Abstract
Class switch DNA recombinations change the constant (C) region of the antibody heavy (H) chain expressed by a B cell and thereby change the antibody effector function. Unusual tandemly repeated sequence elements located upstream of H chain gene exons have long been thought to be important in the targeting and/or mechanism of the switch recombination process. We have deleted the entire switch tandem repeat element (Sμ) from the murine μ H chain gene. We find that the Sμ tandem repeats are not required for class switching in the mouse immunoglobulin H-chain locus, although the efficiency of switching is clearly reduced. Our data demonstrate that sequences outside of the Sμ tandem repeats must be capable of directing the class switch mechanism. The maintenance of the highly repeated Sμ element during evolution appears to reflect selection for a highly efficient switching process rather than selection for a required sequence element.
Keywords: gene rearrangement, B lymphocyte, heavy chain, class switching, immunoglobulin isotype
Introduction
Antibody class switching is a process allowing B cells expressing IgM to give rise to cells that produce IgG, IgA, or IgE. The different classes of Ig molecules are defined by the constant region of the H chain. Class switching allows B cells to change effector function against a foreign antigen without losing antigen receptor specificity and takes place via DNA recombination events that involve looping out and deletion of DNA sequences (for reviews, see references 1 2 3). Unlike V(D)J recombination, which is targeted by specific sequence elements 4, no clear consensus sites for class switch recombination are apparent. The switch recombination mechanism appears to involve stretches of tandemly repeated DNA sequences, called switch (S) regions, which are located upstream of each Ig H chain constant region gene except δ.
The roles of S regions in class switching are not clear, but evidence suggests that the tandemly repeated elements are used as the sites for cleavage 5, as well as being involved in the joining reaction 6 7 8. Studies of integrated or extrachromosomal switch substrates 9 10 11 12 13 14 15 16 have suggested that S regions may be sufficient for targeting switch recombination and that two S regions are required for switch recombination to take place. Finally, transcribed S region tandem repeats have been found to form RNA–DNA complexes, and these have been suggested to play a role in switch recombination 17 18 19 20.
In all species that have been analyzed, switch recombination sites in H chain genes are found within or near a tandemly repeated S region. Furthermore, all identified breakpoints from DNA circles excised during switching in normal mouse splenocytes have been found within the repeated S elements 21 22 23. However, when chromosomal breakpoints in a variety of cell types were analyzed 7 24, it was found that some mapped outside the tandemly repeated elements, especially for the Sμ element. Some breakpoints found in DNA circles that arise from an apparent class switching process in a lymphoma cell line are also found outside of the Sμ region 24. These data suggest that the function of and requirement for the Sμ tandem repeats is not clearly understood.
In the mouse, the Sμ element consists of repeated (GAGCT)nGGGGT sequences, where n varies from one to seven in different repeats, but has an average value of three. To directly test whether Sμ is required for antibody class switching, we have removed all the Sμ tandem repeats from the mouse Igh locus to determine the impact of this deletion on the switching process. We find that the Sμ tandem repeats are not required for class switching in the mouse Igh locus, although the efficiency of switching is reduced. The maintenance of the highly repeated Sμ element during evolution appears, therefore, to reflect selection for a highly efficient switching process rather than selection for a required sequence element.
Materials and Methods
Targeting of Sμ in Embryonic Stem Cells.
The targeting construct is shown in Fig. 1. The 5′ homology region is a 1.4-kb segment upstream of Sμ bounded by EcoRI and HindIII sites. The 3′ homology region is a 4.6-kb segment downstream of Sμ bounded by HindIII and KpnI sites. The neo/loxP cassette (a gift from Dr. F. Alt, Harvard Medical School, Boston, MA) was inserted in between the two homology regions. The targeting construct was used in standard gene targeting approaches to obtain chimeric mice that carried the targeted allele. Chimeric mice that transmitted the mutation were mated to Cre recombinase transgenic mice (a gift of Dr. J. Chen, Massachusetts Institute of Technology, Cambridge, MA) in order to remove the neomycin cassette. This provided the ΔSμ allele that has one loxP site replacing Sμ.
Figure 1.
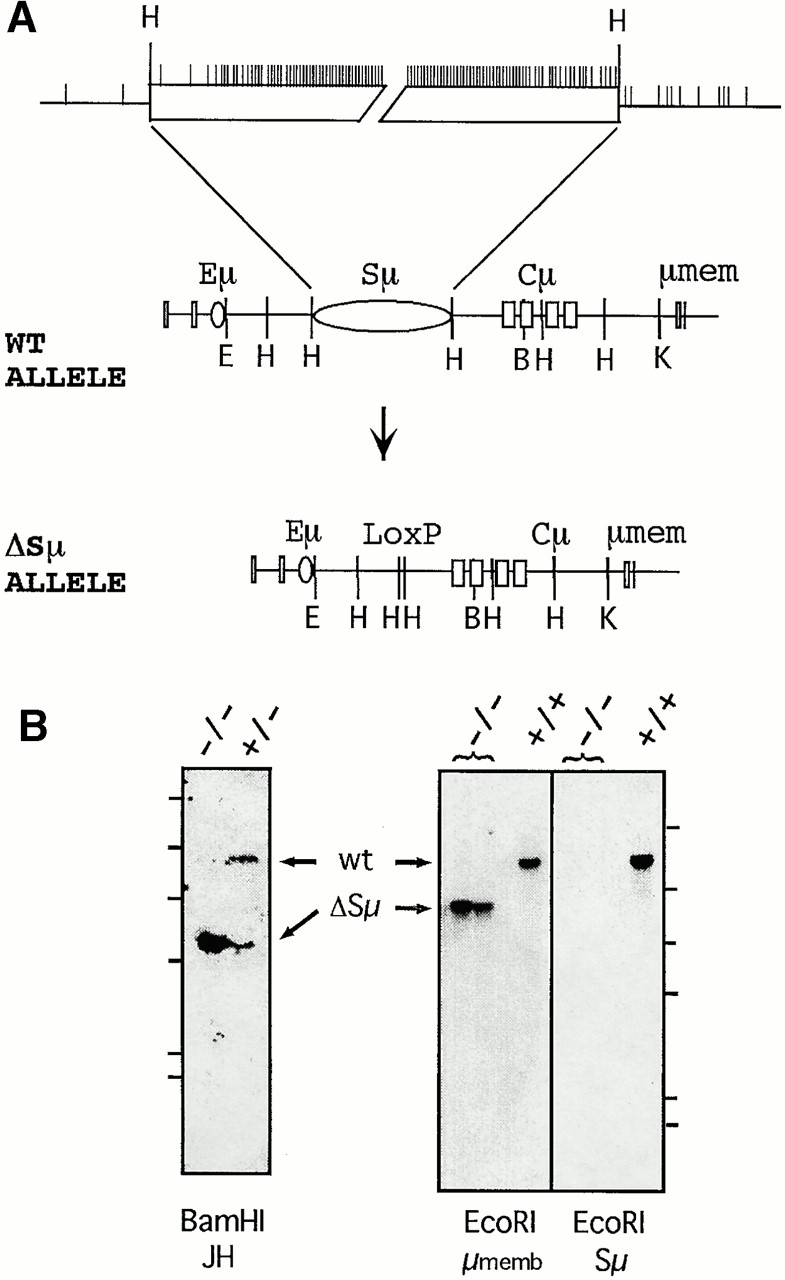
Generation of the ΔSμ mice. (A) Diagram of Sμ tandem repeats and map of the JH-Cμ intron. Eμ, intronic enhancer; Cμ, μ constant region exons; μmem, μ membrane exons; E, EcoRI; H, HindIII; B, BamHI; K, KpnI. In the expanded view of Sμ, each vertical line represents either a GAGCT or GGGGT sequence and the two HindIII sites represent the pair of sites flanking Sμ in the diagram of the wild-type (WT) allele. (B) Southern blot analyses of knockout mice. Genomic DNAs from mice with the indicated phenotypes were digested and hybridized. The left blot contains BamHI digests and was hybridized with pJ11 (reference 52), a 1.8-kb BamHI-EcoRI fragment containing JH3 and JH4. Sizes of wild-type (wt) and ΔSμ alleles are ∼8 and 5.3 kb, respectively. The right blot contains EcoRI digests and was hybridized sequentially with, first, a 900-bp KpnI-XbaI probe containing the μ membrane exons and, second, pM2-20 (reference 52), a 1.8-kb probe containing ∼1.4-kb of Sμ tandem repeat sequences. Sizes of the wild-type and ΔSμ alleles are ∼12 and 8.9 kb, respectively. Positions of λ HindIII DNA fragments are indicated. μmemb, μ membrane exons.
