7408.1986.tb05567.x. [DOI] [PubMed] [Google Scholar]
- 7.Tzipori S, Griffiths J K. Adv Parasitol. 1998;40:5–36. doi: 10.1016/s0065-308x(08)60116-5. [DOI] [PubMed] [Google Scholar]
- 8.Griffiths J K, Balakrishnan R, Widmer G, Tzipori S. Infect Immun. 1998;66:3874–3883. doi: 10.1128/iai.66.8.3874-3883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambudkar S V, Gottesman M M. Methods Enzymol. 1998;292:1–853. doi: 10.1016/s0076-6879(98)92025-0. [DOI] [PubMed] [Google Scholar]
- 10.Borst P, Schinkel A H. Trends Genet. 1997;13:217–222. doi: 10.1016/S0168-9525(97)01112-8. [DOI] [PubMed] [Google Scholar]
- 11.Keppler D, Leier I, Jedlitschky G. Biol Chem. 1997;378:787–791. [PubMed] [Google Scholar]
- 12.Deeley R G, Cole S P C. Semin Cancer Biol. 1997;8:193–204. doi: 10.1006/scbi.1997.0070. [DOI] [PubMed] [Google Scholar]
- 13.Evers R, Kool M, van Deemter L, Janssen H, Calafat J, Oomen L C, Paulusma C C, Oude Elferink R P, Baas F, Schinkel A H, et al. J Clin Invest. 1998;101:1310–1319. doi: 10.1172/JCI119886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Germann U A. Eur J Cancer. 1996;32A:927–944. doi: 10.1016/0959-8049(96)00057-3. [DOI] [PubMed] [Google Scholar]
- 15.Loe D W, Deeley R G, Cole S P C. Eur J Cancer. 1996;32A:945–957. doi: 10.1016/0959-8049(96)00046-9. [DOI] [PubMed] [Google Scholar]
- 16.Devidas S, Guggino W B. J Bioenerg Biomembr. 1997;29:443–451. doi: 10.1023/a:1022430906284. [DOI] [PubMed] [Google Scholar]
- 17.Upton S J, Tilley M, Nesterenko M V, Brillhart D B. FEMS Microbiol Lett. 1994;118:45–50. doi: 10.1111/j.1574-6968.1994.tb06801.x. [DOI] [PubMed] [Google Scholar]
- 18.Perkins M E, Wu T W, Le Blancq S M. Antimicrob Agents Chemother. 1998;42:843–848. doi: 10.1128/aac.42.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasquez J R, Gooze K, Kim K, Gut J, Peterson C, Nelson R. Mol Biochem Parasitol. 1996;79:153–165. doi: 10.1016/0166-6851(96)02647-3. [DOI] [PubMed] [Google Scholar]
- 20.Spano F, Putignani L, Crisanti A, Sallicandro P, Morgan U M, Le Blancq S M, Tchack L, Tzipori S, Widmer W. J Clin Microbiol. 1998;36:3255–3259. doi: 10.1128/jcm.36.11.3255-3259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perkins M E, Volkman S, Wirth D F, Le Blancq S M. Mol Biochem Parasitol. 1997;87:117–122. doi: 10.1016/s0166-6851(97)00053-4. [DOI] [PubMed] [Google Scholar]
- 22.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 23.Le Blancq S M, Khramtsov N V, Zamani F, Upton S J, Wu T W. Mol Biochem Parasitol. 1997;90:463–478. doi: 10.1016/s0166-6851(97)00181-3. [DOI] [PubMed] [Google Scholar]
- 24.Cheng S, Fockler C, Barnes W M, Higuchi R. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genetics Computer Group. Program Manual for the Wisconsin Package, Version 10.0. Madison, WI: Genetics Computer Group; 1999. [Google Scholar]
- 26.Caccio S, La Rosa G, Pozio E. Mol Biochem Parasitol. 1997;89:307–311. doi: 10.1016/s0166-6851(97)00122-9. [DOI] [PubMed] [Google Scholar]
- 27.Taglicht D, Michaelis S. Methods Enzymol. 1998;292:130–162. doi: 10.1016/s0076-6879(98)92012-2. [DOI] [PubMed] [Google Scholar]
- 28.Tusnády G E, Simon I. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- 29.Riordan J R, Rommens J M, Kerem B, Alon N, Rozmahel R, Grzelczak Z, Zielenski J, Lok S, Plavsic N, Chou J L, et al. Science. 1989;245:1066–1073. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- 30.Ishikawa T, Li Z S, Lu Y P, Rea P A. Biosci Rep. 1997;17:189–207. doi: 10.1023/a:1027385513483. [DOI] [PubMed] [Google Scholar]
