Abstract
The receptor tyrosine kinase Flt3 plays an important role in proliferation and survival of hematopoietic stem and progenitor cells. Although some post-receptor signaling events of Flt3 have been characterized, the involvement of the Janus kinase/signal transducer and activator of transcription (Jak/Stat) pathway in Flt3 signaling has not been thoroughly evaluated. To this aim, we examined whether Flt3 activates the Jak/Stat pathway in Baf3/Flt3 cells, a line stably expressing human Flt3 receptor. Stat5a, but not Stats 1–4, 5b, or 6, was potently activated by Flt3 ligand (FL) stimulation. Interestingly, FL did not activate any Jaks. Activation of Stat5a required the kinase activity of Flt3. A selective role for Stat5a in the proliferative response of primary hematopoietic progenitor cells to FL was documented, as FL did not act on progenitors from marrows of Stat5a−/− mice, but did stimulate/costimulate proliferation of these cells from Stat5a+/+, Stat5b−/−, and Stat5b+/+ mice. Thus, Stat5a is essential for at least certain effects of FL. Moreover, our data confirm that Stat5a and Stat5b are not redundant, but rather are at least partially distinctive in their function.
Keywords: Flt3, signal transducer and activator of transfection 5, signal transduction, hematopoiesis, gene knockout
Introduction
Flt3 ligand (FL) is a potent cytokine that acts synergistically with a wide range of CSFs and ILs to stimulate proliferation and differentiation of hematopoietic stem and progenitor cells 1 2 3 4. It also enhances survival of progenitor cells 5 6. Its receptor, Flt3, belongs to the type III receptor tyrosine kinases (RTKs) that also include receptors for CSF-1, Steel (or stem cell) factor (SLF), and platelet-derived growth factor (PDGF; reference 7). This family has five immunoglobulin-like domains in their extracellular region and an intracellular tyrosine kinase made up of an ATP-binding loop and a catalytic domain separated by a kinase insert domain. In vivo and in vitro studies have shown that FL and Flt3 play important roles in proliferation and differentiation of both multipotent stem and lymphoid progenitor cells 1 2 3 4 8 9 10. Mice lacking Flt3 receptor have normal mature hematopoietic populations; however, they exhibit reduced numbers of primitive B lymphoid progenitors and multipotent stem cells 11.
Before the cloning of Flt3 ligand, the capacity of the Flt3 receptor to transmit a signal was studied by constructing chimeric receptors between the cytoplasmic and transmembrane domains of Flt3 and the extracellular domains of either the c-kit 12 or CSF-1 receptor 13 14 15. Using this strategy, some early signaling events and substrate specificities of Flt3 receptor were characterized 13 15 16. The chimeric receptor transduced mitogenic signals through association with and/or phosphorylation of a number of cytoplasmic proteins, including RAS GTPase–activating protein, phospholipase C-γ, Vav, Grb2, Shc, and the p85 subunit of phosphatidylinositol 3 kinase. p85 has been shown to bind tyrosine 958 (YQNM) in the COOH-terminal tail of murine Flt3 15 16. Recently, we found that p85 does not bind directly to human Flt3, which lacks a potential p85-binding site in the COOH terminus 17. In addition, FL stimulation of the human Flt3 receptor resulted in phosphorylation of Src homology 2 (SH2)-containing inositol-5-phosphatase (SHIP) and a 100-kD protein in monocytic THP-1 cells 17; phosphorylation of Shc and Cbl in myeloid cells 18; and SH2-containing tyrosine phosphatase, SHIP, and Cbl-b in pro-B cells 17 19 20. However, whether Janus kinase/signal transducer and activator of transcription (Jak/Stat) proteins are involved in Flt3 signaling has not been clearly studied.
In this study, we examined whether Flt3 activates the Jak/Stat pathway in a murine IL-3–dependent hematopoietic cell line Baf3, which stably expresses full-length human Flt3 receptor. We found that, unlike most RTKs, Flt3 potently activates only Stat5a without activation of any Jaks. This activation of Stat5a by Flt3 was also observed in COS-7 cells cotransfected with Flt3 receptor and Stat5 expression vectors. Moreover, FL did not stimulate/costimulate proliferation of primary hematopoietic progenitor cells from marrows of mice functionally deleted in Stat5a (−/−) but it did act normally on Stat5a littermate control (+/+), Stat5b (−/−), and Stat5b (+/+) progenitor cells. These results demonstrate that Stat5a, but not Stat5b, plays a critical role in mediating FL effects.
Materials and Methods
Cytokines and Antibodies.
Recombinant human FL and murine SLF were provided by Immunex. Recombinant preparations of murine IL-3, GM-CSF, M-CSF, and G-CSF were purchased from R&D Systems. Rabbit polyclonal anti-Flt3, anti-Stat5a, anti-Stat5b, anti-Stat3, anti-Stat6, and anti-Tyk2 antibodies were purchased from Santa Cruz Biotechnology, Inc. Rabbit polyclonal anti-Stat5a, anti-Stat5b, anti-Jak1, anti-Jak2, and anti-Jak3 antibodies and antiphosphotyrosine mAb (4G10) were obtained from Upstate Biotechnology. Anti-Stat1, anti-Stat2, and anti-Stat4 antibodies were from Transduction Laboratories. AG490 and AG1296 were from Calbiochem-Novabiochem.
Cell Culture, DNA Constructs, and Transfection.
The murine IL-3–dependent hematopoietic cell line Baf3/Flt3, a subline of Baf3 transduced with the human Flt3 receptor gene, was cultured as previously described 17. Before cytokine stimulation, Baf3/Flt3 cells were washed with PBS and starved in serum-free RPMI 1640 overnight. COS-7 and HEK293 cells were cultured in DMEM containing 10% fetal bovine serum and 100 U/ml penicillin and streptomycin.
The cDNA encoding human Flt3 (provided by Dr. Steward Lyman, Immunex, Seattle, WA) was subcloned into the expression vector pCDNA3 (Invitrogen). Mutants of Flt3 that lack either COOH terminus (Flt3-ΔCT; amino acids 950–993) or both COOH terminus and the second kinase domain (Flt3-ΔCT/TKII; amino acids 782–993) were constructed by PCR-based mutagenesis. The cDNAs encoding murine Stat5a and Stat5b were obtained from Dr. X.-Y. Fu (Yale University, New Haven, CT). Transfection of COS-7 and 293 cells was performed using SuperFect reagent (Qiagen) according to the manufacturer's protocol. For cytokine stimulation experiments, COS-7 or 293 cells were serum-starved for 24 h and then stimulated with FL (100 ng/ml).
Immunoprecipitation and Immunoblotting.
After starvation, cells were suspended in serum-free media (1–2 × 107 cells/ml) and stimulated at 37°C with human FL or murine IL-3 in the absence of any serum. Cell extracts were prepared and subjected to immunoprecipitation and immunoblotting with the antibodies indicated as previously described 20.
Electrophoretic Mobility Shift Assay.
Nuclear extracts from untreated and FL-treated cells were prepared as described elsewhere 21. For the EMSA, 20,000 cpm of 32P-labeled γ-activated sequence motif corresponding to the β-casein gene promoter (5′-AGATTTCTAGGAATTCAATCC-3′) was incubated with 20 μg of nuclear extract proteins in 20 μl of binding buffer containing 10 mM Tris, pH 7.5, 50 mM NaCl, 1 mM dithiothreitol, 1 mM EDTA, 0.05% NP-40, 2 μg poly(dI·dC):poly(dI·dC), and 10% glycerol at room temperature for 30 min and then resolved on 4% polyacrylamide gels containing 0.5× TBE (1× TBE is 89 mmol/liter Tris borate and 1 mmol/liter EDTA, pH 8) and 2.5% glycerol. Gels were run at 4°C in 0.5× TBE at 20 V/cm, dried, and autoradiographed. Oligonucleotide competition was performed by preincubating nuclear extracts with the cold probe (50-fold excess) and poly(dI·dC):poly(dI·dC) for 30 min at 4°C before the addition of labeled probe. For supershifts, samples were preincubated for 1 h at 4°C with the indicated antibodies.
