Abstract
In Saccharomyces cerevisiae most chitin is synthesized by Chs3p, which deposits chitin in the lateral cell wall and in the bud-neck region during cell division. We have recently found that addition of glucosamine (GlcN) to the growth medium leads to a three- to fourfold increase in cell wall chitin levels. We compared this result to the increases in cellular chitin levels associated with cell wall stress and with treatment of yeast with mating pheromone. Since all three phenomena lead to increases in precursors of chitin, we hypothesized that chitin synthesis is at least in part directly regulated by the size of this pool. This hypothesis was strengthened by our finding that addition of GlcN to the growth medium causes a rapid increase in chitin synthesis without any pronounced change in the expression of more than 6,000 genes monitored with Affymetrix gene expression chips. In other studies we found that the specific activity of Chs3p is higher in the total membrane fractions from cells grown in GlcN and from mutants with weakened cell walls. Sucrose gradient analysis shows that Chs3p is present in an inactive form in what may be Golgi compartments but as an active enzyme in other intracellular membrane-bound vesicles, as well as in the plasma membrane. We conclude that Chs3p-dependent chitin synthesis in S. cerevisiae is regulated both by the levels of intermediates of the UDP-GlcNAc biosynthetic pathway and by an increase in the activity of the enzyme in the plasma membrane.
Chitin is a linear polysaccharide composed of β(1→4)-linked N-acetylglucosamine (GlcNAc) residues. In the yeast Saccharomyces cerevisiae, chitin is an important component of the cell wall and septum. Three chitin synthases, Chs1p, Chs2p, and Chs3p, have the same polymerizing activity but deposit chitin at different times and at different locations during the cell cycle. Chs1p is thought to be a repair enzyme that adds chitin to the birth scar on the daughter cell at the end of cytokinesis. Chs2p is responsible for synthesis of chitin in the primary septum. Chs3p deposits chitin as a ring at the base of an emerging bud and is retained by the mother cell (bud scar) after cell division. Chs3p-generated chitin is also deposited in the lateral wall (reviewed in references 7 and 21). When the cell wall is weakened by mutations (“cell wall stress”) (41, 42) or otherwise modified, as in treatment with mating pheromones (39, 46), chitin is deposited in the lateral wall as a reinforcing polymer.
Chs3p synthesizes about 90% of the chitin in S. cerevisiae. Levels of Chs3p are virtually unaltered during the yeast life cycle (9). However, temporal changes in its subcellular location result from being secreted to and endocytosed from the plasma membrane. Chs3p transits through the endoplasmic reticulum/Golgi secretory pathway to the plasma membrane early in the formation of a daughter cell. Once the daughter cell is full size, Chs3p is retrieved by endocytosis into “chitosomes,” intracellular vesicles related to the trans-Golgi network and early endosomes (22, 59). Several proteins—Chs4p (Skt5p), Chs5p, Chs6p, and Chs7p—regulate Chs3p enzymatic activity and trafficking through the endoplasmic reticulum to the plasma membrane (reviewed in references 7 and 44). Loss of any of these proteins results in a reduction of the chitin concentrationin the cell wall to levels comparable to those observed when Chs3p itself is absent. Chs4p serves a dual role of binding with Chs3p to form an active complex and localizing the active complex to the bud-neck region by also binding to the septin ring through Bni4p (13). The resulting synthesis of a ring of chitin reinforces the bud-neck region during cell division. For vegetatively grown cells, the amount of Chs4p is limiting and therefore impacts chitin synthesis by its availability to complex with Chs3p (38). Chs7p is involved specifically in the exit of Chs3p from the endoplasmic reticulum (51). Chs5p and Chs6p have been identified as components required for transport of secretory and/or endocytic vesicles to the plasma membrane (45, 52, 60). The clathrin AP-1 complex has recently been shown to also be important in the process of retrieval of Chs3p by endocytosis and its recycling into the secretory pathway (52).
Information concerning the molecular activation and trafficking of Chs3p to the sites where it produces chitin is emerging, but details remain to be elucidated. Data from our laboratory and other groups show that chitin levels in S. cerevisiae increase in response to (i) treatment of mating-type a cells with α-factor, a mating pheromone (39, 46; this study), probably as a result of modifications of cell wall architecture in preparation for mating; (ii) mutations resulting in impairment of cell wall integrity, e.g., gas1, fks1, kre6, mnn9, and knr4 mutations (16, 28, 40, 41, 42; this study); or (iii) addition of glucosamine (GlcN) to the growth medium, probably as a result of an increased intracellular pool of metabolites (2; this study).
Chitin passes through the plasma membrane to the extracellular cell wall by the polymerizing activities of chitin synthases with UDP-GlcNAc as a substrate. Biosynthesis of UDP-GlcNAc from glucose, however, takes place in the cytosol. Fructose-6-phosphate is converted to GlcN-6-phosphate (GlcN-6-P) by GlcN-6-P synthase (encoded by GFA1), which is then acetylated by GlcN-6-P acetyltransferase (encoded by GNA1) to GlcNAc-6-P, followed by the conversion of the latter to GlcNAc-1-P, a reaction catalyzed by acetylglucosamine phosphomutase (encoded by AGM1 [PCM1]). The synthesis of UDP-GlcNAc from GlcNAc-1-P and UTP is catalyzed by UDP-GlcNAc pyrophosphorylase (encoded by UAP1 [QRI1]). Where chitin levels have been shown to increase following α-factor treatment, transcriptional profiling has revealed that GFA1 and AGM1 are up-regulated; GNA1 and UAP1 transcript levels remain unchanged. All four genes encoding enzymes involved in UDP-GlcNAc biosynthesis are up-regulated to various degrees during meiosis and sporulation, when new chitin is synthesized for the spore wall (YPD Database [http:www.incyte.com/control/researchproducts/insilico/proteome];KEGG Metabolic Pathways [www.genome.ad.jp/kegg/pathway/map/map00530.html]). It is now recognized that this pathway is highly regulated in yeast and in higher eukaryotes, since UDP-GlcNAc serves as a donor nucleotide sugar for at least four groups of compounds: N-linked glycans, glycosylphosphatidylinositol (GPI)-anchored proteins, chitin, and glycolipids. Treatment of yeast cells with α-factor causes a three- to fivefold increase in the chitin level, associated with an increase in the level of the UDP-GlcNAc precursor pool (39, 46). Gfa1p, the first enzyme in the pathway, is a key step in UDP-GlcNAc biosynthesis; it is regulated at the transcriptional and posttranscriptional levels. Its activity increases in the yeast Candida albicans during hyphal growth (36) and in S. cerevisiae during mating, which correlates with an increase in chitin formation (54). The enzyme is inhibited by UDP-GlcNAc in C. albicans (36), Drosophila melanogaster (20), and bacteria (26). Gfa1p activity is regulated by a protein kinase(s); the protein kinase A-dependent phosphorylated form of Gfa1p appears to have a higher activity than the unphosphorylated protein (20, 57). Protein phosphatase I, encoded in Saccharomyces by GLC7, is a strong repressor of GFA1 transcription. When Glc7p activity is blocked, GFA1 transcription increases (57).
In this paper we report our recent findings on the factors that contribute to the regulation of chitin synthesis. We studied a number of single and double mutants, which elevate or decrease chitin levels, and examined the effect on chitin levels of addition of GlcN or α-factor to the growth medium. We show here that there is a direct correlation between Gfa1p activity, the pool of metabolic intermediates, and chitin synthesis. Finally, since the increase in chitin levels associated with treatment of wild-type cells with GlcN is similar to the increase in chitin levels associated with the cell wall stress response, we investigated the relevant whole-genome transcription responses.
MATERIALS AND METHODS
Yeast strains used in these studies.
Strains of S. cerevisiae either were made by the Saccharomyces Genome Deletion Project (55; www-deletion.stanford.edu/cgi-bin/deletion/search3.pl) and are available from Research Genetics, a division of Invitrogen (San Diego, Calif.), or were from our stock collection as described in Table 1. Mating, sporulation, and tetrad dissection were carried out by standard methods. Due to the poor sporulation of diploids derived from the Research Genetics strains, conditions were modified as described by L. Riles and M. Curtiss in “Sporulation Method for BY4743” (www-sequence.stanford.edu/group/yeast_deletion_project/spo.html). Media used were YPD medium (1% Bacto Yeast Extract, 2% Bacto Peptone, 2% glucose) and SD minimal medium (0.7% Bacto Yeast Nitrogen Base without amino acids, 0.2% SCM supplement [Bufferad, Inc., Lake Bluff, Ill.], 2% glucose) with or without 1.5% agar.
TABLE 1.