- 31.Tusnády G E, Bakos E, Váradi A, Sarkadi B. FEBS Lett. 1997;402:1–3. doi: 10.1016/s0014-5793(96)01478-0. [DOI] [PubMed] [Google Scholar]
- 32.Bakos E, Evers R, Szakacs G, Tusnády G E, Welker E, Szab K, de Haas M, van Deemter L, Borst P, Váradi A, et al. J Biol Chem. 1998;273:32167–32175. doi: 10.1074/jbc.273.48.32167. [DOI] [PubMed] [Google Scholar]
- 33.Belinsky M G, Bain L J, Balsara B B, Testa J R, Kruh G D. J Natl Cancer Inst. 1998;90:1735–1741. doi: 10.1093/jnci/90.22.1735. [DOI] [PubMed] [Google Scholar]
- 34.Lee K, Belinsky M G, Bell D W, Testa J R, Kruh G D. Cancer Res. 1998;58:2741–2747. [PubMed] [Google Scholar]
- 35.Bakos E, Hegedus T, Hollo Z, Welker E, Tusnády G E, Zaman G J R, Flens M J, Varadi A, Sarkadi B. J Biol Chem. 1996;271:12322–12326. doi: 10.1074/jbc.271.21.12322. [DOI] [PubMed] [Google Scholar]
- 36.Kast C, Gros P. J Biol Chem. 1997;272:26479–26487. doi: 10.1074/jbc.272.42.26479. [DOI] [PubMed] [Google Scholar]
- 37.Kast C, Gros P. Biochemistry. 1998;37:2305–2313. doi: 10.1021/bi972332v. [DOI] [PubMed] [Google Scholar]
- 38.Li Z S, Szczypka M, Lu Y P, Thiele D J, Rea P A. J Biol Chem. 1996;271:6509–6517. doi: 10.1074/jbc.271.11.6509. [DOI] [PubMed] [Google Scholar]
- 39.Li Z-S, Lu Y-P, Zhen R G, Szczypka M, Thiele D J, Rea P A. Proc Natl Acad Sci USA. 1997;94:42–47. doi: 10.1073/pnas.94.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trager W, Rozario C, Shio H, Williams J, Perkins M E. Infect Immun. 1992;60:4656–4661. doi: 10.1128/iai.60.11.4656-4661.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beckers C J, Dubremetz J F, Mercereau-Puijalon O, Joiner K A. J Cell Biol. 1994;127:947–961. doi: 10.1083/jcb.127.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lingelbach K, Joiner K A. J Cell Sci. 1998;111:1467–1475. doi: 10.1242/jcs.111.11.1467. [DOI] [PubMed] [Google Scholar]
- 43.Bannantine J P, Rockey D D, Hackstadt T. Mol Microbiol. 1998;28:1017–1026. doi: 10.1046/j.1365-2958.1998.00867.x. [DOI] [PubMed] [Google Scholar]
- 44.Twentyman P R. Biochem Pharmacol. 1992;43:109–117. doi: 10.1016/0006-2952(92)90668-9. [DOI] [PubMed] [Google Scholar]
- 45.Demeule M, Wenger R M, Beliveau R. J Biol Chem. 1997;272:6647–6652. doi: 10.1074/jbc.272.10.6647. [DOI] [PubMed] [Google Scholar]
- 46.Bell A, Wernli B, Franklin R M. Biochem Pharmacol. 1994;48:495–503. doi: 10.1016/0006-2952(94)90279-8. [DOI] [PubMed] [Google Scholar]
- 47.Kocken C H, van der Wel A, Rosenwirth B, Thomas A W. Exp Parasitol. 1996;84:439–443. doi: 10.1006/expr.1996.0132. [DOI] [PubMed] [Google Scholar]
- 48.Silverman J A, Hayes M L, Luft B J, Joiner K A. Antimicrob Agents Chemother. 1997;41:1859–1866. doi: 10.1128/aac.41.9.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole S P C, Bhardwaj G, Gerlach J H, Mackie J E, Grant C E, Almquist K C, Stewart A J, Kurz E U, Duncan A M V, Deeley R G. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 50.Szczypka M S, Wemmie J A, Moye-Rowley W S, Thiele D J. J Biol Chem. 1994;269:22853–22857. [PubMed] [Google Scholar]
- 51.Hipfner D R, Almquist K C, Leslie E M, Gerlach J H, Grant C E, Deeley R G, Cole S P. J Biol Chem. 1997;272:23623–23630. doi: 10.1074/jbc.272.38.23623. [DOI] [PubMed] [Google Scholar]
- 52.Akabas M H, Cheung M, Guinamard R. J Bioenerg Biomembr. 1997;29:453–463. doi: 10.1023/a:1022482923122. [DOI] [PubMed] [Google Scholar]