Mice.
Mice functionally deleted (−/−) in Stat5a 22 and Stat5b 23 were from fifth generation backcrosses to the C57BL/6 background. These mice were generated from heterozygous matings. Some of the characteristics of immune cells from these mice have been described previously 24 25. As controls, cells from age- and sex-matched littermate Stat5a+/+ and Stat5b+/+ mice were used.
Hematopoietic Progenitor Cell Assay.
Unseparated bone marrow (5 × 104 cells/ml) obtained from femurs of Stat5a−/−, Stat5a+/+, Stat5b−/−, and Stat5b+/+ mice were plated in 0.3% agar (Difco) culture medium in the presence of 10% heat-inactivated fetal bovine serum (Hyclone) in the absence and presence of varying concentrations of human FL; murine SLF; murine GM-CSF; murine M-CSF; human G-CSF; the combination of SLF with GM-CSF, M-CSF, or G-CSF; or the combination of FL with SLF, GM-CSF, M-CSF, or G-CSF. Plates were incubated at 5% CO2 and lowered (5%) O2 for 7 d in a humidified atmosphere and scored for colonies deriving from granulocyte and/or macrophage progenitor cells 26. Three plates were scored per point per experiment.
Flow Cytometric Analysis.
After lysis of red blood cells, mouse bone marrow cells were subjected to negative selection using lineage-specific antibodies (anti-Gr1, anti-CD11b, anti-CD3, and anti-B220 antibodies from PharMingen) and complement to remove granulocytes, macrophages, and mature T and B cells. Then the cells were stained with fluorochrome-conjugated antibodies (anti–Flt3–PE and anti–c-kit–FITC from PharMingen) and analyzed using FACScan® with CELLQuest software (Becton Dickinson).
Results
FL Preferentially Activates Stat5a over Stat5b in Baf3/Flt3 Cells.
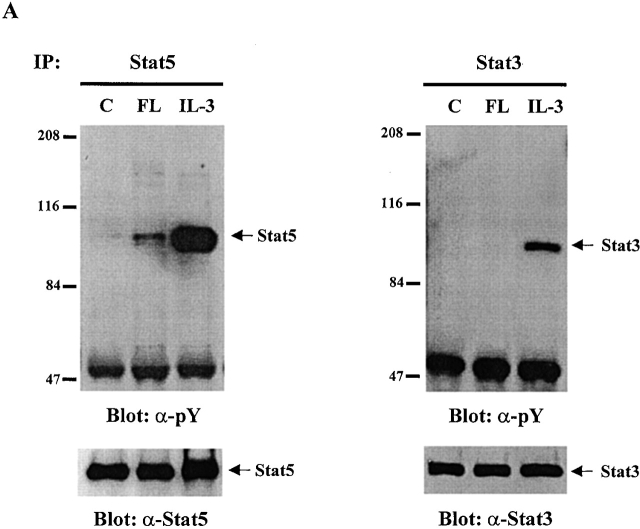
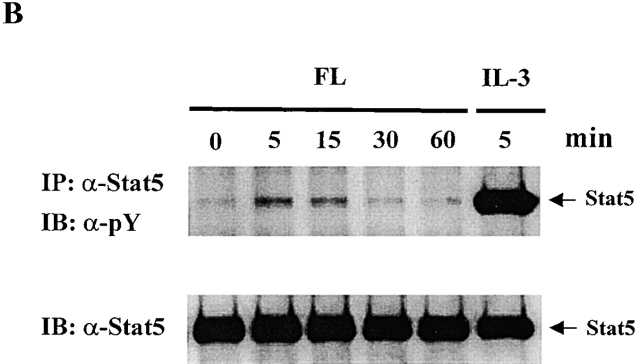
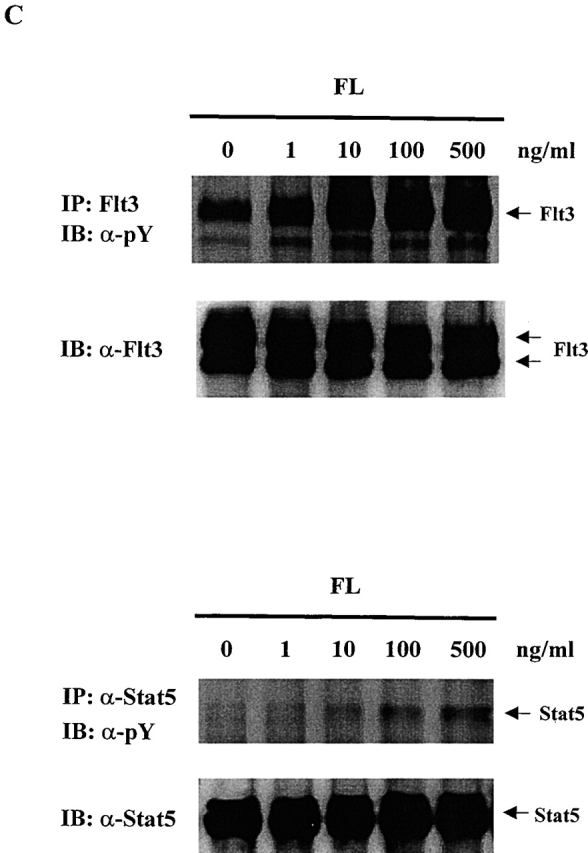
To test whether any Stat proteins are involved in Flt3 signaling, we used Baf3/Flt3 cells transduced with full-length Flt3 cDNA. This subline stably expresses Flt3 receptor on the cell surface and proliferates in response to human FL 20. Among the Stat proteins we examined (Stat1, 2, 3, 4, 5, and 6), only Stat5 was tyrosine phosphorylated by FL stimulation, while IL-3 induced tyrosine phosphorylation of both Stat3 and Stat5 (Fig. 1 A). Tyrosine phosphorylation of Stat5 induced by FL was transient and reached a maximal level at 5 min (Fig. 1 B). It was weak compared with IL-3 stimulation. Fig. 1 C shows the dose–response of tyrosine phosphorylation of Flt3 and Stat5 induced by FL stimulation. We also checked Stat1, 2, 4, and 6. Although they are expressed in Baf3/Flt3 cells, none of them were tyrosine phosphorylated by FL stimulation (data not shown). These results demonstrate that unlike most RTKs, Flt3 selectively activates Stat5.
Figure 1.
FL induces tyrosine phosphorylation of Stat5 in Baf3/Flt3 cells. (A) Growth factor-starved Baf3/Flt3 cells were stimulated with FL (100 ng/ml) or IL-3 (10 ng/ml) for 5 min. Stat5 and Stat3 were immunoprecipitated from cell lysates and immunoblotted with antiphosphotyrosine antibody. (B) Stat5 is transiently tyrosine phosphorylated by FL stimulation. Growth factor–starved Baf3/Flt3 cells were stimulated with FL for various periods of time or with IL-3 for 5 min. Stat5 was immunoprecipitated from cell lysates and immunoblotted with antiphosphotyrosine antibody. (C) Dose–response of tyrosine phosphorylation of Flt3 and Stat5. Growth factor–starved Baf3/Flt3 cells were stimulated with different dose of FL for 5 min. Flt3 and Stat5 were immunoprecipitated from cell lysates and immunoblotted with antiphosphotyrosine antibody. The same membranes were stripped and reblotted with different antibodies as shown. Results shown are from one representative of two to three experiments. pY, antiphosphotyrosine antibody.
