Abstract
A key feature of B and T lymphocyte development is the generation of antigen receptors through the rearrangement and assembly of the germline variable (V), diversity (D), and joining (J) gene segments. However, the mechanisms responsible for regulating developmentally ordered gene rearrangements are largely unknown. Here we show that the E2A gene products are essential for the proper coordinated temporal regulation of V(D)J rearrangements within the T cell receptor (TCR) γ and δ loci. Specifically, we show that E2A is required during adult thymocyte development to inhibit rearrangements to the γ and δ V regions that normally recombine almost exclusively during fetal thymocyte development. The continued rearrangement of the fetal Vγ3 gene segment in E2A-deficient adult thymocytes correlates with increased levels of Vγ3 germline transcripts and increased levels of double-stranded DNA breaks at the recombination signal sequence bordering Vγ3. Additionally, rearrangements to a number of Vγ and Vδ gene segments used predominately during adult development are significantly reduced in E2A-deficient thymocytes. Interestingly, at distinct stages of T lineage development, both the increased and decreased rearrangement of particular Vδ gene segments is highly sensitive to the dosage of the E2A gene products, suggesting that the concentration of the E2A proteins is rate limiting for the recombination reaction involving these Vδ regions.
Keywords: E2A; rearrangement; T cell receptor, γ and δ
The ability of lymphocytes to respond to a vast array of antigens is dependent on the generation of unique surface receptors with diverse binding specificities. The antigen receptors are assembled during lymphocyte development from germline variable (V), diversity (D), and joining (J) gene segments by the process of V(D)J recombination (1). Products of recombination activating gene 1 (Rag-1)1 and Rag-2 recognize and cleave DNA at recombination signal sequences (RSS) that flank all rearranging gene segments (2–4). The restricted expression of Rag-1 and Rag-2 to B and T cells accounts for the lymphoid specificity of the recombinase (3, 5–7). However, within the lymphoid lineages, the process of V(D)J recombination is ordered and developmentally regulated at a number of different levels. For example, Ig V region genes are fully rearranged only in B lymphocytes and TCR V region genes are rearranged only in T lymphocytes. In addition, assembly of the V region genes is regulated in a developmentally stage-specific manner within developing B and T lineage cells. That is, most differentiating B cells assemble Ig heavy chain V regions before light chain V regions, and developing T cells rearrange β chain V regions before α chain V regions (8– 10). During the assembly of the Ig heavy chain and the TCR β chain, the D to J rearrangement step normally occurs before the assembly of the V regions (9, 11). These types of regulations are proposed to be affected by altering the accessibility of the substrate gene segments to the recombinase (12, 13).
Control of V(D)J recombination can also be regulated at the level of V gene usage. During murine B cell development, the 3′-most V gene segments are preferentially used in pre-B cells in fetal liver and adult bone marrow, whereas mature B cell populations use a wide range of VH segments (14–17). A similar level of regulation exists in the rearrangement of the TCR γ and δ genes. During thymic ontogeny, the γ and δ V gene rearrangements occur in waves. Vγ3 and Vγ4, which are most proximal to the Jγ gene segments, are the most frequently rearranged Vγ gene segments during early fetal thymic development (18–21). Rearrangements to Vγ3 and Vγ4 peak at approximately embryonic day 15 and decline thereafter. In the adult thymus, Vγ3 is rarely rearranged. In contrast, rearrangements to the more 5′ V regions, such as Vγ2, begin late in fetal development and predominate in the adult (22). Within the δ locus, Vδ1 rearrangements predominate during early fetal thymic development, but are rare in the adult, whereas Vδ5 usage begins later and predominates in the adult (23, 24). Thus, within the γ and δ loci there is a regulated switch in the use of V gene segments between fetal and adult thymocyte populations. It is likely that the regulation of this developmental switch is controlled, in part, by changes in gene segment accessibility and/or selective recruitment of the recombination enzymes.
Several studies have suggested that transcriptional promoters and enhancers play important roles in the regulation of VDJ recombination by modulating the accessibility of the gene segments to the recombination machinery (13, 25). However, evidence for the involvement of specific transcription factors in this type of regulation is lacking. The basic helix-loop-helix (bHLH) family of transcriptional regulatory proteins has been implicated in the regulation of Ig and TCR gene rearrangement based on the ability of these proteins to bind to and activate transcription from the Ig and TCR gene enhancers (26–28). The E2A gene products, E12 and E47, are broadly expressed members of the HLH family of transcription factors and play important functional roles in the early stages of both B and α/β T lymphocyte development. In the absence of E2A activity, B lymphocytes are blocked at a stage before the initiation of Ig gene rearrangements (29–31). Ectopic expression of either E12 or E47 in the E2A-deficient background allows the activation of several B lineage–restricted genes (32). Interestingly, E2A-deficient mice display an incomplete block at a developmentally similar stage of α/β T cell development (33). A specific role for E2A in B lineage development is also inferred from experiments demonstrating the induction of B cell–specific traits upon overexpression of E47 in cell lines. Ectopic expression of E47 in non-B cell lines results in the activation of a number of B lineage– specific genes, including Rag-1, TdT, and λ5 (34–37). Overexpression of E47 in a pre-T cell line leads to the induction of IgH DJ rearrangements as well (36).
Here we describe a functional role for the E2A gene products during the development of the γ/δ T lymphocytes. The absence of E2A results in the impaired development of the γ/δ T cells found in the secondary lymphoid organs and the intraepithelial layers of the intestine. Rearrangements to the V regions used most predominately by these γ/δ T cells are significantly reduced in E2A-deficient thymocytes. In contrast, skin intraepithelial γ/δ T cells develop normally in E2A-deficient mice, and rearrangements to Vγ3 and Vδ1, the V regions used exclusively by the skin γ/δ T cells, are present at wild-type levels in the E2A-deficient fetal thymus. Remarkably, both Vγ3 and Vδ1 continue to rearrange coordinately in the E2A-deficient adult thymus to an adult configuration of D and J segments, whereas developing thymocytes in wild-type adult thymocytes rarely use these V regions. Interestingly, the data indicate that the regulation of rearrangement to specific Vδ gene segments is dosage sensitive, as mice heterozygous for E2A show significant alterations in rearrangement levels. We propose that the concentration of the E2A proteins is a key factor in regulating, both positively and negatively, the rearrangement of several Vδ and Vγ gene segments.
Materials and Methods
Gene Targeting.
Targeting of the E2A gene has been described previously (29).
Flow Cytometric Analysis.
Isolation of γ/δ T cells from the intestine and skin has been described previously (38). Intestinal cells were stained with anti-γ/δ TCRbiotin (streptavidin-PE) and anti-α/β TCRcy-chrome and were gated on the basis of forward and side scatter. Skin cells were stained with anti-Thy1.1biotin, anti-CD3PE (145-2C11), and anti-Vγ3FITC (536) and were gated on the basis of Thy1 expression and forward scatter. Spleen, thymus, and lymph node cells were stained with anti-CD4PE (RM4-5), anti-CD8aPE (53-6.7), and anti-γ/δ TCRFITC (GL3). All antibodies were obtained from PharMingen.
Rearrangement Southern Blot.
Genomic DNA was prepared from thymocytes by DNAzol. 10 μg DNA was digested with EcoRI, electrophoresed on a 0.8% agarose gel, and transferred to Nytran (Schleicher & Schuell). The blot was hybridized with probe 4, then stripped and reprobed sequentially with the Vδ5-, Vδ1-, and Vδ4-specific probes (39).
Rearrangement PCR.
500 ng of adult or E18-19 fetal thymus genomic DNA (isolated using DNAzol; GIBCO BRL) was analyzed by PCR in a 25 μl reaction volume containing 100 ng of each primer in 10 mM Tris, pH 8.3, 50 mM KCl, and 2 mM MgCl2. All Vγ PCRs were performed for 20 cycles of 1 min at 94°C, 1 min at 58°C, and 1 min at 72°C. All Vδ PCRs were performed for 23 cycles of 1 min at 94°C, 1 min at 61°C, and 1 min at 72°C. The Vγ3(L3), Vγ2(L2), Jγ1, Vγ5, Vδ1, and Jδ2 primers have been described previously (22, 40, 41). Other primers used are as follows: Vδ5, 5′-TGCACGTACAATGCGGATTCTCCAA; and Jδ1, 5′-AGTCACTTGGGTTCCTTGTCC. For the V-D intermediate rearrangement PCR, the following reverse primers were used: Dδ1 rev-2, 5′-GACCTCGTCTACTGGGGCTC; and Dδ2 rev-2, 5′-CAGCAAGTGGAGGTCATATCTT. For the D-J intermediate PCR, the DR6 primer described previously was used in combination with the Jδ1 primer (42). A control reaction was performed using the following p53 primers: FOR, 5′-TATACTCAGAGCCGGCCT; and REV, 5′-ACAGCGTGGTACCTTAT. The p53 PCR was performed for 17 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C. 10 μl of each PCR reaction was run on a 2.2% Nusieve (FMC Corp.) gel and blotted to Nytran. Blots were hybridized with gene-specific probes.
Ligation-mediated PCR.
Adult thymus genomic DNA was prepared by incubation of thymocytes overnight at 55°C with 50 μg/ml proteinase K in 50 mM Tris, pH 8.0, 100 mM EDTA, 100 mM NaCl, and 1% SDS. The DNA was phenol/chloroform extracted, chloroform extracted, and precipitated with 2 vol of ethanol. The DNA was washed in 70% ethanol and dissolved in ddH2O. 3 μg DNA was ligated to annealed BW-1/BW-2 linkers for 12–14 h before inactivation of the ligation reaction as described previously (43). 12 rounds of PCR were performed on 1/20 of the ligation reaction using 100 ng of the BW-1H primer and 100 ng of the following locus-specific primer: Vγ3EXT, 5′-GGGAGTGGATGGAGATGGAAACAGGGC, in 10 mM Tris, pH 8.3, 50 mM KCl, and 2 mM MgCl2; 1/25 of the first PCR reaction was used in a second 25 μl PCR reaction. 27–32 rounds of a second PCR were performed with the BW-1H primer and the following locus-specific primer: Vγ3INT, 5′-GTCACTTGGCTTTTCTGGGCTCCAGCT. The Vγ3 PCR was performed for 30 s at 94°C and 1 min 30 s at 66°C. Primers and PCR conditions for the JH2 ligation-mediated (LM)-PCR have been described previously (43). 10 μl of each second round PCR reaction was run on a 2.5% Nusieve gel, transferred to Nytran, and hybridized with gene-specific probes.
