Abstract
Leukotriene B4 (LTB4) is a potent chemoattractant and activator of both granulocytes and macrophages. The actions of LTB4 appear to be mediated by a specific G protein–coupled receptor (GPCR) BLT1, originally termed BLT (Yokomizo, T., T. Izumi, K. Chang, Y. Takuwa, and T. Shimizu. 1997. Nature. 387:620–624). Here, we report the molecular cloning of a novel GPCR for LTB4, designated BLT2, which binds LTB4 with a Kd value of 23 nM compared with 1.1 nM for BLT1, but still efficiently transduces intracellular signaling. BLT2 is highly homologous to BLT1, with an amino acid identity of 45.2%, and its open reading frame is located in the promoter region of the BLT1 gene. BLT2 is expressed ubiquitously, in contrast to BLT1, which is expressed predominantly in leukocytes. Chinese hamster ovary cells expressing BLT2 exhibit LTB4-induced chemotaxis, calcium mobilization, and pertussis toxin–insensitive inhibition of adenylyl cyclase. Several BLT1 antagonists, including U 75302, failed to inhibit LTB4 binding to BLT2. Thus, BLT2 is a pharmacologically distinct receptor for LTB4, and may mediate cellular functions in tissues other than leukocytes. BLT2 provides a novel target for antiinflammatory therapy and promises to expand our knowledge of LTB4 function. The location of the gene suggests shared transcriptional regulation of these two receptors.
Keywords: BLT, cloning, gene cluster, leukotriene, low-affinity receptor
Introduction
Leukotriene B4 (LTB4; 5[S],12[R]-dihydroxy-6,14-cis-8,10-trans-eicosatetraenoic acid) is a metabolite of arachidonic acid and is one of the most potent activators of granulocytes and macrophages 1 2 3. LTB4 binds to a specific G protein–coupled receptor (GPCR) named BLT and activates the Gi and G16 classes of G proteins 4 to inhibit adenylyl cyclase and activate phospholipase C. Exposure to LTB4 induces adhesion of granulocytes to endothelial cells, degranulation of the lysosomal enzymes, generation of superoxide, and transmigration of granulocytes, all important in the host defense against foreign organisms. Overproduction of LTB4 is involved in inflammatory diseases including psoriasis 5, bronchial asthma 6, rheumatoid arthritis 7, inflammatory bowel diseases 8, and ischemic renal failure 9. Mice lacking leukotriene production are deficient in their response to some acute and chronic inflammatory stimuli 10 11 12 13. Therefore, BLT antagonists are under development as potent antiinflammatory drugs 14 15 16 17. BLT antagonists have most recently been reevaluated as immunosuppressive agents for allograft rejection 18 19 20. We recently reported the characterization of cDNA encoding human BLT1 (originally termed BLT) isolated from HL-60 cells, and showed that BLT1 mRNA is highly expressed in leukocytes, and to a much lesser extent in the other tissues 21. BLT1s isolated from mouse 22 23, guinea pig 24 25 and rat 26 are highly homologous to hBLT1, with amino acid identities of >75% 27. During the course of the analysis of the genomic structures of human and mouse BLT1, we identified a novel gene encoding a putative GPCR with structural similarity to BLT1. Surprisingly, this receptor shows specific binding for LTB4 and activates multiple intracellular signaling pathways when expressed in mammalian cells. In this study, we describe the molecular cloning of this novel LTB4 receptor and demonstrate its specificity, expression, and function. Further, we show that the receptor has pharmacological properties different from those of BLT1. In an accompanying paper, we report that BLT2 open reading frame (ORF) is present in the promoter region of BLT1 28, weaving these two receptors tightly at the genomic and functional level.
Materials and Methods
Isolation of Genomic Clones Containing Human and Mouse BLT1.
Genomic libraries from human and mouse were screened by plaque hybridization. 106 clones from human genomic library (Human Lymphocyte Genomic Library; Stratagene), and mouse library (129SV Mouse Genomic Library; Stratagene) were lifted to Hybond N+ nylon membranes (Amersham Pharmacia Biotech) and screened with [32P]dCTP-labeled ORF of hBLT1 and 800 bp of expressed sequence tag (EST) clones encoding mouse BLT1 (sequence data available from EMBL/GenBank/DDBJ under accession no. AA028322), respectively. Hybridization was carried out in a hybridization buffer containing 6× SSC, 10× Dehnhart's solution, 0.5% SDS, and 100 μg/ml single-stranded salmon sperm DNA at 65°C overnight. The membranes were washed in 2× SSC, 0.1% SDS, followed by washing in 0.5× SSC, 0.1% SDS at 25°C. Tertiary screening gave three human and two mouse clones, which were analyzed by Southern blotting. DNA sequencing was done using an automated DNA sequencer (model 373A; Applied Biosystems) and LI-COR 4000LS (Aloka).
Northern Blotting.
Human multiple-tissue Northern blots (CLONTECH Laboratories, Inc.) were hybridized with [32P] dCTP-labeled ORF of hBLT2 and human β-actin cDNA in ExpressHyb hybridization solution (CLONTECH Laboratories, Inc.) for 18 h (BLT2) or 1 h (β-actin) at 68°C. The membranes were washed in 0.1× SSC, 0.1% SDS for 2 h at 65° C and subjected to autoradiography.
Construction of Expression Vectors for hBLT2.
Two expression vectors for wild-type and hemagglutinin (HA)-tagged hBLT2 were constructed. The inserts were amplified from the genomic clone containing a full-length BLT2 by PCR with sense (5′-CGGGATCCCGCCATGTCGGTCTGCTACCGT-3′ and 5′-CGGGATCCCGCCATGTACCCCTACGACGTGCCCGACTACGCCTCGGTCTGCTACCGTCC-3′ for wild-type and HA, respectively) and antisense (5′-GGAATTCAAAGGTCCCATTCCGG-3′) primers, digested with BamHI and EcoRI, and subcloned into pcDNA3 vector (Invitrogen). The plasmids for wild-type and HA-tagged BLT2 were designated as phBLT2 and pHA-hBLT2, respectively. Entire sequences of the inserts were determined on both strands for unexpected misincorporations.
Cell Culture and Transfection.
HEK 293 and Chinese hamster ovary (CHO) cells were cultured in DMEM and Ham's F12, respectively, supplemented with 10% FCS (Sigma-Aldrich), 100 IU/ml penicillin, and 100 μg/ml streptomycin. For transient expression, HEK 293 cells on 15-cm plates were transfected by lipofection using 20 μg of plasmid DNA and Lipofectamine Plus (Life Technologies) according to the manufacturer's protocol. After 3 d, the cells were harvested and sonicated in a sonication buffer containing 20 mM Tris-HCl, pH 7.4, 0.25 M sucrose, 10 mM MgCl2, 2 mM EDTA-Na2, 2 mM PMSF, and 1 μM pepstatin-A. After centrifugation at 12,000 g for 10 min at 4°C, the remaining supernatants were further centrifuged at 105,000 g for 60 min. The resulting pellets were used for binding assay as membrane fractions. The concentrations of the protein were determined by a method of Bradford using protein assay (Bio-Rad Laboratories). For stable expression of HA-tagged BLT2, CHO-K1 cells were transfected with pHA-hBLT2 by lipofection using Transfectam (Life Technologies) and selected with 1 mg/ml G418. 18 resistant clones were isolated by limiting dilution and examined for the expression of the receptor protein. The cells were fixed with PBS(−) containing 0.5% paraformaldehyde for 5 min on ice and blocked with PBS(−) containing 2% FCS. The cells were bound with 10 μg/ml anti-HA antibody (clone CA12-5; Eastman Kodak Co.) in PBS(−) containing 2% FCS for 1 h, followed by staining with 500× FITC-anti–mouse IgG (Zymed Laboratories) for 30 min. The cells were washed twice with PBS(−) and analyzed with a flow cytometer (Epics XL; Beckman Coulter). Three lines of the cells (HA-12, -13, and -14) with high expression were selected, maintained in Ham F12, 10% FCS, and 0.3 mg/ml G418, and used for the further analyses.
3H–LTB4 Binding Assay.
The membrane fractions of HEK 293 cells were examined for 3H–LTB4 binding. The binding mixture (100 μl) contained membrane fractions and various concentrations of 3H–LTB4 with or without unlabeled LTB4 in binding buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 10 mM NaCl, and 0.05% BSA). For determination of the nonspecific binding, at least 1,000 times concentration of unlabeled LTB4 was used. The mixtures were incubated at room temperature for 60 min with agitation, followed by rapid filtration through GF/C filters (Packard Instrument Co.) and washing with ∼3 ml of binding buffer. The radioactivities of the filters were determined with a scintillation counter (Top Count; Packard Instrument Co.).