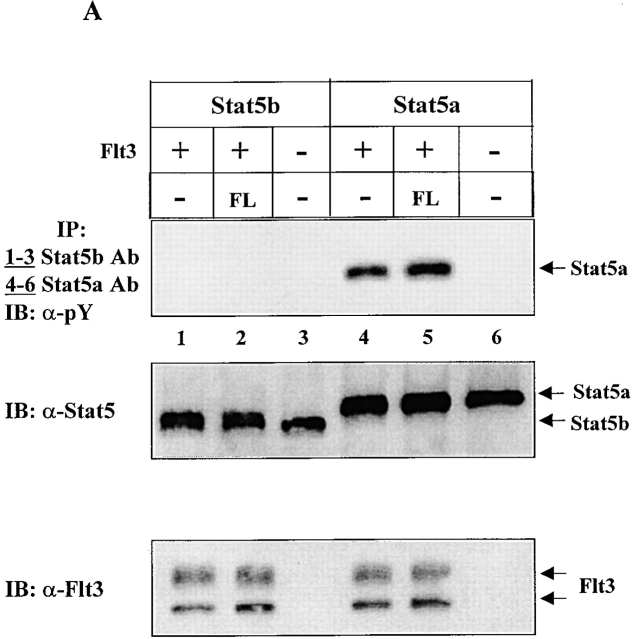
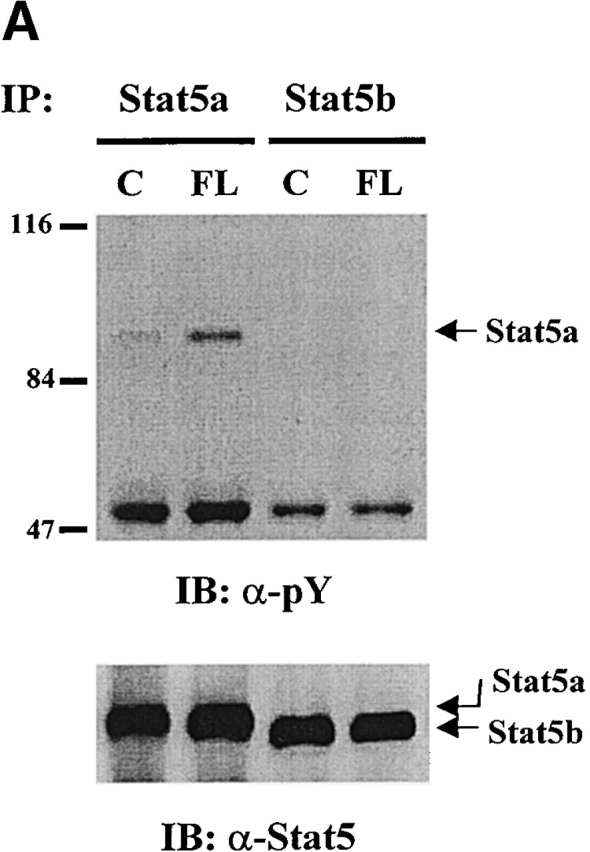
Two different Stat5 genes, encoding Stat5a and Stat5b and sharing >90% identity at the amino acid level, have been identified 27 28 29 30. Both Stat5a and Stat5b are activated by many cytokines and growth factors. The targeted knockout of the individual genes in mice has suggested that they play essential and at least partially overlapping roles in the physiological responses associated with several cytokines 22 23 24 25 31. Despite their homology, there is evidence suggesting that Stat5a and Stat5b may be differentially activated 32. Since the anti-Stat5 antibody we used in Fig. 1 recognizes both Stat5a and Stat5b, we assessed whether FL activated both or only one of the Stat5 proteins. By using specific anti-Stat5a and -Stat5b antibodies, we found that FL induced tyrosine phosphorylation of Stat5a but not Stat5b (Fig. 2 A). IL-3 induced tyrosine phosphorylation of both Stat5a and Stat5b in Baf3/Flt3 cells (data not shown).
Figure 2.
FL preferentially activates Stat5a in Baf3/Flt3 cells. (A) Growth factor–starved Baf3/Flt3 cells were stimulated with FL for 5 min. Stat5a and Stat5b were immunoprecipitated from cell lysates and immunoblotted with anti-phosphotyrosine antibody (pY). The same membrane was stripped and reblotted with anti-Stat5 antibody that recognizes both forms of Stat5. Results shown are from one representative of three experiments. (B) FL activates Stat5a DNA binding activity. Growth factor–starved Baf3/Flt3 cells were either untreated (control) or stimulated with FL for 10 min. Nuclear extracts from these cells were incubated with 32P-labeled probe and subjected to EMSA. Oligonucleotide competition was performed by preincubating nuclear extracts with the cold probe. For supershifts, nuclear extracts were preincubated with the indicated anti-Stat5a or anti-Stat5b antibodies. The arrow indicates the DNA/Stat5 complex. Results shown are from one representative of two experiments.
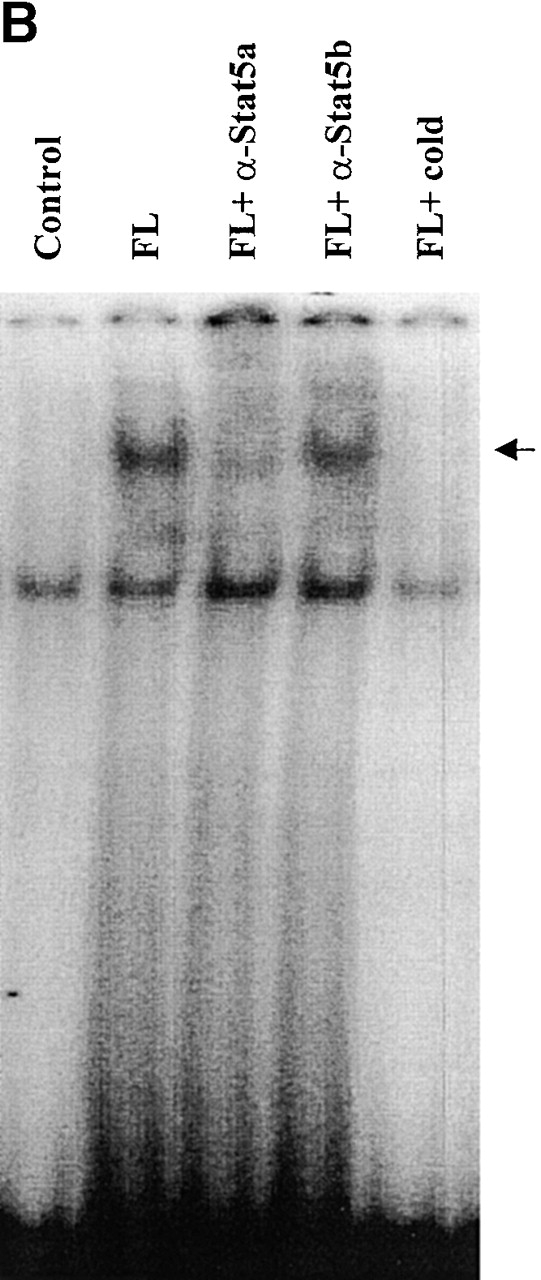
We next investigated if FL could stimulate Stat5a DNA binding activity by EMSAs using a γ-activated sequence motif from the β-casein gene promoter, which is known to bind to both Stat5a and Stat5b. As shown in Fig. 2 B, FL stimulation induced a DNA binding complex that was clearly supershifted by anti-Stat5a antibody, but anti-Stat5b antibody had a much smaller effect. This is consistent with the tyrosine phosphorylation pattern of Stat5 and suggests that gel shift assays tend to be more sensitive than phosphotyrosine blots. These results demonstrate that FL preferentially activates Stat5a in Baf3/Flt3 cells.
FL Does Not Activate Jaks.
