Abstract
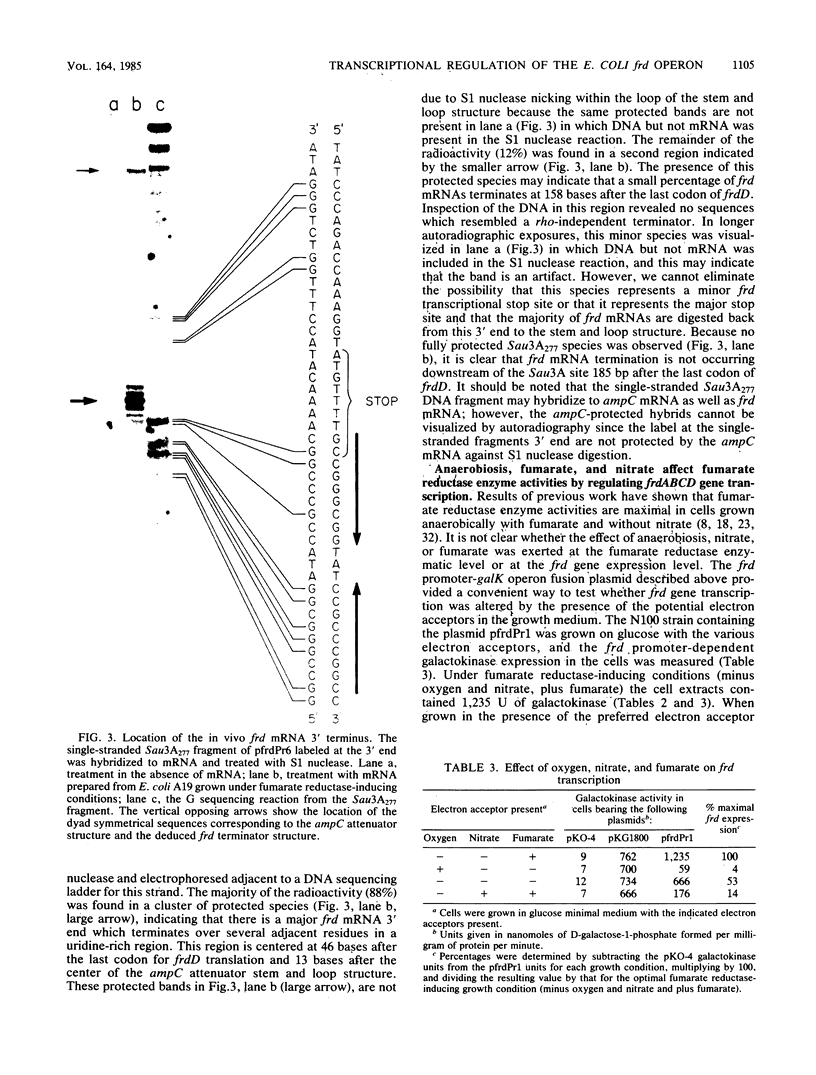
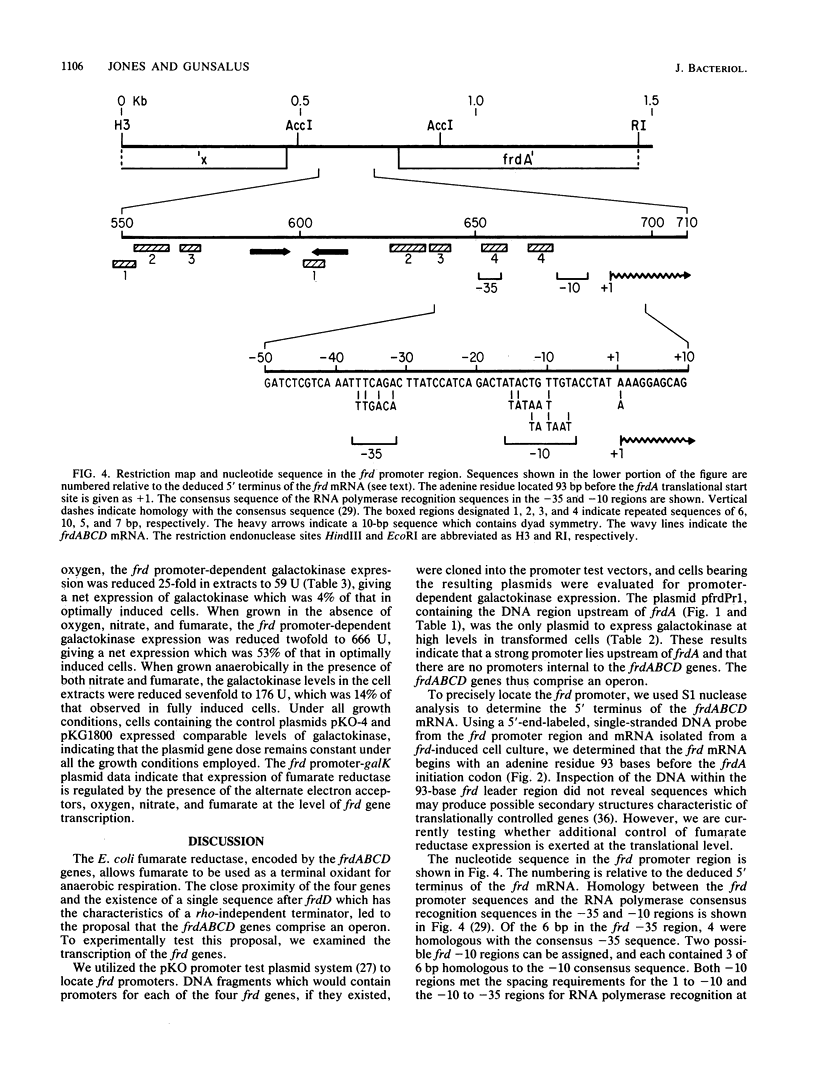
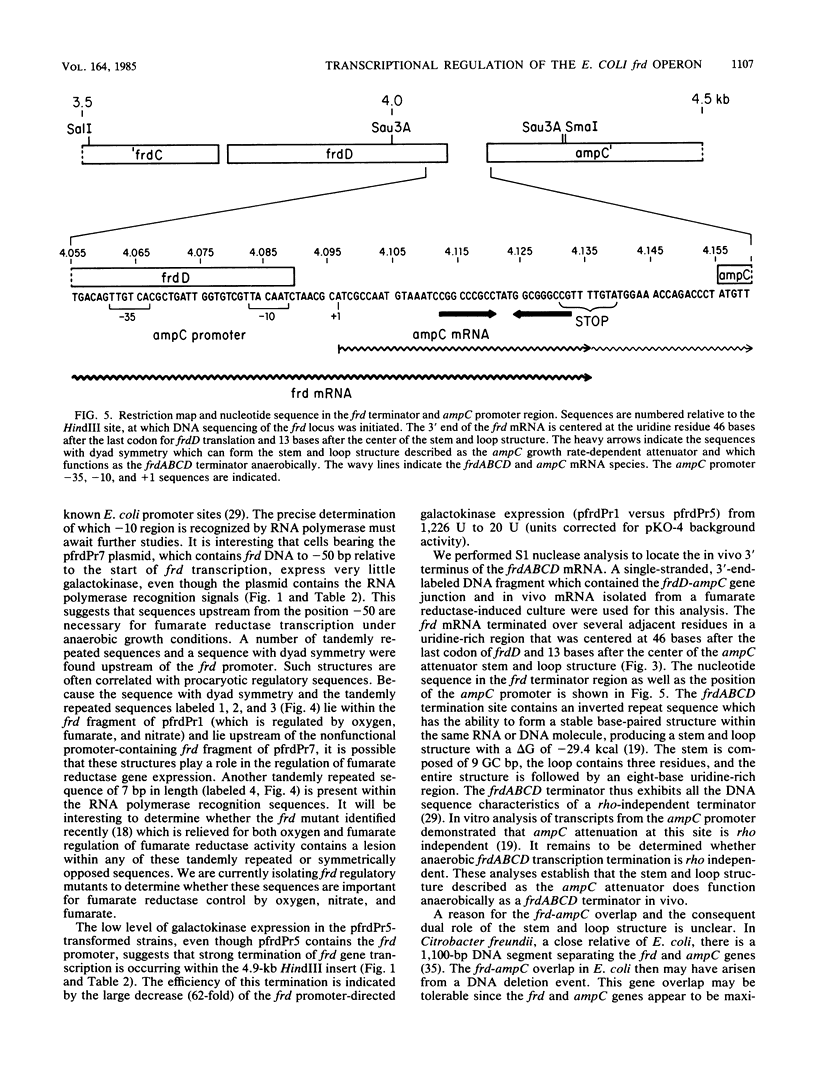
The fumarate reductase enzyme complex allows Escherichia coli to grow anaerobically with fumarate as a terminal electron acceptor for oxidative phosphorylation when the preferred compounds oxygen and nitrate are not available. We used the pKO promoter test vectors to identify a single promoter for the frdABCD genes which encode fumarate reductase. Expression of galactokinase from the frd promoter-galK operon fusion plasmid was repressed by oxygen and by nitrate and was induced by fumarate, indicating that frd gene expression is regulated at the transcriptional level by these terminal electron acceptors. S1 nuclease analysis, using a single-stranded DNA probe from the frd promoter region and mRNA isolated from a fumarate reductase-induced culture, revealed that the frd mRNA transcript initiates with an adenine residue 93 bases prior to the start of frdA translation. No promoters internal to the frd genes were revealed with the plasmid promoter screening system. S1 nuclease analysis revealed that the frd mRNA terminates in a uridine-rich region centered at 46 bases after the last codon of frdD. A stem and loop structure previously described as the growth rate-dependent attenuator for the linked ampC gene precedes the frd mRNA terminus. This result confirms the proposal that the stem and loop structure serves the dual role of a frd terminator anaerobically and an ampC attenuator aerobically. The four frd genes encoding the subunits of the fumarate reductase complex thus comprise an operon which is regulated at the transcriptional level in response to the cellular availability of the alternate electron acceptors oxygen, nitrate, and fumarate.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H. Autoregulation of the Escherichia coli crp gene: CRP is a transcriptional repressor for its own gene. Cell. 1983 Jan;32(1):141–149. doi: 10.1016/0092-8674(83)90504-4. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilous P. T., Weiner J. H. Dimethyl sulfoxide reductase activity by anaerobically grown Escherichia coli HB101. J Bacteriol. 1985 Jun;162(3):1151–1155. doi: 10.1128/jb.162.3.1151-1155.