Measurement of Intracellular Calcium Concentration.
The CHO cells were loaded with 10 μM Fura-2 AM (Dojin) in Hepes-Tyrode's BSA buffer (25 mM Hepes-NaOH, pH 7.4, 140 mM NaCl, 2.7 mM KCl, 1.0 mM CaCl2, 12 mM NaHCO3, 5.6 mM d-glucose, 0.37 mM NaH2PO4, 0.49 mM MgCl2, and 0.1% [wt/vol] fatty acid–free BSA; Fraction V) at 37°C for 2 h. The cells were washed twice and resuspended in Hepes-Tyrode's BSA buffer at the density of 106 cells/ml. 0.5 ml of the cell suspension was applied to a CAF-100 system (Jasco), and 5 μl of ligand solution in ethanol (for eicosanoids) or in PBS (for chemokines) was added. Intracellular Ca2+ concentration was measured by the ratio of emission fluorescence of 500 nm by excitation at 340 and 380 nm.
Measurement of cAMP.
The cells were seeded on 96-well plates (20,000 cells per well) and cultured for another 36 h. The medium was replaced to 100 μl of Hepes-Tyrode's BSA buffer containing 1 mM IBMX (3-isobutyl-1-methylxanthine), and incubated at 37°C for 20 min. The reaction was initiated by adding 100 μl of the ligand solution, and after 30 min of incubation, the reaction was terminated by adding 25 μl of lysis reagent 1A included in Biotrak cAMP enzyme immunoassay system (Amersham Pharmacia Biotech). The cAMP contents in 5 μl of the aliquots were determined using the assay kit according to the manufacturer's protocol.
Chemotaxis Assay.
CHO cells expressing hBLT1 or hBLT2 were examined for their chemotactic responses as described previously 21.
Presentation of the Data.
All figures shown contain representative data from at least two independent experiments with similar results.
Results
Cloning of Human and Mouse BLT2.
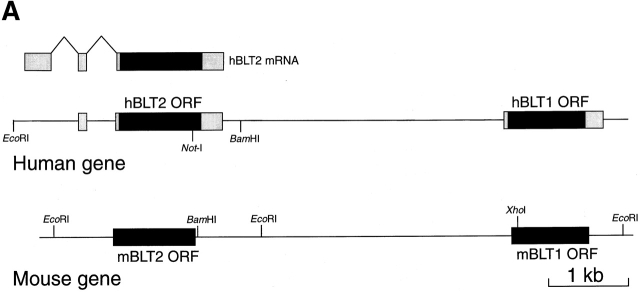
During the course of the analysis of human and mouse BLT1 genes, we identified an ORF encoding a putative seven-transmembrane–type receptor with sequence similarities to BLT1. We designate the new gene BLT2. BLT1 and BLT2 genes are located within 10 kbp of each other both in humans and mice (Fig. 1 A). Because a NotI site is present in the ORF of hBLT2, we could not isolate full-length cDNA clones for hBLT2 from cDNA libraries (data not shown). However, we found several EST clones containing polyadenylation signals followed by poly A tails, or exon/intron junctions upstream of the ORF of hBLT2 (Fig. 1 B). The ORFs of human and mouse BLT2 encode proteins with 358 and 360 amino acids, respectively (Fig. 1 C), which contain several consensus GPCR sequences 29. These include 38N in the transmembrane domain (TM)-1; 38D in TM-2; 146W and 155P in TM-4; 237W, 239P, and 244N in TM-6; 284N, 285P, and 288Y in TM-7; and 94C and 168C, which may be joined by a disulfide bond. The amino acid identity between human and mouse BLT2 was 92.7%. A blast search showed that hBLT2 is highly homologous to human and mouse BLT1, with the amino acid identities of 45.2 and 44.6%, respectively. The similarity between BLT1 and BLT2 was high in the putative TMs, especially in TM-2, -3, and -7. Although BLT2 also showed significant homology to an orphan GPCR (GPR25 [30]) and a recently-cloned chemoattractant receptor–homologous molecule expressed on TH2 cells (CRTH2 [31, 32]), the homology was much lower (<31%) than that for BLT1.
Figure 1.
Cloning of human and mouse BLT2. (A) Structures of human and mouse genomic DNAs containing BLT1 and BLT2. In the human gene, transcribed segments are indicated by open boxes, and putative ORFs are indicated by filled boxes. (B) Primary structure of hBLT2 cDNA and deduced amino acid sequence. Putative TMs are boxed. An in-frame-stop codon (TAG) is indicated by number signs at the 5′ untranslated region of the putative ORF. The positions of introns are shown by the nucleotide numbers of introns. The incomplete polyadenylation signal (AATACA) is underlined. (C) Sequence alignment of BLT1 and BLT2 from humans and mice. Asterisks indicate amino acids conserved among four receptors. Dots indicate amino acids conserved among three receptors. The putative TMs of hBLT2 predicted from a Kyte-Doolittle hydrophobicity analysis are lined and labeled as I–VII. These sequence data are available from EMBL/GenBank/DDBJ under accession nos. AB029892 and AB029893.

Tissue Distribution of BLT2.
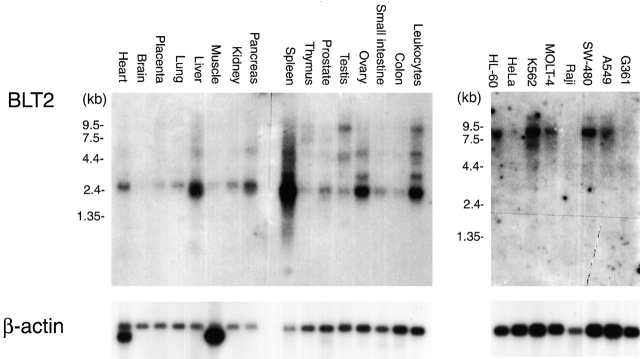
Searching EST database revealed that BLT2 mRNA is expressed in various human tissues, including skeletal muscle, heart, lung, and mammalian gland. Northern blot analyses showed that BLT2 mRNA is expressed most abundantly in spleen, followed by liver, ovary, and leukocytes, with weak signals detected in most human tissues (Fig. 2). The size of the major transcript is 2.5 kb in these tissues, but longer transcripts are also detected. The results show that BLT2 expression is distinct from BLT1, which is expressed almost exclusively in leukocytes 21. BLT2 mRNA is also present in several human cell lines, including undifferentiated HL-60, K562, MOLT-4, SW-480, and A549 cells, but the size of these transcripts is much longer than those of the human tissues (Fig. 2).
Figure 2.
Northern blot analyses of BLT2 mRNA in various human tissues and cells. Human multiple-tissue Northern blot filters (2 μg poly-A RNA/lane; CLONTECH Laboratories, Inc.) were hybridized with [32P]dCTP-labeled ORF of hBLT2 or human β-actin cDNA (CLONTECH Laboratories, Inc.).
Characterization of hBLT2 as a Pharmacologically Distinct LTB4 Receptor.
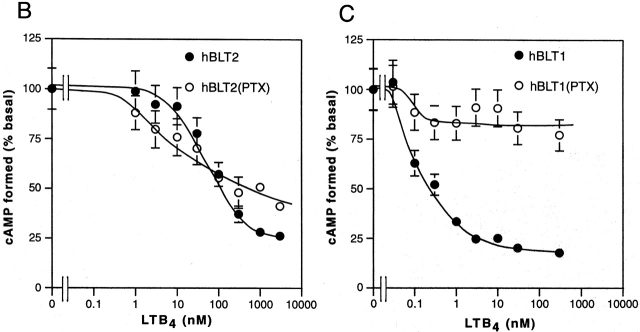
Because sequence similarities between BLT1 and BLT2 suggested that BLT2 is also a receptor for eicosanoids, we examined 3H–LTB4 binding to the membrane fractions of HEK 293 cells transiently transfected with an hBLT2 expression construct. We chose HEK 293 cells for transfection because they showed no specific binding for 3H–LTB4 at concentrations of up to 5 nM (data not shown). Membrane fractions of HEK 293 cells expressing hBLT2 showed a specific and saturable LTB4 binding, whereas those of empty vector-transfected cells did not (Fig. 3 A). Scatchard analysis revealed that the Kd value of hBLT2 for LTB4 was 22.7 nM (Fig. 3B and Fig. C), which was 20-fold higher than that of hBLT1 (1.1 nM; data not shown). We next examined the inhibition of 3H–LTB4 binding by two distinct BLT1 antagonists and various eicosanoids, using the membrane fractions of HEK 293 cells transfected with hBLT1 or hBLT2. Although 3H–LTB4 binding to hBLT1 was inhibited by both ONO 4057 and U 75302, 3H–LTB4 binding to hBLT2 was not inhibited by U 75302 (Fig. 3D and Fig. E). Several other BLT antagonists also failed to inhibit LTB4 binding to BLT2 (data not shown). 3H–LTB4 binding to BLT2 was competed with 5 μM LTB4 (100%) or 20-hydroxy LTB4 (78.6 ± 4.4% compared with the competition by LTB4). 20-carboxy-LTB4, lipoxin (LX)A4, LXB4, or 5-oxo-eicosatetraenoic acid slightly (<40%) competes with 5 nM 3H–LTB4 binding to BLT2 at the concentration of 5 μM. These results clearly show that BLT2 is a pharmacologically distinct receptor from BLT1 21.