Yeast strains
| Strain | Genotype | Reference or source |
|---|---|---|
| PRY242 | MATaura3-552 lys2-801 his3Δ200 trp1Δ1 leu2-3.112 | 36a |
| PRY488 | MATaura3-552 lys2-801 his3Δ200 trp1Δ1 leu2-3,112 cts1::TRP1 | 36a |
| PRY88.1D | MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 leu2Δ1 | 40 |
| PRY533 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 leu2Δ1 chs3::LEU2 | 40 |
| PRY408 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 leu2Δ1 chs6::TRP1 | 40 |
| OCY6 | MATaura3-52 lys2-801 ade2-101 his3Δ200 trp1Δ1 leu2Δ1 kre5::HIS3 | 8a |
| BY4741 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| BY4742 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| BY4743 | MATa/α (BY4741/BY4742) | Research Genetics |
| 2020 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chs1::kanMX4 | Research Genetics |
| 3175 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chs2::kanMX4 | This work |
| 13175 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chs2::kanMX4 | This work |
| 3160 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chs3::kanMX4 | Research Genetics |
| 13160 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chs3::kanMX4 | Research Genetics |
| 3087 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chs4::kanMX4 | Research Genetics |
| 13087 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chs4::kanMX4 | Research Genetics |
| 5239 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chs5::kanMX4 | Research Genetics |
| 15239 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chs5::kanMX4 | Research Genetics |
| 1324 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chs6::kanMX4 | Research Genetics |
| 11324 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chs6::kanMX4 | Research Genetics |
| 2835 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chs7::kanMX4 | Research Genetics |
| 12835 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chs7::kanMX4 | Research Genetics |
| 5251 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 fks1::kanMX4 | Research Genetics |
| 15251 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 fks1::kanMX4 | Research Genetics |
| 897 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gas1::kanMX4 | Research Genetics |
| 10897 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 gas1::kanMX4 | Research Genetics |
| 5574 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kre6::kanMX4 | Research Genetics |
| 15574 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 kre6::kanMX4 | Research Genetics |
| 2778 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 mnn9::kanMX4 | Research Genetics |
| 277 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 anp1::kanMX4 | Research Genetics |
| 7116 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 van1::kanMX4 | Research Genetics |
| 4406 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 och1::kanMX4 | Research Genetics |
| 14406 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 och1::kanMX4 | Research Genetics |
| 5882 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 knr4::kanMX4 | Research Genetics |
| 15882 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 kur4::kanMX4 | Research Genetics |
| 14317 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 kre2::kanMX4 | Research Genetics |
| 3792 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pmt1::kanMX4 | Research Genetics |
| 13792 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pmt1::kanMX4 | Research Genetics |
| 385 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 pmt2::kanMX4 | Research Genetics |
| 10385 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 pmt2::kanMX4 | Research Genetics |
| 1974 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 kex2::kanMX4 | Research Genetics |
| 4572 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 chc1::kanMX4 | Research Genetics |
| 14572 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 chc1::kanMX4 | Research Genetics |
| 14797 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 clc1::kanMX4 | Research Genetics |
| 11969 | MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 end4::kanMX4 | Research Genetics |
| 24954 | MATa/α GFA1/gfa1::kanMX4 (BY4743) | Research Genetics |
| 174.1C | MATahis3Δ1 leu2Δ0 ura3Δ0 gfa1::kanMX4 | This work |
| 174.1D | MATahis3Δ1 leu2Δ0 ura3Δ0 gfa1::kanMX4 | This work |
| 184.4A | MATα his3Δ1 leu2Δ0 lys2Δ0 met15Δ ura3Δ0 gas1::kanMX4 chs5::kanMX4 | This work |
| 184.4B | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 gas1::kanMX4 chs5::kanMX4 | This work |
| 183.1C | MATα his3Δ1 leu2Δ0 ura3Δ0 gas1::kanMX4 chs6::kanMX4 | This work |
| 183.3C | MATahis3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 gas1::kanMX4 chs6::kanMX4 | This work |
| 175.2C | MATα his3Δ1 leu2Δ0 lys2Δ0 met15Δ ura3Δ0 fks1::kanMX4 chs6::kanMX4 | This work |
| 177.1D | MATα his3Δ1 leu2Δ0 ura3Δ0 fks1::kanMX4 chs6::kanMX4 | This work |
α-Factor induction.
MATa haploid strains were shaken overnight in YPD medium at 30°C, diluted 1:100 into fresh YPD medium (optical density at 600 nm [OD600], 0.1), and grown 4 h to an OD of 0.4 to 0.5, at which time the medium was adjusted to contain 5 μM α-factor (Sigma-Aldrich, St. Louis, Mo.). At hourly intervals after the addition of α-factor, an aliquot of cells was harvested as described below to determine the chitin content.
Measurement of the chitin content of cells.
The Morgan-Elson method (29) for colorimetric determination of GlcNAc was adapted for microplate readers in measurements of cellular chitin levels. Cultures for chitin determination were made from those initially grown to stationary phase in liquid YPD medium and then diluted 1:100 in fresh medium and incubated at 30°C with shaking for 18 to 22 h. Typically, 1 ml of culture was spun in a tared microcentrifuge tube and then washed once with 1 ml of water, and all residual liquid was removed from the pellet to yield 10 to 25 mg (wet weight) of cells. The cells were suspended in 0.5 ml of 6% KOH and heated at 80°C for 90 min. Samples were centrifuged at 20,000 × g for 10 min, and the supernatant was discarded. The pellet was suspended in 1 ml of phosphate-buffered saline and spun again, and the buffer was discarded. Each pellet was suspended in 0.1 ml of McIlvaine's buffer, pH 6.0, and 5 μl of purified Streptomyces plicatus chitinase-63 (5 mg/ml in phosphate-buffered saline) was added to hydrolyze chitin to GlcNAc; samples were incubated for 24 h at 37°C. Ten microliters of 0.27 M sodium borate, pH 9.0, and 10 μl of sample supernatant were combined in 0.2-ml PCR tubes. Samples were heated in a thermocycler (Techne Inc., Princeton, N.J.) to 99.9°C for about 60 s, mixed gently, and incubated further at 99.9°C for 10 min. Immediately upon cooling to room temperature, 100 μl of freshly diluted DMAB solution (Ehrlich's reagent, consisting of 10 g of p-dimethylaminobenzaldehyde in 12.5 ml of concentrated HCl and 87.5 ml of glacial acetic acid, diluted 1:10 with glacial acetic acid) was added to samples, which were incubated at 37°C for 20 min. Seventy-five microliters was transferred to 96-well low-evaporation microtiter plates, and absorbance at 585 nm was recorded. Standard curves were prepared from stocks of 0.2 to 2.0 mM GlcNAc.
Preparation of cell extracts for enzyme measurements.
Yeast cells were grown in standard YPD medium at 30°C and harvested in the logarithmic phase (OD600, 1.5 to 2.0) by centrifugation at 1,800 × g for 10 min. Cells were resuspended in a buffer (60 mM potassium phosphate [pH 7.0], 1 mM EDTA, 1 mM dithiothreitol) in the presence of fungal protease inhibitors (40 μl/20 ml of buffer) (Sigma-Aldrich) and disrupted with 425- to 600-μm-diameter beads (Sigma-Aldrich) in 2-ml flat-bottom screw-cap tubes in a Mini-Bead Beater-8 Cell Disrupter (Biospec Products, Bartlesville, Okla.) at the maximum power in three 1-min cycles, with 4 min on ice between cycles. Broken-cell extracts were either immediately frozen in liquid nitrogen and stored at −80°C for future use or centrifuged at 1,800 × g for 10 min at 4°C. The resulting supernatant was collected and subjected to a 30-min centrifugation at 20,000 × g. The final supernatant was collected and used immediately to assay for Gfa1p activity.
Assay for Gfa1p (EC 2.6.1.16) activity.
The activity of Gfa1p (GlcN-6-P synthase) was assayed as described previously with modifications (19). The reaction mixture contained the following: 15 mM fructose-6-phosphate, 10 mM l-glutamine, 1 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, and 50 mM potassium phosphate (pH 6.5). Addition of 5 to 10 μl of the enzyme to a final volume of 50 μl started enzymatic reactions. Tubes were incubated at 30°C for 30 min, and heating at 100°C for 2 min terminated the reaction. Amounts of GlcN-6-P formed were determined by a modified Morgan-Elson procedure (29). Portions (10 μl) of the reaction mixture were transferred to PCR strip tubes, and 4 μl of 5% acetic anhydride in acetone was added to each tube and incubated for 3 min at room temperature, followed by addition of 14 μl of 0.33 M potassium tetraborate, pH 9.0, and incubation at 99.9°C for 10 min in a thermocycler (Techne Inc.). Color was developed by addition of 140 μl of Ehrlich's reagent and heating for 20 min at 37°C and was recorded at 590 nm. GlcN-6-P (Sigma-Aldrich) was used to generate a standard curve. Assays were performed in triplicate in two independent preparations. The reaction was shown to be linear with respect to time and enzyme concentration. Specific enzyme activity is expressed as micromoles of GlcN-6-P formed per minute per milligram of protein.
Gfa1p overexpression in a gfa1 deletion mutant.
A haploid strain with the gfa1 deletion was obtained by sporulation of a GFA1/gfa1 diploid strain (24954). Because the gfa1 deletion is lethal, progeny (BY4741 gfa1Δ) were identified as auxotrophic for GlcN and were maintained in liquid and solid media supplemented with 5 mM GlcN. A DNA fragment containing the GFA1 gene with 600 bp upstream of the open reading frame (ORF) was amplified by PCR from the yeast genomic DNA by using forward primer 5′-GAGCTCGAATTCGGCGAGTTGTGA-3′ and reverse primer 5′-TTATTCGACGGTAACAGATTTAGCC-3′. The PCR product of 2,936 kb was directly cloned into the high-copy-number yeast vector pYES2.1 TOPO TA (Invitrogen). Standard methods were used for transformation of Escherichia coli and for preparation of plasmid DNA. Competent cells of strain BY4741 gfa1Δ were transformed chemically by using the Frozen-EZ Yeast Transformation II kit (Zymo Research, Orange, Calif.). Positive clones were selected on YPD agar medium, on which only cells expressing functional Gfa1p can grow.
Measurements of intracellular UDP-GlcNAc.
YPD medium (20 ml) was inoculated with overnight cultures to an OD600 of 0.4 to 0.6, and cultures were then grown for 5 to 7 h at 30°C to an OD600 of 1.8 to 2.3. Cells were harvested by centrifugation for 5 min at 1,800 × g at room temperature and were flash-frozen by adding 20 ml of 100% methanol chilled to −20°C. Cells were either used immediately for metabolite extraction or kept frozen in methanol at −80°C (4).
Cells were centrifuged for 10 min at 1,800 × g and −10°C, and the supernatant was removed by aspiration. Metabolites were extracted from cell pellets in 2 ml (for every 20 ml of the original culture) of 1 M formic acid saturated with 1-butanol at 4°C for 1 h. Cells were then centrifuged, and supernatants were collected and lyophilized. Cell pellets were washed twice with water, collected, and weighed to determine wet weights of cells. The lyophilized supernatant was dissolved in 100 μl of 200 mM Tris base (pH was monitored by pH indicator paper, and Tris base was added if needed to obtain a pH of 8). Samples were incubated with 100 U of alkaline phosphatase (New England Biolabs,Beverly, Mass.) overnight at 37°C. Protein was precipitated with 50% ethanol, samples were centrifuged for 10 min at 20,000 × g, and the supernatant was collected and dried by lyophilization. Dried samples were reconstituted in 200 mM citrate phosphate buffer (pH 4.0) in a volume of buffer that was 10 times greater than the wet weight of the cells.
Twenty microliters of each sample was loaded onto a normal-phase, Ultrasphere-amide (-NH2) (4.6- by 250-mm) column (Beckman Coulter, Inc.) and eluted in a narrow gradient (60 to 50% [vol/vol]) of acetonitrile in 5 mM citrate phosphate buffer (pH 4.0) at a flow rate of 0.5 ml/min. Nucleotides and nucleotide sugars were detected at 254 nm. UDP-GlcNAC (Sigma-Aldrich) was used as a standard to calculate the amounts of UDP-GlcNAc in cell extracts.
Preparation of total cell membrane and sucrose gradient fractionation.