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chippaux M., Bonnefoy-Orth V., Ratouchniak J., Pascal M. C. Operon fusions in the nitrate reductase operon and study of the control gene nir R in Escherichia coli. Mol Gen Genet. 1981;182(3):477–479. doi: 10.1007/BF00293938. [DOI] [PubMed] [Google Scholar]
- Chippaux M., Giudici D., Abou-Jaoudé A., Casse F., Pascal M. C. Laboratoire de Chimie Bactérienne C.N.R.S., Marsielle, France. Mol Gen Genet. 1978 Apr 6;160(2):225–229. doi: 10.1007/BF00267485. [DOI] [PubMed] [Google Scholar]
- Cole J. A., Wimpenny J. W. Metabolic pathways for nitrate reduction in Escherichia coli. Biochim Biophys Acta. 1968 Jul 16;162(1):39–48. doi: 10.1016/0005-2728(68)90212-0. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Grundström T., Jaurin B., Robinson J. J., Weiner J. H. Location and nucleotide sequence of frdB, the gene coding for the iron-sulphur protein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982 Aug;126(1):211–216. doi: 10.1111/j.1432-1033.1982.tb06768.x. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Guest J. R. Genetic and physical characterization of lambda transducing phages (lambda frdA) containing the fumarate reductase gene of Escherichia coli K12. Mol Gen Genet. 1980;178(2):409–418. doi: 10.1007/BF00270492. [DOI] [PubMed] [Google Scholar]
- Cole S. T. Nucleotide sequence coding for the flavoprotein subunit of the fumarate reductase of Escherichia coli. Eur J Biochem. 1982 Mar 1;122(3):479–484. doi: 10.1111/j.1432-1033.1982.tb06462.x. [DOI] [PubMed] [Google Scholar]
- Edlund T., Grundström T., Normark S. Isolation and characterization of DNA repetitions carrying the chromosomal beta-lactamase gene of Escherichia coli K-12. Mol Gen Genet. 1979 Jun 7;173(2):115–125. doi: 10.1007/BF00330301. [DOI] [PubMed] [Google Scholar]
- Fimmel A. L., Haddock B. A. Use of chlC-lac fusions to determine regulation of gene chlC in Escherichia coli K-12. J Bacteriol. 1979 Jun;138(3):726–730. doi: 10.1128/jb.138.3.726-730.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundström T., Jaurin B. Overlap between ampC and frd operons on the Escherichia coli chromosome. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1111–1115. doi: 10.1073/pnas.79.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J. R. Partial replacement of succinate dehydrogenase function by phage- and plasmid-specified fumarate reductase in Escherichia coli. J Gen Microbiol. 1981 Feb;122(2):171–179. doi: 10.1099/00221287-122-2-171. [DOI] [PubMed] [Google Scholar]