Figure 3.
Binding of 3H–LTB4 to the membrane fractions of HEK 293 cells transiently transfected with hBLT1 or hBLT2. (A) Binding of 5 nM 3H–LTB4 to the membrane fractions (20 μg of protein) from HEK 293 cells transfected with control vector (Vector), BLT1 expression vector (hBLT1), or BLT2 expression vector (hBLT2). Total binding (black columns) and nonspecific binding (white columns) are presented (mean ± SD, n = 3). d.p.m., disintegration per minute. (B and C) Binding isotherms (B) and Scatchard analysis (C) of 3H–LTB4 binding to membrane fractions of HEK 293 cells transfected with hBLT2. In B, total binding (□), nonspecific binding (○), and specific binding (•) are presented (mean ± SD, n = 3). (D and E) Inhibition of 5 nM 3H–LTB4 binding to the membrane fractions (20 μg of protein) of HEK 293 cells transfected with hBLT2 (•) or hBLT1 (○) by two BLT antagonists, (F) ONO 4057 and (G) U 75302 (mean ± SD, n = 3).
Intracellular Signaling of BLT2.
To examine whether the binding of LTB4 to BLT2 transduces intracellular signaling, we established several lines of CHO cells stably expressing BLT2 (CHO-hBLT2) and compared LTB4-induced cellular effects to those obtained with CHO cells expressing hBLT1 (CHO-hBLT1 [21]). CHO-K1 cells were transfected with an expression vector for HA-tagged hBLT2, selected with G418, and examined for expression of the tagged receptor using an anti-HA antibody. 18 clones were picked by limiting dilution, and 3 representative clones with high HA expression were selected (data not shown). As each of these three clones showed LTB4-induced calcium mobilization and chemotaxis, we chose one clone as a representative and analyzed the LTB4-induced intracellular signaling in more detail.
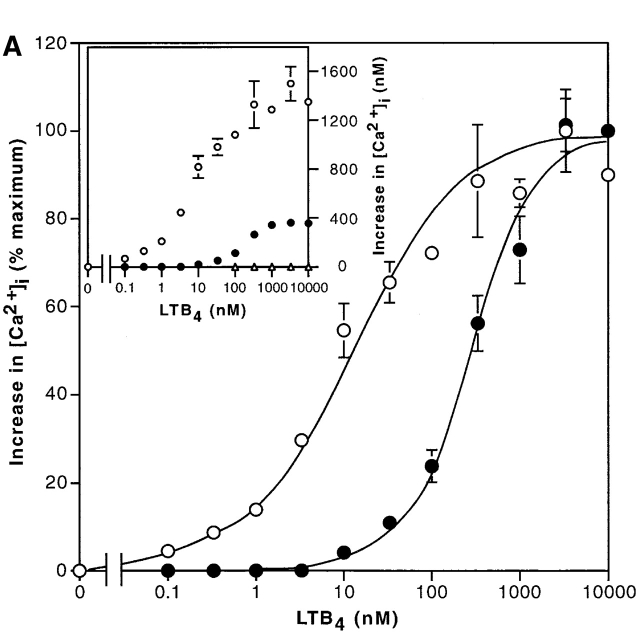
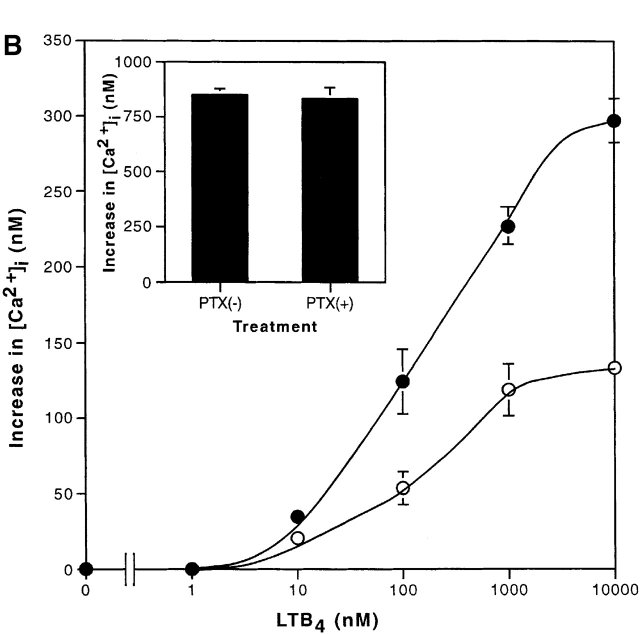
Fig. 4 A shows the increases in intracellular calcium concentrations induced by LTB4 in CHO-hBLT1 and CHO-hBLT2 cells. In both cells, LTB4 increased intracellular calcium in a dose-dependent manner, but the maximum increase in CHO-hBLT2 cells was only one third of that in CHO-hBLT1 cells (Fig. 4 A, inset). The dose response curve for LTB4 in CHO-hBLT2 cells was right shifted from that in CHO-hBLT1 cells by 2 orders of magnitude (Fig. 4 A). Other ligands, including leukotriene C4, leukotriene D4, leukotriene E4, 5(S)-hydroxyeicosatetraenoic acid (HETE), 5(R)-HETE, 12(S)-HETE, 12(R)-HETE, 15(S)-HETE, 15(R)-HETE, 5-oxo-eicosatetraenoic acid, LXA4, LXB4, IL-8, C5a (a component of complement), and FMLP, were not able to induce any significant change in intracellular calcium in CHO-hBLT2 cells at the concentrations of up to 1 μM (data not shown). 20-hydroxy-LTB4, which partially inhibited 3H–LTB4 binding to BLT2, did not induce any change in intracellular calcium in CHO-hBLT2 at 1 μM (data not shown). To determine the subtypes of G protein(s) responsible for calcium increase by BLT2, we pretreated CHO-hBLT2 cells with 100 ng/ml pertussis toxin (PTX) for 12 h and examined the intracellular calcium response. Pretreatment of the cells with PTX diminished by half the response in CHO-hBLT2 cells (Fig. 4 B). PTX pretreatment of these cells did not affect the intracellular calcium increases induced with 2 U/ml thrombin (Fig. 4 B, inset), but completely abolished LTB4-induced cell migration (see Fig. 6 B) in CHO-hBLT2 cells. Therefore, we conclude that the calcium response mediated by hBLT2 involves both PTX-sensitive and -insensitive G protein(s) in CHO cells.
Figure 4.
Calcium mobilization in CHO-hBLT1 and CHO-hBLT2 cells by LTB4. (A) Increases in intracellular calcium after exposure to various concentrations of LTB4 were measured in CHO-hBLT1 (○) and CHO-hBLT2 (•) cells, and were represented as percentages of the maximum responses. The inset graph shows absolute values of increase in intracellular calcium concentrations (mean ± SD, n = 3). (B) Effects of PTX pretreatment on LTB4-induced increases in intracellular calcium concentrations in CHO-hBLT2 cells. The cells were pretreated with 100 ng/ml PTX (○) or vehicle (•) for 12 h. The inset shows increases in intracellular calcium concentrations, evoked by 2 U/ml thrombin, that were not affected by PTX pretreatment (mean ± SD, n = 3).
Figure 6.