Reverse-transcription PCR.
Total RNA was prepared from thymocytes isolated from 4–6-wk-old E2A-deficient mice and heterozygous littermates by TriZOL (GIBCO BRL). 10 μg of total thymocyte RNA was DNase treated, and 3 μg of the DNase-treated RNA was reverse transcribed as described previously (29). The PCR reaction was run on a 2.5% Nusieve gel, transferred to Nytran, and hybridized with gene-specific probes. The actin and Vγ3 primers and PCR conditions have been described previously (22, 32).
Results
Impaired Development of γ/δ T Lymphocytes in E2A-deficient Mice.
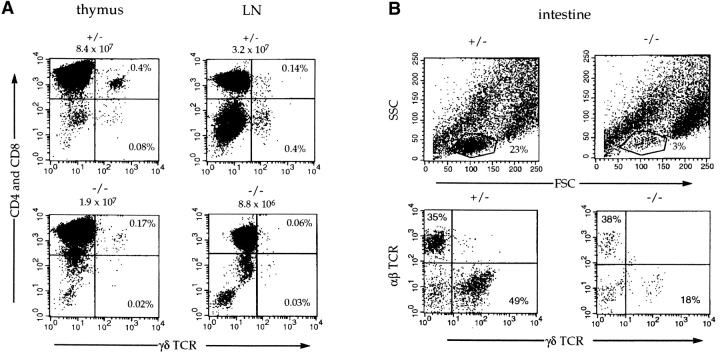
The E2A proteins are expressed at high levels in prothymocytes and are important in the development of committed α/β T cells from uncommitted progenitors (33, 44). Since both α/β and γ/δ T lymphocytes develop from the same progenitor thymocytes, we analyzed E2A-deficient mice for the presence of γ/δ T cells by flow cytometry (45–47). E2A-deficient mice display significantly reduced numbers of γ/δ T lymphocytes in the thymus, spleen, and lymph node compared with their heterozygous littermates (Fig. 1 A, and data not shown). In the thymus and lymph nodes, γ/δ T cell numbers are reduced from 8- to 40-fold (Fig. 1 A). Similarly, the number of γ/δ T cells is decreased ∼20-fold in the intestinal epithelium of E2A-deficient mice (Fig. 1 B). Surprisingly, mice that lacked E2A showed almost normal numbers of skin intraepithelial lymphocytes, suggesting that the E2A gene products differentially control γ/δ T cell development (Fig. 1 C).
Figure 1.
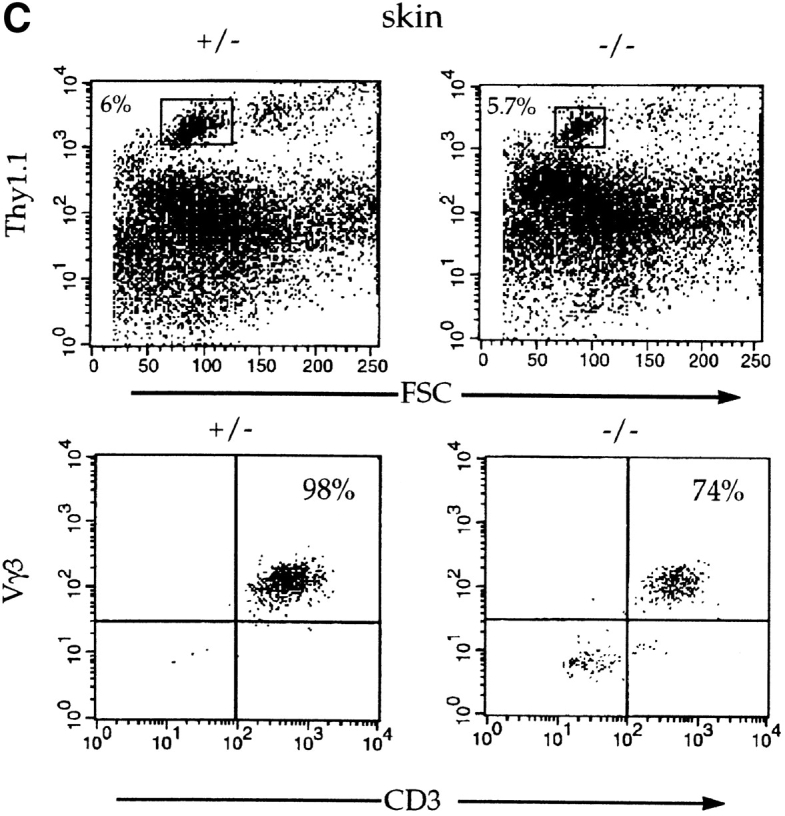
Flow cytometric analysis of γ/δ T cell populations in E2A-deficient mice. Flow cytometric analysis of thymus and lymph node (LN) cells from a 6-wk-old E2A-deficient mouse and a heterozygous littermate (A). Cells were stained with anti-CD4PE, anti-CD8PE, and anti–γ/δ TCRFITC. The number shown above each FACS® profile represents the total number of cells isolated from that tissue. The percentages of γ/δ cells are indicated. The total number of γ/δ cells is decreased 10–20-fold in the thymus and lymph nodes of E2A-deficient mice. (B) Intestinal T cells from an E2A-deficient mouse and a heterozygous littermate were stained with anti–α/β TCRcy-chrome and anti–γ/δ TCRbiotin (streptavidin-PE). The total number of intestinal lymphocytes (top) is decreased approximately sevenfold in the E2A-deficient mice (3 vs. 23%). Bottom panels show the α/β versus γ/δ staining of the lymphocytes gated in the top panels with the percentages of each population indicated. (C) Cells isolated from the skin of an E2A-deficient mouse and a heterozygous littermate were stained with anti-Thy1.1biotin (streptavidin–Red 670), anti-CD3PE, and anti-Vγ3FITC. T cells were gated on the basis of forward scatter and Thy1.1 expression (top), and the gated cells were analyzed for the expression of CD3 and Vγ3 (bottom).
E2A-deficient Mice Display an Altered Pattern of Vδ Gene Usage.
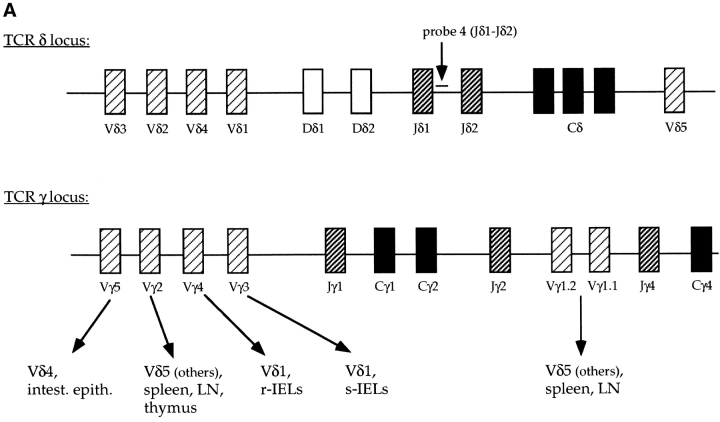
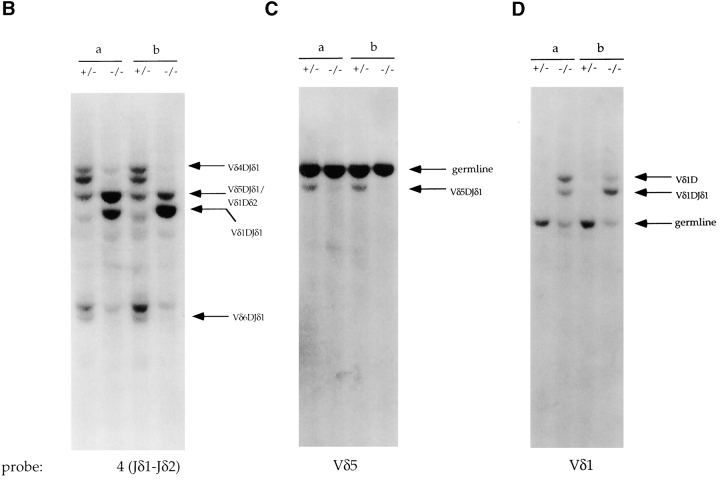
γ/δ T lymphocytes can be divided into two broad subtypes which differ in the type of V region used, their junctional diversity, and their ability to home to specific sites (48; Fig. 2 A). Skin intraepithelial lymphocytes represent one subtype, while γ/δ T cells in the secondary lymphoid organs and the intestinal epithelium are members of the second subtype (48). The first γ/δ T cells to develop in the thymus express an invariant receptor composed of γ and δ chains that have used exclusively Vγ3 and Vδ1 (19, 20, 40, 49–51). These γ/δ cells migrate to the skin (50). γ/δ T cells that populate the secondary lymphoid organs express variable receptors using predominately the γ2, γ1, and δ5 V regions, and intestinal γ/δ cells express variable receptors often using Vγ5 and Vδ4 (23, 41, 52, 53; Fig. 2 A). Rearrangements to these V regions begin late in fetal development and predominate in the adult (48). Since the development of each subtype of γ/δ cells is associated with specific γ and δ rearrangements, the development of only a subset of γ/δ T cells in the E2A-deficient mice suggests that γ and/or δ rearrangements may be affected. Committed α/β T lymphocytes retain previously rearranged δ genes in both chromosomal and extrachromosomal DNA (39, 47, 54). Thus, the frequency of specific Vδ rearrangements can be analyzed in total thymus DNA despite the fact that 95% of the cells are committed to the α/β T cell lineage. To determine the relative frequency of Vδ gene usage, we analyzed adult thymus DNA from E2A-deficient mice and heterozygous littermates by Southern blotting. As a probe, we used a genomic DNA fragment, termed probe 4(Jδ1-Jδ2), which is located between the Jδ1 and Jδ2 gene segments and allows for the detection of TCR δ genomic rearrangements (39; Fig. 2 A). Southern blot analysis with radiolabeled probe 4(Jδ1-Jδ2) identified a series of bands present in both heterozygous controls (Fig. 2 B). Several of these bands have been previously identified and are indicated by arrows (23, 39). Interestingly, thymus DNA from the E2A-deficient mice gave a distinctly different pattern of DNA fragments hybridizing to probe 4 (Fig. 2 B). Many of the bands that are clearly visible in the wild-type DNA samples are nearly undetectable in the E2A-deficient DNA samples. However, there are two strongly hybridizing fragments in the E2A-deficient DNAs, one of which is undetectable in wild-type DNAs (Fig. 2 B, fragment indicated by a bent arrow). To determine the identity of the bands, the blot was rehybridized with Vδ5-, Vδ4-, and Vδ1-specific probes (Fig. 2, C and D, and data not shown). Bands corresponding to Vδ5DJδ and Vδ4DJδ rearrangements were detectable in both heterozygous DNAs but were virtually absent in the E2A-deficient DNA samples (Fig. 2 C, and data not shown). These data demonstrate that the predominate δ rearrangements in a wild-type adult thymus, which involve the δ5 and δ4 V regions, are significantly underrepresented in thymocyte DNA derived from E2A-deficient mice.