Immunizations and Immunoassays.
Mice were immunized with 100 μg/ml p-phenylarsonate-KLH (Ars-KLH) in CFA (GIBCO BRL), and boosted two times with Ars-KLH in incomplete adjuvant. For hybridoma production, animals were boosted with a final injection of 100 μg/ml Ars-KLH in PBS 3 d before fusion.
Total IgG titers were determined by ELISA on plates coated with 4 μg/ml rat anti–mouse IgG (Zymed Laboratories) and blocked with PBS/3% BSA (Sigma-Aldrich). Samples were applied and detected with 0.5 μg/ml biotin-labeled rat anti–mouse IgG (Zymed Laboratories) and 0.5 μg/ml alkaline phosphatase–streptavidin (Boehringer). Plates were developed with 20 μg/ml of p-nitrophenyl phosphate (Sigma-Aldrich) and analyzed on a Dynatech MR700 plate reader.
For quantitation of the different isotypes in the sera, the same basic protocol described for the total IgG ELISA was used with the following modifications: plates were coated with unlabeled goat anti–mouse IgG3, IgG1, IgG2b, and IgA (all from Southern Biotechnology Associates, Inc.). Detecting antibodies used were: goat anti–mouse IgG3, goat anti–mouse IgG1, goat anti–mouse IgG2b, and (for IgA) both goat anti–mouse κ and λ (all from Southern Biotechnology Associates, Inc.). Total IgM was determined by ELISA using the same basic protocol as above with goat anti-IgM as the coating reagent and rat anti-IgM as the detecting reagent.
Surface IgM staining was done on splenocytes from wild-type and ΔSμ mice using FITC-goat anti–mouse IgM (Southern Biotechnology Associates, Inc.). Samples were collected on a FACScan™ (Becton Dickinson) and analyzed using FloJo (Treestar) analysis software.
In Vitro Switching and Digestion Circularization PCR.
T cell–depleted splenocytes were cultured at 2 × 105/ml in 5-ml cultures with LPS alone to induce IgG3, or with cytokines added to induce IgG1 (IL-4, 800 U/ml), IgG2b (dextran sulfate, 30 μg/ml), or IgG2a (IFN-γ, 10 U/ml). After 4 d, cells were acid treated (50 mM sodium acetate, pH 5.2, 85 mM sodium chloride, 5 mM potassium chloride, 1% FCS) for 2 min on ice to remove Ig bound to Fc receptors, and washed and stained for FACS® analyses with FITC-goat anti–mouse IgM and PE-goat F(ab′)2 anti–mouse IgG1, IgG3, IgG2a, or IgG2b (Southern Biotechnology Associates, Inc.).
Digestion circularization (DC)-PCR is described briefly here and in detail elsewhere 25 26. Genomic DNAs from LPS and LPS plus IL-4–induced cultures were digested with EcoRI (2 μg/100 μl) overnight, ligated overnight at a concentration of 180 ng/μl, and then dialyzed against distilled water. PCR primers are as published for acetylcholine receptor (AchR) and IgG1 25 and IgG2b 10. Twofold dilutions of ligated DNA and a plasmid standard for AchR, P2AO 25, were analyzed to demonstrate that the amount of product depends on the amount of input DNA in a linear fashion. For quantitation, [α-32P]dCTP was incorporated, and the amounts of Sμ-Sγ1 and Sμ-Sγ2b PCR products were normalized to the amount of AchR product to control for the efficiency of digestion and circularization.
Filter Hybridization.
Northern blots were prepared and analyzed as described previously 27 using a 0.7-kb EcoRI-HindIII probe containing the Iμ exon. For Southern blots, genomic DNA was digested, electrophoresed, and transferred to nitrocellulose (Sartorius). The membranes were UV cross-linked and hybridized (1× SSC, 1× Blotto, 0.1% SDS) with the indicated probes (48 h at 65°C). The blots were washed sequentially with 3× SSC, 5 mM EDTA, 0.1% SDS, 1× Blotto, and 50 μg/ml salmon sperm DNA; 0.2× SSC, 1.25 mM EDTA, and 0.1% SDS; and 0.1× SSC, 1.25 mM EDTA, and 0.1% SDS (all at 65°C), and autoradiographed.
Sμ/Sγ1 Junction Analysis.
PCR and sequencing of recombination junctions was generally as described previously 28. Nested μ primers were 5′-AAGTTGAGGATTCAGCCGAAACTGGAGAGG-3′ (MUSIGCD07 nos. 3434–3463) followed by 5′-GCTTGAGTAGTTCTAGTTTCC-3′ (MUSICD07 nos. 3502–3522). The sequence of the nested set of γ1 primers was 5′-CAATTAGCTCCTGCTCTTCTGTGG-3′ (D78344 nos. 8464–8488) followed by 5′-TCTAATCTGCCCCTGTTCCTCTACAACTAC-3′ (D78344 nos. 8365–8395). PCR products were directly sequenced or cloned and sequenced using a TA cloning kit (Invitrogen). Switch junctions in ΔSμ and wild-type mice were analyzed for mutation frequency. Mutations in ΔSμ junctions were identified by comparison with μ and γ1 germline sequences that were determined by compiling sequences from several independent PCR clones. These germline sequences (derived from the 129 strain) were found to be identical to the corresponding published BALB/c sequences. Mutations in junction regions were confirmed from two independent PCR clones. The frequencies of mutation per basepair in the donor (5′) and acceptor (3′) S regions were determined by dividing the total number of mutations by the total number of basepairs in the sequences. Mutation frequencies in wild-type junctions were similarly calculated using published sequences 7 8.
Results
Mutant Mice that Lack Sμ.
The first step in the creation of the Sμ knockout was defining which sequences to remove from the JH-Cμ intron. The goal was to remove all of the tandem repeats without deleting elements required for transcription of the locus. The upstream border of the Sμ deletion was set at the second HindIII site downstream of Eμ (MUSIGCD07 base no. 4995, sequence data available from Genbank/EMBL/DDBJ under accession no. J00440), leaving intact important elements such as the μ enhancer (Eμ), matrix attachment regions 29, and the Iμ exon (Fig. 1). The downstream border was set at the HindIII site ∼1.2 kb upstream of Cμ (MUSIGCD09 base no. 1208, sequence data available from Genbank/EMBL/DDBJ under accession no. J00442, leaving a region of DNA indicated to be important in the transcription of Ig H chain transgenes 27 30. This deletion removes all of the Sμ tandem repeat region. As indicated in Fig. 1 A, some GAGCT or GGGGT sequences remain in the JH-Cμ intron after the deletion of the Sμ element. One GAGCT is left in the intron between Eμ and the 5′ border of the deletion, while 11 additional nontandemly spaced GAGCT sequences are located between the end of the deletion and Cμ. Also, a single GGGGT is ∼150 bp upstream of the deletion. Gene targeting was used to produce mice carrying the ΔSμ deletion, which was confirmed by Southern blotting (Fig. 1 B).
Serum IgG Antibodies in ΔSμ Mice.