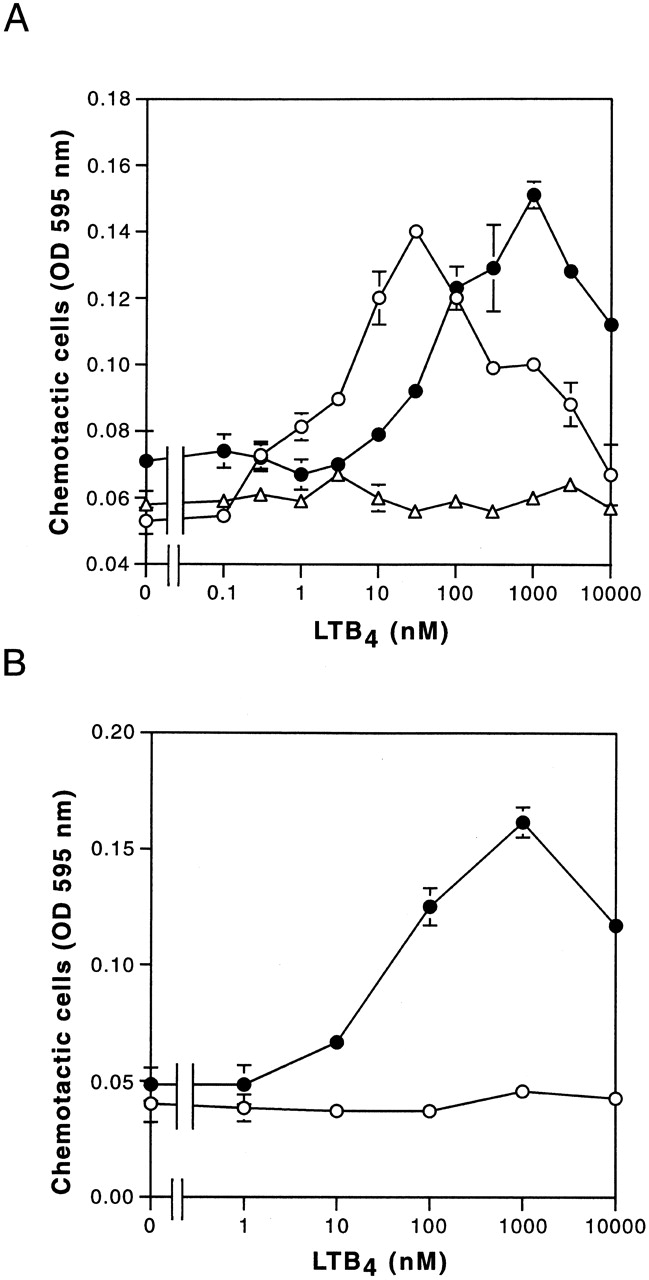
LTB4-induced cell migration in CHO-hBLT1 and CHO-hBLT2 cells. (A) Dose dependency of LTB4-induced cell migration was measured in CHO-hBLT1 (○), CHO-hBLT2 (•), and CHO vector (▵) cells (mean ± SE, n = 4). (B) Effects of PTX pretreatment (100 ng/ml, 12 h) on LTB4-induced cell migration in CHO-hBLT2 cells (mean ± SE, n = 4).
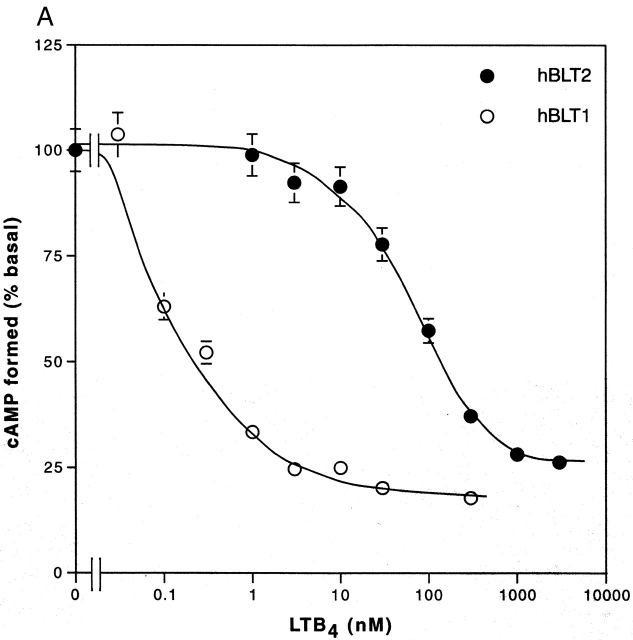
We next examined the effects of LTB4 stimulation on adenylyl cyclase activity in these cells by measuring the cAMP accumulation. In the absence of forskolin, which activates adenylyl cyclases, no increase in cAMP concentration was observed either in CHO-hBLT1 or CHO-hBLT2 cells (data not shown). On the other hand, LTB4 inhibited 50 μM forskolin-activated adenylyl cyclase activities in a dose-dependent manner in both CHO-hBLT1 and CHO-hBLT2 cells. The IC50 value of LTB4 for inhibiting adenylyl cyclase activities in CHO-hBLT2 cells (80 nM; Fig. 5 A) was higher than that of CHO-hBLT1 cells (0.1 nM; Fig. 5 A). Pretreatment of the cells with 100 ng/ml PTX for 12 h had markedly different effects on LTB4-induced adenylyl cyclase inhibition. Although PTX pretreatment abolished ∼80% of LTB4-induced adenylyl cyclase inhibition in CHO-hBLT1 cells, the same pretreatment had a negligible effect on cAMP accumulation in CHO-hBLT2 cells (Fig. 5B and Fig. C). These results indicate that BLT2 uses Gz, a PTX-insensitive G protein 33 34, in inhibiting adenylate cyclases.
Figure 5.
Cyclic AMP accumulation in forskolin-treated CHO-hBLT1 and CHO-hBLT2 cells. (A) LTB4 inhibits 50 μM forskolin–induced adenylyl cyclase activities in both CHO-hBLT1 (○) and CHO-hBLT2 (•) cells. Forskolin-induced cAMP levels in the absence of LTB4 were 44.2 ± 4.5 pmol/well in CHO-hBLT2 and 112.0 ± 11.8 pmol/well in CHO-hBLT1 cells (mean ± SE, n = 4). (B) The effects of PTX pretreatment (100 ng/ml, 12 h) on cAMP accumulation in CHO-hBLT2 cells. The 50 μM forskolin–induced cAMP levels in the absence of LTB4 were 44.2 ± 4.5 pmol/well in untreated cells and 48.3 ± 4.9 pmol/well in PTX-treated cells (mean ± SE, n = 4). (C) The effects of PTX pretreatment (100 ng/ml, 12 h) on cAMP accumulation in CHO-hBLT1 cells. The 50 μM forskolin–induced cAMP levels in the absence of LTB4 were 112.0 ± 11.8 pmol/well in untreated cells and 71.0 ± 7.3 pmol/well in PTX-treated cells (mean ± SE, n = 4).
LTB4 is a potent chemoattractant of granulocytes and macrophages, and CHO cells migrate toward LTB4 when transfected with BLT1 21. Examining the chemotactic activities of LTB4 mediated by BLT2 using CHO-hBLT2 cells, both CHO-hBLT1 and CHO-hBLT2 cells showed potent chemotactic activities, with bell-shaped dose response curves (Fig. 6 A). The optimum concentration of LTB4 needed for chemotaxis in CHO-hBLT2 cells was higher than that of CHO-hBLT1 cells by 2 orders of magnitude, enhancing further the idea that hBLT2 is a low-affinity but still potent chemotactic receptor for LTB4. As shown in Fig. 6 B, chemotaxis of CHO-hBLT2 cells induced by LTB4 was abolished by PTX, showing that PTX-sensitive G protein(s) are absolutely required for LTB4-induced cell migration through BLT1 and BLT2.
Discussion
LTB4 is one of the most potent chemoattractants for leukocytes, and is unique because it is a lipid mediator biosynthesized from membrane phospholipids by the actions of cytosolic phospholipase A2 35 36, 5-lipoxygenase, and LTA4 hydrolase 3 37. LTB4–BLT interaction plays important roles in host defense mechanism and inflammatory diseases. Mice lacking in leukotriene production are insensitive to some inflammatory stimuli, and mice overexpressing BLT1 exhibit enhanced responses to infections and lung ischemic-reperfusion injury 38. Numerous biochemical and pharmacological studies indicated that high-affinity and LTB4-specific receptors exist in membranes of neutrophils, macrophages, eosinophils, and T cells. Although many antagonists for LTB4 receptor are under development, their main target has been directed to the high-affinity receptor for LTB4, BLT1. There are also reports of low-affinity binding protein(s) for LTB4 in human granulocytes 39, murine spleen 15, and guinea pig alveolar and peritoneal eosinophils 40 41. Some investigators have speculated that the high-affinity receptor mediates chemotaxis for LTB4, and the low-affinity receptor mediates LTB4-induced secretary and oxidase-activation responses 42. Therefore, we have had only suggestive information on a low-affinity LTB4 receptor.
Identification of a Novel GPCR, BLT2, in a Gene Cluster with BLT1.