Figure 2.
E2A-deficient mice display an altered pattern of Vδ gene rearrangements. (A) Schematic diagram of the TCR δ and γ loci showing the relative location of the V, D, and J gene segments. The arrows in the γ locus indicate the Vδ region that normally pairs with that Vγ and the site to which the cells expressing that specific TCR home. s-IEL and r-IEL, skin intraepithelial lymphocytes and reproductive tract intraepithelial lymphocytes, respectively. (B–D) Southern blot analysis of EcoRI- digested total thymus DNA from two E2A-deficient mice and their heterozygous littermates to determine the types of Vδ rearrangements detectable. (B) The blot hybridized with probe 4 which is located between Jδ1 and Jδ2 (A, δ locus). The band marked by the bent arrow is detectable only in the E2A-deficient DNA samples. The probe 4(Jδ1-Jδ2) blot was stripped and rehybridized sequentially with a Vδ5-specific (C) or Vδ1-specific (D) probe. Vδ5DJδ1 rearrangements are present in the control DNA samples (C, arrow). The Vδ5 germline fragment is also indicated. Wild-type DNAs show only the germline band when hybridized with a Vδ1 probe (D), but two rearrangements are detectable in the E2A-deficient DNAs. Based on the size of the two rearranged bands (∼8.8 and 9.5 kb) and the fact that they also hybridize with probe 4, they likely represent Vδ1D intermediate rearrangements and Vδ1DJδ1 rearrangements.
The flow cytometric data showed that the γ/δ T cells in the skin, which express a TCR composed exclusively of Vγ3 and Vδ1, are readily detectable at close to wild-type levels in E2A-null mutant mice (Fig. 1 C). Vδ1 rearrangements occur predominately during fetal development, and a significant proportion of Vδ1 rearrangements use the Jδ2 region, which results in the deletion of the DNA hybridizing to probe 4(Jδ1-Jδ2) (24, 41, 48; Fig. 2 A). Thus, Vδ1 rearrangements are not normally detectable in wild-type adult thymus DNA (23). Consistent with these data, only the germline band is visible in the heterozygous DNA samples when hybridized with a Vδ1-specific probe (Fig. 2 D). Surprisingly, the Vδ1 probe hybridized to two DNA fragments in the E2A-deficient DNA samples (Fig. 2 D). These Vδ1-specific rearrangements comigrate with the two predominate fragments recognized by probe 4(Jδ1-Jδ2) (Fig. 2, B and D). Based on their sizes, the bands likely represent Vδ1DJδ1 rearrangements and Vδ1Dδ2 intermediate rearrangements (23). These data indicate that rearrangements involving Vδ1, which are normally not detectable in wild-type mice, make up the majority of the rearrangements in adult E2A-deficient thymocytes. The appearance of Vδ1 rearrangements and the lack of Vδ5 and Vδ4 rearrangements indicate that a deficiency in E2A leads to a deregulation of V region usage at the TCR δ locus in adult thymocytes.
Absence of E2A Leads to Alterations in Both Vδ and Vγ Gene Rearrangements.
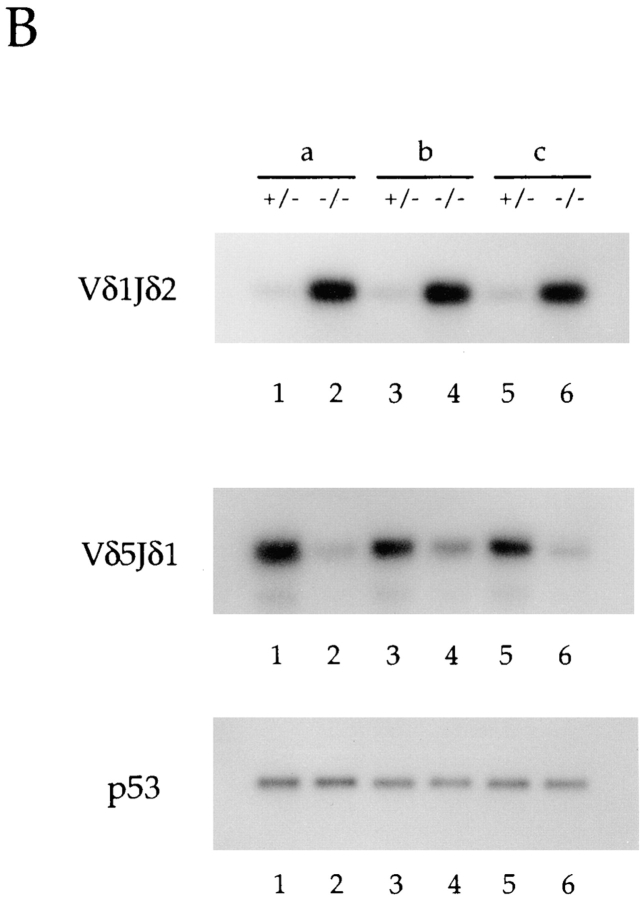
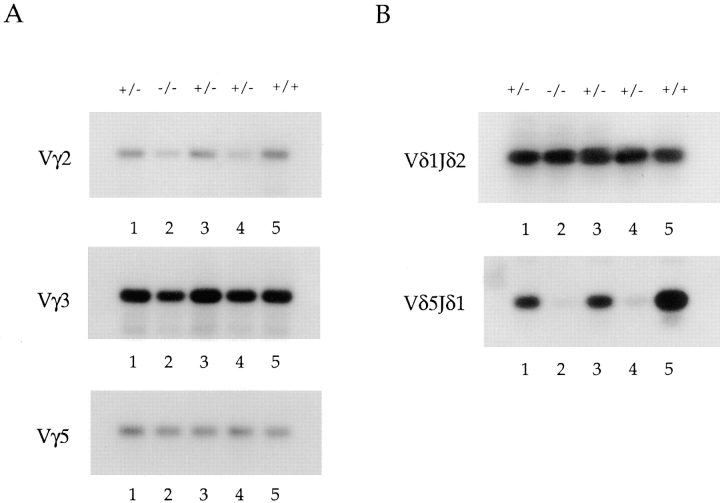
To confirm the δ rearrangement data described above and to examine the level of γ gene rearrangements, we analyzed total thymus DNA for Vγ and Vδ rearrangements by PCR using forward primers specific for the V regions and reverse primers recognizing the J gene segments. Only upon rearrangement are the primers brought into sufficient proximity to allow PCR amplification. The data shown are derived from three independent sets of littermates, designated as a, b, and c (Fig. 3, A and B). As expected, all E2A-deficient mice analyzed displayed reduced levels of Vδ5DJ rearrangements, although the decreases varied from 3- to 50-fold (Fig. 3 B). As predicted from the Southern blot analysis, Vδ1 rearrangements were increased ∼10–50-fold in thymus DNA derived from E2A-deficient mice compared with control littermates (Fig. 3 B).
Figure 3.
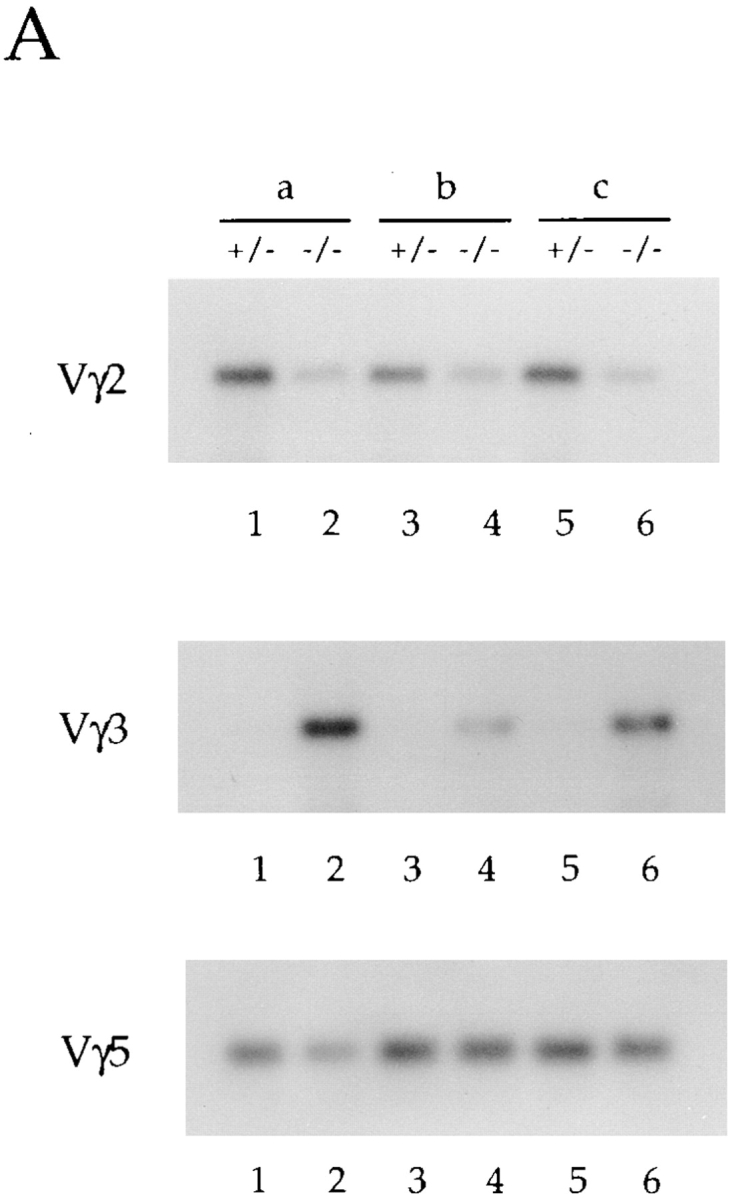
Differential usage of the Vγ and Vδ gene segments in E2A-deficient mice. (A and B) PCR analysis of total thymus DNA from adult (6-wk-old) E2A-deficient mice and their heterozygous littermates to determine the relative level of Vγ (A) and Vδ (B) gene rearrangements. Forward primers were specific for each V region. The Jγ1 reverse primer used recognizes all Jγ gene segments, whereas the reverse Jδ primers are specific for either Jδ1 or Jδ2. The δ PCRs were performed with both reverse primers, and the results were identical. A control reaction was performed with p53 primers. Shown are three independent knockouts (designated a, b, and c) and their heterozygous littermates.