Yeast cells were inoculated from the fresh overnight cultures and grown in 400 ml of YPD medium (supplemented with 23 mM GlcN where indicated) at 30°C with vigorous shaking to an OD600 of 1.5 to 2.0. Cells were harvested by centrifugation at 1,800 × g and 4°C, washed in ice-cold breaking buffer (50 mM Tris buffer [pH 7.5]-1 mM EDTA), and resuspended in 30 ml of breaking buffer with 65 μl of fungal protease inhibitor cocktail (Sigma-Aldrich). Cells were mechanically disrupted by being subjected to high pressure (1,200 lb/in2) three times in a French press (Spectronic Instruments, Rochester, N.Y.). The broken-cell extract was then centrifuged for 10 min at 1,800 × g (4°C) to remove cell walls and unbroken cells. The supernatant was collected, and the total membrane fraction was isolated by centrifugation for 2 h at 100,000 × g. The pellet was resuspended in approximately 1 ml of 50 mM Tris buffer (pH 7.5); protein concentration was measured by the method of Lowry et al. (32). The total membrane fraction was centrifuged for 10 min at 13,000 × g to separate the plasma membrane (pellet) from the subcellular organelles (supernatant). One milliliter of the supernatant (10 to 15 mg of protein) was loaded on a 25-ml discontinuous sucrose gradient (12.5 to 50%) and centrifuged for 3 h at 100,000 × g (55.2 Ti rotor; Beckman). One-milliliter fractions were collected from the top of the gradient, protein content was determined with a Bradford reagent, and chitin synthase 3 (CSIII) activity was determined as described below.
Determination of CSIII (Chs3p) activity.
CSIII activity was measured by a colorimetric assay in 96-well microtiter plates as recently described by Lucero et al. (33).
Gene expression analysis.
An overnight culture of parental strain BY4741 grown in YPD medium was diluted 50-fold in fresh YPD medium and grown on an orbitalshaker for 2 or 3 h; then GlcN was added to a final concentration of 15 mM. Cells were grown in GlcN for 1 to 2 h to an OD600 of 1.6 to 1.8 and were harvested by centrifugation. Alternatively, cell cultures grown in GlcN (15 mM) overnight were diluted 1:50 into fresh YPD medium supplemented with GlcN, allowed to grow to mid-log phase (OD600, 1.6 to 1.8), and harvested by centrifugation. The fks1 mutant strain (5251) was grown similarly in YPD medium.
Total RNA was isolated by a hot phenol procedure (47). Target probe preparation was carried out according to the Affymetrix (Santa Clara, Calif.) Gene Expression technical manual. Briefly, first-strand and double-stranded cDNA were synthesized from total RNA (10 to 40 μg) (SuperScript II RT). cDNA was converted to biotinylated cRNA by in vitro transcription containing T7 RNA polymerase (Enzo Biochem). The cRNA product was cleaned up on RNeasy spin columns (Qiagen, Valencia, Calif.) followed by spectrophotometric quantitation (UV λ = 260 nm) and by gel electrophoresis. About 20 μg of cRNA was fragmented to yield 35- to 200-base fragments before hybridization. RNA probes were hybridized to the entire yeast genome microarray (YG-S98; Affymetrix) for 16 h at 45°C. After being washed in a nonstringent and a stringent wash buffer, the probe arrays were stained with streptavidin phycoerythrin in the GeneChip Fluidics Station and scanned with the Affymetrix GeneChip Scanner according to the manufacturer's instructions. Following data acquisition, the scanned images were quantified by using Microarray Suite 5.0 (MAS 5.0) software (Affymetrix) yielding a signal intensity for each probe on the GeneChip. The signal intensities from the 22 probes for each gene were then used to determine an overall expression level, a detection confidence score, and a present-or-absent call according to algorithms implemented in MAS 5.0 software. The arrays were then linearly scaled to an average expression level of 500 U on each chip in MAS 5.0. For each gene, the fold change and statistical significance of differential expression were calculated. The fold change was calculated using the average signal from the two groups.
Other methods.
Protein levels were determined on microtiter plates by using the bicinchoninic acid protein reagent (Pierce Biotech. Inc., Rockford, Ill.) or by the Bradford method (Bio-Rad Laboratories, Hercules, Calif.) when samples containing high sucrose concentrations were assayed. Gel electrophoresis was carried out on sodium dodecyl sulfate-10% polyacrylamide gels under reducing conditions. Protein was transferred to polyvinylidene difluoride membranes, blocked in 5% milk, and probed with a polyclonal antibody against a 17-mer peptide from the N terminus of Chs3p, monoclonal antibodies to Pep12p, Vma2p, and Pho8p (Molecular Probes, Eugene, Oreg.), and a polyclonal antibody against Pma1p (a gift from R. Serrano, Universidad Politecnica de Valencia-CSIC, Valencia, Spain). Western blots were developed with Chemiluminescence Reagent Plus (Perkin-Elmer, Boston, Mass.). Films were scanned in a Fluor-S MultiImager (Bio-Rad) and subjected to densitometry, and quantitation was carried out with Quantity One software (Bio-Rad). GDPase (Gda1p) activity was measured as previously described (56).
RESULTS
Chitin synthesis in response to cell wall stress.
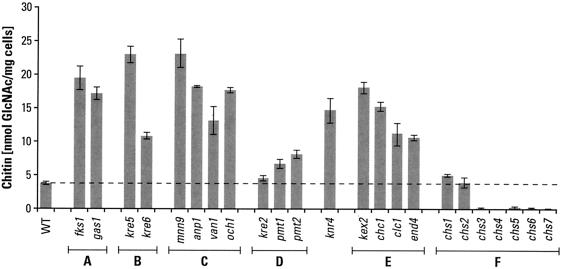
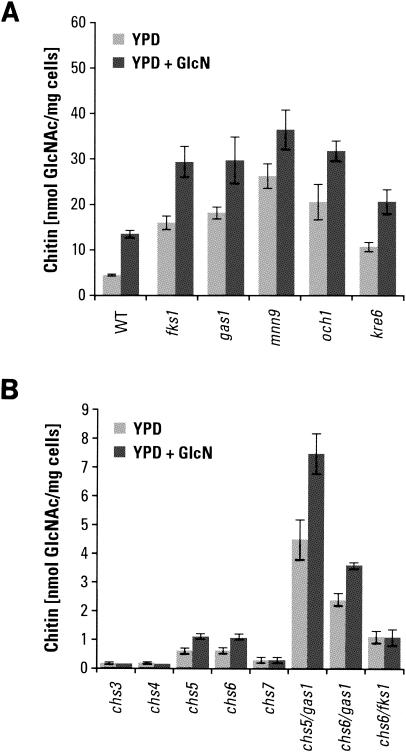
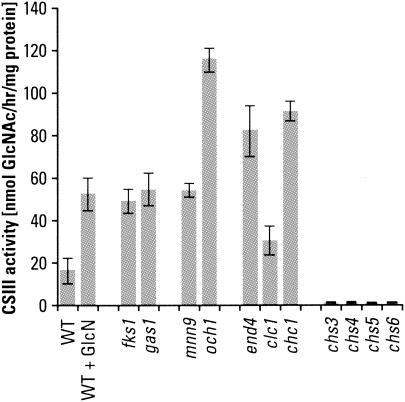
We have begun a survey of full-ORF-deletion strains for their chitin content, concentrating principally on those available through Research Genetics. This has minimized genetic background effects and made possible a ranking of the effects of gene loss on chitin synthesis. It has also avoided complications that would remain undefined if strains had genes mutated by other means. Work in several laboratories, including our own, has shown that mutations that affect cell wall structure and integrity lead to extensive deposition of chitin in the lateral wall (11, 28, 41, 42; this study). Some typical results for chitin levels in cells with defects in glucan or mannan synthesis or in cell wall cross-linking are shown in Fig. 1. It is also apparent from this preliminary survey that deletions of genes that impact endocytosis and/or retention of Chs3p in intracellular vesicles yield strains that have high chitin contents. This suggests that Chs3p is being retained at the plasma membrane and/or that more Chs3p is being directed to the plasma membrane in these strains. It is also reasonable to consider that cells respond to cell wall stress by redirecting their intracellular pools of Chs3p. The knr4 deletion is not categorized in Fig. 1 because the function of Knr4p is not well defined. An extensive list of chitin measurements in other Saccharomyces mutants may be found on our laboratory website (http://dentalschool.bu.edu/research/molecular-faculty/supplemental.html).
FIG. 1.
Effects of gene deletions on chitin content. Deletions of individual genes produce specific changes in the chitin contents of S. cerevisiae strains. Genes are grouped into functional categories A (β1,3-glucan synthesis), B (β1,6-glucan synthesis), C (N glycosylation), D (O glycosylation), E (endocytosis), and F (chitin synthesis). Cells were cultured in YPD medium at 30°C for 18 to 22 h. Strains studied were BY4741 and BY4742 (wild type [WT]), 5251 and 15251 (fks1), 897 and 10897 (gas1), OCY6 (kre5), 5574 and 15574 (kre6), 2778 (mnn9), 277 (anp1), 7116 (van1), 4406 and 14406 (och1), 14317 (kre2), 3792 and 13792 (pmt1), 385 and 10385 (pmt2), 5882 and 15882 (knr4), 1974 (kex2), 4572 and 14572 (chc1), 14797 (clc1), 11969 (end4), 2020 (chs1), 3175 and 13175 (chs2), 3160 and 13160 (chs3), 3087 and 13087 (chs4), 5239 and 15239 (chs5), 1324 and 11324 (chs6), and 2835 and 12835 (chs7). Data are averages of two chitin determinations for each genotype.
Genomewide transcriptional profile of the fks1Δ strain.
Although our major effort in transcriptional profiling was concerned with the study of GlcN supplementation (see below), we also carried out a briefer profiling of the fks1 strain and compared our results with those reported in other publications (Table 2). Jung and Levin (24) used miniarray filters to compare expression profiles of 6,144 ORFs encoded by the yeast genome in response to the cell wall integrity pathway. Rather than using a cell wall mutation to create stress conditions, they compared wild type cells with cells bearing a gain-of-function allele of MKK1, which encodes a key-enzyme in the cell wall integrity pathway (1, 30) (see Discussion). Terashima et al. (50) used high-density gene microarrays to identify up-regulation of genes in the fks1 strain. They then fused the 800 bp of the 5′ noncoding region from each gene to E. coli lacZ, introduced plasmids containing these constructs into wild-type cells and fks1 mutant cells, and monitored β-galactosidase expression in the two strains. In addition, Hughes et al. (23) constructed a reference database (Hughes compendium) in which they analyzed the global-genome responses to 300 different mutations and chemical treatments. This set of data (available at http://www.transcriptome.ens.fr/ymgv) was especially relevant to our studies, because the analysis was performed with Research Genetics strains and Affymetrix chips.