Our interest in understanding the molecular mechanisms that regulate the transcription of BLT1 (originally termed BLT) gene led us to isolate several genomic clones containing BLT1 gene from humans and mice. The precise structure of the hBLT1 gene and the mechanism by which the BLT1 gene is regulated is published in an accompanying paper 28. During the course determining the nucleotide sequences of these genomic clones, we discovered an ORF for a putative seven-transmembrane receptor (BLT2) that is similar to BLT1 (Fig. 1a and Fig. b). Human and mouse BLT2 genes encode proteins of 358 and 360 amino acids, respectively (Fig. 1 C), similar to hBLT1 (352 amino acids). The amino acid identity of hBLT1 and hBLT2 is 45.2%, and that of human and mouse BLT2 is 92.7%. The amino acid identity of BLT2 between human and mouse is higher than BLT1 between two species (78.6%), suggesting that BLT2 has been conserved during evolution and must therefore play an important role. We conclude that BLT2 is not a pseudogene based on the following criteria: (a) Northern blotting showed that mRNA expression is seen in various tissues and cells in humans; (b) the primary structures of human and mouse BLT2 are well conserved, with the amino acid identity of 92.7%; (c) the structures of genomic clones containing BLT1 and BLT2 are similar both in humans and mice; (d) there are no frame shifts in either human or mouse BLT2 ORF; and (e) many EST clones encoding BLT2 have been deposited in the EMBL/GenBank/DDBJ database. hBLT2 showed a significant homology to a recently cloned chemoattractant receptor–homologous molecule expressed on TH2 cells 31 32 and an orphan GPCR, GPR25 30, with amino acid identities of 30.5 and 28.5%, respectively. A recently cloned receptor for cysyteinyl leukotrienes, Cys-LT1, showed 28% amino acid identity to hBLT1 43 44. Thus, BLT2 is the second member for a LTB4 receptor family.
hBLT1 was reported to be located on human chromosome 14q11.2-q12 45, and our present finding shows that the ORF for hBLT2 is located at 3 kbp upstream of hBLT1 ORF (Fig. 1 A). Similar gene clusters of GPCRs have been reported for the FMLP receptor, its related receptors 46, and C5a receptor on chromosome 19q13.3-q13.4 47, and for CXCR1 and 2, the receptors for IL-8, on 2q34-q35 48 49 50. β2 and α1 adrenergic receptors are also closely linked on human chromosome 5q32-q34, and β1 and α2 adrenergic receptors are both located on human chromosome 10q24-q26 51. Formyl peptide receptor (FPR)-like (FPRL) 1–LXA4 receptor (AXLR), which is a neighboring receptor to the FMLP specific receptor (FPR1), was reported to respond to lipoxin A4 at nM 52 and the acute phase protein serum amyloid A at μM 53. It was shown recently that MHC binding peptide and synthetic peptide MMK-1 bind to FPRL1/ALXR at sub-μM concentration 54. In the case of IL-8 receptors, CXCR1 is a high-affinity and specific receptor for IL-8, and CXCR2 is a less selective receptor for IL-8, which also binds growth regulatory oncogene (GRO)-α, -β, -γ and neutrophil-activating peptide (NAP)-2 55. These receptor clusters appear to be generated by gene duplication, with the degree of homology among the members of the cluster indicative of the timing of the duplication during evolution 50. The amino acid identity between hBLT1 and 2 (45.2%) is much lower than that between CXCR1 and 2 (77%) and between FPR1 and FPRL1/ALXR (72%), suggesting that the duplication event that generated BLT2 occurred earlier than the events generating the other receptor clusters. The reasons why these receptors for chemoattractants form clusters are not yet clear, but may become evident as more information is gathered. Surprisingly, the promoter region of BLT1 is localized in the ORF of BLT2. This, to our knowledge, is the first mammalian example of “promoter in ORF,” which is reported in bacteria 56 and bacteriophages 57.
Characterization of BLT2 as a Low-Affinity Receptor for LTB4.
We first examined whether or not BLT2 can recognize LTB4. A binding assay using membrane fractions from HEK 293 cells exogenously transfected with the receptor cDNA showed that BLT2 is a low-affinity receptor for LTB4 (Fig. 3A–E). The Kd value was 22.7 nM, which is higher than that of BLT1 21 22 23 24 25 26. High- and low-affinity LTB4 binding sites were reported in human 42 58 and rabbit 59 neutrophils, differentiated HL-60 cells 60 61, guinea pig peritoneal eosinophils 41, and murine spleen membrane 62. The Kd values of these low-affinity binding sites for LTB4 were reported to be between 70 and 580 nM, similar to the value obtained from the transfection study using BLT2 (Fig. 3B and Fig. C). Thus, BLT2 appears to be a low-affinity binding site for LTB4. The high expression of BLT2 in spleen on Northern blots (Fig. 2) also supports this speculation, as spleen is the tissue where the low-affinity receptor is most well characterized 14 15 62. Studies using BLT antagonists showed that BLT2 is a pharmacologically novel receptor for LTB4, as binding of LTB4 to BLT2 was not inhibited by a classical BLT antagonist, U 75302 (references 21, 63; Fig. 3D and Fig. E).
Signaling from BLT2.
We next established several lines of CHO cells stably expressing BLT2 to examine intracellular signaling. CHO cells were transfected with an expression construct for HA-tagged BLT2 and were selected with culture media containing G418, and stable clones were isolated. All three cell lines selected for high HA expression responded to LTB4 with calcium mobilization and chemotaxis, as opposed to cells transfected with the empty vector, which did not respond. In calcium mobilization assays, IC50 values for LTB4 in CHO-hBLT1 and CHO-hBLT2 cells were 10 nM and 300 nM, respectively (Fig. 4 A). Calcium increases in CHO-hBLT1 cells were not inhibited by PTX pretreatment 21, and cotransfection of BLT1 cDNA with G16α, but not with G11α, increased LTB4-induced d-myo-inositol 1, 4, 5-triphosphate (IP3) accumulation in Cos-7 cells 4. LTB4–BLT1 interaction appears to activate G16α protein, leading to the activation of phospholipase C. On the other hand, about half of the calcium increase by LTB4 through BLT2 was inhibited by pretreatment of PTX (Fig. 4 B), suggesting that different types and/or different ratios of G proteins are activated by binding of LTB4 to BLT2. Differences in the maximum responses in LTB4-induced calcium mobilizations in CHO-BLT1 and CHO-BLT2 cells (Fig. 4 A, inset) also support this hypothesis. As the low-affinity binding of LTB4 to BLT2 raised the possibility of the other ligands for this receptor, we examined calcium mobilization in CHO-BLT2 cells by various eicosanoids and chemoattractants. Other than LTB4 (discussed in Results), none of the ligands tested showed positive responses up to 1 μM, whereas 100 nM LTB4 showed a clear calcium increase (Fig. 4).
With respect to inhibition of adenylyl cyclase, PTX pretreatment had different effects on these two related receptors (Fig. 5 A). LTB4-mediated inhibition of adenylyl cyclase in CHO-hBLT1 cells is completely PTX sensitive, but in CHO-hBLT2 it is hardly affected by PTX pretreatment (Fig. 5B and Fig. C), suggesting that BLT1 and BLT2 use different subtype(s) of Gi-like G proteins. Among Gi family members with inhibitory effects on adenylyl cyclases, only Gzα is insensitive to PTX, as it lacks the Cys residue at position (−4) from the COOH terminus 33 34. Therefore, BLT2 may use Gzα to inhibit adenylyl cyclase in transfected CHO cells. On the other hand, LTB4-induced chemotactic activities of CHO-hBLT1 and CHO-hBLT2 cells were completely PTX sensitive (Fig. 5 B). Thus BLT2, like BLT1, uses at least three classes of G proteins (Gi, Gq-like, and Gz) for signaling.
BLT2, a Possible Therapeutic Target.