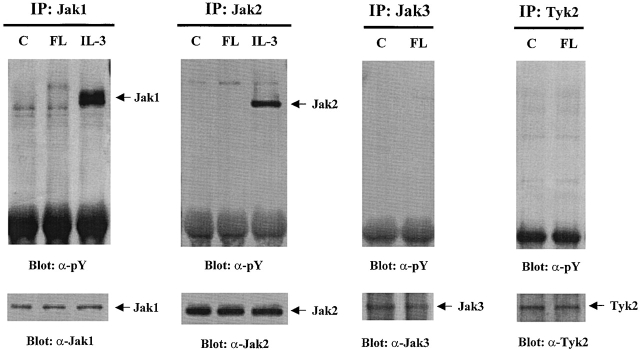
Since Stat5a is activated by FL stimulation, we tested whether Jaks are activated by FL. Jak1, Jak2, Jak3, and Tyk2 were immunoprecipitated from cells treated with either FL or IL-3, separated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and blotted with antiphosphotyrosine antibody. IL-3 has been reported to activate both Jak1 and Jak2 33 34 35. As shown in Fig. 3, although IL-3 induced rapid tyrosine phosphorylation of Jak1 and Jak2, none of the Jaks were activated by FL. These results show that FL does not activate Jaks and suggests that activation of Stat5a is likely mediated by other tyrosine kinases, possibly the Flt3 receptor.
Figure 3.
Jaks are not activated by FL stimulation in Baf3/Flt3 cells. Growth factor–starved Baf3/Flt3 cells were stimulated with FL (100 ng/ml) or IL-3 (10 ng/ml) for 5 min. Jak1, Jak2, Jak3, and Tyk2 were immunoprecipitated from cell lysates and immunoblotted with antiphosphotyrosine antibody (pY). The same membranes were stripped and reblotted with different antibodies as shown in the figure. Results shown are from one representative of three experiments.
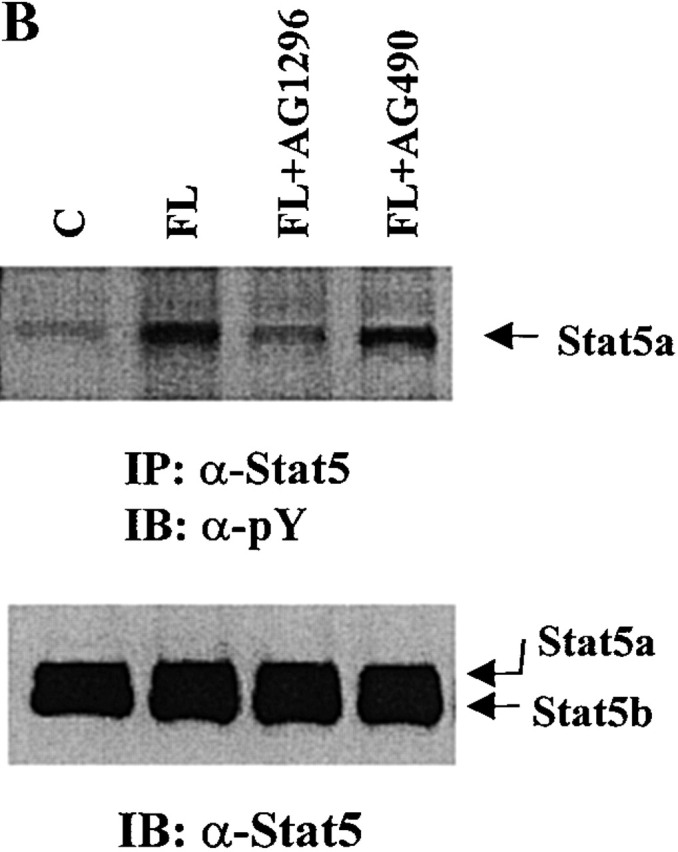
Activation of Stat5a Requires the Kinase Activity of Flt3.
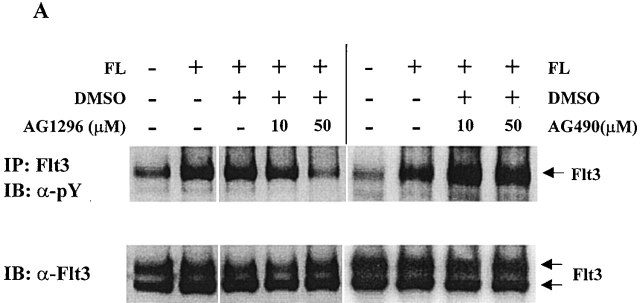
Since Stat5a is tyrosine phosphorylated after FL stimulation and none of the Jaks is activated by FL stimulation, it is possible that Flt3 may directly phosphorylate Stat5a. To test this hypothesis, we first used type III receptor tyrosine kinase inhibitor AG1296, which has been shown to specifically inhibit SLF and PDGF signaling 36. As a control, we used AG490, which inhibits the kinase activity of Jak2 and Jak3 37 38. As shown in Fig. 4 A, AG1296 at 50 μM diminished tyrosine phosphorylation of Flt3 to the control level, whereas AG490 did not have an effect. After Baf3/Flt3 cells were pretreated with these two inhibitors, Stat5 was immunoprecipitated from cells treated with FL and tyrosine phosphorylation of Stat5 was determined by Western blot analysis with antiphosphotyrosine antibody. As shown in Fig. 4 B, tyrosine phosphorylation of Stat5a was decreased close to control levels by AG1296, but not by AG490. This is consistent with the results that Jaks were not activated by FL stimulation and suggests that the tyrosine kinase activity of Flt3 is required for activating Stat5a.
Figure 4.
Inhibition of tyrosine phosphorylation of Flt3 and Stat5a by AG1296, but not by AG490. Baf3/Flt3 cells were pretreated with either control diluent DMSO or AG1296 and AG490 for 1 h at 37°C and washed once with PBS. Then the cells were treated with FL for 5 min. Flt3 (A) and Stat5a (B) were immunoprecipitated from cell lysates and immunoblotted with antiphosphotyrosine antibody (pY). The same membranes were stripped and reblotted with different antibodies as shown in the figure. In B, AG1296 and AG490 were used at 50 μM. Results shown are from one representative of two experiments.
Next, we transiently transfected Flt3 cDNA with either Stat5a or Stat5b cDNA into COS-7 and HEK293 cells and examined whether Flt3 could induce tyrosine phosphorylation and DNA binding activity of Stat5. After serum-starvation for 24 h, the transfected cells were stimulated with human FL (100 ng/ml) for 5 min. Stat5a and Stat5b were immunoprecipitated from cell lysates and tyrosine phosphorylation was analyzed by immunoblotting with antiphosphotyrosine antibody. As shown in Fig. 5 A, no phosphorylation of either Stat5a or Stat5b was detected in COS-7 cells that were mock transfected. Cotransfection of Flt3 and Stat5a cDNA resulted in strong phosphorylation of Stat5a. Addition of FL slightly increased phosphorylation of Stat5a. In contrast, the phosphorylation of Stat5b was very weak, and could only be detected after longer exposure when coexpressed with Flt3. Control immunoblots showed that equivalent levels of Flt3 were expressed. Similar results were obtained when using 293 cells (data not shown). We then evaluated the DNA binding activity of Stat5 by EMSAs. As shown in Fig. 5 B, no DNA binding activities were detected in COS-7 cells that were transfected with vector control plus Stat5a or Stat5b alone. Cotransfection of Flt3 cDNA with Stat5a resulted in high levels of a DNA complex that was supershifted by anti-Stat5a antibody. Although Stat5a and Stat5b were expressed at similar levels (data not shown), cotransfection of Flt3 with Stat5b yielded only a weaker DNA binding activity. These results confirmed that Flt3 preferentially activated Stat5a.
Figure 5.