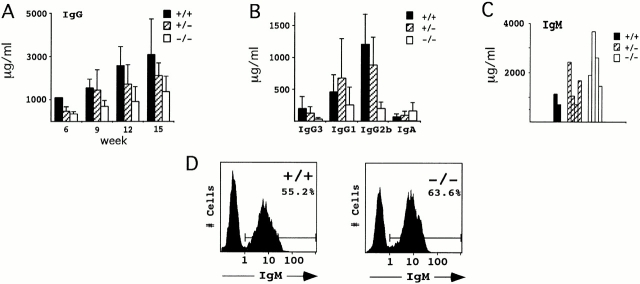
Once the ΔSμ deletion was bred to homozygosity, we bled the animals at various time points to determine whether the mice had detectable IgG titers. Sera from the mice were analyzed at weeks 6, 9, 12, and 15. The results show that ΔSμ animals have the ability to produce IgG, although the levels appear to be reduced approximately twofold relative to wild-type mice (Fig. 2 A). We also analyzed the serum isotype profile of these mice at week 15 (Fig. 2 B). It appears that ΔSμ mice have a slight reduction in IgG1 and IgG3, a relatively large reduction in IgG2b, and no decrease in IgA. When IgM titers were checked on a subset of these week 15 sera, an increase in the amount of serum IgM in the ΔSμ mice was noted (Fig. 2 C). However, when surface IgM of splenocytes was examined by flow cytometry, both C57BL/6 and ΔSμ mice had approximately equivalent numbers of IgM expressing B cells and the same levels of surface IgM expression (Fig. 2 D). Northern blot analyses of unstimulated and LPS-stimulated splenocytes also showed identical levels of spliced Iμ germline transcripts in ΔSμ and wild-type mice (data not shown). These results suggest that the Sμ deletion has little or no effect on early B cell differentiation or on the accessibility of the Iμ-Cμ locus.
Figure 2.
Analysis of IgM and IgG production in unimmunized ΔSμ mice and their littermate controls. (A) Animals were bled and total IgG titers were determined for three wild-type, six heterozygous, and eight mutant mice at weeks 6, 9, 12, and 15. (B) Isotype profile of serum from wild-type, heterozygous, and mutant mice were analyzed at week 15. (C) Serum IgM titers at week 15 were determined on a subset of wild-type, heterozygous, and mutant mice. (D) Surface IgM staining of wild-type and mutant mice was examined by flow cytometry using a FITC-conjugated monoclonal antibody reactive with mouse IgM.
Switching of ΔSμ B Cells in Culture.
To determine if the differences in serum isotype profiles of the ΔSμ mice were due to an intrinsic difference in the B cells, we polyclonally stimulated splenic B cells from the mice in vitro for 4 d and measured the amount of switching to the IgG subclasses by surface staining of Ig isotypes and flow cytometry (representative data shown in Fig. 3). The results (summarized in Table ) extend the serological data and show that switching by the ΔSμ mice is partially reduced for all isotypes analyzed: two- to threefold for IgG1, IgG3, and IgG2a, and fivefold for IgG2b.
Figure 3.
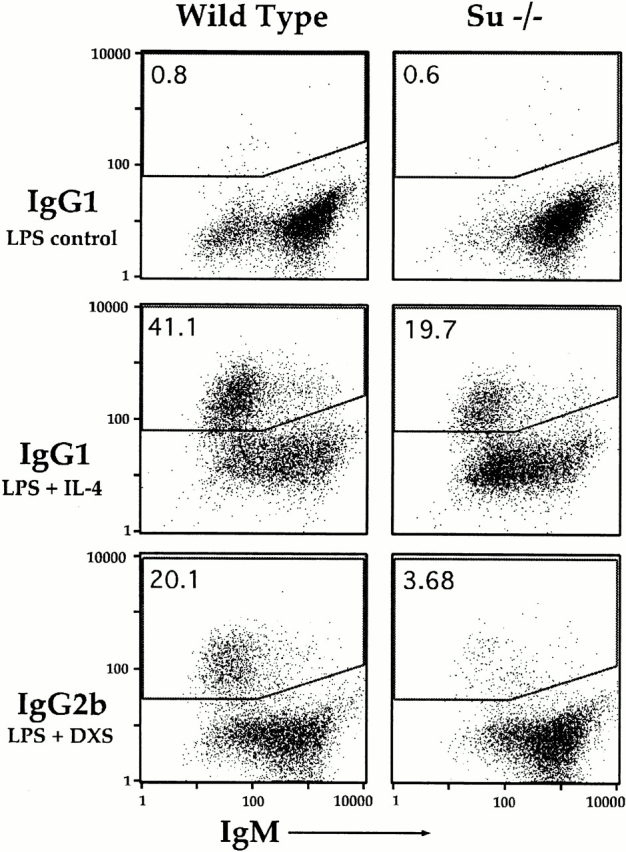
Reduced, but easily detectable IgG expression on B cells from ΔSμ mice. IgG2b is more severely affected than IgG1 by the deletion of Sμ tandem repeats. Splenic B cells from wild-type or ΔSμ mice were cultured with either LPS plus IL-4 to induce IgG1 or with LPS plus dextran sulfate (DXS) to induce IgG2b. On day 4 the cells were stained for surface IgG and IgM and analyzed by flow cytometry. The percentage of cells in the upper gate (indicated in the top right corner of each panel) includes cells positive for IgG alone and for IgG and IgM together. The gates for IgG1 were set on wild-type cells treated with LPS alone, which does not induce switching to IgG1, and then were adjusted for the other isotypes to optimize separation of the populations. Gates for wild-type and ΔSμ cells were always the same for a given isotype.
Table 1.
FACS® Analysis of In Vitro Switching on Day 4 by Splenic B Cells from ΔSμ and Wild-type Littermates
| Percent switched cells | |||||
|---|---|---|---|---|---|
| Exp. 1 | Exp. 2 | Percent WT | |||
| IgG1 | WT | LPS | 1.9 | 0.6 | |
| LPS plus IL-4 | 24.7 | 41.1 | |||
| ΔSμ | LPS | 1.4 | 0.3 | ||
| LPS plus IL-4 | 19.6 | 19.8 | 63 ± 15* | ||
| IgG3 | WT | LPS | 13.4 | 9.8 | |
| LPS plus DxSO4 | 10.7 | 11.6 | |||
| ΔSμ | LPS | 6.3 | 4.5 | ||
| LPS plus DxSO4 | 5.2 | 4.1 | 44.2 ± 2* | ||
| IgG2b | WT | LPS | 7.6 | 9.1 | |
| LPS plus DxSO4 | 23.9 | 19.7 | |||
| ΔSμ | LPS | 3.3 | 1.2 | ||
| LPS plus DxSO4 | 5.7 | 3.3 | 24.3 ± 12* | ||
| IgG2a | WT | LPS plus IFN-γ | ND | 11.0 | |
| ΔSμ | LPS plus IFN-γ | ND | 3.6 | 33 | |
Exp., experiment; ND, not determined. The percentages of cells expressing the switched isotypes are shown for two experiments (Exp. 1 and 2). For each isotype, average values for ΔSμ switching as percentages of WT are also shown.
*±SEM.