The differential V gene usage observed in the δ locus in E2A-deficient mice was evident in the γ locus as well. PCR analysis using specific Vγ forward primers in conjunction with a Jγ reverse primer demonstrated significant decreases in the levels of Vγ2 rearrangements in E2A-deficient adult thymus DNAs (Fig. 3 A). The decreases in the level of Vγ2 rearrangements from 10 independent E2A- deficient mice ranged from 2- to 10-fold. Within any one E2A-deficient thymus, rearrangements to Vδ5 were generally more significantly decreased than rearrangements to Vγ2. Analysis of E2A-deficient thymus DNA for recombination to the Vγ5 gene segment demonstrated that Vγ5 rearrangements are not significantly affected by the absence of E2A (Fig. 3 A). These data demonstrate that the proper V(D)J rearrangement of particular V gene segments in both the γ and δ locus requires the E2A gene products.
The TCR γ V gene segment, Vγ3, recombines coordinately with Vδ1 during early embryogenesis. To determine whether Vγ3 rearrangements, like those of Vδ1, are increased in adult thymocytes derived from E2A-deficient mice, genomic DNA was analyzed by PCR using a primer specific for Vγ3 (Fig. 3 A). Remarkably, rearrangements to Vγ3 are also significantly increased in DNA isolated from E2A-deficient thymocytes (Fig. 3 A). Thus, the absence of E2A leads to a coordinate increase in the rearrangement of the γ and δ V gene segments normally used predominately during early fetal development to an adult configuration of D and J segments. These data suggest that during adult thymocyte development, the E2A gene products act as both positive and negative regulators of γ and δ V gene usage, and are essential for the temporally ordered recombination of the TCR γ and δ loci.
Altered Frequency of Vδ and Vγ Rearrangements in E2A- deficient Fetal Thymi.
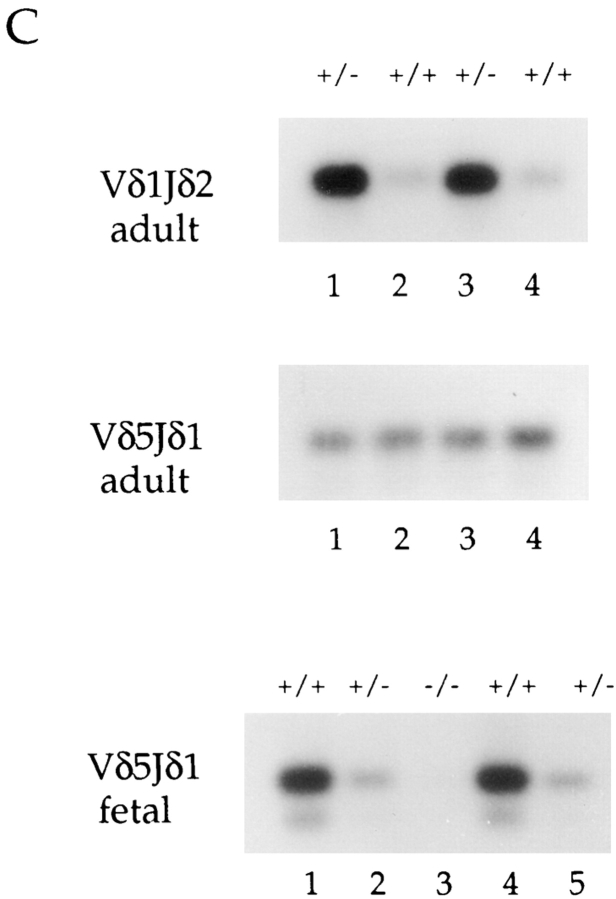
Because the fetal and adult thymus differ with respect to the V regions predominately rearranged and expressed, we analyzed DNA from E19 fetal thymocytes for the presence of γ and δ rearrangements. As in the adult, Vγ5 rearrangements were present at relatively normal levels in DNA derived from E2A-deficient fetal thymi, whereas Vγ2 rearrangements were decreased an average of three- to fourfold (Fig. 4 A). Additionally, we observed a striking decrease in the level of Vδ5 rearrangements in the E2A-deficient fetal thymus DNAs compared with the wild-type littermates (∼ 50-fold; Fig. 4 B). Surprisingly, we consistently detected three- to fourfold decreases in the level of Vδ5 rearrangements in fetal thymus DNAs derived from mice heterozygous for E2A compared with wild-type littermates (Fig. 4, B and C). Thus, the absence of the E2A gene products during fetal thymocyte development appears to have a more significant effect on rearrangement to the δ5 V region than to the γ2 V region. In addition, the frequency of rearrangement to the δ5 V gene segment during fetal thymocyte development is sensitive to the dosage of the E2A proteins.
Figure 4.
PCR analysis of V gene rearrangements in E2A-deficient fetal thymus DNAs. Vγ (A) and Vδ (B) gene rearrangements from E19 fetal thymus DNAs. All fetal thymus DNA samples shown are from the same litter. (C) PCR analysis of Vδ5 and Vδ1 gene rearrangements from two independent adult (6-wk-old) E2A heterozygous mice and their wild-type littermates. Vδ5 rearrangements from five E19 fetal thymocyte DNAs are shown for comparison. The number of PCR cycles used for the adult Vδ1 rearrangements in C was increased (compared with Fig. 3 B) in order to enhance the signal from the heterozygotes.
Vγ3 and Vδ1 rearrangements, which are virtually undetectable in the adult thymus, predominate during fetal thymic development. As analyzed by PCR, the frequency of usage of these V regions was similar in fetal thymus DNA derived from E2A-deficient and control mice (Fig. 4, A and B). These data suggest that the E2A gene products play multiple and distinct roles in regulating γ and δ V(D)J recombination. The E2A gene products are nonessential in the initiation of Vδ1 and Vγ3 rearrangements early in fetal thymic development. However, the presence of the E2A gene products is required at the later stages of thymocyte differentiation to inhibit the usage of Vγ3 and Vδ1, which are normally dormant in wild-type mice. Additionally, activity of the E2A gene products is important for initiating the proper rearrangement of the δ5 and γ2 V regions.
The observation that the frequency of Vδ5 usage in fetal thymus development is dependent on the dosage of the E2A proteins led us to examine more carefully whether V region usage during adult thymocyte development is correspondingly dosage sensitive. Thus, we analyzed DNAs from wild-type and heterozygous littermates by PCR to determine the relative levels of V gene rearrangements. Unlike in the fetal thymus DNAs, Vδ5 rearrangements were comparable in adult thymocyte DNA derived from E2A heterozygous and wild-type mice (Fig. 4 C). Similarly, rearrangements involving all other V regions were unaffected in E2A heterozygous mice compared with wild-type littermates, with the exception of Vδ1 (Fig. 4 C, and data not shown). Unexpectedly, we found that the frequency of Vδ1 gene usage is increased dramatically in E2A heterozygous mice compared with wild-type littermates (Fig. 4 C). Therefore, the proper inhibition of Vδ1 rearrangement during adult thymus development is additionally dosage dependent. In summary, the data indicate that the activity and concentration of the E2A gene products in thymocytes undergoing site-specific recombination are key factors in inducing proper Vδ5 rearrangement during fetal thymus development and for inhibiting rearrangements to Vδ1 during adult thymocyte development.
E2A Is Required at the Initial Step of δ Gene Recombination.
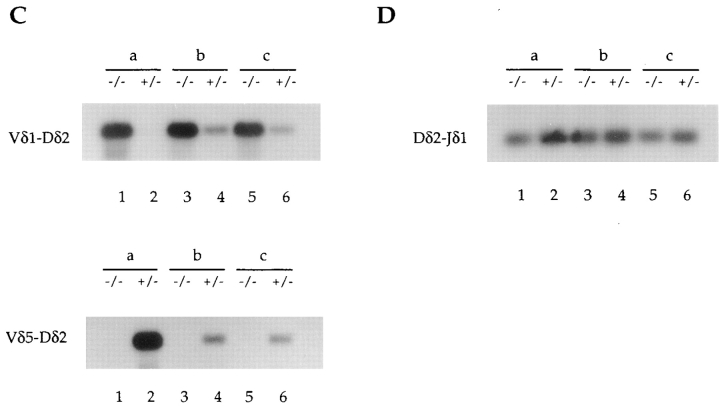
The process of V(D)J recombination is tightly regulated and proceeds in well-defined stages. During the assembly of the Ig heavy chain and the TCR β chain, the D to J rearrangement step normally precedes the assembly of the V regions (9, 11). However, recombination at the δ locus is unique in that V to D rearrangement normally represents the initial recombination event and precedes rearrangement to the downstream J regions (24). As described above, the end products of the recombination reaction involving Vδ5 are significantly decreased in adult thymus DNA from E2A-deficient mice, whereas those involving Vδ1 are strikingly increased. To further define the stage at which the recombination events involving these V regions are deregulated, we analyzed total thymus DNA from E2A-deficient mice and wild-type controls for the presence of rearrangement intermediates. V to D rearrangement intermediates can be detected using a V region–specific forward primer and a reverse primer located 3′ of the Dδ2 coding sequence (Fig. 5 A). Since fully rearranged VDJ products result in the deletion of the DNA hybridizing to the reverse 3′ Dδ2 primer, only intermediate V-D rearrangements can be PCR amplified (Fig. 5 A). Like the fully rearranged products, Vδ5-Dδ2 intermediate rearrangement products are present at significantly reduced levels in E2A-deficient adult thymus DNA (Fig. 5 C). Likewise, the Vδ1-Dδ2 intermediate rearrangements are increased to a similar extent as the fully rearranged Vδ1DJ products in thymus DNA lacking E2A (Fig. 5 C). Similar results were obtained using a reverse primer located 3′ of Dδ1 (data not shown). Interestingly, the presence of Vδ1Dδ1 rearrangements in the E2A-deficient thymus demonstrates that these Vδ1 rearrangements are not from residual fetal cells, as fetally derived Vδ1 rearrangements use exclusively the Dδ2 gene segment. These data suggest that the deregulation of TCR δ V to D rearrangement is not D region specific.