- HIRSCH C. A., RASMINSKY M., DAVIS B. D., LIN E. C. A FUMARATE REDUCTASE IN ESCHERICHIA COLI DISTINCT FROM SUCCINATE DEHYDROGENASE. J Biol Chem. 1963 Nov;238:3770–3774. [PubMed] [Google Scholar]
- Ingledew W. J., Poole R. K. The respiratory chains of Escherichia coli. Microbiol Rev. 1984 Sep;48(3):222–271. doi: 10.1128/mr.48.3.222-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kuritzkes D. R., Lin E. C. Escherichia coli mutant with altered respiratory control of the frd operon. J Bacteriol. 1985 Mar;161(3):1023–1028. doi: 10.1128/jb.161.3.1023-1028.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaurin B., Grundström T., Edlund T., Normark S. The E. coli beta-lactamase attenuator mediates growth rate-dependent regulation. Nature. 1981 Mar 19;290(5803):221–225. doi: 10.1038/290221a0. [DOI] [PubMed] [Google Scholar]
- Jones H. M., Brajkovich C. M., Gunsalus R. P. In vivo 5' terminus and length of the mRNA for the proton-translocating ATPase (unc) operon of Escherichia coli. J Bacteriol. 1983 Sep;155(3):1279–1287. doi: 10.1128/jb.155.3.1279-1287.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritzkes D. R., Zhang X. Y., Lin E. C. Use of phi(glp-lac) in studies of respiratory regulation of the Escherichia coli anaerobic sn-glycerol-3-phosphate dehydrogenase genes (glpAB). J Bacteriol. 1984 Feb;157(2):591–598. doi: 10.1128/jb.157.2.591-598.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambden P. R., Guest J. R. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976 Dec;97(2):145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- Lemire B. D., Robinson J. J., Weiner J. H. Identification of membrane anchor polypeptides of Escherichia coli fumarate reductase. J Bacteriol. 1982 Dec;152(3):1126–1131. doi: 10.1128/jb.152.3.1126-1131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmeier E., Hagen D. S., Dickie P., Weiner J. H. Cloning and expression of fumarate reductase gene of Escherichia coli. Can J Biochem. 