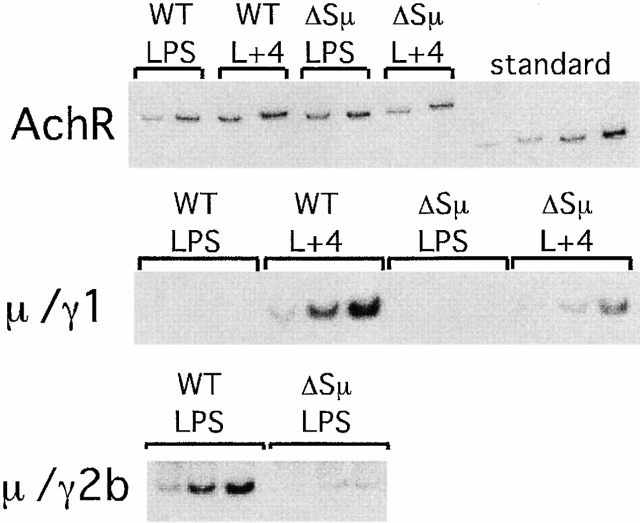
To determine whether the class switching detected in the ΔSμ mice is due to a DNA recombination event occurring within the Sμ intron and to obtain a semiquantitative estimate for the level of switch recombination in these mice, we analyzed DNA from in vitro–activated B cells by DC-PCR 25 26 31. Using one primer upstream of Sμ and one primer downstream of either Sγ1 or Sγ2b, this technique yields a single size band that can be quantified and is independent of the location of switch recombination junctions. An Sμ/Sγ1 product was clearly detectable in DNA from ΔSμ B cells stimulated with LPS plus IL-4, but not with LPS alone (Fig. 4), and was reduced fourfold relative to wild-type in both of two independent PCRs. In the experiment shown, the reduction in IgG1 switching as assayed by cytometry was twofold (Table , experiment 2). The Sμ/Sγ2b PCR product from LPS-stimulated ΔSμ B cells was reduced by an average of ninefold relative to wild-type in two independent PCR assays, compared with eightfold reduction assayed by cytometry. Thus, decreases in IgG expression observed by flow cytometry are concordant with the reduction in switch recombination in the ΔSμ B cells as determined by DC-PCR.
Figure 4.
DC-PCR demonstrates switch recombination at the DNA level is reduced in B cells from ΔSμ mice stimulated with LPS or LPS plus IL-4 (L+4). DNA template levels were adjusted by normalizing to the amount of AchR DC-PCR product and then twofold dilutions of adjusted levels of input or a plasmid standard were amplified for AchR, and for Sμ/Sγ1 and Sμ/Sγ2b recombination, with the incorporation of [α-32P]dCTP. The fold reductions relative to wild-type (WT) from two independent PCR reactions were: IgG1, 3.6 and 3.65; IgG2b, 9.7 and 8.5.
Switch Recombination Sites in IgG-Producing ΔSμ Hybridomas.
To characterize the nature of switch recombination events in ΔSμ mice, we produced IgG1-expressing hybridomas from animals immunized with Ars-KLH. DNA fragments containing recombination sites were PCR-amplified from hybridoma DNAs. Approximately 30% of the hybridomas yielded PCR fragments. Fragments from 10 hybridomas were sequenced to locate the switch joining sites. Sequenced fragments were compared with germline JH-Cμ sequences and germline Cγ1 flanking sequences to locate the sites of recombination.
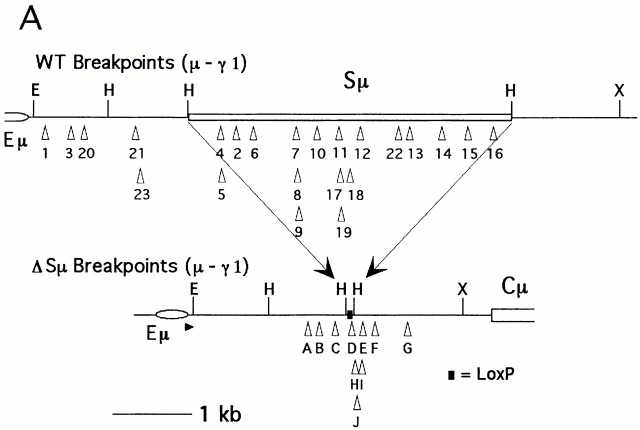
As shown in Fig. 5 A, the recombination sites in the ΔSμ JH-Cμ intron are found at a variety of locations; ∼2 kb separate the most 5′ and 3′ sites. In wild-type mice, most switch sites in IgG1-producing cells occur within Sμ; for those sites that are outside of Sμ, most occur in the 5′ flanking sequence with a much smaller percentage in the 3′ flanking sequence. In the ΔSμ mice, most μ switch sites in IgG1-producing cells are in the 3′ flanking sequence with a smaller proportion in the 5′ flanking sequence. Although our data set is limited, there does not appear to be any shared motif at the sites of recombination. Only one site is within a GAGCT sequence, but all sites are located <250 bp from the nearest GAGCT.
Figure 5.
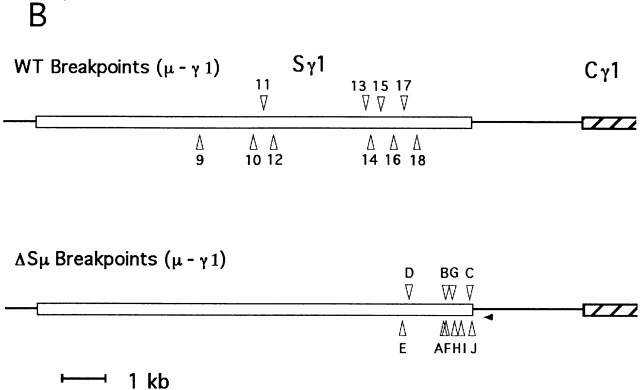
(A) ΔSμ and wild-type (WT) recombination junctions in the JH –Cμ intron. The wild-type junctions pictured are all from μ − γ1 recombinations and were derived from published sequences. Junctions nos. 1 and 2 are from reference 53, nos. 3 and 4 from reference 7, nos. 5–7 from reference 54, no. 8 from reference 21, nos. 9–19 from reference 23, nos. 20 and 21 from reference 55, no. 22 from reference 56, and no. 23 from reference 57. ΔSμ recombination junctions were generated by PCR from IgG1-producing hybridomas. Sites were from hybridomas 16B3 (A), 18A4 (B), 8B5 (C), 8A2 (D), 18B6 (E), 18B2 (F), 9A6 (G), 8C4 (H), 4D5 (I), and 16B5 (J). Only the relevant restriction sites are shown on the map. E, EcoRI; H, HindIII; X, XbaI. Location of the PCR primer used to generate ΔSμ junctions is represented by small arrowheads. (B) ΔSμ and wild-type (WT) switch junctions upstream of Cγ1. The wild-type junctions are all μ −γ1 recombinations and are derived from (reference 55). The wild-type junctions shown here are a subset of the group shown in A because we could not accurately place all the breakpoints within the γ1 tandem repeats. ΔSμ junctions were generated by PCR from IgG1-producing hybridomas; the small arrowheads represents the location of the PCR primer used.
The sites of switch recombination within the Sγ1 region of the mutant mice are shown in Fig. 5 B and these are compared with analogous sites in wild-type animals. The γ1 recombination sites observed for ΔSμ hybridomas are clearly clustered to the 3′ end of Sγ1 relative to the γ1 sites in wild-type mice. However, because the ΔSμ sites were from PCR amplifications, whereas the published wild-type sites were generally obtained by cloning genomic DNA fragments, we believe that this clustering may be due to smaller fragments being more readily amplified by PCR. Shorter PCR products will mostly exhibit recombination sites located at the 3′end of the long (10 kb) Sγ1 element. On the other hand, there is no indication that the use of PCR has affected our analysis of the μ switch recombination sites in ΔSμ JH-Cμ intron. PCR effects would result in μ switch sites clustered at the 5′ end of the JH-Cμ intron. Instead, the actual observed μ switch sites (Fig. 5 A) are located toward the 3′ end of this intron.
The DNA sequences surrounding the recombination sites in H chain genes that have undergone class switching frequently display single basepair mutations. These mutations have been suggested to reflect a role for error-prone DNA synthesis in the switching mechanism 6 32. Mutations are seen surrounding the switch sites in ΔSμ mice and, within the μ sequences, the frequency of mutation (1.8%) is similar to that previously reported for wild-type mice (1.4%). However, within the γ1 sequences flanking the switch junctions in ΔSμ mice there appears to be an elevated level of mutation (5.5%) relative to the levels found in analogous flanking S regions in wild-type mice (1.3%). This elevated level is mainly due to a rather large number of mutations found in 3 of the 11 cloned recombination sites that have been analyzed (Fig. 6).