Figure 5.
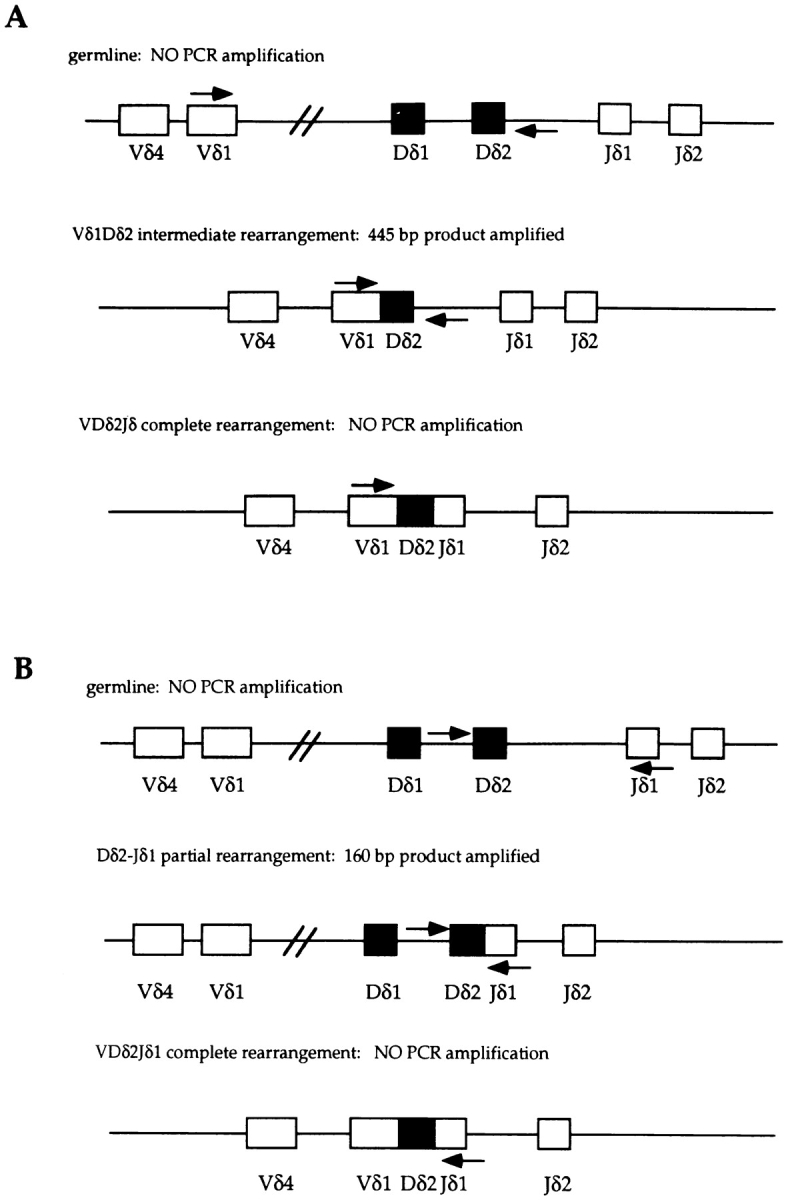
Detection of δ gene intermediate rearrangement products in E2A-deficient mice and their heterozygous littermates. (A) Schematic diagram of the δ locus showing the relative location of the forward and reverse primers used to detect V-D intermediate rearrangement products. A fully rearranged V-D-J allele results in the deletion of the reverse primer, which is located 3′ of Dδ2. As a result, only the intermediate V-D rearrangements can be PCR amplified. (B) Diagram showing the location of the PCR primers used in the detection of D-J intermediates. Rearrangement of any V region to Dδ2 results in the deletion of the forward PCR primer. Thus, only D-J intermediates can be PCR amplified. (C) Vδ1-Dδ2, Vδ5-Dδ2, and Dδ2-Jδ1 PCR analysis of total thymus DNA from three independent 6-wk-old E2A-deficient mice (designated a, b, and c) and their heterozygous littermates.
Although recombination at the δ locus normally involves an initial V to D rearrangement step, the Dδ2 and Jδ1 elements have been shown previously to be involved in Dδ2-Jδ1 and Dδ1-Dδ2-Jδ1 intermediate rearrangements which do not involve V gene rearrangement (54, 55). To examine the efficiency of D to J joining, we analyzed total thymus DNA by PCR using a forward primer located 5′ of the Dδ2 RSS and a reverse primer specific for Jδ1 (Fig. 5 B). Since rearrangement of any gene segment to Dδ2 results in the deletion of the DNA hybridizing to the forward primer, only Dδ2-Jδ1 intermediates can be PCR amplified (Fig. 5 B). As shown, Dδ2-Jδ1 and Dδ1-Jδ1 intermediate rearrangements are present at normal levels in adult thymus DNA derived from E2A-deficient mice (Fig. 5 D, and data not shown). Thus, E2A plays an important role in regulating the rearrangement of the V gene segments within the TCR δ locus, but is dispensable for normal Dδ-Jδ rearrangement.
Increases in Vγ3 Gene Usage Correlate with Increases in the Level of Double-strand Breaks at the Vγ3 RSS.
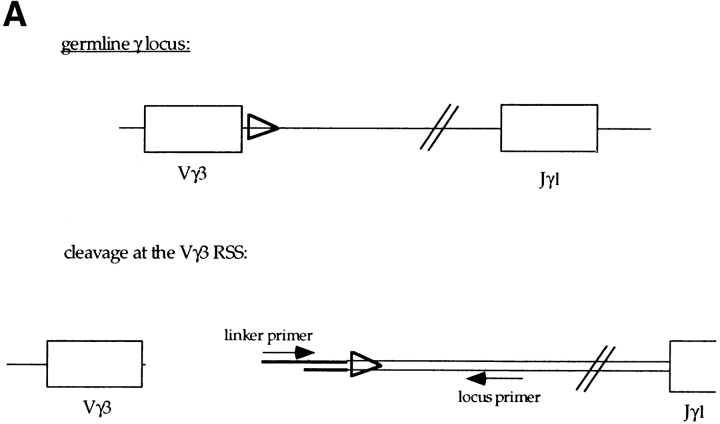
The recombination process that assembles the functional γ and δ TCR genes involves cleavage at the RSS that border the coding segments and then joining of the coding ends (56, 57). The RAG-1 and RAG-2 proteins were previously shown to be sufficient for cleavage of the V(D)J recombination signal on extrachromosomal and in vitro substrates (3, 4, 58). As described above, rearrangement to several Vγ and Vδ gene segments is deregulated in the absence of E2A. In adult E2A-deficient mice, the most dramatic phenotype observed is the increased rearrangement to Vγ3 and Vδ1. To determine whether the increased usage of Vγ3 reflected an increase in the cleavage of the Vγ3 RSS, we assayed for the presence of double-stranded breaks at the RSS 3′ of Vγ3 using LM-PCR (Fig. 6 A). Double-stranded breaks at Vγ3 were absent in E2A heterozygous and wild-type adult thymus DNA, but could be detected in the E2A-deficient thymus DNA (Fig. 6 B). In contrast, both the E2A-deficient DNA and the control DNA displayed similar levels of broken ends at the RSS 5′ of JH2 (Fig. 6 B). These data suggest that the E2A gene products inhibit rearrangement of the γ3 V region by modulating the accessibility of γ3 gene segments to the recombination machinery.
Figure 6.
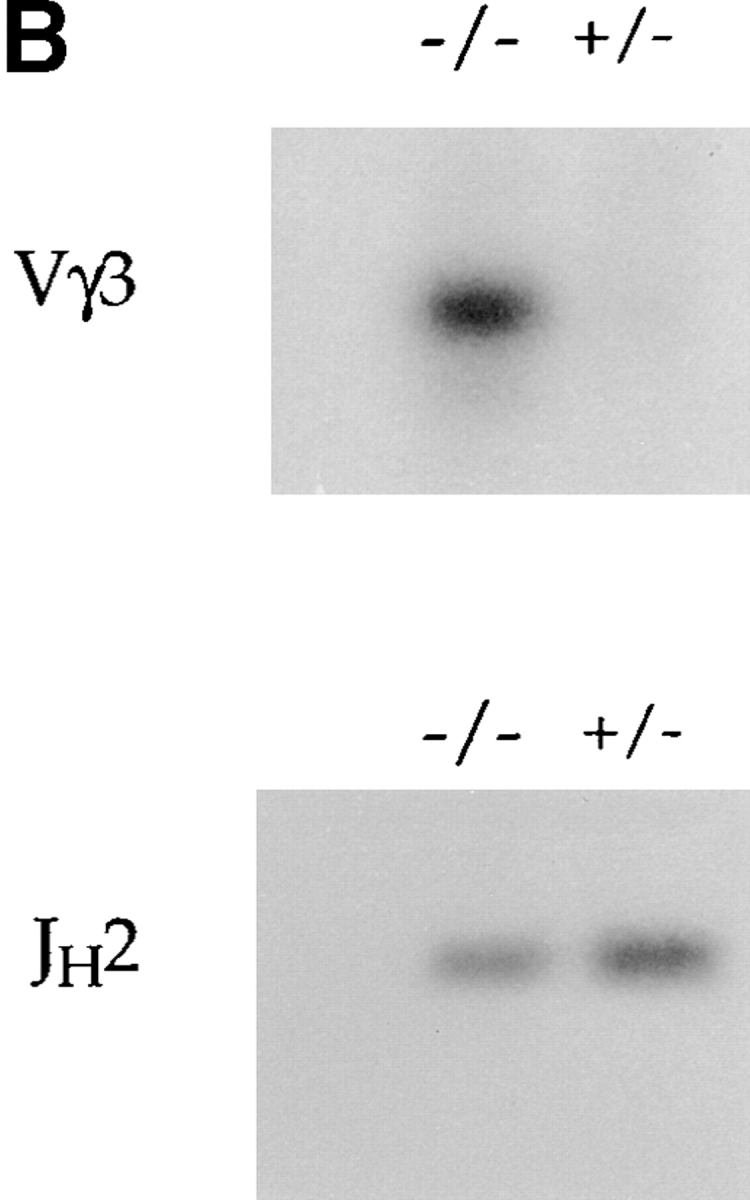
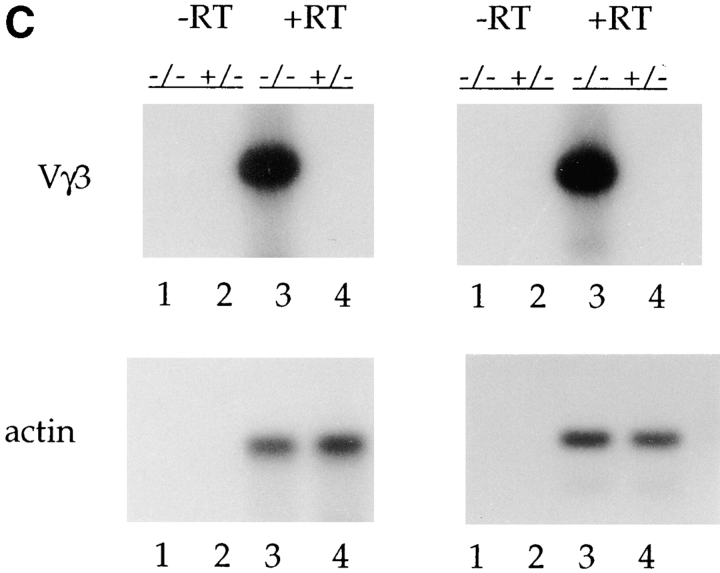
Analysis of the level of double-stranded DNA breaks at the Vγ3 and JH2 RSS and the level of Vγ3 germline transcripts in E2A-deficient mice. (A) Schematic diagram of the linker-ligation assay for detecting broken ends at the Vγ3 RSS. The thick line represents the ligated BW-1/BW-2 linker. (B) Total thymus DNA from an E2A-deficient mouse and a heterozygous littermate was analyzed for broken ends at the RSS 3′ of Vγ3 and 5′ of JH2 using LM-PCR. The same linker-ligated DNA was used for each of the PCR reactions. (C) Total thymocyte RNA from adult E2A-deficient mice and heterozygous littermates was analyzed by RT-PCR for the presence of Vγ3 sterile transcripts. Positive control RT-PCR reactions were performed with β-actin primers.