TABLE 2.
Effects of cell wall stress response on gene expression
| ORFa | Gene name | Log2 fold change in
expression (fks1 strain/WT)b in the
indicated study:
|
Log2 fold change in expression with 50 nM α-factor for 120 minc | Function | ||
|---|---|---|---|---|---|---|
| This study | Hughes et al. (23) | Terashima et al. (50) | ||||
| Cell wall proteins | ||||||
| YKL163W** | PIR3 | 2.6 | 2.3 | —d | 1.1 | Similar to Pir1p/Hsp150/Pir3 family |
| YLR194C** | 2.6 | 2.8 | 2.7 | 3.1 | Cell wall protein, GPI anchored | |
| YGR189C** | CRH1 | 0.9 | 1.8 | 1.8 | 1.0 | Cell wall protein, GPI anchored |
| YDR055W** | PST1 | 1.3 | 1.3 | 1.3 | 0.7 | Cell wall protein, GPI anchored |
| YKL096W** | CWP1 | 0.8 | 3.6 | 1.1 | 2.2 | Cell wall mannoprotein |
| Y1R039C | YPS6 | 1.6 | 1.1 | — | 1.3 | Aspartyl protease. GPI anchored |
| YLR121C* | YPS3 | 2.2 | 3.0 | 2.7 | 2.9 | Aspartyl protease, GPI anchored |
| Cell wall synthesis | ||||||
| YGR032W | FKS2 | 2.9 | 0.7 | 1.5 | 2.9 | Beta-1,3-glucan synthase |
| YKR061W* | KTR2 | 2.0 | 1.3 | 0.8 | 2.4 | Mannosyltransferase of KRE2 family |
| YGR166W | KRE11 | 1.9 | 1.5 | — | 1.7 | Beta-1,6-glucan synthesis |
| YNL192W | CHS1 | 1.2 | 0.7 | 1.3 | 2.0 | Chitin synthase 1 |
| YMR238W* | DFG5 | 1.0 | 1.1 | — | 1.3 | PIP binding protein |
| YBR023C** | CHS3 | 0.7 | 0.6 | 0.8 | 1.5 | Chitin synthase 3 |
| YHR142W** | CHS7 | 0.6 | 0.6 | 1.0 | 0.5 | Chitin synthase 7 |
| Environmental stress response | ||||||
| YPR005C | HAL1 | 1.7 | 1.1 | — | 2.0 | Ion homestasis |
| YGL248W | PDE1 | 1.0 | 0.6 | — | 1.5 | Cyclic AMP phosphodiesterase |
| YHR006W | STP2 | 1.5 | 0.8 | — | 0.9 | Transcription factor, Msn2/Msn4 dependent |
| YGL184C | STR3 | 1.9 | 0.9 | — | 0.9 | Cystathione beta-lyase |
| Cell signaling | ||||||
| YKL161C** | 3.5 | 3.1 | — | 2.6 | Ser/Thr kinase | |
| YHR030C** | SLT2 | 2.6 | 1.6 | 1.5 | 2.6 | Ser/Thr mitogen-activated protein kinase family |
| YPL089C | RLM1 | 1.5 | 1.2 | — | 1.6 | Transcription factor |
| YOR208W** | PTP2 | 1.4 | 1.0 | 0.8 | 1.6 | Protein tyrosine phosphatase |
| YNL289W | PCL1 | 0.8 | 0.0 | 1.5 | 0.1 | G1/S cyclin |
| YKR013W*** | PRY2 | 1.0 | 1.0 | 0.8 | 2.0 | Cell surface signaling |
| Mating | ||||||
| YIL117C** | PRM5 | 3.3 | 1.7 | — | 2.8 | Mating, cell wall stress |
| YBR296C | PHO89 | 2.9 | 1.7 | — | 2.4 | Sodium symporter |
| YDR085C*** | AFR1 | 2.6 | 1.7 | — | 4.5 | Shmoo formation |
| YEL060C | PRB1 | 1.5 | 2.0 | — | 2.8 | α-Factor receptor-degrading protease |
| Other proteins | ||||||
| YHR209W | 4.5 | 2.9 | — | 2.7 | Putative methyltransferase | |
| YAL053W* | 1.6 | 1.3 | 1.4 | 1.6 | Unknown function | |
| YOR306C*** | MCH5 | 1.9 | 0.7 | — | 1.3 | Permease |
| YBR071W* | 1.3 | 1.0 | 0.8 | −0.3 | Unknown function | |
| YPL221W* | BOP1 | 1.2 | 1.1 | 0.8 | 0.8 | Unknown function |
| YPL088W*** | 3.2 | 1.7 | — | 4.0 | Putative alcohol dehydrogenase | |
| YKR091W | SRL3 | 3.5 | 1.7 | — | 2.1 | Unknown function |
| YKL104C* | GFA1 | 1.0 | 0.9 | 0.6 | 2.0 | Glutamine-fructose-6-P aminotransferase |
| YPL163C | SVS1 | 0.7 | 1.1 | 1.3 | −0.7 | Unknown function |
These proteins were found in the YPD Database. A complete list is available at http://dentalschool.bu.edu/research/molecular-faculty/supplemental.html. *, Rlm1p binding site(s) in a promoter; **, Rlm1p induced; ***, Rlm1p transcription coregulation.
WT, wild type.
From the study of Roberts et al. (43).
—, not done.
The most noticeable effect in our studies and those of others (24, 49) is the strong induction of cell wall integrity genes, which either are directly induced by the stress-related transcription factor Rlm1p (PIR3, YLR194C, CRH1, CWP1, CHS3, CHS7, YKL161C, SLT2, PTP2, PRM5, and PST1), have a putative Rlm1p binding site(s) (YPS3, KTR2, DFG5, YAL053W, YBR071W, BOP1, and GFA1), or are transcriptionally coregulated with other proteins involved in aging, the cell cycle, the cell wall, the cytoskeleton, glycolysis, or endocytosis (SRL3, AFR1, PRY2, and MCH5) (YPD Database) (Table 2). RLM1 itself is also activated in our studies. Although details differ, probably as a result of the very different methodologies used, the above studies give similar pictures (Table 2). All studies show a strong induction of genes for cell wall proteins such as PIR3, CRH1, CWP1, and YLR194C. Our study, that of Terashima et al. (50), and the Hughes compendium (23) also show very strong induction of YPS3, which encodes a GPI-anchored aspartyl protease and, to a lesser extent, induction of the related gene YPS6. All studies also demonstrate the expected induction of the alternative glucan synthase gene FKS2 and chitin synthase genes. It is interesting that both CHS3 and CHS7 are induced in the fks1 strain. Chs7p has been shown to be responsible for the export of Chs3p from the endoplasmic reticulum, and increased Chs3p levels at the cell surface require induction of both genes (51). Our results and those of Terashima et al. (50) also show induction of KTR2, a mannosyltransferase gene associated with stress and pseudohyphal growth (34). Among the interesting genes in this “other proteins” category are PST1, encoding another GPI-anchored cell wall protein, and PTP2, encoding a phosphatase which probably is involved in maintaining homeostasis in the cell wall integrity pathway (35). In addition, our data confirm the transcriptional up-regulation of GFA1, a key regulatory enzyme in the UDP-GlcNAc biosynthetic pathway (see below), in agreement with other reports (28, 50). Interestingly, treatment of cells with the α-factor pheromone for 120 min (Hughes compendium) similarly up-regulates most of the genes identified in Table 2; only two genes (YBR071W and SVS1) are clearly down-regulated by pheromone treatment. This supports the idea that cell wall remodeling induced by pheromones may in fact occur partly in response to cell wall stress (25).
Role of metabolic precursors in the regulation of chitin synthesis.
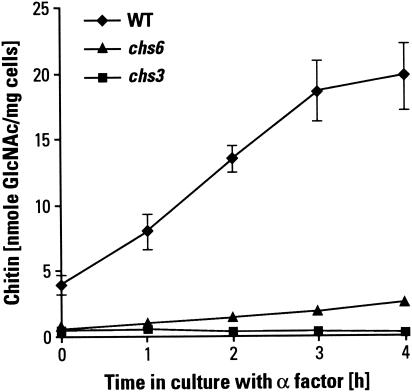
The concept that metabolic intermediates leading to UDP-GlcNAc play an important regulatory role in chitin synthesis was developed from the early studies of Cabib and coworkers, who showed that in vitro, chitin synthesis was strongly stimulated by GlcNAc (6). Seminal in vivo evidence came from studies by Schekman and Brawley (46) and Orlean et al. (39) of the extra chitin synthesis that occurs when mating-type a cells are treated with the pheromone α-factor, a phenomenon probably related to chitin synthesis in response to cell wall stress. Orlean et al. (39) found that treatment with α-factor led to a rapid four- to fivefold increase in the pool of soluble chitin precursors, followed by a three- to fivefold increase in the rate of chitin synthesis. In our studies we observed that wild-type cells treated with the α-factor for 3 h had about a fivefold increase in chitin content. No such effect was observed in a chs3 strain, where the chitin level remained unchanged. When chs6 cells were exposed to α-factor, chitin levels increased slightly over those in untreated cells, suggesting that there is an alternative, Chs6p-independent chitin deposition in the lateral cell wall (Fig. 2).
FIG. 2.
Effect of α-factor on chitin content. Addition of 5 μM α-factor to MATa cells cultured in liquid YPD medium at 30°C causes a rapid increase in the level of chitin, which is synthesized by Chs3p. Three wild-type (WT) strains (PRY242, PRY488, and PRY88.1D) were analyzed, and their chitin contents were averaged. These were compared to the responses of a chs3Δ strain (PRY533) and a chs6Δ strain (PRY408).
Transcription profiling studies of cells treated with α-factor have shown substantial increases in the transcription of GFA1 and AGM1 mRNAs, encoding two of the four enzymes in the UDP-GlcNAc biosynthetic pathway. An increase in the transcription of all four genes accompanies meiosis and sporulation, when chitin synthesis is required for spore wall formation (48). Special attention has recently focused on the regulatory role of Gfa1p (28).
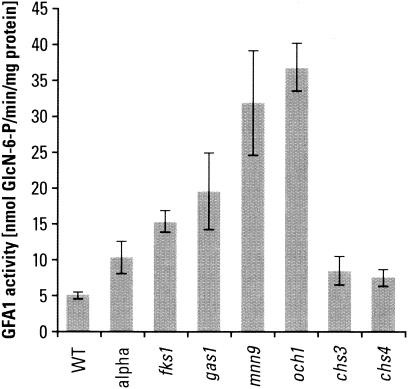
Gfa1p activity in wild-type and mutant strains.