A unique characteristic of BLT2 is its ubiquitous expression, with the highest mRNA levels in spleen, followed by liver, ovary, and peripheral leukocytes (Fig. 2). In ovary, LTB4 was reported to mediate ovulation in rat 64 and rabbit 65, without affecting the production of estradiol and progesterone from the ovarian cells. The biological roles of LTB4 in liver are unknown, but the liver expresses the LTB4-producing enzyme LTA4 hydrolase 66 67 and LTB4-degrading enzymes LTB4 20-hydroxylase 68 69, LTB4 12-hydroxydehydrogenase 70, and β-oxidizing enzymes 71. The liver also expresses high amounts of peroxisome proliferator-activated factor (PPAR)α, a reported nuclear receptor for LTB4, which controls transcription of various genes involved in lipid metabolism 72 73. Therefore, it is reasonable to consider that LTB4 has some as yet unidentified roles in the liver function that are mediated by BLT2. Northern blotting shows that human peripheral leukocytes express both BLT1 and BLT2 (Fig. 2; reference 21), which agrees well with a previous report of the coexistence of high- and low-affinity LTB4 receptors in these cells 39. Some investigators have speculated that the high-affinity receptor mediates chemotaxis for LTB4 and the low-affinity receptor mediates LTB4-induced degranulation and superoxide anion generation 42. Transfection studies using BLT1 or BLT2 or both will enable us to precisely analyze and distinguish the roles of two receptors in vitro. We should again emphasize that LTB4 binding to BLT2 is not inhibited by most of the previously developed BLT antagonists, suggesting that BLT2 could be a therapeutic target for novel drugs related to contraception and immunosuppression. The lack of significant effects of the current BLT antagonists in some studies may be due to the lack of their effects on BLT2. Further study will be needed to clarify the biological roles of LTB4 through BLT2, especially in relation to the spleen, and in ovaries and the liver. Although LTB4 was the best ligand among those tested for hBLT2, we should pay attention to the possible existence of better ligand(s) for BLT2 than LTB4.
In conclusion, we have identified a second leukotriene B4 receptor, BLT2, from humans and mice. The BLT2 gene closely located to the BLT1 gene in both humans and mice. In an accompanying paper 28, we detail the presence of the ORF for BLT2 localized in the promoter region of BLT1. BLT2 is a low-affinity and pharmacologically novel receptor for LTB4, activating different G protein(s) to increase intracellular calcium, inhibit adenylyl cyclase, and stimulate chemotaxis. These findings may lead us to identify other members of LTB4 receptor family, and provide new important clues concerning the physiological and pathophysiological roles of LTB4.
Acknowledgments
We thank D. Saffen and D. Wong (The University of Tokyo) for critically reading this manuscript, and S. Ishii, N. Uozumi, M. Taniguchi, S. Kato, and K. Takeyama (The University of Tokyo) for discussion.
This work was supported in part by grants-in-aid from the Ministry of Education, Science, Sports, and Culture and Human Science Foundation, and by grants from the Yamanouchi Foundation for Metabolic Disorders, the Uehara Memorial Foundation, and the Cell Science Research Foundation.
Footnotes
Abbreviations used in this paper: ALXR, lipoxin A4 receptor; CHO, Chinese hamster ovary; EST, expressed sequence tag; FPR, formyl peptide receptor; FPRL, formyl peptide receptor–like; GPCR, G protein–coupled receptor; HETE, hydroxyeicosatetraenoic acid; LTB4, leukotriene B4; LX, lipoxin; ORF, open reading frame; PTX, pertussis toxin; TM, transmembrane domain.
References
- Samuelsson B., Dahlén S.E., Lindgren J.Å., Rouzer C.A., Serhan C.N. Leukotrienes and lipoxinsstructures, biosynthesis, and biological effects. Science. 1987;237:1171–1176. doi: 10.1126/science.2820055. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Wolfe L.S. Arachidonic acid cascade and signal transduction. J. Neurochem. 1990;55:1–15. doi: 10.1111/j.1471-4159.1990.tb08813.x. [DOI] [PubMed] [Google Scholar]
- Serhan C.N., Haeggstrom J.Z., Leslie C.C. Lipid mediator networks in cell signalingupdate and impact of cytokines. FASEB J. 1996;10:1147–1158. doi: 10.1096/fasebj.10.10.8751717. [DOI] [PubMed] [Google Scholar]
- Gaudreau R., Gouill C.L., Metaoui S., Lemire S., Stankova J., Rola-Pleszczynski M. Signalling through the leukotriene B4 receptor involves both alpha i and alpha 16, but not alpha q or alpha 11 G-protein subunits. Biochem. J. 1998;335:15–18. doi: 10.1042/bj3350015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen L., Kragballe K., Ziboh V.A. Significance of leukotriene-A4 hydrolase in the pathogenesis of psoriasis. Skin. Pharmacol. 1997;10:169–177. doi: 10.1159/000211501. [DOI] [PubMed] [Google Scholar]
- Turner C.R., Breslow R., Conklyn M.J., Andresen C.J., Patterson D.K., Lopez A.A., Owens B., Lee P., Watson J.W., Showell H.J. In vitro and in vivo effects of leukotriene B4 antagonism in a primate model of asthma. J. Clin. Invest. 1996;97:381–387. doi: 10.1172/JCI118426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R.J., Pettipher E.R., Koch K., Farrell C.A., Breslow R., Conklyn M.J., Smith M.A., Hackman B.C., Wimberly D.J., Milici A.J. Leukotriene B4 plays a critical role in the progression of collagen-induced arthritis. Proc. Natl. Acad. Sci. USA. 1995;92:517–521. doi: 10.1073/pnas.92.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon P., Stenson W.F. Enhanced synthesis of leukotriene B4 by colonic mucosa in inflammatory bowel disease. Gastroenterology. 1984;86:453–460. [PubMed] [Google Scholar]
- Noiri E., Yokomizo T., Nakao A., Izumi T., Fujita T., Kimura S., Shimizu T. A novel in vivo approach showing the chemotactic activity of leukotriene B4 in acute renal ischemic-reperfusion injury. Proc. Natl. Acad. Sci. USA. 2000;97:823–828. doi: 10.1073/pnas.97.2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths R.J., Smith M.A., Roach M.L., Stock J.L., Stam E.J., Milici A.J., Scampoli D.N., Eskra J.D., Byrum R.S., Koller B.H., McNeish J.D. Collagen-induced arthritis is reduced in 5-lipoxygenase–activating protein–deficient mice. J. Exp. Med. 1997;185:1123–1129. doi: 10.1084/jem.185.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrum R.S., Goulet J.L., Griffiths R.J., Koller B.H. Role of the 5-lipoxygenase–activating protein (FLAP) in murine acute inflammatory responses. J. Exp. Med. 1997;185:1065–1075. doi: 10.1084/jem.185.6.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.-S., Sheller J.R., Johnson E.N., Funk C.D. Role of leukotriene revealed by targeted disruption of the 5-lipoxygenase gene. Nature. 1994;372:179–182. doi: 10.1038/372179a0. [DOI] [PubMed] [Google Scholar]
- Goulet J.L., Snouwaert J.N., Latour A.M., Coffman T.M., Koller B.H. Altered inflammatory responses in leukotriene-deficient mice. Proc. Natl. Acad. Sci. USA. 1994;91:12852–12856. doi: 10.1073/pnas.91.26.12852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.T., Froelich L.L., Boyd R.J., Schrementi J.P., Saussy D.L., Jr., Schultz R.M., Sawyer J.S., Sofia M.J., Herron D.K., Goodson T., Jr. Pharmacologic actions of the second-generation leukotriene B4 receptor antagonist LY293111in vitro studies. J. Pharmacol. Exp. Ther. 1999;288:286–294. [PubMed] [Google Scholar]
- Showell H.J., Conklyn M.J., Alpert R., Hingorani G.P., Wright K.F., Smith M.A., Stam E., Salter E.D., Scampoli D.N., Meltzer S. The preclinical pharmacological profile of the potent and selective leukotriene B4 antagonist CP-195543. J. Pharmacol. Exp. Ther. 1998;285:946–954. [PubMed] [Google Scholar]