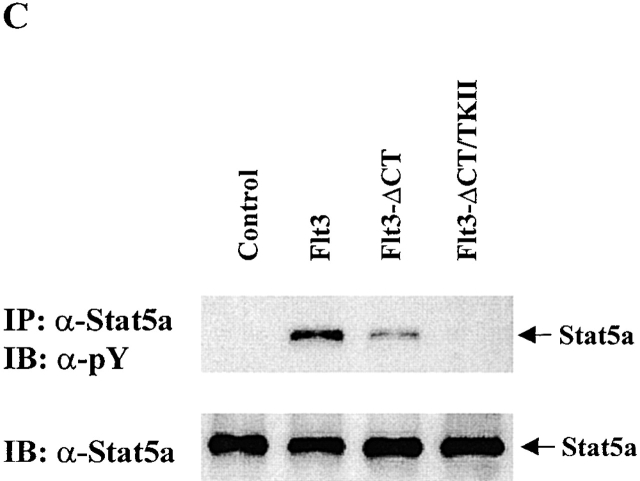
Flt3 preferentially activates Stat5a in COS7 cells. (A) COS-7 cells were transfected with Stat5a, Stat5b, Flt3, or control vector in the combinations shown. After stimulation with either medium control or FL, cells were lysed and Stat5a or Stat5b was immunoprecipitated from cell lysates and immunoblotted with antiphosphotyrosine antibody. The same membrane was stripped and reblotted with anti-Stat5 antibody that recognizes both Stat5a and Stat5b. The expression of Flt3 was confirmed by immunoblot with anti-Flt3 antibody on total cell lysates. (B) Nuclear extracts from COS7 cells transfected with Stat5a, Stat5b, Flt3, or vector control in the combination shown were incubated with 32P-labeled probe and subjected to EMSA. Oligonucleotide competition was performed by preincubating nuclear extracts with the cold probe. For supershifts, nuclear extracts were preincubated with the anti-Stat5a or -Stat5b antibodies. The arrow indicates the DNA–Stat5 complex. (C) COS-7 cells were transfected with Stat5a plus control vector, Flt3, Flt3-ΔCT, or Flt3-ΔCT/TKII, as shown. The tyrosine phosphorylation of Stat5a was examined by immunoprecipitation and immunoblotting with antiphosphotyrosine antibody. The same membrane was stripped and reblotted with anti-Stat5a antibody. pY, antiphosphotyrosine antibody; SS, supershift.

Although expression of wild-type Flt3 in COS-7 cells led to phosphorylation of Stat5a, mutant Flt3 lacking the second kinase domain did not induce phosphorylation of Stat5a, and mutant Flt3 lacking the COOH terminus induced less phosphorylation of Stat5a (Fig. 5 C). Wild-type and mutants of Flt3 were expressed at similar levels as measured by flow cytometry (data not shown). These results demonstrate that phosphorylation of Stat5a requires tyrosine kinase activity of Flt3. Thus, Stat5a is either a direct substrate of Flt3, or Flt3 activates an endogenous tyrosine kinase that in turn phosphorylates Stat5a, or both.
Myeloid Progenitors from Stat5a−/− Mice Do Not Respond to the Stimulating/Costimulating Effects of FL.
Use of cells from mice functionally deleted in genes for certain intracellular molecules has allowed the elucidation of the roles of these gene-encoded proteins in blood cell regulation 39. To this end, we evaluated the responsiveness in vitro of myeloid progenitor cells from femurs of Stat5a−/− and Stat5a+/+ mice to the proliferation effects of FL. By itself, FL is a weak stimulator of the proliferation of myeloid progenitor cells. However, in combination with GM-CSF, M-CSF, or G-CSF, it is a potent costimulating molecule that acts synergistically with these CSFs to enhance the number and size of colonies 4.
Bone marrow cells from Stat5a−/− and littermate control Stat5a+/+ mice were plated in the absence and presence of varying concentrations of FL, SLF, GM-CSF, M-CSF, or G-CSF, or varying concentrations of SLF or FL with each other, or varying concentrations of each of the CSFs (Fig. 6). FL by itself had minimal activity on stimulation of colony formation of Stat5a+/+ progenitors and little or no stimulation of Stat5a−/− progenitors. No significant differences (P > 0.05) were noted in response of Stat5a−/− and Stat5a+/+ progenitors to stimulation by SLF, GM-CSF, M-CSF, or G-CSF, or to the synergistic effects of 50 ng/ml SLF plus GM-CSF, M-CSF, or G-CSF. However, the costimulating synergistic effects of 100 ng/ml FL with SLF, GM-CSF, M-CSF, or G-CSF seen with cells from Stat5a+/+ mice (P < 0.001) were not apparent with cells from Stat5a−/− mice. The effects of FL in combination with the different concentrations of SLF or CSFs on Stat5a−/− cells were equivalent to the single actions of these concentrations of SLF or CSFs. Similar differences were noted using 10 ng/ml FL or SLF with varying concentrations of CSFs (data not shown). Greater than 90% of colonies stimulated with M-CSF with or without FL or SLF were composed purely of macrophages. Greater than 90% of colonies stimulated with G-CSF with or without FL or SLF were composed of neutrophilic granulocytes. Colonies stimulated with GM-CSF with or without FL or SLF were >50% composed of granulocytes and macrophages with the remainder being pure granulocyte or pure macrophage colonies.
Figure 6.

FL stimulates/costimulates proliferation of myeloid progenitor cells from marrows of Stat5a+/+ but not Stat5a−/− mice. Results are shown as the average percentage of control of cells incubated with 50 ng/ml SLF plus 10 ng/ml GM-CSF for a total of six experiments each. The control colony numbers upon which the percent of control are based were 52 ± 2 (mean ± 1 SEM), 70 ± 3, 89 ± 4, 60 ± 4, 152 ± 3, and 132 ± 2 for Stat5a−/− cells and 62 ± 3, 87 ± 3, 101 ± 8, 71 ± 2, 70 ± 2, and 55 ± 3 for Stat5a+/+ cells.
To evaluate the specificity of these effects, similar comparative experiments were done with myeloid progenitor cells from Stat5b−/− and their littermate control Stat5b+/+ mice (Fig. 7). No significant differences (P > 0.05) were noted in the responses of Stat5b−/− and Stat5b+/+ cells to the individual effects of FL, SLF, GM-CSF, M-CSF, G-CSF, or to the combined effects of SLF or FL with each other or with any of the CSFs. At the same time these experiments were done, experiments were also done with Stat5a−/− and Stat5a+/+ cells with results comparable to those seen in Fig. 6.
Figure 7.

FL stimulates/costimulates proliferation of myeloid progenitor cells from marrows of Stat5b+/+ and Stat5b−/− mice to the same extent. Results are shown as the average percentage of control of cells incubated with 50 ng/ml SLF plus 10 ng/ml GM-CSF for a total of two experiments each. The control colony numbers upon which the percentage of control are based were 66 ± 2 and 93 ± 1 for Stat5b−/− cells, and 93 ± 2 and 74 ± 3 for Stat5b+/+ cells.
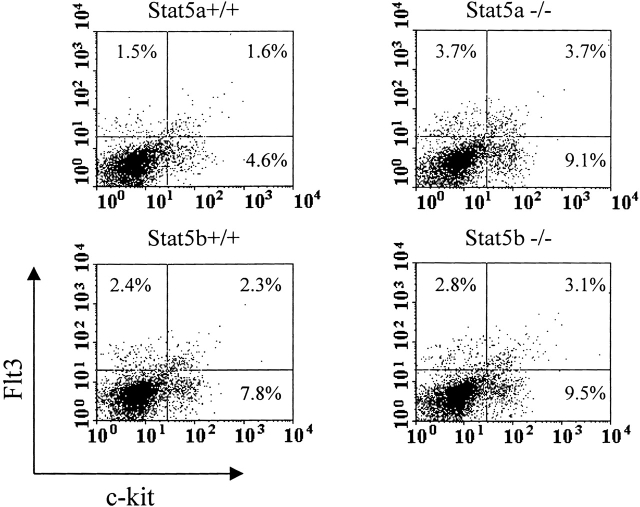
These results demonstrate the critical role played by Stat5a, but not Stat5b, in myeloid progenitor cell responsiveness to the stimulating/costimulating effects of FL. To make sure that the differences noted between Stat5a−/− cells compared with those of Stat5a+/+, Stat5b−/−, and Stat5b+/+ cells were not due to differential expression of Flt3 on the cells, protein levels of Flt3 were assessed by flow cytometry on the surface of bone marrow cells that were depleted of lineage-positive cells. (Fig. 8). Expression of Flt3 on Stat5a−/− c-kit+Lin− cells, a population highly enriched for myeloid progenitor cells 40, was at least as high as on the Stat5a+/+, Stat5b+/+, and Stat5b−/− cells. Thus, the expression levels of Flt3 on the cells from these mice could not explain the nonresponsiveness of Stat5a−/− progenitors to stimulation/costimulation by FL.