1981 Mar;59(3):158–164. doi: 10.1139/o81-023. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKenney K., Shimatake H., Court D., Schmeissner U., Brady C., Rosenberg M. A system to study promoter and terminator signals recognized by Escherichia coli RNA polymerase. Gene Amplif Anal. 1981;2:383–415. [PubMed] [Google Scholar]
- Newman B. M., Cole J. A. The chromosomal location and pleiotropic effects of mutations of the nirA+ gene of Escherichia coli K12: the essential role of nirA+ in nitrite reduction and in other anaerobic redox reactions. J Gen Microbiol. 1978 May;106(1):1–12. doi: 10.1099/00221287-106-1-1. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Ruch F. E., Kuritzkes D. R., Lin E. C. Use of lac operon fusions to isolate Escherichia coli mutants with altered expression of the fumarate reductase system in response to substrate and respiratory controls. Biochem Biophys Res Commun. 1979 Dec 28;91(4):1365–1370. doi: 10.1016/0006-291x(79)91217-8. [DOI] [PubMed] [Google Scholar]
- Salser W., Gesteland R. F., Bolle A. In vitro synthesis of bacteriophage lysozyme. Nature. 1967 Aug 5;215(5101):588–591. doi: 10.1038/215588a0. [DOI] [PubMed] [Google Scholar]
- Spencer M. E., Guest J. R. Isolation and properties of fumarate reductase mutants of Escherichia coli. J Bacteriol. 1973 May;114(2):563–570. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J. H., Dickie P. Fumarate reductase of Escherichia coli. Elucidation of the covalent-flavin component. J Biol Chem. 1979 Sep 10;254(17):8590–8593. [PubMed] [Google Scholar]
- Yamamoto T., Murayama S. Y., Sawai T. Cloning and expression of the gene(s) for cephalosporinase production of Citrobacter freundii. Mol Gen Genet. 1983;190(1):85–91. doi: 10.1007/BF00330328. [DOI] [PubMed] [Google Scholar]
- Yanofsky C. Attenuation in the control of expression of bacterial operons. Nature. 1981 Feb 26;289(5800):751–758. doi: 10.1038/289751a0. [DOI] [PubMed] [Google Scholar]
- Zalkin H., Yanofsky C., Squires C. L. Regulated in vitro synthesis of Escherichia coli tryptophan operon messenger ribonucleic acid and enzymes. J Biol Chem. 1974 Jan 25;249(2):465–475. [PubMed] [Google Scholar]