Figure 6.
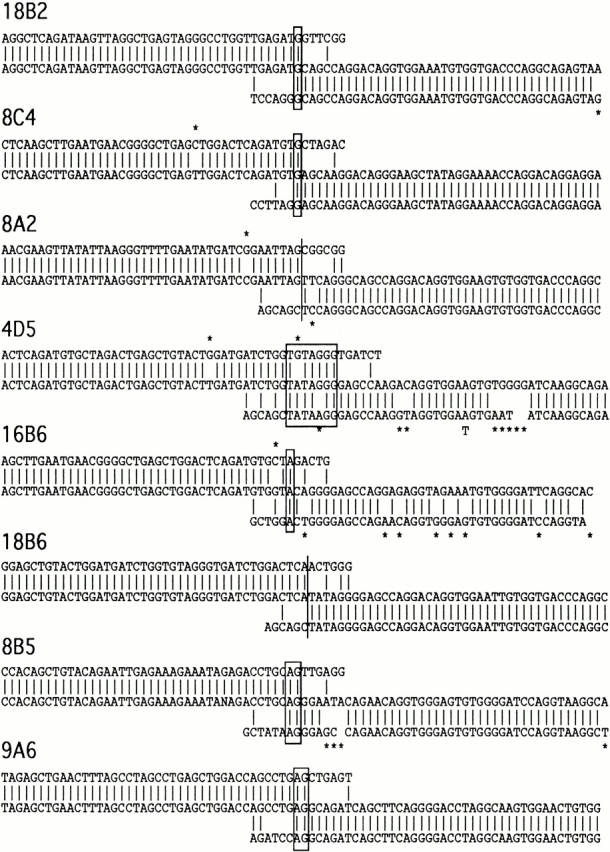
Sequence analysis of ΔSμ IgG1 hybridoma switch recombination junctions. Approximately 80 bp of each clone is shown, 40 bp upstream and 40 bp downstream of each breakpoint. The sequence on top is the corresponding germline μ sequence, and the sequence on the bottom is the germline γ1 sequence. A small vertical line is drawn between bases if they are identical. The switch junctions are shown as either a large vertical line if there are not any shared base pairs, or as a box if there are shared base pairs. Junctions are assigned according to the following criteria: if there is a base change close to the potential breakpoint it must be followed by at least one base that is the same as germline; if there are two or more base changes they must be followed by at least the same number of germline bases. Base changes are marked with an asterisk, insertions have no corresponding germline sequence, and deletions have the corresponding germline base placed below the germline sequence.
Hybridomas from ΔSμ Mice Frequently Have Switch Recombination at Only One μ Allele.
The panel of ΔSμ IgG1-producing hybridomas was analyzed by Southern blot hybridization to assess the recombination events that had occurred during class switching in immunized ΔSμ B cells. The hybridomas in this panel were isolated as clones from soft agar but were not re-cloned after isolation; thus, a fraction of the hybridomas might represent multiple clones. This fraction appears to be small because only one of the hybridomas displays more than two B cell–derived Igh alleles when the allele derived from the X63.Ag8 tumor fusion parent is discounted.
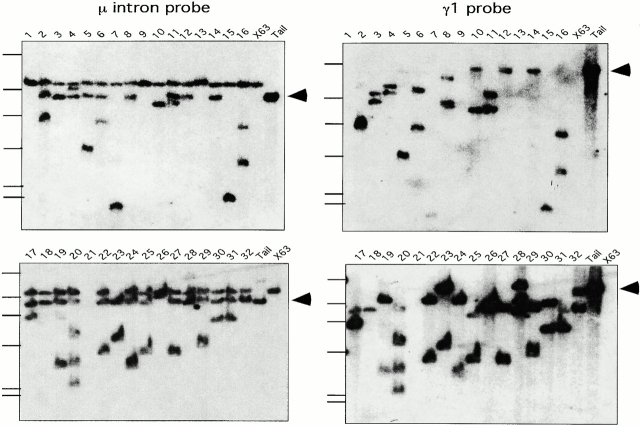
Hybridoma DNAs were analyzed both with 5′ Sμ probes (Fig. 7, left) and γ1 probes (Fig. 7 right). Consistent with the sequenced PCR clones described above, recombined DNA fragments that hybridize with both 5′ Sμ and Sγ1 probes are found in almost all of the analyzed hybridomas, including some of those that did not yield detectable fragments in PCR assays. The inability to obtain PCR products from some hybridomas is likely due to a long distance between the μ and γ1 primers used in the amplification. The large size of some recombined bands, together with the known size of the ΔSμ JH-Cμ intron, suggests that these bands reflect Sγ1 recombination sites located further 5′ in the Sγ1 element.
Figure 7.
Southern blots of ΔSμ IgG1 hybridomas. DNAs from 32 hybridomas were digested with EcoRI and hybridized. The blots on the left were hybridized with a 700-bp EcoRI-HindIII fragment located just 3′ of the Eμ enhancer (pJ14c; reference 53). On the right, the same blots were probed with a 10-kb EcoRI fragment containing the Sγ1 S region (pγ1EH10.0; reference 58). The germline bands are marked with an arrowhead and the sizes are ∼8.9 kb for the μ intron and 16 kb for the γ1 intron. Positions of Hind III fragments of λ phage DNA are indicated for each gel.
The most notable finding from the Southern blot analyses of ΔSμ IgG1-producing hybridomas is that most of the hybrids display germline JH-Cμ alleles (indicated by the arrowheads in Fig. 7). This result indicates that only one of the two JH-Cμ alleles in these B cells has undergone switch recombination. In contrast, three separate studies of wild-type mice showed that 80–100% of IgG hybridomas displayed switch recombinations at both μ alleles 33 34 35. Because the ΔSμ hybridomas produce IgG1, it appears that the nonproductive chromosome is the one that has not undergone switching within the μ region. On the other hand, most of the ΔSμ hybridomas have undergone recombination at both Sγ1 regions as indicated by the small number of germline Sγ1 bands among the hybridoma panel (Fig. 7). Therefore, the lack of switching in the ΔSμ JH-Cμ intron indicates a defect in the ability of the μ locus to participate in switch recombination rather than an overall defect in switch recombination of the nonproductive chromosome.
Discussion
Sμ Is Not Required for Class Switch Recombination.
The tandemly repeated S region sequences are the most unique sequence features found within the DNA regions known to be involved in class switch recombination. Since their discovery 36, the tandem repeats have been the focus of investigations into the targeting and mechanism of class switching. These repetitive sequences have been suggested to be the sites of DNA breakage for switch recombination 5 7, and transfection experiments using switch constructs have suggested that S regions may be sufficient to direct switch recombination 9 10 11 12 13 14 15 16. However, our analyses of the ΔSμ mice, in which all the tandem repeats in the JH-Cμ intron have been deleted, demonstrate that the Sμ element is not required for antibody class switching to occur.
Tandem Repeats Appear to be Important for Efficient Switching of Both Igh Alleles.
In normal mice, >90% of hybridomas that exhibit switching on a productive H chain allele also exhibit switch recombination on the nonproductive allele, and these are frequently to the same isotype 34 37 38. There is no evidence to suggest that the switch mechanism can distinguish between the productive and nonproductive alleles in a B cell; therefore, this efficient recombination of both alleles has probably been evolutionarily selected to ensure that the productive allele undergoes class switching when the cell is appropriately stimulated.
In the ΔSμ knockout mouse, however, a large percentage of the hybridomas that have undergone switching exhibit recombination only at the functional μ allele. In these cells, the nonfunctional μ allele does not appear to have undergone any recombination event. This result suggests that during immune responses in ΔSμ mice some cells that have been stimulated to switch might recombine only a nonfunctional μ allele, and some may fail to recombine either μ allele. This notion would appear to be consistent with the observed increased IgM production in the ΔSμ mice. Thus, the efficiency of switching that is provided by the presence of the tandem repeats appears to increase the probability that B cells participating in an immune response undergo switching.