Figure 8.
Flt3 receptor expression on the surface of bone marrow cells of Stat5a+/+, Stat5a−/−, Stat5b+/+, and Stat5b−/− mice as analyzed by flow cytometry. Mouse bone marrow cells were depleted of lineage-positive cells and stained with anti–Flt3–PE and anti–c-kit–FITC antibodies. Large granulocytes and debris were gated out on the basis of their light scatter pattern. The quadrants were set up based on isotype control antibodies. The numbers shown on the two-dimensional dot plots are percentages of single positive cells (Flt3+ or c-kit+) and double positive cells (Flt3+c-kit+).
Discussion
In this study we have found that FL preferentially activates Stat5a over Stat5b, and that myeloid progenitors from the bone marrow of Stat5a+/+, Stat5b+/+, and Stat5b−/−, but not from Stat5a−/−, mice respond to the stimulating/costimulating effects of FL. These results demonstrate that Stat5a plays a critical role in mediating FL effects. Moreover, this adds additional evidence to the literature showing the nonoverlapping effects of Stat5a and Stat5b.
Among the different Stat proteins examined, only Stat5a was potently tyrosine phosphorylated by FL stimulation. FL did not seem to activate any Janus kinase as measured by phosphotyrosine immunoblotting (Fig. 3). Longer stimulation with FL (30 min) did not result in any phosphorylation of Jak1 and Jak2 (data not shown). This is in contrast to most growth factors whose receptors are tyrosine kinases, such as PDGF, SLF, and M-CSF (or CSF-1). PDGF, SLF, and M-CSF have been shown to activate multiple Jak and Stat proteins. M-CSF induces tyrosine phosphorylation of Jak1, Tyk2, and Stat1, 3, and 5; PDGF activates Jak1, Jak2, Jak3, and Stat1, 3, 5, and 6 41 42 43 44; and SLF activates Jak2 and Stat1, 3, and 5 45 46. These growth factors activate both Stat5a and Stat5b. Our results suggest that FL differs from these RTKs in that it activates only Stat5a without an apparent activation of Jaks.
Recently, a group reported that a tandem-duplicated form of Flt3 constitutively activates Stat5 and mitogen-activated protein (MAP) kinase, whereas FL stimulation leads to activation of MAP kinase but not Stat5 activation in Baf3 and 32D cells expressing wild-type Flt3 47. In that study 47, it was not specified whether Hayakawa et al. were dealing with Stat5a or Stat5b or both Stat5 proteins. Our data show that FL stimulation of wild-type Flt3 preferentially activates Stat5a, and our in vivo data strongly suggest that Stat5a, but not Stat5b, plays a critical and potentially physiological role in mediating the synergistic effects of FL. Although the same cell line, Baf3, was used, the discrepancy between the study by Hayakawa et al. and ours may be due to different levels of Flt3 expression and the assays used.
Stat5a and Stat5b are two highly related transcription factors encoded by two different genes. Stat5a and Stat5b are activated by a broad range of cytokines and growth factors. Although most cytokines and growth factors activate both Stat5a and Stat5b, there are some studies showing that Stat5a and Stat5b can be differentially activated and regulate different sets of genes. For example, GM-CSF preferentially activates Stat5b, but not Stat5a, in human neutrophils, whereas IFN-α and -γ predominantly activate Stat5a in promonocytic U937 cells 48 49. Insulin preferentially activates Stat5b, but not Stat5a, via a Jak2-independent signaling pathway in kym-1 rhabdomyosarcoma cells 50. Not only can Stat5a and Stat5b be differentially activated, they also have distinctive functions. In support of this, Stat5a−/− and Stat5b−/− mice have different phenotypes 22 23 24 25 51. Our findings that FL preferentially activates Stat5a in both Baf3/Flt3 and COS7 cells and that myeloid progenitors from the marrow of Stat5a+/+, Stat5b+/+, and Stat5b−/−, but not from Stat5a−/−, mice respond to the stimulating/costimulating effects of FL provide another example of differential activation of Stat5 protein. Moreover, our results strongly suggest that Stat5a, but not Stat5b, plays a critical role in myeloid progenitor cell responsiveness to the stimulating/costimulating effects of FL.
The Jak/Stat pathway has been well characterized in cytokine signaling. In the type I cytokine receptor family, ligand stimulation induces rapid activation of Jaks which then phosphorylate Stat proteins. In contrast, in tyrosine kinase receptors, the role of Jak proteins in Stat activation is not clearly defined. Some RTKs like PDGFR and epidermal growth factor receptor (EGFR), although they can activate Jaks, activate Stat proteins in a Jak-independent fashion 43 52. In contrast, the activity of the intrinsic RTK is absolutely required for Stat activation by EGF or PDGF. A mutant PDGFR, which lacks kinase activity, is unable to stimulate Stat1 and Stat3 activation in response to PDGF stimulation 43. Stat1 can directly interact with EGF, PDGF, and SLF receptors and these receptors can phosphorylate Stat1 in vitro 46 53 54. EGFR can also directly phosphorylate Stat3 in vitro 55. These results suggest that Stat proteins may be direct substrates of receptor tyrosine kinases. Our findings that the kinase activity of Flt3 is required for Stat5a activation supports this hypothesis. However, our results do not rule out the possibility that Flt3 may activate an endogenous tyrosine kinase that in turn phosphorylates Stat5a. Regardless of the mechanism, our data confirm that Stat5a and Stat5b are not redundant, but rather are at least partially distinctive in their functions.
Acknowledgments
We would like to thank Dr. Stewart Lyman for Baf3 and Baf3/Flt3 cells; Dr. X.-Y. Fu (for Stat5a and Stat5b cDNA; and Dr. Mark Kaplan and Charlie Mantel for reading the manuscript and for their comments.
This work was supported by US Public Health Service grants R01 HL56416 and R01 DK53674 to H.E. Broxmeyer.
Footnotes
Abbreviations used in this paper: FL, Flt3 ligand; Jak, Janus kinase; PDGF, platelet-derived growth factor; RTK, receptor tyrosine kinase; SLF, Steel factor; Stat, signal transducer and activator of kinase.