The first reaction in the chitin synthesis pathway is the formation of GlcN-6-P from fructose-6-phosphate and glutamine, catalyzed by Gfa1p. In an extensive study, Lagorce et al. (28) demonstrated that the chitin metabolic pathway is impressively hierarchical, dominated by the cellular Gfa1p level. Some of our own measurements of Gfa1p enzymatic activity in wild-type and mutant strains are shown in Fig. 3. In agreement with the results of Lagorce et al. (28), we find that while the correlation between the cell wall stress-related transcriptional activation of GFA1 and the rise in chitin synthesis is impressive, it is not strictly quantitative, suggesting that additional factors are involved. It is interesting that the chs3Δ and chs4Δ strains, which are unable to synthesize ring or lateral wall chitin, still express a somewhat increased level of Gfa1p activity, possibly reflecting cell wall stress and increased GFA1 transcription even in the absence of extensive chitin synthesis. It is possible that accumulation of intermediates as a result of lack of chitin synthesis may moderate the level of the enzyme activity.
FIG. 3.
Gfa1p activity in wild-type (WT) and mutant strains. The specific activity of Gfa1p was measured in a soluble, cytosolic fraction of cells grown in YPD medium at 30°C to mid-log phase. Strains studied were BY4741, 5251 (fks1), 897 (gas1), 2778 (mnn9), 4406 (och1), 3160 (chs3), and 3087 (chs4). Cells were treated with 5 μM α-factor for 4 h. Cell extracts were prepared in three independent experiments, and activity measurements were carried out in duplicate.
If the Gfa1p or the precursor pool plays a direct or indirect role in chitin synthesis, then increasing the enzyme and/or precursor pool by overexpressing GFA1 should give rise to extra chitin deposition. In fact, we found that when GFA1 was expressed on a multicopy plasmid with its own promoter, cellular chitin levels increased from the normal level of 4 to 5 nmol of GlcNAc per mg of cells to 14 nmol, which directly correlated with the ∼4- to 5-fold increase in Gfa1p activity under these conditions. These results are in agreement with those of Lagorce et al. (28), who also made the observation that simultaneous overexpression of GFA1, CHS3, and CHS7 leads to about the same amount of chitin synthesis as when GFA1 alone is overexpressed.
Chitin synthesis response to GlcN supplementation.
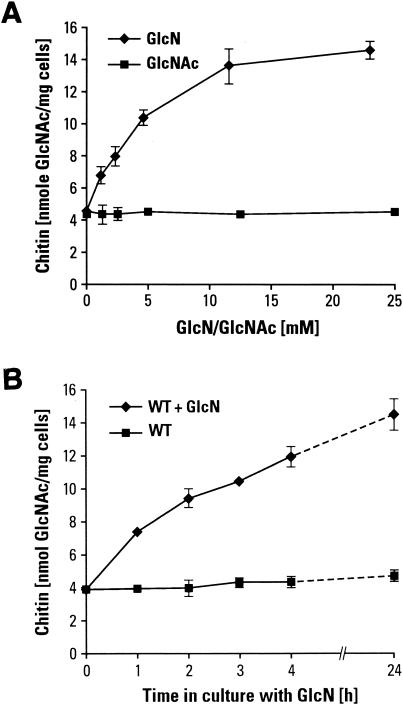
The increase in chitin synthesis that occurs in response to GFA1 overexpression is probably not a response to cell wall stress. It may represent a bypass of the stress response, i.e., stress may normally lead first to stimulation of GFA1, which in turn stimulates chitin synthesis directly or indirectly (see Discussion for an analysis of this hypothesis). If this is the case, then increasing the GlcN-6-P pool by simply adding GlcN to the growth medium might also lead to increased chitin synthesis. It has been shown that GlcN can readily be taken up and phosphorylated by S. cerevisiae (2). Typical results are shown in Fig. 4A. The chitin content of cells treated with GlcN (0 to 23 mM in YPD medium) increases from 4 to 5 nmol of GlcNAc per mg (wet weight) to about 14 nmol. GlcN concentrations higher than 23 mM in the medium have a toxic effect on cells. We also measured the rate of chitin synthesis in yeast cells exposed to 15 mM GlcN over time (Fig. 4B). There was no apparent lag in chitin synthesis, as chitin levels nearly doubled in the first hour. After 4 h, chitin content approached its new steady-state level, as found for cells grown overnight with GlcN. Upon examination by fluorescence microscopy using Calcofluor to stain chitin, large budded cells as well as mother cells showed increased fluorescence in their lateral walls following culture in 15 mM GlcN (data not shown).
FIG. 4.
(A) Effect of GlcN addition on chitin level. Addition of GlcN to wild-type cells cultured in YPD medium at 30°C for 18 to 22 h caused a concentration-dependent increase in chitin levels. Addition of GlcNAc had no effect on chitin levels. Each data point is the combined average of the chitin determinations for strains BY4741 and BY4742. (B) The rate of chitin synthesis of a wild-type (WT) strain (BY4741) was measured during growth in YPD medium at 30°C in the presence and absence of 15 mM GlcN. Aliquots of cells were withdrawn at 1-h intervals for chitin determinations.
Cells grown overnight with 15 mM GlcN usually have wet weights 21 to 25% lower than parallel WT cultures (data not shown). In order to determine the source of this difference, we measured the cell growth rate by OD600, performed cell counts, and also measured the dry weight of cells. ODs and numbers of cells were comparable in cells grown in the presence and absence of GlcN, indicating that 15 mM GlcN does not inhibit the rate of cell growth. Furthermore, the dry weights of these two sets were comparable, suggesting that the difference in the wet weights should probably be attributed to water content. Thus, for reasons we do not yet understand, cells grown in the presence of GlcN appeared to retain less water.
GlcN is presumed to enter cells by way of hexose transport systems and to be phosphorylated by hexokinase(s). The resulting GlcN-6-P is a normal intermediate in the chitin synthesis pathway, and the increased levels of metabolic intermediates must initiate the increased chitin formation observed. GlcNAc, on the other hand, either is not transported into the cell or is not phosphorylated.
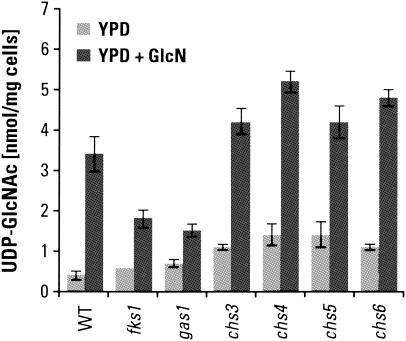
GlcN supplementation also increased chitin levels in various cell wall mutants, as shown in Fig. 5A. In some cases the amount of chitin increased to nearly 10 times the normal wild-type level. In these cases, chitin constituted as much as 20% of the cell wall mass. We have not noted any decrease in the viability or growth rate of these cells. Considering also the minimal amount of chitin in, for example, a chs3Δ strain (relative to the maximum observed upon GlcN supplementation), it is clear that S. cerevisiae readily adapts to a >50-fold change in cell wall chitin levels.
FIG. 5.
(A) Effect of GlcN on chitin contents of wild-type (WT) and mutant strains. Mutant strains that already had elevated chitin levels showed increased chitin synthesis upon addition of GlcN. Cells were cultured in YPD medium plus 15 mM GlcN at 30°C for 18 to 22 h before being collected for chitin determinations. Strains studied were BY4741 (wild type), 5251 (fks1), 897 (gas1), OCY6 (mnn9), 4406 (och1),and 5574 (kre6). Data are averages of two chitin determinations for each genotype. (B) Effects of GlcN on chitin synthesis for single- and double-mutant strains defective in the activity or localization of Chs3p were measured. Strains studied were 3160 (chs3), 3087 (chs4), 5239 (chs5), 1324 (chs6), 2835 (chs7), 184.4A and 184.4B (chs5 gas1), 183.1C and 183.3C (chs6 gas1), and 175.2C and 177.1D (chs6 fks1). Data are averages of two chitin determinations for each genotype.
Chitin levels in chs3Δ, chs4Δ, and chs5Δ mutants and some double-mutant strains are shown in Fig. 5B. It is obvious that the increase in chitin synthesis brought about by addition of GlcN to the medium is mediated by Chs3p, since chs3 and chs4 deletion mutants produce almost no chitin, whether cells are exposed to GlcN or not. Previous results (40) suggested that the increased lateral wall chitin synthesis associated with an fks1 glucan synthase mutation could “bypass” loss of Chs6p, a factor required for targeting of chitosomes to the plasma membrane. For reasons that are still unclear, we were unable to restore a chs6 mutant phenotype to the chs6 fks1 mutant strain used in that study by making a chs6/chs6 FKS1/fks1 diploid. We have therefore turned to studies with standard Research Genetics deletion strains (Table 1) for clarification. As can be seen, the presence of the fks1 deletion with the chs6 deletion causes chitin levels to increase somewhat over those with chs6Δ alone; similarly, the gas1 deletion produces a larger increase. The stimulation of chitin formation by the gas1 mutation is even more pronounced in a chs5Δ strain. The level of chitin synthesis in these double mutants is still consistent with an active role for Chs5p and Chs6p in lateral wall chitin synthesis, as reported recently by Carotti et al. (8).
Cellular UDP-GlcNAc levels.
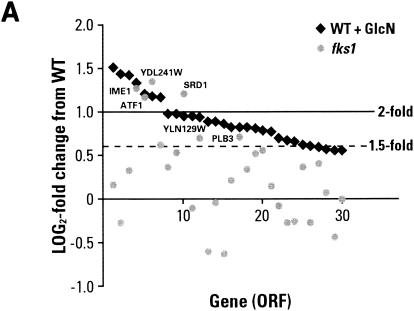
To further explore the role of the precursor pool in chitin synthesis by Chs3p, the levels of UDP-GlcNAc in several cell wall mutants were quantitated. Soluble components were extracted from the cytosol of logarithmically growing cells with formic acid. Extracted metabolites were incubated with alkaline phosphatase to remove phosphomonoester intermediates. This treatment reduces the complexity of the high-pressure liquid chromatographyprofile, since diester-linked metabolites are limited to nucleotide sugars and a few coenzymes. Figure 6 shows the levels of UDP-GlcNAc in cells grown in complete medium alone and with added GlcN. In each case the level was measured three to five times, and the values were consistent. Surprisingly, similar levels of UDP-GlcNAc were also found in cultures grown to saturation overnight.