Potential Roles for Sμ in Switch Recombination.
Class switch recombination occurs by a type of nonhomologous end joining (NHEJ) as indicated by the involvement of DNA protein kinase and Ku in the process 39 40 41, and by the absence of any extensive homology surrounding the joining sites 7. Switch recombination can be divided into three major steps: targeting, initiation (cleavage), and resolution (rejoining; reference 42). The Sμ tandem repeats could act at one or more of these steps. Targeting of isotype switching appears to be relatively intact in the ΔSμ mice. The expected isotypes are produced upon in vitro stimulation with cytokines, and the locations of the switch junctions in both the JH-Cμ intron and in Sγ1 appear consistent with normal targeting of switch recombination. Thus, it seems more likely that Sμ has a role in either switch initiation or resolution.
Our results provide some data suggesting that Sμ tandem repeats could be involved during the resolution phase of switch recombination. ΔSμ mice and mismatch repair (MMR)-deficient mice exhibit switching defects that have similar isotype profiles 26 28. It has been proposed that the MMR protein Msh2 is involved in end processing that occurs after formation of switch double strand breaks but before ligation 26 28. Therefore, the similar defects in isotype expression could suggest that Msh2 and Sμ both affect switch resolution. However, Msh2 and Sμ do not appear to impinge on switch recombination in the identical manner because ΔSμ mice do not exhibit the focus of μ switch sites within GAGCT sequences that has been reported in Msh2 knockout animals 28. A defect in switch resolution might also result in increased mutations around switch sites; our limited data from ΔSμ switch junctions show potential increases in mutation frequency that would be consistent with this suggestion.
The Sμ element could certainly also be important in the initiation of switch recombination. The observation that many ΔSμ B cells have Igh alleles that are germline in the JH-Cμ intron but rearranged at γ1 suggests that initiation is intact at Sγ1, whereas it is decreased at μ. One proposed role for S regions in the initiation of class switching is as the site of double strand breaks; this role is supported by the detection of strand breaks within Sγ3 in B cells undergoing switching 5. Although no site for break induction has yet been identified in Sμ, it is likely that some sites are located within Sμ and it is possible that the loss of these sites is responsible for the decrease in switching. Furthermore, NHEJ is important in the resolution step of switch recombination and is generally considered to be sequence independent. Therefore, the presence or absence of specific Sμ sequences would seem less likely to affect the efficiency of NHEJ during resolution, suggesting that Sμ might be more likely to be important for switch initiation. However, because little is known about the nature of the DNA ends generated during switch initiation, or about possible processing of these ends before DNA joining, it may well be that Sμ sequences play a role at both the initiation and the resolution of switch breakpoints.
Locations of ΔSμ Switch Sites Suggest that Sequences 3′ of Sμ Might Be Important for Switch Recombination.
Because Sμ is not required for isotype switching, it appears that sequences located outside of Sμ must be capable of directing the switch recombination mechanism. RNA transcription and splicing have been shown to be important in S region function 43 44 45 46, although they cannot be sufficient for specific targeting of the switch mechanism and must be coupled with some additional sequence or structural information. Perhaps targeting is focused by sequences located between Eμ and Sμ or between Sμ and Cμ; both of these regions have not yet been tested by deletional mutagenesis.
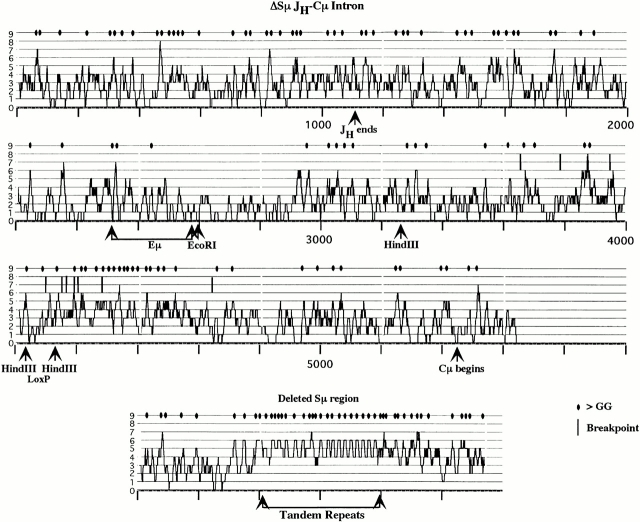
Sμ, as well as other S regions, is made up of tandemly repeated sequences that exhibit relatively high levels of G nucleotide residues and GGG or GGGG sequences on the nontranscribed DNA strand. Fig. 8 diagrams the location of G-rich segments and GGG sequences for both the wild-type Sμ region and the ΔSμ JH-Cμ intron. A segment of ∼200–300 bp located 3′ of the loxP site in the ΔSμ intron exhibits G richness, clusters of GGG sequences, and some isolated GAGCT sequences (Fig. 1 and Fig. 8). This 3′ element does not exhibit the repetitive pattern of G-richness that is apparent in the 3,000-bp wild-type Sμ, part of which is shown at the bottom of Fig. 8. This 200–300 bp 3′ region has not been considered part of Sμ because there are no tandem repeats present and the frequency of switch recombination sites in wild-type mice within this element is lower than within Sμ. Indeed, the frequency of recombination sites within the 3′ element in wild-type mice is lower than the frequencies within other JH-Cμ regions that exhibit no GAGCT sequences (see wild-type breakpoints summarized in Fig. 5 A). The recombinational activity of this 3′ region has never been tested in switch substrate experiments. Nevertheless, the 200–300-bp element is found centrally situated in the region where ΔSμ switch sites occur. Perhaps this region is important in the switch recombination process.
Figure 8.
Analysis of the frequency of G nucleotides in the ΔSμ JH-Cμ intron. The top three graphs wrap around to form a continuous sequence from JH to the beginning of Cμ in the ΔSμ mouse. The bottom graph represents a portion of the sequence that has been removed in the ΔSμ mouse; however, ∼2.5 kb of tandem repeats is not included in this graph because the sequence is not known. The analysis was done by counting the number of G residues for nucleotide nos. 1–10 (numbers not shown), and plotting the value on the graph. This was repeated for nucleotide nos. 2–11, nos. 3–12, etc. Essentially, a peak in the graph indicates a stretch of DNA with numerous G residues. A random sequence would have 2.5 G residues per 10 nucleotides. An oval on the top line of the graph represents runs of G that are three nucleotides or greater. The positions of the recombination sites in the ΔSμ IgG1 hybridomas are indicated by a horizontal line between the 7th and 8th line of the graph. Relevant restriction sites and features of the intron are indicated.
It has been hypothesized that S regions might function in class switching by forming a special secondary structure that starts the recombinational process 5 47 48 49 50. The proposed mechanisms all involve single-stranded S region DNA. Transcription of tandemly repeated S regions has been shown to lead to stable RNA–DNA complexes and single-stranded S region sequences which could play an important role in class switch recombination 17 18 19 20.
It is not known what causes RNA–DNA complexes to form in transcribed S regions, but extensive polypurine or GGG stretches have been suggested to play a role. In ΔSμ mice, the JH-Cμ intron exhibits considerably fewer of these types of sequence elements than do the Sμ tandem repeats. However, it is possible that the complexes can still form in the ΔSμ JH-Cμ intron, perhaps using the shorter stretches of GGG clusters remaining. The reduction in ΔSμ switching might be due to a decrease in the efficiency of formation or the stability of the complex in vivo.