References
- Lyman S.D., James L., Vanden Bos T., de Vries P., Brasel K., Gliniak B., Hollingsworth L.T., Picha K.S., McKenna H.J., Splett R.R. Molecular cloning of a ligand for the flt3/flk-2 tyrosine kinase receptora proliferative factor for primitive hematopoietic cells. Cell. 1993;75:1157–1167. doi: 10.1016/0092-8674(93)90325-k. [DOI] [PubMed] [Google Scholar]
- Lyman S.D., James L., Johnson L., Brasel K., de Vries P., Escobar S.S., Downey H., Splett R.R., Beckmann M.P., McKenna H.J. Cloning of the human homologue of the murine flt3 liganda growth factor for early hematopoietic progenitor cells. Blood. 1994;83:2795–2801. [PubMed] [Google Scholar]
- Hannum C., Culpepper J., Campbell D., McClanahan T., Zurawski S., Bazan J.F., Kastelein R., Hudak S., Wagner J., Mattson J. Ligand for FLT3/FLK2 receptor tyrosine kinase regulates growth of haematopoietic stem cells and is encoded by variant RNAs. Nature. 1994;368:643–648. doi: 10.1038/368643a0. [DOI] [PubMed] [Google Scholar]
- Broxmeyer H.E., Lu L., Cooper S., Ruggieri L., Li Z.H., Lyman S.D. Flt3 ligand stimulates/costimulates the growth of myeloid stem/progenitor cells. Exp. Hematol. 1995;23:1121–1129. [PubMed] [Google Scholar]
- Veiby O.P., Jacobsen F.W., Cui L., Lyman S.D., Jacobsen S.E. The flt3 ligand promotes the survival of primitive hemopoietic progenitor cells with myeloid as well as B lymphoid potential. Suppression of apoptosis and counteraction by TNF-alpha and TGF-beta. J. Immunol. 1996;157:2953–2960. [PubMed] [Google Scholar]
- Takahira H., Lyman S.D., Broxmeyer H.E. Flt3 ligand prolongs survival of CD34+++ human umbilical cord blood myeloid progenitors in serum-depleted culture medium. Ann. Hematol. 1996;72:131–135. doi: 10.1007/s002770050150. [DOI] [PubMed] [Google Scholar]
- Rosnet O., Birnbaum D. Hematopoietic receptors of class III receptor-type tyrosine kinases. Crit. Rev. Oncog. 1993;4:595–613. [PubMed] [Google Scholar]
- Jacobsen S.E., Okkenhaug C., Myklebust J., Veiby O.P., Lyman S.D. The FLT3 ligand potently and directly stimulates the growth and expansion of primitive murine bone marrow progenitor cells in vitrosynergistic interactions with interleukin (IL) 11, IL-12, and other hematopoietic growth factors. J. Exp. Med. 1995;181:1357–1363. doi: 10.1084/jem.181.4.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna H.J., de Vries P., Brasel K., Lyman S.D., Williams D.E. Effect of flt3 ligand on the ex vivo expansion of human CD34+ hematopoietic progenitor cells. Blood. 1995;86:3413–3420. [PubMed] [Google Scholar]
- Lyman S.D., Jacobsen S.E. c-kit ligand and Flt3 ligandstem/progenitor cell factors with overlapping yet distinct activities. Blood. 1998;91:1101–1134. [PubMed] [Google Scholar]
- Mackarehtschian K., Hardin J.D., Moore K.A., Boast S., Goff S.P., Lemischka I.R. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 1995;3:147–161. doi: 10.1016/1074-7613(95)90167-1. [DOI] [PubMed] [Google Scholar]
- Lyman S.D., James L., Zappone J., Sleath P.R., Beckmann M.P., Bird T. Characterization of the protein encoded by the flt3 (flk2) receptor-like tyrosine kinase gene. Oncogene. 1993;8:815–822. [PubMed] [Google Scholar]
- Dosil M., Wang S., Lemischka I.R. Mitogenic signalling and substrate specificity of the Flk2/Flt3 receptor tyrosine kinase in fibroblasts and interleukin 3-dependent hematopoietic cells. Mol. Cell. Biol. 1993;13:6572–6585. doi: 10.1128/mcb.13.10.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroc N., Rottapel R., Rosnet O., Marchetto S., Lavezzi C., Mannoni P., Birnbaum D., Dubreuil P. Biochemical characterization and analysis of the transforming potential of the FLT3/FLK2 receptor tyrosine kinase. Oncogene. 1993;8:909–918. [PubMed] [Google Scholar]
- Rottapel R., Turck C.W., Casteran N., Liu X., Birnbaum D., Pawson T., Dubreuil P. Substrate specificities and identification of a putative binding site for PI3K in the carboxy tail of the murine Flt3 receptor tyrosine kinase. Oncogene. 1994;9:1755–1765. [PubMed] [Google Scholar]
- Casteran N., Rottapel R., Beslu N., Lecocq E., Birnbaum D., Dubreuil P. Analysis of the mitogenic pathway of the FLT3 receptor and characterization in its C terminal region of a specific binding site for the PI3′ kinase. Cell. Mol. Biol. 1994;40:443–456. [PubMed] [Google Scholar]
- Zhang S., Broxmeyer H.E. p85 subunit of Pi3 kinase does not bind to human Flt3 receptor, but associates with Shp2, Ship, and a tyrosine-phosphorylated 100-kDa protein in Flt3 ligand-stimulated hematopoietic cells. Biochem. Biophys. Res. Commun. 1999;254:440–445. doi: 10.1006/bbrc.1998.9959. [DOI] [PubMed] [Google Scholar]
- Lavagna-Sevenier C., Marchetto S., Birnbaum D., Rosnet O. FLT3 signaling in hematopoietic cells involves CBL, SHC and an unknown P115 as prominent tyrosine-phosphorylated substrates. Leukemia. 1998;12:301–310. doi: 10.1038/sj.leu.2400921. [DOI] [PubMed] [Google Scholar]
- Lavagna-Sevenier C., Marchetto S., Birnbaum D., Rosnet O. The CBL-related protein CBLB participates in FLT3 and interleukin-7 receptor signal transduction in pro-B cells. J. Biol. Chem. 1998;273:14962–14967. doi: 10.1074/jbc.273.24.14962. [DOI] [PubMed] [Google Scholar]
- Zhang S., Mantel C., Broxmeyer H.E. Flt3 signaling involves tyrosyl-phosphorylation of SHP-2 and SHIP and their association with Grb2 and Shc in Baf3/Flt3 cells. J. Leukoc. Biol. 1999;65:372–380. doi: 10.1002/jlb.65.3.372. [DOI] [PubMed] [Google Scholar]
- Sadowski H.B., Gilman M.Z. Cell-free activation of a DNA-binding protein by epidermal growth factor. Nature. 1993;362:79–83. doi: 10.1038/362079a0. [DOI] [PubMed] [Google Scholar]
- Liu X., Robinson G.W., Wagner K.U., Garrett L., Wynshaw-Boris A., Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- Udy G.B., Towers R.P., Snell R.G., Wilkins R.J., Park S.H., Ram P.A., Waxman D.J., Davey H.W. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc. Natl. Acad. Sci. USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H., Liu X.W., Wynshaw-Boris A., Rosenthal L.A., Imada K., Finbloom D.S., Hennighausen L., Leonard W.J. An indirect effect of Stat5a in IL-2-induced proliferationa critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity. 1997;7:691–701. doi: 10.1016/s1074-7613(00)80389-1. [DOI] [PubMed] [Google Scholar]
- Imada K., Bloom E.T., Nakajima H., Horvath-Arcidiacono J.A., Udy G.B., Davey H.W., Leonard W.J. Stat5b is essential for natural killer cell–mediated proliferation and cytolytic activity. J. Exp. Med. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, S., and H.E. Broxmeyer. 1996. Measurement of interleukin-3 and other hematopoietic growth factors, such as GM-CSF, G-CSF, M-CSF, erythropoietin and the potent co-stimulating cytokines Steel factor and Flt-3 ligand. Curr. Prot. Immunol. Suppl. 18:6.4.1–6.4.12. [DOI] [PubMed]
- Mui A.L., Wakao H., O'Farrell A.M., Harada N., Miyajima A. Interleukin-3, granulocyte-macrophage colony stimulating factor and interleukin-5 transduce signals through two STAT5 homologs. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:1166–1175. doi: 10.1002/j.1460-2075.1995.tb07100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Robinson G.W., Gouilleux F., Groner B., Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc. Natl. Acad. Sci. USA. 1995;92:8831–8835. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M., Erdjument-Bromage H., Kreider B.L., Xia M., Quelle F., Basu R., Saris C., Tempst P., Ihle J.N., Schindler C. Interleukin-3 signals through multiple isoforms of Stat5. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:1402–1411. doi: 10.1002/j.1460-2075.1995.tb07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.X., Mietz J., Modi W.S., John S., Leonard W.J. Cloning of human Stat5B. Reconstitution of interleukin-2-induced Stat5A and Stat5B DNA binding activity in COS-7 cells. J. Biol. Chem. 1996;271:10738–10744. [PubMed] [Google Scholar]