Increased Vγ3 Germline Transcripts in E2A-deficient Adult Thymus.
Several studies have demonstrated that gene rearrangement is correlated with prior transcription of the unrearranged genes. Indeed, there is a striking correlation between the level of gene rearrangement and the level of expression of the unrearranged genes (59–65). In fact, production of sterile transcripts has been postulated to direct recombination through the alteration of gene segment accessibility. In the γ locus, decreases in Vγ3 rearrangements correlate with decreases in the level of Vγ3 germline transcripts (22). To determine whether the increased cleavage of the γ3 V region is paralleled by increases in Vγ3 germline transcripts, we analyzed total thymus RNA from E2A-deficient mice and wild-type littermates by reverse-transcription (RT)-PCR. Consistent with the presence of γ3 rearrangements, Vγ3 sterile transcripts were readily detectable in the thymus of the E2A-deficient mice, but were absent from the thymus of control littermates (Fig. 6 C). Thus, the absence of the E2A proteins during adult thymocyte development allows for the continued transcription of the unrearranged γ3 V gene segment, likely rendering this V region accessible to the recombination machinery.
Discussion
The E2A gene products have the ability to bind to the enhancer elements within the Ig heavy and light chain genes as well as the TCR α and β genes (66–69). The ability of these enhancer elements to promote lineage-specific activation of recombination suggests that the E2A gene products might be involved in regulating Ig and TCR gene rearrangements (70–73). However, whether the E2A proteins are important for recombination of the Ig or TCR α and β loci is still unclear. Here we have examined the rearrangement efficiency of the two other TCR genes, the γ and δ genes, within E2A-deficient α/β T cells. Committed α/β T cells, which represent >90% of total thymocytes, retain previously rearranged γ and δ genes in both chromosomal and extrachromosomal DNA (39, 47, 54). Importantly, committed α/β T lymphocytes are selected and expanded on the basis of β chain rearrangements, and not on the basis of γ and δ rearrangements (74–76). Thus, the relative level of Vγ and Vδ gene rearrangements in total thymus DNA reflects the frequency of Vγ and Vδ gene usage during double-negative thymocyte development. The data presented here demonstrate that the E2A gene products are critical regulators of γ and δ V(D)J rearrangement during fetal and adult thymocyte development. First, they are required during fetal development to allow efficient rearrangement of at least one TCR Vδ gene segment and to a lesser extent a TCR Vγ gene segment. Second, the presence of the E2A proteins during adult thymocyte development is required to prevent the inappropriate rearrangement of V regions that primarily recombine in fetal thymocytes. The decreased usage of particular V gene segments correlates well with the deficiency of the γ/δ subpopulations expressing TCRs that use those V regions. Thus, the E2A gene products, which are required at two distinct stages of γ/δ T cell development, regulate the development of the γ/δ subpopulations through the ability to influence V gene usage during the recombination process.
Positive Regulation of V(D)J Recombination by the E2A Gene Products.
E2A-deficient mice have significantly reduced levels of rearrangements to particular V regions, including Vδ5 and Vγ2. There are various ways in which the bHLH proteins might positively regulate VDJ rearrangement. Several studies have suggested that transcriptional enhancers play important roles in modulating the accessibility of the gene segments to the recombination machinery, and E2A protein binding sites have been identified in the enhancer element of the δ locus (13, 25, 77, 78). However, others have shown that the TCR δ enhancer is important for J segment accessibility (79). In transgenic mice lacking the δ enhancer, V to D rearrangement is intact, but V-D to J rearrangement is inhibited (79). Since this phenotype is clearly distinct from that observed in the E2A-deficient mice, it is unlikely that bHLH proteins are functioning through the TCR δ enhancer.
The data described here raise the possibility that bHLH proteins play a role in the initiation of germline transcription. Indeed, there are several E box elements in the Vγ2 upstream sequences but not in the Vγ3 upstream sequences (Raulet, D., personal communication). However, we have examined for the presence of Vγ2 germline transcripts in E2A-deficient and wild-type thymocytes and have not found a tight correlation between the level of Vγ2 rearrangements and the level of Vγ2 germline transcripts (Bain, G., unpublished observations). An alternative mechanism is that bHLH proteins directly target the recombinase to the RSS. We note the presence of consensus E box binding sites in the linker sequences separating the heptamer and nonamer of almost all Vγ and Vδ gene segments. E2A and HEB proteins bind to these sites with relatively high affinity using nuclear extracts derived from thymocytes (Bain, G., and C. Murre, unpublished results). To determine how the E2A proteins positively regulate V(D)J recombination, it will be important to examine the functional significance of the E box binding sites in the promoter and spacer regions by mutational analysis using knock-in strategies, and to determine the transcriptional activity of particular V region promoters in the absence of all E box binding activity.
Negative Regulation of V(D)J Recombination by the E2A Proteins.
Normally, Vγ3 and Vδ1 rearrange coordinately during early thymic development and give rise to γ/δ T cells that migrate to the epithelium of the skin (20, 48). In thymocytes that develop in the adult mouse, rearrangements to these V regions occur only at a very low frequency (23). Interestingly, we show here that both Vγ3 and Vδ1 gene rearrangements are significantly overrepresented in DNA derived from E2A-deficient adult thymi. Thus, the presence of E2A is essential in order to properly regulate the ordered rearrangement of these TCR loci in adult thymocytes.
The question then becomes, how does E2A restrict the usage of the δ1 and γ3 V regions in wild-type thymocytes? As discussed above, positive regulation of recombination to the δ5 and γ2 V gene segments is impaired in E2A-deficient mice. It is conceivable that the differences in V region usage observed in the E2A-deficient thymus are simply the result of competition between V gene segments. That is, if Vδ5 and Vγ2 are prevented from rearranging efficiently in the absence of E2A, then Vγ3 and Vδ1 rearrangements, which don't require E2A, might continue to rearrange and be overrepresented. However, we consider it unlikely that a competition model could explain the data observed, particularly within the δ locus, for the following reasons. First, thymus DNAs isolated from adult E2A heterozygous mice show normal levels of Vδ5 rearrangements, but at the same time display significantly increased levels of Vδ1 rearrangements compared with wild-type littermates (Fig. 4 C). Thus, we observe increased recombination to the δ1 V region in the absence of a decrease in rearrangement to the δ5 V region. In addition, Vδ5 rearrangements are significantly decreased in fetal thymus DNAs derived from E2A-deficient and E2A heterozygous mice despite the fact that the Vδ1 rearrangements in these mice occur at a similar frequency as in the wild-type fetal thymus (Fig. 4 B). Taken together, we favor a model in which the E2A gene products normally function in restricting the accessibility of Vδ1 and Vγ3 to the recombinase.
Previous studies have identified a repressor element located in the Vγ3-Vγ4 intergenic region that functions on heterologous promoters in transient transfection assays (80). It has been proposed that the activation of this repressor element during adult thymocyte development renders the γ3 gene segment inaccessible to the recombinase (80). Several consensus protein binding sites have been identified in the repressor region; however, no E2A protein binding sites are present. Nevertheless, it will be important to examine whether E2A activity is required, indirectly, for the Vγ3-Vγ4 repressor activity. Additionally, it will be interesting to determine whether a similar repressor element exists in the δ locus. Taken together, we favor a model in which the absence of the E2A proteins during adult thymocyte development allows for the continued transcription of unrearranged fetal V gene segments, rendering continued access to the recombination machinery.
Dosage-dependent Regulation of V(D)J Recombination by the E2A Proteins.