RNA–DNA complexes are an intriguing mechanism for targeting switch recombination; however, there is no direct evidence that demonstrates a role for these complexes in class switch recombination. In fact, switching in some species appears to involve sequences that are not G rich or even purine rich 50 51. Although switching processes could differ in different species, it is also possible that RNA–DNA complexes are not actually needed for switching or that the sequence features that promote complex formation are not yet understood. The location of switch sites in ΔSμ mice do, however, suggest that the 200–300-bp element in the ΔSμ JH-Cμ intron could contain the minimal sequences required for switching. The long Sμ tandem repeat element found in a variety of animal species appears to be present for optimal efficiency of the switch recombination process.
Acknowledgments
We thank Naomi Rosenberg for helpful comments on the manuscript and Jørgen Selsing for help with sequence analyses.
Supported by National Institutes of Health grants AI23283 and AI42108 (J. Stavnezer), AI24465 and AI42569 (E. Selsing), and AI07077 (T.M. Luby).
Footnotes
Abbreviations used in this paper: Ach, acetylcholine; Ars, p-phenylarsonate; DC, digestion circularization; NHEJ, nonhomologous end joining; S, switch.
References
- Gritzmacher C.A. Molecular aspects of heavy chain class switching. Crit. Rev. Immunol. 1989;9:173–200. [PubMed] [Google Scholar]
- Stavnezer, J. 1996. Antibody class switch. In Advances in Immunology. Vol. 61. Academic Press, Inc., San Diego, CA. 79–146. [DOI] [PubMed]
- Stavnezer J. Molecular processes that regulate class switching. Curr. Top. Microbiol. Immunol. 2000;245:127–168. doi: 10.1007/978-3-642-59641-4_6. [DOI] [PubMed] [Google Scholar]
- Sakano H., Huppi K., Heinrich G., Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- Wuerffel R.A., Du J., Thompson R.J., Kenter A.L. Ig Sγ3 DNA-specific double strand breaks are induced in mitogen-activated B cells and are implicated in switch recombination. J. Immunol. 1997;159:4139–4144. [PubMed] [Google Scholar]
- Dunnick W., Wilson M., Stavnezer J. Mutations, duplication and deletion of recombined switch regions suggest a role for DNA replication in the immunoglobulin heavy chain switch. Mol. Cell. Biol. 1989;9:1850–1856. doi: 10.1128/mcb.9.5.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W., Hertz G.Z., Scappino L., Gritzmacher C. DNA sequences at immunoglobulin switch region recombination sites. Nucleic Acids Res. 1993;21:365–372. doi: 10.1093/nar/21.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J., Ye Z., Shanmugam A., Kenter A. Analysis of immunoglobulin Sγ3 recombination breakpoints by PCRimplications for the mechanism of isotype switching. Nucleic Acids Res. 1997;25:3066–3073. doi: 10.1093/nar/25.15.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels G.A., Lieber M.R. Starnd specificity in the transcriptional targeting of recombination at immunoglobulin switch sequences. Proc. Natl. Acad. Sci. USA. 1995;92:5625–5629. doi: 10.1073/pnas.92.12.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballantyne J., Henry D., Muller J., Briere F., Snapper C., Kehry M., Marcu K. Efficient Recombination of a switch substrate retrovector in CD40-activated B lymphocytesimplications for the control of CH gene switch recombination. J. Immunol. 1998;161:1336–1347. [PubMed] [Google Scholar]
- Leung H., Maizels N. Transcriptional regulatory elements stimulate recombination in extrachromosomal substrates carrying immunoglobulin switch-region sequences. Proc. Natl. Acad. Sci. USA. 1992;89:4154–4158. doi: 10.1073/pnas.89.9.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung H., Maizels N. Regulation and targeting of recombination in extrachromosomal substrates carrying immunoglobulin switch region sequences. Mol. Cell. Biol. 1994;14:1450–1458. doi: 10.1128/mcb.14.2.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita K., Tashiro J., Tomita S., Lee C.-G., Honjo T. Target specificity of immunoglobulin class switch recombination is not determined by the nucleotide sequences of S regions. Immunity. 1998;9:849–858. doi: 10.1016/s1074-7613(00)80650-0. [DOI] [PubMed] [Google Scholar]
- Petry K., Siebenkotten G., Christine R., Hein K., Radbruch A. An extrachromosomal switch recombination substrate reveals kinetics and substrate requirements of switch recombination in primary murine B cells. Int. Immunol. 1999;11:753–763. doi: 10.1093/intimm/11.5.753. [DOI] [PubMed] [Google Scholar]
- Stavnezer J., Bradley S.P., Rousseau N., Pearson T., Shanmugam A., Waite D.J., Rogers P.R., Kenter A.L. Switch recombination in a transfected plasmid occurs preferentially in a B cell line that undergoes switch recombination of its chromosomal Ig heavy chain genes. J. Immunol. 1999;163:2028–2040. [PubMed] [Google Scholar]
- Shanmugam A., Shi M.J., Yauch L., Stavnezer J., Kenter A.L. Evidence for class-specific factors in immunoglobulin isotype switching. J. Exp. Med. 2000;191:1365–1380. doi: 10.1084/jem.191.8.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaban M.E., Griffin J.A. Induction of RNA-stabilized DNA conformers by transcription of an immunoglobulin switch region. Nature. 1990;348:342–344. doi: 10.1038/348342a0. [DOI] [PubMed] [Google Scholar]
- Reaban M.E., Lebowitz J., Griffin J.A. Transcription induces the formation of a stable RNA.DNA hybrid in the immunoglobulin alpha switch region. J. Biol. Chem. 1994;269:21850–21857. [PubMed] [Google Scholar]
- Daniels G.A., Lieber M.R. RNA:DNA complex formation upon transcription of immunoglobulin switch regionsimplications for the mechanism and regulation of class switch recombination. Nucleic Acids Res. 1995;23:5006–5011. doi: 10.1093/nar/23.24.5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M., Alt F.W. Transcription-induced cleavage of immunoglobulin switch regions by nucleotide excision repair nucleases in vitro. J. Biol. Chem. 2000;275:24163–24172. doi: 10.1074/jbc.M003343200. [DOI] [PubMed] [Google Scholar]
- Matsuoka M., Yoshida K., Maeda T., Usuda S., Sakano H. Switch circular DNA formed in cytokine-treated mouse splenocytesevidence for intramolecular DNA deletion in immunoglobulin class switching. Cell. 1990;62:135–142. doi: 10.1016/0092-8674(90)90247-c. [DOI] [PubMed] [Google Scholar]
- von Schwedler U., Jack H.-M., Wabl M. Circular DNA is a product of immunoglobulin class switch rearrangement. Nature. 1990;345:452–456. doi: 10.1038/345452a0. [DOI] [PubMed] [Google Scholar]
- Iwasato T., Shimizu A., Honjo T., Yamagishi H. Circular DNA is excised by immunoglobulin class switch recombination. Cell. 1990;62:143–149. doi: 10.1016/0092-8674(90)90248-d. [DOI] [PubMed] [Google Scholar]
- Lee C., Kondo S., Honjo T. Frequent but biased class switch recombination in the Sμ flanking regions. Curr. Biol. 1998;8:227–230. doi: 10.1016/s0960-9822(98)70087-9. [DOI] [PubMed] [Google Scholar]
- Chu C.C., Paul W.E., Max E.E. Quantitation of immunoglobulin μ-γ1 heavy chain switch region recombination by a digestion-circularization polymerase chain reaction method. Proc. Natl. Acad. Sci. USA. 1992;89:6978–6982. doi: 10.1073/pnas.89.15.6978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader C., Edelmann W., Kucherlapati R., Stavnezer J. Reduced isotype switching in splenic B cells from mice deficient in mismatch repair enzymes. J. Exp. Med. 1999;190:323–330. doi: 10.1084/jem.190.3.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdardottir D., Sohn J., Kass J., Selsing E. Regulatory regions 3′ of the immunoglobulin heavy chain intronic enhancer differentially affect expression of a heavy chain transgene in resting and activated B cells. J. Immunol. 1995;154:2217–2225. [PubMed] [Google Scholar]