- Teglund S., McKay C., Schuetz E., van Deursen J.M., Stravopodis D., Wang D., Brown M., Bodner S., Grosveld G., Ihle J.N. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93:841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- Yamashita H., Xu J., Erwin R.A., Farrar W.L., Kirken R.A., Rui H. Differential control of the phosphorylation state of proline-juxtaposed serine residues Ser725 of Stat5a and Ser730 of Stat5b in prolactin-sensitive cells. J. Biol. Chem. 1998;273:30218–30224. doi: 10.1074/jbc.273.46.30218. [DOI] [PubMed] [Google Scholar]
- Silvennoinen O., Witthuhn B.A., Quelle F.W., Cleveland J.L., Yi T., Ihle J.N. Structure of the murine Jak2 protein-tyrosine kinase and its role in interleukin 3 signal transduction. Proc. Natl. Acad. Sci. USA. 1993;90:8429–8433. doi: 10.1073/pnas.90.18.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouro T., Kikuchi Y., Kanazawa H., Hirokawa K., Harada N., Shiiba M., Wakao H., Takaki S., Takatsu K. Critical proline residues of the cytoplasmic domain of the IL-5 receptor alpha chain and its function in IL-5-mediated activation of JAK kinase and STAT5. Int. Immunol. 1996;8:237–245. doi: 10.1093/intimm/8.2.237. [DOI] [PubMed] [Google Scholar]
- Callus B.A., Mathey-Prevot B. Interleukin-3-induced activation of the JAK/STAT pathway is prolonged by proteasome inhibitors. Blood. 1998;91:3182–3192. [PubMed] [Google Scholar]
- Kovalenko M., Gazit A., Bohmer A., Rorsman C., Ronnstrand L., Heldin C.H., Waltenberger J., Bohmer F.D., Levitzki A. Selective platelet-derived growth factor receptor kinase blockers reverse sis-transformation. Cancer Res. 1994;54:6106–6114. [PubMed] [Google Scholar]
- Meydan N., Grunberger T., Dadi H., Shahar M., Arpaia E., Lapidot Z., Leeder J.S., Freedman M., Cohen A., Gazit A. Inhibition of acute lymphoblastic leukaemia by a Jak-2 inhibitor. Nature. 1996;379:645–648. doi: 10.1038/379645a0. [DOI] [PubMed] [Google Scholar]
- Kirken R.A., Erwin R.A., Taub D., Murphy W.J., Behbod F., Wang L., Pericle F., Farrar W.L. Tyrphostin AG-490 inhibits cytokine-mediated JAK3/STAT5a/b signal transduction and cellular proliferation of antigen-activated human T cells. J. Leukoc. Biol. 1999;65:891–899. doi: 10.1002/jlb.65.6.891. [DOI] [PubMed] [Google Scholar]
- Broxmeyer H.E. Regulation of growth factor signaling as assessed by gene deletion and gene transduction. In: Zon L.I., editor. Developmental Biology of Hematopoiesis. Oxford University Press; Oxford: 2000. pp. 247–257. [Google Scholar]
- Reid S., Ritchie A., Boring L., Gosling J., Cooper S., Hangoc G., Charo I.F., Broxmeyer H.E. Enhanced myeloid progenitor cell cycling and apoptosis in mice lacking the chemokine receptor, CCR2. Blood. 1999;93:1524–1533. [PubMed] [Google Scholar]
- Novak U., Harpur A.G., Paradiso L., Kanagasundaram V., Jaworowski A., Wilks A.F., Hamilton J.A. Colony-stimulating factor 1-induced STAT1 and STAT3 activation is accompanied by phosphorylation of Tyk2 in macrophages and Tyk2 and JAK1 in fibroblasts. Blood. 1995;86:2948–2956. [PubMed] [Google Scholar]
- Novak U., Mui A., Miyajima A., Paradiso L. Formation of STAT5-containing DNA binding complexes in response to colony-stimulating factor-1 and platelet-derived growth factor. J. Biol. Chem. 1996;271:18350–18354. doi: 10.1074/jbc.271.31.18350. [DOI] [PubMed] [Google Scholar]
- Vignais M.L., Sadowski H.B., Watling D., Rogers N.C., Gilman M. Platelet-derived growth factor induces phosphorylation of multiple JAK family kinases and STAT proteins. Mol. Cell. Biol. 1996;16:1759–1769. doi: 10.1128/mcb.16.4.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel B.K., Wang L.M., Lee C.C., Taylor W.G., Pierce J.H., LaRochelle W.J. Stat6 and Jak1 are common elements in platelet-derived growth factor and interleukin-4 signal transduction pathways in NIH 3T3 fibroblasts. J. Biol. Chem. 1996;271:22175–22182. doi: 10.1074/jbc.271.36.22175. [DOI] [PubMed] [Google Scholar]
- Linnekin D., Mou S., Deberry C.S., Weiler S.R., Keller J.R., Ruscetti F.W., Longo D.L. Stem cell factor, the JAK-STAT pathway and signal transduction. Leuk. Lymphoma. 1997;27:439–444. doi: 10.3109/10428199709058310. [DOI] [PubMed] [Google Scholar]
- Deberry C., Mou S., Linnekin D. Stat1 associates with c-kit and is activated in response to stem cell factor. Biochem. J. 1997;327:73–80. doi: 10.1042/bj3270073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa F., Towatari M., Kiyoi H., Tanimoto M., Kitamura T., Saito H., Naoe T. Tandem-duplicated Flt3 constitutively activates STAT5 and MAP kinase and introduces autonomous cell growth in IL-3-dependent cell lines. Oncogene. 2000;19:624–631. doi: 10.1038/sj.onc.1203354. [DOI] [PubMed] [Google Scholar]
- Meinke A., Barahmand-Pour F., Wohrl S., Stoiber D., Decker T. Activation of different Stat5 isoforms contributes to cell-type-restricted signaling in response to interferons. Mol. Cell. Biol. 1996;16:6937–6944. doi: 10.1128/mcb.16.12.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shami A., Mahanna W., Naccache P.H. Granulocyte-macrophage colony-stimulating factor-activated signaling pathways in human neutrophils. Selective activation of Jak2, Stat3, and Stat5b. J. Biol. Chem. 1998;273:1058–1063. doi: 10.1074/jbc.273.2.1058. [DOI] [PubMed] [Google Scholar]
- Storz P., Doppler H., Pfizenmaier K., Muller G. Insulin selectively activates STAT5b, but not STAT5a, via a JAK2-independent signalling pathway in kym-1 rhabdomyosarcoma cells. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1999;464:159–163. doi: 10.1016/s0014-5793(99)01689-0. [DOI] [PubMed] [Google Scholar]
- Park S.H., Liu X., Hennighausen L., Davey H.W., Waxman D.J. Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression. Impact of STAT5a gene disruption. J. Biol. Chem. 1999;274:7421–7430. doi: 10.1074/jbc.274.11.7421. [DOI] [PubMed] [Google Scholar]
- Leaman D.W., Pisharody S., Flickinger T.W., Commane M.A., Schlessinger J., Kerr I.M., Levy D.E., Stark G.R. Roles of JAKs in activation of STATs and stimulation of c-fos gene expression by epidermal growth factor. Mol. Cell. Biol. 1996;16:369–375. doi: 10.1128/mcb.16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okimoto T., Kohno K., Kuwano M., Gopas J., Kung H.F., Ono M. Regulation of epidermal growth factor receptor by activated H-ras and V-myc oncogenes in mouse Balb/3T3 cellspossible roles of AP-1. Oncogene. 1996;12:1625–1633. [PubMed] [Google Scholar]
- Choudhury G.G., Ghosh-Choudhury N., Abboud H.E. Association and direct activation of signal transducer and activator of transcription 1α by platelet-derived growth factor receptor. J. Clin. Invest. 1998;101:2751–2760. doi: 10.1172/JCI1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park O.K., Schaefer T.S., Nathans D. In vitro activation of Stat3 by epidermal growth factor receptor kinase. Proc. Natl. Acad. Sci. USA. 1996;93:13704–13708. doi: 10.1073/pnas.93.24.13704. [DOI] [PMC free article] [PubMed] [Google Scholar]