An intriguing result of the data presented here is that the regulation of δ5 rearrangement during fetal thymic development appears to be highly sensitive to the dosage of E2A. We would like to consider the possibility that the concentration of E2A molecules in a double-negative T cell actively undergoing Vδ5 rearrangement is limiting to the recombination process. Site-specific recombination in B lymphocytes is controlled in such a way that if the initial VH to DJH rearrangement results in the production of a functional heavy chain, a signal is sent to shut down the heavy chain rearrangement process. As a result, an individual B cell expresses only one functional heavy chain. However, if the initial rearrangement is nonproductive, an additional VH to DJH rearrangement can be initiated on the other allele (9). This model predicts that VH gene rearrangement occurs only on one allele at a time. These data raise the question whether a limiting concentration of some factor is important in regulating this aspect of V(D)J recombination. The results described here indicate that the concentration of E2A proteins present in fetal thymocytes undergoing TCR δ rearrangements is rate limiting. Thus, it is conceivable that, within any one T lymphocyte, the level of E2A is such that only one allele of the TCR δ locus is undergoing rearrangement. In addition, these data suggest that the level of E2A activity is significantly lower in fetal thymocytes and relatively higher in adult thymocytes. Although E2A transcript levels are comparable between fetal and adult thymus RNAs, Id-2 transcript levels are threefold higher in fetal thymocytes compared with adult thymocytes (Bain, G., and C. Murre, unpublished data). Since the Id-2 protein can function as a negative regulator of E2A activity, it is likely that E2A activity is significantly lower during fetal thymocyte development.
Like the TCR γ and δ loci, the murine Ig heavy chain locus displays preferential rearrangement of particular V gene segments. For example, B cell precursors characteristically use a restricted set of VH segments, with the 3′ V gene segments being preferentially rearranged, whereas mature B cell populations use a wide range of VH segments (14–17). There is a dosage-dependent effect of E2A on differentiation through the B cell lineage as well, since mice heterozygous for E2A have ∼50% of the wild-type numbers of progenitor B cells (31, 81). These data raise the question of whether the E2A gene products control Ig gene rearrangements in a similar dosage-dependent fashion as described here for the TCR δ locus.
Acknowledgments
We thank Drs. F. Livak and D. Schatz (Yale University, New Haven, CT) for providing the δ Southern probes, and Dr. D. Raulet (University of California, Berkeley, CA) for providing Vγ2 and Vγ3 plasmids.
This work was supported by grants from the National Institutes of Health (to C. Murre and W.L. Havran), the Council for Tobacco Research, and the Malinkrodt Foundation (to C. Murre).
Abbreviations used in this paper
- bHLH
basic helix-loop-helix
- LM
ligation-mediated
- RAG
recombination activating gene
- RSS
recombination signal sequence(s)
- RT
reverse transcription
References
- 1.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 2.Schatz D, Oettinger M, Schlissel M. V(D)J recombination: molecular biology and regulation. Annu Rev Immunol. 1992;10:359–383. doi: 10.1146/annurev.iy.10.040192.002043. [DOI] [PubMed] [Google Scholar]
- 3.Oettinger M, Schatz D, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 4.McBlane J, van Gent D, Ramsden D, Romeo C, Cuomo C, Gellert M, Oettinger M. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 5.Mombaerts P, Iacomini J, Johnson RS, Herrup K, Tonegawa S, Papaioannou VE. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 6.Shinkai Y, Rathbun G, Lam K-P, Oltz EM, Stewart V, Mendelsohn M, Charron J, Datta M, Young F, Stall AM, Alt FW. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell. 1992;68:855–867. doi: 10.1016/0092-8674(92)90029-c. [DOI] [PubMed] [Google Scholar]
- 7.Schatz D, Oettinger M, Baltimore D. The V(D)J recombination activating gene, Rag-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Alt FW. Gene rearrangement and B cell development. Curr Opin Immunol. 1993;5:194–200. doi: 10.1016/0952-7915(93)90004-c. [DOI] [PubMed] [Google Scholar]
- 9.Alt F, Yancopoulos G, Blackwell T, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO (Eur Mol Biol Organ) J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kisielow P, von Boehmer H. Development and selection of T cells: facts and puzzles. Adv Immunol. 1995;58:87–209. doi: 10.1016/s0065-2776(08)60620-3. [DOI] [PubMed] [Google Scholar]
- 11.Strominger J. Developmental biology of T cell receptors. Science. 1989;244:943–950. doi: 10.1126/science.2658058. [DOI] [PubMed] [Google Scholar]
- 12.Stanhope-Baker P, Hudson K, Shaffer A, Constantinescu A, Schlissel M. Cell type-specific chromatin structure determines the targeting of V(D)J recombinase activity in vitro. . Cell. 1996;85:887–897. doi: 10.1016/s0092-8674(00)81272-6. [DOI] [PubMed] [Google Scholar]
- 13.Sleckman B, Gorman J, Alt F. Accessibility control of antigen-receptor variable-region gene assembly: role of cis-acting elements. Annu Rev Immunol. 1996;14:459–481. doi: 10.1146/annurev.immunol.14.1.459. [DOI] [PubMed] [Google Scholar]
- 14.Yancopoulos G, Malynn B, Alt F. Developmentally regulated and strain-specific expression of murine VH gene families. J Exp Med. 1988;168:417–435. doi: 10.1084/jem.168.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu G, Paige C. VH gene family utilization in colonies derived from B and pre-B cells detected by the RNA colony blot assay. EMBO (Eur Mol Biol Organ) J. 1986;5:3475–3481. doi: 10.1002/j.1460-2075.1986.tb04672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marshall A, Wu G, Paige C. Frequency of VH81X usage during B cell development. Initial decline in usage is independent of Ig heavy chain cell surface expression. J Immunol. 1996;156:2077–2084. [PubMed] [Google Scholar]
- 17.Dildrop R, Krawinkel U, Winter E, Rajewsky K. VH-gene expression in murine lipopolysaccharide blasts distributes over the nine known VH-gene groups and may be random. Eur J Immunol. 1985;15:1154–1156. doi: 10.1002/eji.1830151117. [DOI] [PubMed] [Google Scholar]
- 18.Raulet D. The structure, function, and molecular genetics of the γ/δ T cell receptor. Annu Rev Immunol. 1989;7:175–207. doi: 10.1146/annurev.iy.07.040189.001135. [DOI] [PubMed] [Google Scholar]
- 19.Ito K, Bonneville M, Takagaki Y, Nakanishi N, Kanagawa O, Krecko E, Tonegawa S. Different γ/δ T cell receptors are expressed on thymocytes at different stages of development. Proc Natl Acad Sci USA. 1989;86:631–635. doi: 10.1073/pnas.86.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Havran W, Allison J. Developmentally ordered appearance of thymocytes expressing different T cell antigen receptors. Nature. 1988;335:443–445. doi: 10.1038/335443a0. [DOI] [PubMed] [Google Scholar]
- 21.Garman R, Doherty P, Raulet D. Diversity, rearrangement and expression of murine T cell gamma genes. Cell. 1986;45:733–742. doi: 10.1016/0092-8674(86)90787-7. [DOI] [PubMed] [Google Scholar]
- 22.Goldman J, Spencer D, Raulet D. Ordered rearrangement of variable region genes of the T cell receptor γ locus correlates with transcription of the unrearranged genes. J Exp Med. 1993;177:729–739. doi: 10.1084/jem.177.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwashima M, Green A, Davis M, Chien Y. Variable region (Vδ) gene segment most frequently utilized in adult thymocytes is 3′ of the constant (Cδ) region. Proc Natl Acad Sci USA. 1988;85:8161–8165. doi: 10.1073/pnas.85.21.8161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chien Y, Iwashima M, Wettstein D, Kaplan K, Elliott J, Born W, Davis M. T cell receptor δ gene rearrangements in early thymocytes. Nature. 1987;330:722–727. doi: 10.1038/330722a0. [DOI] [PubMed] [Google Scholar]
- 25.Schlissel M, Stanhope-Baker P. Accessibility and the developmental regulation of V(D)J recombination. Semin Immunol. 1997;9:161–170. doi: 10.1006/smim.1997.0066. [DOI] [PubMed] [Google Scholar]
- 26.Nelsen B, Sen R. Regulation of immunoglobulin gene transcription. Int Rev Cytol. 1992;133:121–148. doi: 10.1016/s0074-7696(08)61859-8. [DOI] [PubMed] [Google Scholar]
- 27.Murre C, Bain G, van Dijk MA, Engel I, Furnari BA, Massari ME, Matthews JR, Quong MW, Rivera RR, Stuiver MH. Structure and function of helix-loop-helix proteins. Biochim Biophys Acta. 1994;1218:129–135. doi: 10.1016/0167-4781(94)90001-9. [DOI] [PubMed] [Google Scholar]
- 28.Kadesch T. Helix loop helix proteins in the regulation of immunoglobulin gene transcription. Immunol Today. 1992;13:31–36. doi: 10.1016/0167-5699(92)90201-h. [DOI] [PubMed] [Google Scholar]
- 29.Bain G, Maandag E, Izon D, Amsen D, Kruisbeek A, Weintraub B, Krop I, Schlissel M, Feeney A, van Roon M, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 30.Sun X-H. Constitutive expression of the Id1gene impairs mouse B cell development. Cell. 1994;79:893–900. doi: 10.1016/0092-8674(94)90078-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhuang Y, Soriano P, Weintraub H. The helix-loop-helix gene E2A is required for B cell formation. Cell. 1994;79:875–884. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 32.Bain G, Robanus EC, Maandag, te Riele HP, Feeney A, Sheehy A, Schlissel M, Shinton S, Hardy R, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 33.Bain G, Engel I, Robanus EC, Maandag, te Riele HP, Voland J, Sharp L, Chun J, Huey B, Pinkel D, Murre C. E2A deficiency leads to abnormalities in αβ T-cell development and to the rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kee B, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix-loop-helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi J, Shen C-P, Radomska H, Eckhardt L, Kadesch T. E47 activates the Ig-heavy chain and TdT loci in non-B cells. EMBO (Eur Mol Biol Organ) J. 1996;15:5014–5021. [PMC free article] [PubMed] [Google Scholar]