FIG. 6.
UDP-GlcNAc levels in wild-type (WT) and mutant strains grown in the presence of GlcN. UDP-GlcNAc was quantitated from mid-log-phase cultures of the wild-type strain BY4741 and mutant strains 5251 (fks1), 897 (gas1), 3160 (chs3), 3087 (chs4), 5239 (chs5), and 1324 (chs6) grown in YPD medium at 30°C. GlcN was added to 23 mM. Each measurement was done three to five times, and the standard deviation was determined.
An obvious question is whether chitin synthesis is proportional to, or is in any way “driven” by, the cellular UDP-GlcNAc level. The data show that this is not the case. Although we and Lagorce et al. (28) found that chitin synthesis is proportional to the cellular level of Gfa1p, the enzyme responsible for the formation of GlcN-6-P, it is clear that UDP-GlcNAc concentrations are increased to only moderately higher levels in mutant strains that are making more than twice as much chitin as wild-type cells. More dramatically, when chitin synthesis is driven not by Gfa1p but by added GlcN, the level of UDP-GlcNAc is actually lowest in cells that show the highest rate of synthesis. It is thus clear that the UDP-GlcNAc concentration alone does not control the rate of chitin formation. In summary, since chitin synthesis is proportional to Gfa1p but not to UDP-GlcNAc concentrations, it is likely that either Gfa1p itself or another GlcN metabolite plays a role in the activation and/or localization of Chs3p. However, the recent results of Valdivia et al. (52) show that control of Chs3p targeting may be complex and subject to secondary mutations. These investigators demonstrated that mutations in proteins of the clathrin AP-1 complex allow extensive bypassing of the Chs6p requirement for lateral wall chitin synthesis.
Genomewide transcription profiling of cells exposed to GlcN.
Another obvious question was whether treatment of cells with GlcN would lead to transcription of any of the genes found to be “turned on” in strains with mutations affecting cell wall structure, since in both cases there is a major increase in the synthesis of lateral cell wall chitin. Also, we wanted to know whether GlcN simply produces—either directly or indirectly—a cell wall stress. In that case, the transcription profile following addition of GlcN would be similar to that seen, for example, in the fks1 mutant. We designed two experiments. In one, the cells were grown first in YPD medium and then for 1 to 2 h after supplementation with GlcN (to 15 mM). In the other, cells were grown in the presence of GlcN overnight, diluted in fresh medium containing 15 mM GlcN, and again grown to mid-log phase (these are referred to below as cells exposed to “steady-state” conditions). Analysis of genomewide expression was carried out in triplicate in all cases, and the average intensities of the mRNA signals showing a significant change are reported in Table 3.
TABLE 3.
Effects of GlcN addition on gene expressiona
| ORF | Gene name | Function | Expression level
(U) under the following
condition:
|
Log2 ratio (GlcN added/ no GlcN) | Expression level (U) with steady- state GlcN | Log2 ratio (steady-state GlcN/no GlcN) | |
|---|---|---|---|---|---|---|---|
| No GlcN | GlcN added after 2 h | ||||||
| Up-regulated genes | |||||||
| Genes involved in mating, cell cycle arrest, sporulation | |||||||
| YJR094C | IME1 | Transcription factor | 93 ± 17 | 113 ± 8 | 0.3 | 263 ± 50 | 1.5 |
| YJR086W | STE18 | GTPase activator | 87 ± 25 | 113 ± 26 | 0.4 | 198 ± 15 | 1.2 |
| YGR049W | SCM4 | Putative cell cycle control | 202 ± 7 | 195 ± 58 | 0 | 390 ± 68 | 1.0 |
| YBL016W | FUS3 | Mitogen-activated protein kinase | 180 ± 38 | 193 ± 61 | 0.1 | 355 ± 38 | 1.0 |
| Metabolic genes | |||||||
| YOR377W | ATF1 | Alcohol dehydrogenase; lipids, sterols | 143 ± 19 | 141 ± 34 | 0 | 386 ± 100 | 1.4 |
| YBR085W | AAC3 | ADP/ATP transporter mitochondria | 223 ± 29 | 177 ± 14 | −0.3 | 537 ± 150 | 1.3 |
| Other genes | |||||||
| YIL080W | Unknown function | 87 ± 15 | 106 ± 45 | 0.3 | 217 ± 55 | 1.3 | |
| YLR040C | Possible cell wall mannoproteins | 172 ± 66 | 176 ± 44 | 0 | 395 ± 45 | 1.2 | |
| YCR018C | SRD1 | Protein involved in pre-rRNA processing | 111 ± 15 | 107 ± 30 | 0 | 248 ± 20 | 1.1 |
| YNL129W | Unknown function | 102 ± 14 | 165 ± 43 | 0.7 | 200 ± 36 | 1.0 | |
| Down-regulated genes | |||||||
| Energy generation, aerobic respiration | |||||||
| YJR048W | CYC1 | Cytochrome c | 1,282 ± 362 | 1,166 ± 579 | −0.1 | 387 ± 29 | −1.7 |
| YGL187C | COX4 | Cytochrome c oxidase | 1,839 ± 379 | 1,486 ± 502 | −0.3 | 660 ± 74 | −1.5 |
| YLR304C | ACO1 | Aconitate hydratase | 3,590 ± 750 | 3,069 ± 1,158 | −0.2 | 1,378 ± 450 | −1.4 |
| YLR214W | FRE1 | Flavocytochrome | 237 ± 54 | 138 ± 53 | −0.8 | 92 ± 14 | −1.3 |
| YOR161C | Mitochondrial protein | 698 ± 87 | 550 ± 109 | −0.3 | 280 ± 166 | −1.3 | |
| YKL141W | SDH3 | Succinate dehydrogenase complex | 1,889 ± 232 | 1,804 ± 596 | 0 | 936 ± 41 | −1.0 |
| YJR077C | MIR1 | Mitochondrial phosphate carrier | 5,060 ± 1,152 | 4,831 ± 1,606 | 0 | 2,473 ± 331 | −1.0 |
| YLR395C | COX8 | Cytochrome c oxidase | 2,838 ± 569 | 2,356 ± 960 | −0.3 | 1,455 ± 200 | −1.0 |
| YFR033C | QCR6 | Ubiquinol-cytochrome c reductase | 787 ± 91 | 660 ± 100 | −0.3 | 383 ± 25 | −1.0 |
| Amino acid metabolism | |||||||
| YLR303W | MET17 | Acetylhomoserine sulfhydrylase | 1,046 ± 314 | 645 ± 362 | −0.7 | 152 ± 62 | −2.7 |
| YDR380W | ARO10 | Putative indole-3-pyruvate decar- boxylase | 400 ± 35 | 524 ± 97 | 0.4 | 129 ± 18 | −1.6 |
| YER119C | Amino acid permease | 180 ± 11 | 169 ± 37 | −0.1 | 81 ± 27 | −1.2 | |
| YCL064C | CHA1 | Serine/threonine deaminase | 579 ± 119 | 594 ± 123 | 0 | 273 ± 81 | −1.1 |
| YHR137W | ARO9 | Aromatic amino acid amino- transferase | 947 ± 135 | 1,459 ± 111 | 0.6 | 463 ± 97 | −1.0 |
| Other metabolic genes | |||||||
| YBR093C | PHO5 | Acid phosphatase | 3,357 ± 650 | 1,187 ± 386 | −1.5 | 728 ± 509 | −2.2 |
| YDR281C | PHM6 | Phosphate metabolism | 456 ± 32 | 189 ± 28 | −1.3 | 124 ± 84 | −1.9 |
| YPR184W | GDB1 | Glycogen debranching enzyme | 421 ± 54 | 253 ± 99 | −0.7 | 212 ± 69 | −1.0 |
| Cell stress | |||||||
| YBR244W | GPX2 | Glutathione peroxidase | 216 ± 20 | 245 ± 137 | 0.2 | 88 ± 43 | −1.3 |
| YNL241C | ZWF1 | Glucose-6-phosphate dehydroge- enase | 1,330 ± 68 | 1,099 ± 138 | −0.3 | 675 ± 190 | −1.0 |
| Other | |||||||
| YOL155C | Similar to glucan 1,4-glucosidase | 7,062 ± 107 | 4,911 ± 554 | −0.5 | 1,810 ± 225 | −1.9 | |
| YML091C | RPM2 | RNA processing/modification | 236 ± 37 | 230 ± 98 | 0 | 73 ± 38 | −1.7 |
| YPL198W | RPL7B | Ribosomal protein L7 | 1,898 ± 201 | 677 ± 252 | −1.5 | 658 ± 536 | −1.5 |
| YHR136C | SPL2 | Inhibitor of cyclin-dependent protein kinase | 1,005 ± 75 | 697 ± 72 | −0.5 | 334 ± 300 | −1.6 |
| YBR230C | Unknown function | 1,478 ± 95 | 1,018 ± 342 | −0.5 | 654 ± 184 | −1.2 | |
Experiments were carried out in triplicate. P ≤ 0.02; t ≥ 3.7; fold change, ≥2.0. A complete list is available at http://dentalschool.bu.edu/research/molecular-faculty/supplemental.html.
Microarray chips used in these studies (GeneChip Yeast Genome S98 Array; Affymetrix) contained approximately 6,200 S. cerevisiae ORFs recognized by the Saccharomyces Genome Database and an additional 600 putative ORFs identified by other methods, such as SAGE (serial analysis of gene expression) analysis. In order to determine how gene expression is differentially altered, we applied the t test, where we assumed a P value of ≤0.02. We selected those genes which had a t of ≥3.7 and also a ≥1.5-fold change from the signal measured for wild-type cells cultured in YPD medium without GlcN. Of nearly 7,000 ORFs, only 105 met the above criteria (complete data sets are available at the National Center for Biotechnology Information under GEO series number GSE441 and on our website [http://dentalschool.bu.edu/research/molecular-faculty/supplemental.html]). Table 3 contains the data set for genes that showed a ≥2-fold change.Surprisingly, we found fewer up-regulated than down-regulated genes. It is somewhat surprising that GlcN has such a moderate effect on the transcription profile, considering its strong stimulation of chitin synthesis. Only four genes (URA4, RIB4, GLN1, and YNL129W) were up-regulated both 2 h after GlcN addition and in the steady state (see our website).