- Kishikawa K., Tateishi N., Maruyama T., Seo R., Toda M., Miyamoto T. ONO-4057, a novel, orally active leukotriene B4 antagonisteffects on LTB4-induced neutrophil functions. Prostaglandins. 1992;44:261–275. doi: 10.1016/0090-6980(92)90002-b. [DOI] [PubMed] [Google Scholar]
- Taylor B.M., Crittenden N.J., Bruden M.N., Wishka D.G., Morris J., Richards I.M., Sun F.F. Biological activity of leukotriene B4 analogsinhibition of guinea pig eosinophil migration in vitro by the 2,6-disubstituted pyridine analogs U-75,302 and U-75,485. Prostaglandins. 1991;42:211–224. doi: 10.1016/0090-6980(91)90111-r. [DOI] [PubMed] [Google Scholar]
- Spurney R.F., Ibrahim S., Butterly D., Klotman P.E., Sanfilippo F., Coffman T.M. Leukotrienes in renal transplant rejection in rats. Distinct roles for leukotriene B4 and peptideleukotrienes in the pathogenesis of allograft injury. J. Immunol. 1994;152:867–876. [PubMed] [Google Scholar]
- Weringer E.J., Perry B.D., Sawyer P.S., Gilman S.C., Showell H.J. Antagonizing leukotriene B4 receptors delays cardiac allograft rejection in mice. Transplantation. 1999;67:808–815. doi: 10.1097/00007890-199903270-00005. [DOI] [PubMed] [Google Scholar]
- Morita H., Takeda K., Yagita H., Okumura K. Immunosuppressive effect of leukotriene B4 receptor antagonist in vitro. Biochem. Biophys. Res. Commun. 1999;264:321–326. doi: 10.1006/bbrc.1999.1523. [DOI] [PubMed] [Google Scholar]
- Yokomizo T., Izumi T., Chang K., Takuwa Y., Shimizu T. A G-protein-coupled receptor for leukotriene B4 that mediates chemotaxis. Nature. 1997;387:620–624. doi: 10.1038/42506. [DOI] [PubMed] [Google Scholar]
- Martin V., Ronde P., Unett D., Wong A., Hoffman T.L., Edinger A.L., Doms R.W., Funk C.D. Leukotriene binding, signaling, and analysis of HIV coreceptor function in mouse and human leukotriene B4 receptor-transfected cells. J. Biol. Chem. 1999;274:8597–8603. doi: 10.1074/jbc.274.13.8597. [DOI] [PubMed] [Google Scholar]
- Huang W.W., Garcia-Zepeda E.A., Sauty A., Oettgen H.C., Rothenberg M.E., Luster A.D. Molecular and biological characterization of the murine leukotriene B4 receptor expressed on eosinophils. J. Exp. Med. 1998;188:1063–1074. doi: 10.1084/jem.188.6.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda K., Yokomizo T., Izumi T., Shimizu T. cDNA cloning and characterization of guinea-pig leukotriene B4 receptor. Biochem. J. 1999;342:79–85. [PMC free article] [PubMed] [Google Scholar]
- Boie Y., Stocco R., Sawyer N., Greig G.M., Kargman S., Slipetz D.M., O'Neill G.P., Shimizu T., Yokomizo T., Metters K.M., Abramovitz M. Characterization of the cloned guinea pig leukotriene B4 receptorcomparison to its human orthologue. Eur. J. Pharmacol. 1999;380:203–213. doi: 10.1016/s0014-2999(99)00514-2. [DOI] [PubMed] [Google Scholar]
- Toda A., Yokomizo T., Masuda K., Nakao A., Izumi T., Shimizu T. Cloning and characterization of rat leukotriene B4 receptor. Biochem. Biophys. Res. Commun. 1999;262:806–812. doi: 10.1006/bbrc.1999.1284. [DOI] [PubMed] [Google Scholar]
- Yokomizo T., Masuda K., Kato K., Toda A., Izumi T., Shimizu T. Leukotriene B4 receptor. Cloning and intracellular signaling. Am. J. Respir. Crit. Care. Med. 2000;161:S51–S55. doi: 10.1164/ajrccm.161.supplement_1.ltta-11. [DOI] [PubMed] [Google Scholar]
- Kato K., Yokomizo T., Izumi T., Shimizu T. Cell-specific transcriptional regulation of human leukotriene B4 receptor gene. J. Exp. Med. 2000;192:413–420. doi: 10.1084/jem.192.3.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gether U., Kobilka B.K. G protein-coupled receptors. II. Mechanism of agonist activation. J. Biol. Chem. 1998;273:17979–17982. doi: 10.1074/jbc.273.29.17979. [DOI] [PubMed] [Google Scholar]
- Jung B.P., Nguyen T., Kolakowski L.F., Jr., Lynch K.R., Heng H.H., George S.R., O'Dowd B.F. Discovery of a novel human G protein-coupled receptor gene (GPR25) located on chromosome 1. Biochem. Biophys. Res. Commun. 1997;230:69–72. doi: 10.1006/bbrc.1996.5828. [DOI] [PubMed] [Google Scholar]
- Nagata K., Tanaka K., Ogawa K., Kemmotsu K., Imai T., Yoshie O., Abe H., Tada K., Nakamura M., Sugamura K., Takano S. Selective expression of a novel surface molecule by human Th2 cells in vivo. J. Immunol. 1999;162:1278–1286. [PubMed] [Google Scholar]
- Marchese A., Sawzdargo M., Nguyen T., Cheng R., Heng H.H., Nowak T., Im D.S., Lynch K.R., George S.R., O'Dowd B.F. Discovery of three novel orphan G-protein-coupled receptors. Genomics. 1999;56:12–21. doi: 10.1006/geno.1998.5655. [DOI] [PubMed] [Google Scholar]
- Fong H.K., Yoshimoto K.K., Eversole-Cire P., Simon M.I. Identification of a GTP-binding protein alpha subunit that lacks an apparent ADP-ribosylation site for pertussis toxin. Proc. Natl. Acad. Sci. USA. 1988;85:3066–3370. doi: 10.1073/pnas.85.9.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields T.A., Casey P.J. Signalling functions and biochemical properties of pertussis toxin- resistant G-proteins. Biochem. J. 1997;321:561–571. doi: 10.1042/bj3210561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonventre J.V., Huang Z., Taheri M.R., O'Leary E., Li E., Moskowitz M.A., Sapirstein A. Reduced fertility and postischaemic brain injury in mice deficient in cytosolic phospholipase A2 . Nature. 1997;390:622–625. doi: 10.1038/37635. [DOI] [PubMed] [Google Scholar]
- Uozumi N., Kume K., Nagase T., Nakatani N., Ishii S., Tashiro F., Komagata Y., Maki K., Ikuta K., Ouchi Y. Role of cytosolic phospholipase A2 in allergic response and parturition. Nature. 1997;390:618–622. doi: 10.1038/37622. [DOI] [PubMed] [Google Scholar]
- Minami M., Ohno S., Kawasaki H., Rådmark O., Samuelsson B., Jörnvall H., Shimizu T., Seyama Y., Suzuki K. Molecular cloning of a cDNA coding for human leukotriene A4 hydrolase. Complete primary structure of an enzyme involved in eicosanoid synthesis. J. Biol. Chem. 1987;262:13873–13876. [PubMed] [Google Scholar]
- Chiang N., Gronert K., Clish C.B., O'Brien J.A., Freeman M.W., Serhan C.N. Leukotriene B4 receptor transgenic mice reveal novel protective roles for lipoxins and aspirin-triggered lipoxins in reperfusion. J. Clin. Invest. 1999;104:309–316. doi: 10.1172/JCI7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showell H.J., Pettipher E.R., Cheng J.B., Breslow R., Conklyn M.J., Farrell C.A., Hingorani G.P., Salter E.D., Hackman B.C., Wimberly D.J. The in vitro and in vivo pharmacologic activity of the potent and selective leukotriene B4 receptor antagonist CP-105696. J. Pharmacol. Exp. Ther. 1995;273:176–184. [PubMed] [Google Scholar]
- Maghni K., de Brum-Fernandes A.J., Foldes-Filep E., Gaudry M., Borgeat P., Sirois P. Leukotriene B4 receptors on guinea pig alveolar eosinophils. J. Pharmacol. Exp. Ther. 1991;258:784–789. [PubMed] [Google Scholar]
- Sehmi R., Rossi A.G., Kay A.B., Cromwell O. Identification on receptors for leukotriene B4 expressed on guinea-pig peritoneal eosinophils. Immunology. 1992;77:129–135. [PMC free article] [PubMed] [Google Scholar]
- Lin A.H., Ruppel P.L., Gorman R.R. Leukotriene B4 binding to human neutrophils. Prostaglandins. 1984;28:837–849. doi: 10.1016/0090-6980(84)90038-8. [DOI] [PubMed] [Google Scholar]
- Lynch K.R., O'Neill G.P., Liu Q., Im D.S., Sawyer N., Metters K.M., Coulombe N., Abramovitz M., Figueroa D.J., Zeng Z. Characterization of the human cysteinyl leukotriene CysLT1 receptor. Nature. 1999;399:789–793. doi: 10.1038/21658. [DOI] [PubMed] [Google Scholar]
- Sarau H.M., Ames R.S., Chambers J., Ellis C., Elshourbagy N., Foley J.J., Schmidt D.B., Muccitelli R.M., Jenkins O., Murdock P.R. Identification, molecular cloning, expression, and characterization of a cysteinyl leukotriene receptor. Mol. Pharmacol. 1999;56:657–663. doi: 10.1124/mol.56.3.657. [DOI] [PubMed] [Google Scholar]