- Ehrenstein M.R., Neuberger M.S. Deficiency in Msh2 affects the efficiency and local sequence specificity of immunoglobulin class-switch recombinationparallels with somatic hypermutation. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:3484–3490. doi: 10.1093/emboj/18.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockerill P.N., Yuen M.-H., Garrard W.T. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J. Biol. Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- Gram H., Zenke G., Geisse S., Kleuser B., Bürki K. High-level expression of a human immunoglobulin γ1 transgene depends on switch region sequences. Eur. J. Immunol. 1992;22:1185–1191. doi: 10.1002/eji.1830220512. [DOI] [PubMed] [Google Scholar]
- Ballantyne J., Henry D.L., Marcu K.B. Antibody class switch recombinase activity is B cell stage specific and functions stochastically in the absence of ‘targeted accessibility’ control Int Immunol 9 1997. 963 974[published erratum at 9:1773] [DOI] [PubMed] [Google Scholar]
- Dunnick W., Stavnezer J. Copy choice mechanism of immunoglobulin heavy-chain switch recombination. Mol. Cell. Biol. 1990;10:397–400. doi: 10.1128/mcb.10.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottaro A., Young F., Chen J., Serwe M., Sablitzky F., Alt F.W. Deletion of the IgH intronic enhancer and associated matrix-attachment regions decreases, but does not abolish, class switching at the mu locus. Int. Immunol. 1998;10:799–806. doi: 10.1093/intimm/10.6.799. [DOI] [PubMed] [Google Scholar]
- Hummel M., Berry J.K., Dunnick W. Switch region content of hybridomasthe two spleen cell Igh loci tend to rearrange to the same isotype. J. Immunol. 1987;138:3539–3548. [PubMed] [Google Scholar]
- Winter E., Krawinkel U., Radbruch A. Directed Ig class switch recombination in activated murine B cells. EMBO (Eur. Mol. Biol. Organ.) J. 1987;6:1663–1671. doi: 10.1002/j.1460-2075.1987.tb02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M.M., Kim S.K., Hood L.E. DNA sequences mediating class switching in α-immunoglobulins. Science. 1980;209:1360–1365. doi: 10.1126/science.6774415. [DOI] [PubMed] [Google Scholar]
- Radbruch A., Muller W., Rajewsky K. Class switch recombination is IgG1 specific on active and inactive IgH loci of IgG1-secreting B-cell blasts. Proc. Natl. Acad. Sci. USA. 1986;83:3954–3957. doi: 10.1073/pnas.83.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenkotten G., Esser C., Wabl M., Radbruch A. The murine IgG1/IgE class switch program. Eur. J. Immunol. 1992;22:1827–1834. doi: 10.1002/eji.1830220723. [DOI] [PubMed] [Google Scholar]
- Casellas R., Nussenzweig A., Wuerffel R., Pelanda R., Reichlin A., Suh H., Qin X.F., Besmer E., Kenter A., Rajewsky K., Nussenzweig M.C. Ku80 is required for immunoglobulin isotype switching. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manis J.P., Gu Y., Lansford R., Sonoda E., Ferrini R., Davidson L., Rajewsky K., Alt F.W. Ku70 is required for late B cell development and immunoglobulin heavy chain class switching. J. Exp. Med. 1998;187:2081–2089. doi: 10.1084/jem.187.12.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A., Melchers F., Andersson J. The SCID but not the RAG-2 gene product is required for S mu-S epsilon heavy chain class switching. Immunity. 1996;5:319–330. doi: 10.1016/s1074-7613(00)80258-7. [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Lee C.-G., Tashiro J., Muramatsu M., Chen X.-C., Yoshikawa K., Honjo T. Molecular mechanism of immunoglobulin class switch recombination. Cold Spring Harbor Symp. Quant. Biol. 1999;64:217–226. doi: 10.1101/sqb.1999.64.217. [DOI] [PubMed] [Google Scholar]
- Stavnezer-Nordgren J., Sirlin S. Specificity of immunoglobulin heavy chain switch correlates with activity of germline heavy chain genes prior to switching. EMBO (Eur. Mol. Biol. Organ.) J. 1986;5:95–102. doi: 10.1002/j.1460-2075.1986.tb04182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M., Jung S., Radbruch A. Switch transcripts in immunoglobulin class switching. Science. 1995;267:1825–1828. doi: 10.1126/science.7892607. [DOI] [PubMed] [Google Scholar]
- Hein K., Lorenz G.O., Siebenkotten G., Petry K., Christine R., Radbruch A. Processing of switch transcripts is required for targeting of antibody class switch recombination. J. Exp. Med. 1998;188:2369–2374. doi: 10.1084/jem.188.12.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottaro A., Lansford R., Xu L., Zhang J., Rothman P., Alt F.W. S region transcription per se promotes basal IgE class switch recombination but additional factors regulate the efficiency of the process. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:665–674. doi: 10.1002/j.1460-2075.1994.tb06305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr J., Pennell N.M., Shulman M.J. Analysis of a hot spot for DNA insertion suggests a mechanism for Ig switch recombination. J. Immunol. 1996;157:3430–3435. [PubMed] [Google Scholar]
- Dempsey L.A., Sun H., Hanakahi L.A., Maizels N. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D, A role for G-G pairing in immunoglobulin switch recombination. J. Biol. Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Honjo T. Unique and unprecedented recombination mechanisms in class switching. Curr. Opin. Immunol. 2000;12:195–198. doi: 10.1016/s0952-7915(99)00072-2. [DOI] [PubMed] [Google Scholar]
- Mussmann R., Courtet M., Schwager J., Du Pasquier L. Microsites for immunoglobulin switch recombination breakpoints from Xenopus to mammals. Eur. J. Immunol. 1997;27:2610–2619. doi: 10.1002/eji.1830271021. [DOI] [PubMed] [Google Scholar]
- Kitao H., Arakawa H., Kuma K., Yamagishi H., Nakamura N., Furusawa S., Matsuda H., Yasuda M., Ekino S., Shimizu A. Class switch recombination of the chicken IgH chain genesimplications for the primordial switch region repeats. Int. Immunol. 2000;12:959–968. doi: 10.1093/intimm/12.7.959. [DOI] [PubMed] [Google Scholar]
- Marcu K.B., Banerji J., Penncavage N.A., Lang R., Arnheim N. 5′ flanking region of immunoglobulin heavy chain constant region genes displays length heterogeneity in germlines of inbred mouse strains. Cell. 1980;22:187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- Katzenberg D.R., Birshtein B.K. Sites of switch recombination in IgG2b- and IgG2a-producing hybridomas. J. Immunol. 1988;140:3219–3227. [PubMed] [Google Scholar]
- Petrini J., Shell B., Hummel M., Dunnick W. The immunoglobulin heavy chain switchstructural features of gamma 1 recombinant switch regions. J. Immunol. 1987;138:1940–1946. [PubMed] [Google Scholar]
- Iwasato T., Arakawa H., Shimizu A., Honjo T., Yamagishi H. Biased distribution of recombination sites within S regions upon immunoglobulin class switch recombination induced by transforming growth factor beta and lipopolysaccharide. J. Exp. Med. 1992;175:1539–1546. doi: 10.1084/jem.175.6.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick W., Rabbitts T.H., Milstein C. An immunoglobulin deletion mutant with implications for the heavy-chain switch and RNA splicing. Nature. 1980;286:669–675. doi: 10.1038/286669a0. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Matsuoka M., Usuda S., Mori A., Ishizaka K., Sakano H. Immunoglobulin switch circular DNA in the mouse infected with Nippostrongylus brasiliensisevidence for successive class switching from mu to epsilon via gamma 1. Proc. Natl. Acad. Sci. USA. 1990;87:7829–7833. doi: 10.1073/pnas.87.20.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowatt M., Dunnick W. DNA sequence of murine γ1 switch segment reveals novel structural elements. J. Immunol. 1986;136:2674–2683. [PubMed] [Google Scholar]