- 36.Schlissel M, Voronova A, Baltimore D. Helix loop helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 1991;5:1367–1376. doi: 10.1101/gad.5.8.1367. [DOI] [PubMed] [Google Scholar]
- 37.Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- 38.Boismenu R, Havran W. Modulation of epithelial cell growth by intraepithelial γδ T cells. Science. 1994;266:1253–1255. doi: 10.1126/science.7973709. [DOI] [PubMed] [Google Scholar]
- 39.Livak F, Patrie H, Crispe I, Schatz D. In-frame TCR δ gene rearrangements play a critical role in the αβ/γδ T cell lineage decision. Immunity. 1995;2:617–627. doi: 10.1016/1074-7613(95)90006-3. [DOI] [PubMed] [Google Scholar]
- 40.Asarnow D, Kuziel W, Bonyhadi M, Tigelaar R, Tucker P, Allison J. Limited diversity of γδ antigen receptor genes of thy-1+ dendritic epidermal cells. Cell. 1988;55:837–847. doi: 10.1016/0092-8674(88)90139-0. [DOI] [PubMed] [Google Scholar]
- 41.Asarnow D, Goodman T, LeFrancois L, Allison J. Distinct antigen receptor repertoires of two classes of murine epithelium-associated T cells. Nature. 1989;341:60–62. doi: 10.1038/341060a0. [DOI] [PubMed] [Google Scholar]
- 42.Roth D, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 44.Heemskerk M, Blom B, Nolan G, Stegmann A, Bakker A, Weijer K, Res P, Spits H. Inhibition of T cell and promotion of natural killer cell development by the dominant negative helix loop helix factor Id3. J Exp Med. 1997;186:1597–1602. doi: 10.1084/jem.186.9.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petrie H, Scollay R, Shortman K. Commitment to the T-cell receptor αβ or γδ lineages can occur just prior to the onset of CD4 and CD8 expression among immature thymocytes. Eur J Immunol. 1992;22:2185–2188. doi: 10.1002/eji.1830220836. [DOI] [PubMed] [Google Scholar]
- 46.Godfrey D, Kennedy J, Suda T, Zlotnik A. A developmental pathway involving four phenotypically and functionally distinct subsets of CD3−CD4−CD8−triple-negative adult mouse thymocytes defined by CD44 and CD25 expression. J Immunol. 1993;150:4244–4252. [PubMed] [Google Scholar]
- 47.Dudley E, Girardi M, Owens M, Hayday A. αβ and γδ T cells can share a late common precursor. Curr Biol. 1995;5:659–669. doi: 10.1016/s0960-9822(95)00131-x. [DOI] [PubMed] [Google Scholar]
- 48.Raulet D, Spencer D, Hsiang Y-H, Goldman J, Bix M, Liao N-S, Zijlstra M, Jaenisch R, Correa I. Control of γδ T-cell development. Immunol Rev. 1991;120:185–204. doi: 10.1111/j.1600-065x.1991.tb00592.x. [DOI] [PubMed] [Google Scholar]
- 49.Carding S, Kyes S, Jenkinson E, Kingston R, Bottomly K, Owen T, Hayday A. Developmentally regulated fetal thymic and extrathymic T-cell receptor γδ gene expression. Genes Dev. 1990;4:1304–1315. doi: 10.1101/gad.4.8.1304. [DOI] [PubMed] [Google Scholar]
- 50.Lafaille J, DeCloux A, Bonneville M, Takagaki Y, Tonegawa S. Junctional sequences of T cell receptor γδ genes: implications for γδ T cell lineages and for novel intermediate V-(D)-J joining. Cell. 1989;59:859–870. doi: 10.1016/0092-8674(89)90609-0. [DOI] [PubMed] [Google Scholar]
- 51.McVay L, Carding S, Bottomly K, Hayday A. Regulated expression and structure of T cell receptor γ/δ transcripts in human thymic ontogeny. EMBO (Eur Mol Biol Organ) J. 1991;10:83–91. doi: 10.1002/j.1460-2075.1991.tb07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elliot J, Rock E, Patten P, Davis M, Chien Y. The adult T-cell receptor δ-chain is diverse and distinct from that of fetal thymocytes. Nature. 1988;331:627–631. doi: 10.1038/331627a0. [DOI] [PubMed] [Google Scholar]
- 53.Heilig J, Tonegawa S. Diversity of murine gamma genes and expression in fetal and adult T lymphocytes. Nature. 1986;322:836–840. doi: 10.1038/322836a0. [DOI] [PubMed] [Google Scholar]
- 54.Nakajima P, Menetski J, Roth D, Gellert M, Bosma M. V-D-J rearrangements at the T cell receptor δ locus in mouse thymocytes of the αβ lineage. Immunity. 1995;3:609–621. doi: 10.1016/1074-7613(95)90132-9. [DOI] [PubMed] [Google Scholar]
- 55.Zhu C, Bogue M, Lim D-S, Hasty P, Roth D. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 56.Cortes P, Weis-Garcia F, Misulovin Z, Nussenzweig A, Lai J-S, Li G, Nussenzweig M, Baltimore D. In vitro V(D)J recombination: signal joint formation. Proc Natl Acad Sci USA. 1996;93:14008–14013. doi: 10.1073/pnas.93.24.14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lewis S. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 58.Eastman Q, Leu T, Schatz D. Initiation of V(D)J recombination in vitroobeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 59.Blackwell T, Moore M, Yancopoulos G, Suh H, Lutzker S, Selsing E, Alt F. Recombination between immunoglobulin variable region gene segments is enhanced by transcription. Nature. 1986;324:585–590. doi: 10.1038/324585a0. [DOI] [PubMed] [Google Scholar]
- 60.Ferrier P, Krippl B, Blackwell TK, Furley AJW, Suh H, Winoto A, Cook WD, Hood L, Costantini F, Alt FW. Separate elements control DJ and VDJ rearrangement in a transgenic recombination substrate. EMBO (Eur Mol Biol Organ) J. 1990;9:117–125. doi: 10.1002/j.1460-2075.1990.tb08087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lennon G, Perry R. The temporal order of appearance of transcripts from unrearranged and rearranged Ig genes in murine fetal liver. J Immunol. 1990;144:1983–1987. [PubMed] [Google Scholar]
- 62.Martin D, Huang R, LeBien T, VanNess B. Induced rearrangement of κ genes in the BLIN-1 human pre-B cell line correlates with germline J-Cκ and Vκ transcription. J Exp Med. 1991;173:639–645. doi: 10.1084/jem.173.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schlissel MS, Baltimore D. Activation of immunoglobulin kappa gene rearrangement correlates with induction of germline kappa gene transcription. Cell. 1989;58:1001–1007. doi: 10.1016/0092-8674(89)90951-3. [DOI] [PubMed] [Google Scholar]
- 64.Yancopoulos G, Alt FW. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell. 1985;40:271–281. [PubMed] [Google Scholar]
- 65.Yancopoulos G, Blackwell T, Suh H, Hood L, Alt F. Introduced T cell receptor variable region gene segments recombine in pre-B cells: evidence that B and T cells use a common recombinase. Cell. 1986;44:251–259. doi: 10.1016/0092-8674(86)90759-2. [DOI] [PubMed] [Google Scholar]
- 66.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer μE5/κE2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 67.Ho I-C, Yang L-H, Morle G, Leiden J. A T-cell-specific transcriptional enhancer element 3′ of Cα in the human T-cell receptor α locus. Proc Natl Acad Sci USA. 1989;86:6714–6718. doi: 10.1073/pnas.86.17.6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murre C, McCaw PS, Baltimore D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and mycproteins. Cell. 1989;56:777–783. doi: 10.1016/0092-8674(89)90682-x. [DOI] [PubMed] [Google Scholar]
- 69.Takeda J, Cheng A, Mauxion F, Nelson CA, Newberry RD, Sha WC, Sen R, Loh DY. Functional analysis of the murine T-cell receptor β enhancer and characteristics of its DNA-binding proteins. Mol Cell Biol. 1990;10:5027–5035. doi: 10.1128/mcb.10.10.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lauzurica P, Krangel M. Temporal and lineage-specific control of T cell receptor α/δ gene rearrangement by T cell receptor α and δ enhancers. J Exp Med. 1994;179:1913–1921. doi: 10.1084/jem.179.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Capone M, Watrin F, Fernex C, Horvat B, Krippl B, Wu L, Scollay R, Ferrier P. TCRβ and TCRα gene enhancers confer tissue- and stage-specificity on V(D)J recombination events. EMBO (Eur Mol Biol Organ) J. 1993;12:4335–4346. doi: 10.1002/j.1460-2075.1993.tb06118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okada A, Mendelsohn M, Alt F. Differential activation of transcription versus recombination of transgenic T cell receptor β variable region gene segments in B and T lineage cells. J Exp Med. 1994;180:261–272. doi: 10.1084/jem.180.1.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roberts J, Lauzurica P, Krangel M. Developmental regulation of VDJ recombination by the core fragment of the T cell receptor α enhancer. J Exp Med. 1997;185:131–140. doi: 10.1084/jem.185.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mombaerts P, Clarke A, Rudnicki M, Iacomini J, Itohara S, Lafaille J, Wang L, Ichikawa Y, Jaenisch R, Hooper M, Tonegawa S. Mutations in T-cell antigen receptor genes α and β block thymocyte development at different stages. Nature. 1992;360:225–231. doi: 10.1038/360225a0. [DOI] [PubMed] [Google Scholar]
- 75.Mallick C, Dudley E, Viney J, Owen M, Hayday A. Rearrangement and diversity of T cell receptor β chain genes in thymocytes: a critical role for the β chain in development. Cell. 1993;73:513–519. doi: 10.1016/0092-8674(93)90138-g. [DOI] [PubMed] [Google Scholar]
- 76.Shinkai Y, Koyasu S, Nakayama K, Murphy K, Loh D, Reinherz E, Alt F. Restoration of T-cell development in RAG-2 deficient mice by functional TCR transgenes. Science. 1993;259:822–825. doi: 10.1126/science.8430336. [DOI] [PubMed] [Google Scholar]
- 77.Redondo J, Hata S, Brocklehurst C, Krangel M. A T cell specific transcriptional enhancer within the human T cell receptor δ locus. Science. 1990;247:1225–1229. doi: 10.1126/science.2156339. [DOI] [PubMed] [Google Scholar]
- 78.Redondo J, Pfohl J, Krangel M. Identification of an essential site for transcriptional activation within the human T cell receptor δ enhancer. Mol Cell Biol. 1991;11:5671–5680. doi: 10.1128/mcb.11.11.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.McMurry M, Hernandez-Munain C, Lauzurica P, Krangel M. Enhancer control of local accessibility to V(D)J recombinase. Mol Cell Biol. 1997;17:4553–4561. doi: 10.1128/mcb.17.8.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clausell A, Tucker P. Functional analysis of the V gamma 3 promoter of the murine gamma delta T-cell receptor. Mol Cell Biol. 1994;14:803–814. doi: 10.1128/mcb.14.1.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes, E2A, E2-2, and . HEB Mol Cell Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]