- Owman C., Nilsson C., Lolait S.J. Cloning of cDNA encoding a putative chemoattractant receptor. Genomics. 1996;37:187–194. doi: 10.1006/geno.1996.0541. [DOI] [PubMed] [Google Scholar]
- Nomura H., Nielsen B.W., Matsushima K. Molecular cloning of cDNAs encoding a LD78 receptor and putative leukocyte chemotactic peptide receptors. Int. Immunol. 1993;5:1239–1249. doi: 10.1093/intimm/5.10.1239. [DOI] [PubMed] [Google Scholar]
- Perez H.D., Holmes R., Kelly E., McClary J., Andrews W.H. Cloning of a cDNA encoding a receptor related to the formyl peptide receptor of human neutrophils. Gene. 1992;118:303–304. doi: 10.1016/0378-1119(92)90208-7. [DOI] [PubMed] [Google Scholar]
- Mollereau C., Muscatelli F., Mattei M.G., Vassart G., Parmentier M. The high-affinity interleukin 8 receptor gene (IL8RA) maps to the 2q33-q36 region of the human genomecloning of a pseudogene (IL8RBP) for the low-affinity receptor. Genomics. 1993;16:248–251. doi: 10.1006/geno.1993.1167. [DOI] [PubMed] [Google Scholar]
- Morris S.W., Nelson N., Valentine M.B., Shapiro D.N., Look A.T., Kozlosky C.J., Beckmann M.P., Cerretti D.P. Assignment of the genes encoding human interleukin-8 receptor types 1 and 2 and an interleukin-8 receptor pseudogene to chromosome 2q35. Genomics. 1992;14:685–691. doi: 10.1016/s0888-7543(05)80169-7. [DOI] [PubMed] [Google Scholar]
- Ahuja S.K., Ozcelik T., Milatovitch A., Francke U., Murphy P.M. Molecular evolution of the human interleukin-8 receptor gene cluster. Nat. Genet. 1992;2:31–36. doi: 10.1038/ng0992-31. [DOI] [PubMed] [Google Scholar]
- Yang-Feng T.L., Xue F.Y., Zhong W.W., Cotecchia S., Frielle T., Caron M.G., Lefkowitz R.J., Francke U. Chromosomal organization of adrenergic receptor genes. Proc. Natl. Acad. Sci. USA. 1990;87:1516–1520. doi: 10.1073/pnas.87.4.1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiore S., Maddox J.F., Perez H.D., Serhan C.N. Identification of a human cDNA encoding a functional high-affinity lipoxin A4 receptor. J. Exp. Med. 1994;180:253–260. doi: 10.1084/jem.180.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S.B., Gong W., Gao J.L., Shen W., Murphy P.M., Oppenheim J.J., Wang J.M. A seven-transmembrane, G protein–coupled receptor, FPRL1, mediates the chemotactic activity of serum amyloid A for human phagocytic cells. J. Exp. Med. 1999;189:395–402. doi: 10.1084/jem.189.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang N., Fierro I.M., Gronert K., Serhan C.N. Activation of lipoxin A4 receptors by aspirin-triggered lipoxins and select peptides evokes ligand-specific responses in inflammation. J. Exp. Med. 2000;191:1197–1208. doi: 10.1084/jem.191.7.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy P.M., Tiffany H.L. Cloning of complementary DNA encoding a functional human interleukin-8 receptor. Science. 1991;253:1280–1283. doi: 10.1126/science.1891716. [DOI] [PubMed] [Google Scholar]
- Grundstrom T., Jaurin B. Overlap between ampC and frd operons on the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA. 1982;79:1111–1115. doi: 10.1073/pnas.79.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R.H., Foeller C., Bidwell K., Landy A. Site-specific recombination functions of bacteriophage lambdaDNA sequence of regulatory regions and overlapping structural genes for Int and Xis . Proc. Natl. Acad. Sci. USA. 1980;77:2482–2486. doi: 10.1073/pnas.77.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D.W., Gifford L.A., Olson D.M., Goetzl E.J. Transduction by leukotriene B4 receptors of increases in cytosolic calcium in human polymorphonuclear leukocytes. J. Immunol. 1985;135:525–530. [PubMed] [Google Scholar]
- Goldman D.W., Enkel H., Gifford L.A., Chenoweth D.E., Rosenbaum J.T. Lipopolysaccharide modulates receptors for leukotriene B4, C5a, and formyl-methionyl-leucyl-phenylalanine on rabbit polymorphonuclear leukocytes. J. Immunol. 1986;137:1971–1976. [PubMed] [Google Scholar]
- Benjamin C.W., Rupple P.L., Gorman R.R. Appearance of specific leukotriene B4 binding sites in myeloid differentiated HL-60 cells. J. Biol. Chem. 1985;260:14208–14213. [PubMed] [Google Scholar]
- Goldman D.W., Olson D.M., Payan D.G., Gifford L.A., Goetzl E.J. Development of receptors for leukotriene B4 on HL-60 cells induced to differentiate by 1 alpha,25-dihydroxyvitamin D3. J. Immunol. 1986;136:4631–4636. [PubMed] [Google Scholar]
- Showell H.J., Breslow R., Conklyn M.J., Hingorani G.P., Koch K. Characterization of the pharmacological profile of the potent LTB4 antagonist CP-105,696 on murine LTB4 receptors in vitro. Br. J. Pharmacol. 1996;117:1127–1132. doi: 10.1111/j.1476-5381.1996.tb16706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone R.C., Aharony D. Modulation of affinity and density of LTB4 receptors on guinea pig lung membranes by divalent cations and guanine nucleotides. Eur. J. Pharmacol. 1991;206:333–338. doi: 10.1016/0922-4106(91)90118-2. [DOI] [PubMed] [Google Scholar]
- Mikuni M., Yoshida M., Hellberg P., Peterson C.A., Edwin S.S., Brannstrom M., Peterson C.M. The lipoxygenase inhibitor, nordihydroguaiaretic acid, inhibits ovulation and reduces leukotriene and prostaglandin levels in the rat ovary. Biol. Reprod. 1998;58:1211–1216. doi: 10.1095/biolreprod58.5.1211. [DOI] [PubMed] [Google Scholar]
- Yoshimura Y., Nakamura Y., Shiraki M., Hirota Y., Yamada H., Ando M., Ubukata Y., Suzuki M. Involvement of leukotriene B4 in ovulation in the rabbit. Endocrinology. 1991;129:193–199. doi: 10.1210/endo-129-1-193. [DOI] [PubMed] [Google Scholar]
- Ohishi N., Minami M., Kobayashi J., Seyama Y., Hata J., Yotsumoto H., Takaku F., Shimizu T. Immunological quantitation and immunohistochemical localization of leukotriene A4 hydrolase in guinea pig tissues. J. Biol. Chem. 1990;265:7520–7525. [PubMed] [Google Scholar]
- Orning L., Gierse J.K., Fitzpatrick F.A. The bifunctional enzyme leukotriene-A4 hydrolase is an arginine aminopeptidase of high efficiency and specificity. J. Biol. Chem. 1994;269:11269–11273. [PubMed] [Google Scholar]
- Kikuta Y., Miyauchi Y., Kusunose E., Kusunose M. Expression and molecular cloning of human liver leukotriene B4 omega-hydroxylase (CYP4F2) gene. DNA Cell Biol. 1999;18:723–730. doi: 10.1089/104454999315006. [DOI] [PubMed] [Google Scholar]
- Christmas P., Ursino S.R., Fox J.W., Soberman R.J. Expression of the CYP4F3 gene. Tissue-specific splicing and alternative promoters generate high and low Km forms of leukotriene B4 omega-hydroxylase. J. Biol. Chem. 1999;274:21191–21199. doi: 10.1074/jbc.274.30.21191. [DOI] [PubMed] [Google Scholar]
- Yokomizo T., Ogawa Y., Uozumi N., Kume K., Izumi T., Shimizu T. cDNA cloning, expression, and mutagenesis study of leukotriene B4 12-hydroxydehydrogenase. J. Biol. Chem. 1996;271:2844–2850. doi: 10.1074/jbc.271.5.2844. [DOI] [PubMed] [Google Scholar]
- Wheelan P., Murphy R.C. Metabolism of leukotriene B4 in cultured hepatoma cells. Arch. Biochem. Biophys. 1995;321:381–389. doi: 10.1006/abbi.1995.1408. [DOI] [PubMed] [Google Scholar]
- Devchand P.R., Hihi A.K., Perroud M., Schleuning W.D., Spiegelman B.M., Wahli W. Chemical probes that differentially modulate peroxisome proliferator-activated receptor alpha and BLTR, nuclear and cell surface receptors for leukotriene B4 . J. Biol. Chem. 1999;274:23341–23348. doi: 10.1074/jbc.274.33.23341. [DOI] [PubMed] [Google Scholar]
- Devchand P.R., Keller H., Peters J.M., Vazquez M., Gonzalez F.J., Wahli W. The PPARalpha-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]