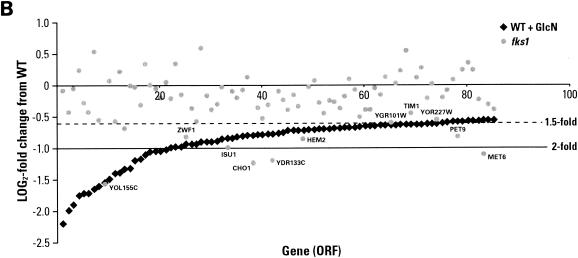
Other genes were virtually unaffected after 2 h of GlcN treatment. The largest functional group of transcripts up-regulated in the steady state represents genes involved in mating, sporulation, and cell cycle arrest. Activation of these genes in the steady state may be a secondary effect of GlcN and an adaptive response to new growth conditions. In the gene transcription pattern there is essentially no overlap (with the exception of IME1, ATF1, YDL241W, SRD1, YNL129W, and PLB3) between the cell wall stress response in fks1Δ strains and exposure to GlcN (Fig. 7A), although in these two cases chitin synthesis is stimulated to about the same extent. Since most of the overlapping genes are involved in mating and sporulation, this may again be an adaptation to steady-state exposure to GlcN rather than a direct impact of GlcN on their transcription.
FIG. 7.
Effects of GlcN addition on transcription of genes that were up-regulated or down-regulated. (A) Genes up-regulated in cells exposed to GlcN in YPD medium (WT + GlcN, steady-state) (P ≤ 0.02; t ≥3.7; fold change [WT + GlcN/WT], ≥1.5) were compared with the same genes regulated in a fks1 mutant strain (fks1 strain/WT). Solid and dashed lines represent a 2- and 1.5-fold change in transcription, respectively. (B) Genes down-regulated in cells treated with GlcN in YPD medium (WT + GlcN, steady-state) (P ≤ 0.02; t ≥ 3.7; fold change [WT + GlcN/WT], ≥1.5) were compared with genes regulated in a fks1 mutant strain (fks1 strain/WT). Dashed and solid lines represent a 1.5- and 2.0-fold repression, respectively.
Steady-state GlcN appears to suppress aerobic respiration, as the largest group of down-regulated genes is composed of mitochondrial respiratory genes. We do not have a rationale for this phenomenon, but in this case also it is predominantly a steady-state effect (Table 3). We found that the cell stress response genes were among the down-regulated genes, reaffirming that GlcN does not “stress” cells. None of the typical environmental stress response identifiers, e.g., Yap1 and Msn2/Msn4, which are activated upon any environmental stress (18), were found in our analysis. As with up-regulated genes, there is little overlap between down-regulated genes in steady-state GlcN-treated wild-type cells and genes down-regulated due to fks1 mutation, with a few exceptions (ZWF1, ISU1, HEM2, YGR101W, TIM1, YOR227W, and PET9) (Fig. 7B). Further experiments are needed to determine whether these overlapped transcripts represent nonspecific “biological noise” or a true effect.
In summary, it appears that GlcN or one of its metabolites must activate chitin synthesis. We assume that the same is true for the chitin synthesis that occurs when Gfa1p is overexpressed. Possible explanations are discussed below. It should be emphasized that although Gfa1p provides GlcN-6-P for UDP-GlcNAc synthesis, this reaction is not required when GlcN is being used as a substrate, and in any case, GFA1 is neither up- nor down-regulated by GlcN addition.
CSIII (Chs3p) enzymatic activity in wild-type and mutant cells.
Although it is clear that precursor molecules play an important role in regulating chitin synthesis, the data above show that UDP-GlcNAc concentrations are not the only controlling factor in chitin biosynthesis. It has been reported previously that CSIII activity increases in fks1 cells (16) and in gas1 cells (53). These reports correlate well with the fact that both strains show elevated levels of chitin. We isolated total cellular membrane fractions and measured CSIII specific activity in wild-type cells and deletion mutants which either had elevated chitin levels (cell wall mutants) or made very little chitin (Fig. 8). In all cases, when the chitin level was elevated, CSIII activity also increased. The same observation was made for cells grown in the presence of GlcN. A study of the time course of activation by GlcN shows little effect in the first hour of exposure but full activation after 2 h (data not shown). Since treatment with GlcN does not lead to extra CHS3 transcription (see above), we assume that the increase is the result of some type of activation and/or redistribution of the enzyme.
FIG. 8.
CSIII activity in wild-type (WT) and mutant strains. CSIII activity was measured in total membrane fractions of WT cells (BY4741) grown in YPD medium at 30°C supplemented with 23 mM GlcN and in deletion mutant strains 5251 (fks1), 897 (gas1), 2778 (mnn9), 4406 (och1), 3160 (chs3), 3087 (chs4), 11969 (end4), 14797 (clc1), 14572 (chc1), 13160 (chs3), 3087 (chs4), 5239 (chs5), and 1324 (chs6). Measurements were done for three independent experiments.
Interestingly, there is a general, although not strictly quantitative, correlation between the amount of cellular chitin and CSIII activity (for comparisons see Fig. 1 and 8). All mutants with elevated chitin content also have higher CSIII activities than wild-type cells. Among mutant strains, however, the amount of elevated chitin is not necessarily proportional to CSIII activity. For example, in comparing mnn9 and och1 N-glycosylation mutants, the CSIII activity found for the och1 mutant is higher than that for the mnn9 mutant, although the chitin content of the former is lower. In all mutants which make little chitin (chs3Δ, chs4Δ, chs5Δ, and chs6Δ mutants), CSIII activity is virtually absent. The low signal probably represents trace activities of other chitin synthases (CSI and CSII). Consistently, our data for strains with chs6 (csd3) deletions have contrasted with those reported by Bulawa (5), who found wild-type activities of CSIII for the csd3 mutant strain. Yeast strains defective in proteins involved in cellular trafficking and endocytosis (end4, clc1, and chc1) have an elevated CSIII activity, suggesting that Chs3p has accumulated at the plasma membrane.
The next question we posed was whether small precursor molecules might activate CSIII. We measured CSIII enzymatic activity in total membrane fractions in vitro in the presence of added GlcN-6-P, GlcNAc-1-P, GlcNAc-6-P, or GalNAc. None of these metabolites had any apparent stimulatory or inhibitory effect on CSIII activity (data not shown). The only stimulator, GlcNAc, is not actually a chitin precursor or primer (6, 14) and is used in the standard assay at the very high concentration of 80 mM. Its two- to fourfold activation of CSIII is seen in wild-type membranes as well as in membranes of cell wall mutants (data not shown). Activity increases linearly over the GlcNAc concentration range of 0 to 40 mM (data not shown). The mechanism of this effect has yet to be determined.
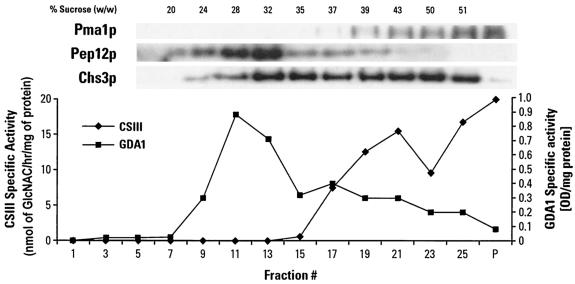
CSIII (Chs3p) activity in subcellular fractions.
Chs3p follows an intricate secretory and endocytotic pathway, which distributes it primarily to the plasma membrane and subcellular compartments. We separated subcellular membranes (devoid of much of the plasma membrane and microsomal fractions due to a precentrifugation step) on a sucrose gradient and then correlated the distribution of Chs3p (by using an anti-Chs3 antibody) with its enzymatic activity. As shown in Fig. 9, which is representative of four experiments, samples were essentially devoid of plasma membrane as indicated by the distribution of the plasma membrane ATPase Pma1p (3). Chs3p was broadly distributed in the gradient as two peaks, similar to findings reported from previous studies (22, 45, 59). While Holthuis et al. (22) did not measure enzyme activity, their resolution of the subcellular distribution of Chs3p was similar to what we have observed. We found no Chs3p in fractions (18 to 20% sucrose) containing the vacuolar protein Vma2p or Pho8p (data not shown). The peak of Chs3p found at sucrose concentrations of 32 to 35% was in subcellular fractions partially colocalizing with the late endosome marker Pep12p and Golgi GDPase (Gda1p). Chs3p was inactive in those subcellular fractions. However, Chs3p was enzymatically active in fractions with higher sucrose concentrations, with the highest specific activity found in a fraction (43% sucrose) with very little of the plasma membrane marker Pma1p. The Chs3p that was enzymatically active had a higher specific activity in the plasma membrane than in its subcellular fraction, as observed by Lucero et al. (33). The distribution we observed likely represents the mixed population of vesicles through which Chs3p transits going to and from the plasma membrane. For wild-type cells treated with GlcN, the amount of Chs3p decreased in the intracellular compartments (our unpublished data) and was probably translocated to the plasma membrane, as reflected by the increased CSIII specific activity shown in Fig. 8.
FIG. 9.
Chs3p localization and activity in subcellular fractions. The total membrane fraction of wild-type cells grown in YPD medium at 30°C and devoid of plasma membrane was separated on a discontinuous (12.5 to 60%) sucrose gradient. Fractions were assayed for CSIII (Chs3p), Gda1p, and GDPase (Gda1p) activities and were probed with antibodies against Chs3p, Pep12p, and Pma1p (upper panel). The remaining pellet (P) contained mostly plasma membrane.
DISCUSSION
We have confirmed and extended the finding that a great many mutations or conditions that produce cell wall weakening or stress lead to extensive deposition of chitin in the lateral cell wall of S. cerevisiae. The effect seems to be limited to situations that activate the cell wall integrity pathway (24, 43). For example, careful study of the high-osmolarity glycerol response has shown that induction of this pathway does not affect chitin synthesis (17). Our genomewide profiling of genes activated by the cell wall integrity pathway is in general agreement with the findings of other studies, showing up-regulation of cell wall proteins, cell wall synthesis genes, and genes of the chitin precursor pathway (23, 24, 50). It is striking that not all the genes of each functional category are turned on and that levels of activation are different for genes within each category. One finding that is consistent with the observed expression is that those genes that were up-regulated have prominent binding sites in their promoters for the transcription factor Rlm1p (YPD Database). Among the genes induced by fks1 stress is PTP2, encoding a phosphatase that modulates pathway activity (35). It will be of interest to see whether deletion of this gene leads to extra cell wall chitin deposition, since the null mutant appears to allow exaggerated and even excessive expression of other stress pathway genes.
Activation of the cell wall stress pathway leads to increased chitin synthesis and to up-regulation of the enzymes that synthesize the metabolic precursors of chitin. However, we and Lagorce et al. (28) have shown that increasing flux through the pathway, even without the application of stress, also leads to increased cell wall chitin synthesis. Lagorce et al. (28) used overexpression of GFA1, the first gene in the metabolic pathway. We have made the same observation and have also shown that chitin synthesis can be activated simply by adding GlcN to the growth medium. That stress is not a factor in this situation has been shown by our survey of genes that are activated and repressed following GlcN treatment. Very few genes are up- or down-regulated in common between cells subjected to weakened-cell-wall stress (fks1Δ mutants) and GlcN-treated cells, and the slight overlap is probably the secondary effect of extra chitin deposition or other minor changes in cell wall metabolism. The fact that cell wall stress is not involved suggests the hypothesis that when stress is applied to the cell, one of the primary responses is activation of the GFA1 metabolic pathway. The proteins or metabolic products of these reactions by themselves (and without other transcriptional changes) are able to increase the rate of chitin synthesis.
In addition to its regulation by cell wall stress, Gfa1p has properties and structural motifs that suggest that it may act in a regulatory manner itself. A global analysis of protein activities using proteome chips (58) has shown that Gfa1p binds in a strong, specific manner to phosphatidyl inositol 3-phosphate [PI(3)P] the phosphoinositide that plays a major role in the activity of intracellular vesicles involved in protein targeting (12). In the absence of either Vps34p or Vps15p, the proteins required to make PI(3)P, missorting of vacuolar and perhaps other proteins occurs. Biochemical studies localizing the Vps34/15 phosphoinositol kinase suggest a functional role for PI(3)P in membrane trafficking from the Golgi apparatus to the endosome and, by extension, a possible role of Gfa1p in these processes. Gfa1p also has a myristoylation site and possibly a VHS domain (15), found in the Vps27, Hrs, and STAM proteins (37). The latter domain is found in proteins associated with endocytosis and/or vesicular trafficking (31). Both of these motifs as well as the PI(3)P binding activity would allow direct participation of Gfa1p in the trafficking of Chs3p. In their extensive study of subcellular localization of the yeast proteome, Kumar et al. (27) found that Gfa1p has both cytoplasmic and “granular” distribution in the cell. Other specific points concerning the hypothesis that stress leads to activation of the GFA1 metabolic pathway and that the protein(s) or metabolic products by themselves play a major role in activating chitin synthesis are summarized briefly below.
(i) The hypothesis as stated may not be the whole story of weakened-cell-wall stress-induced chitin synthesis, since cell wall stress produces about a twofold increase in the expression of CHS3 and CHS7, genes that are not up-regulated by GlcN treatment. It has been demonstrated that increasing the transcription of these two genes may lead to an increase in the level of Chs3p in the cell (51). However, it has not been shown whether overexpression of these two proteins together necessarily leads to increased chitin synthesis. Lagorce et al. (28) found that when Chs3p and Chs7p were overproduced in cells also overproducing Gfa1p, no more chitin was made than in cells overproducing Gfa1p alone. This is consistent, however, with Chs4p (which was not overexpressed) limiting the activity of Chs3p (38). Therefore, the role played by extra transcription of CHS3 and CHS7, although suggestive, is not yet clear. Furthermore, in our studies there is no clear-cut quantitative relationship among in vitro enzymatic activity, stress-related chitin synthesis, and GlcN-stimulated chitin synthesis.
(ii) Given the high levels of chitin synthesized in response to cell wall stress, it might be anticipated that the amount of chitin made would reach a “saturation” level and that addition of GlcN would not bring about further increases. In fact, as shown in Fig. 5A, the increases in chitin levels upon addition of GlcN are roughly the same in wild-type and stressed cells, independent of the level present before GlcN addition. This would suggest that stress and GlcN operate independently. If this is true, it is surprising, since it has seemed probable to us that both have the same basic effect, namely, an increase in the acetylglucosamine phosphate pool. Viewed in another way, if GlcN saturates the soluble precursor pool, then stress must act by other mechanisms to increase chitin synthesis.
Although we and Lagorce et al. (28) found that chitin synthesis is proportional to the cellular level of Gfa1p, the enzyme responsible for the formation of GlcN-6-P, it is clear that UDP-GlcNAc concentrations are increased to only moderately higher levels in mutant strains that are making more than four times as much chitin as wild-type cells. More dramatically, when chitin synthesis is driven not by Gfa1p but by added GlcN, the level of UDP-GlcNAc is actually lowest in cells that show the highest rate of synthesis. It is thus clear that the UDP-GlcNAc concentration alone does not control the rate of chitin formation. In fact, if the intracellular concentration of substrate saturates the enzyme under all conditions (a question to be explored), chitin formation would be independent of the UDP-GlcNAc concentration, barring allosteric and other indirect effects. In summary, since chitin synthesis is proportional to the Gfa1p activity but not to the UDP-GlcNAc concentration, it is likely that either Gfa1p itself (see above) or a GlcN metabolite plays a role in the activity and/or localization of Chs3p.
(iii) Surprisingly, GlcN has only minor effects on gene expression on a whole-genome scale. Only a few genes seem responsive to GlcN treatment, and of these, suppressed genes are prevalent. Down-regulated genes are mostly those of mitochondrial respiration following steady-state GlcN exposure. Thus far, we cannot provide an explanation for this phenomenon. The largest group of up-regulated genes (again only in the steady state) are involved in mating and sporulation. This may well be a secondary effect of accumulated chitin rather than an effect of GlcN itself. Furthermore, upon treatment with GlcN, very few genes show as much as a threefold or higher change; most genes respond at lower levels. From the above we conclude that chitin biosynthesis is regulated by gene expression only to a minor extent and that GlcN likely affects chitin synthesis metabolically without drastically changing gene expression.
(iv) Although GlcN treatment does not increase CHS3 transcription, it does produce a substantial increase in the activity of the enzyme within 2 h. The mechanism of this activation remains to be determined. Changes in cellular location of the enzyme, activation by interaction with Chs4p, or some other type of activation may be involved.
(v) Our vesicle fractionation studies show that both enzymatically active and inactive forms of Chs3p are usually present in the cell. While Chs3p-containing chitosomes have been described (10, 45, 59), clarification is now needed for whether “active chitosomes” and “inactive chitosomes” would be more appropriate terminology. Because Chs3p activity requires interaction with Chs4p, our results also suggest that Chs4p is present in the active chitosome and is formed in a post-Golgi compartment.
(vi) The CHS3 gene, as well as all the genes responsible for its intracellular movement and localization, is required for both stress- and GlcN-induced chitin synthesis. We find we must revise the previous indication that the targeting gene, CHS6, is not required for stress-related chitin synthesis (40). Although there is indeed some bypassing of the chs6 mutation in the chs6 fks1 double mutant, this is reflected as an increase in chitin levels of less than 20% of the chitin made in wild-type cells. Interestingly, the chs6 mutation is more prone to a bypass when combined with gas1 mutation. And finally, when the chs5 deletion is combined with gas1 deletion, the bypass is exemplified by restoration of chitin levels to those of wild-type cells.
(vii) Although it is still possible that the metabolic precursors of chitin, GlcN-6-P, GlcNAc-6-P, and GlcNAc-1-P, may bring about some kind of allosteric activation of Chs3p, we have not yet seen stimulation in vitro. The only effective low-molecular-weight activator of the enzyme to date is GlcNAc, which is not a metabolic intermediate and which does not serve as a chitin primer (14).
Acknowledgments
This work was supported by grants from the National Institutes of Health (GM31318 and AI44070) to P.W.R. C.A.S. was supported by grant AI25780 to Stuart M. Levitz.
We are grateful to John V. Goodman for assistance in producing the figures and tables. We are also grateful to the Microarray Resource Center at Boston University Medical Center for their assistance.
REFERENCES
- 1.Ammerer, G. 1994. Sex, stress and integrity: the importance of MAP kinases in yeast. Curr. Opin. Genet. Dev. 4:90-95. [DOI] [PubMed] [Google Scholar]
- 2.Ballou, C. E., S. K. Maitra, J. W. Walker, and W. L. Whelan. 1977. Developmental defects associated with glucosamine auxotrophy in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 74:4351-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black, M. W., and R. B. Pelham. 2000. A selective transport route from Golgi to late endosomes that requires the yeast GGA proteins. J. Cell Biol. 151:587-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, A., and M. Tuile. 1998. Yeast gene analysis. Methods Microbiol. 26:318-320. [Google Scholar]
- 5.Bulawa, C. E. 1992. CSD2, CSD3, and CSD4, genes required for chitin synthesis in Saccharomyces cerevisiae: the CSD2 gene product is related to chitin synthases and to developmentally regulated proteins in Rhizobium species and Xenopus laevis. Mol. Cell. Biol. 12:1764-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabib, E. 1972. Chitin synthase system from yeast. Methods Enzymol. 28:572-580. [Google Scholar]
- 7.Cabib, E., D. H. Roh, M. Schmidt, L. B. Crotti, and A. Varma. 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276:19679-19682. [DOI] [PubMed] [Google Scholar]
- 8.Carotti, C., L. Ferrario, C. Roncero, M. H. Valdivieso, A. Duran, and L. Popolo. 2002. Maintenance of cell integrity in the gas1 mutant of Saccharomyces cerevisiae requires the Chs3p-targeting and activation pathway and involves an unusual Chs3p localization. Yeast 19:1113-1124. [DOI] [PubMed] [Google Scholar]
- 8a.Castro, O., L. Y. Chen, A. J. Parodi, and C. Abeijon. 1999. Uridine diphosphate-glucose transport into the endoplasmic reticulum of Saccharomyces cerevisiae: in vivo and in vitro evidence. Mol. Biol. Cell 10:1019-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, W. J., B. Santos, A. Duran, and E. Cabib. 1994. Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? Mol. Cell. Biol. 14:7685-7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chuang, J. S., and R. W. Schekman. 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dallies, N., J. Francois, and V. Paquet. 1998. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 14:1297-1306. [DOI] [PubMed] [Google Scholar]
- 12.De Camilli, P., S. D. Emr, P. S. McPherson, and P. Novick. 1996. Phosphoinositides as regulators in membrane traffic. Science 271:1533-1539. [DOI] [PubMed] [Google Scholar]
- 13.DeMarini, D. J., A. E. Adams, H. Fares, C. De Virgilio, G. Valle, J. S. Chuang, and J. R. Pringle. 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139:75-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duran, A., and E. Cabib. 1978. Solubilization and partial purification of yeast chitin synthetase. Confirmation of the zymogenic nature of the enzyme. J. Biol. Chem. 253:4419-4425. [PubMed] [Google Scholar]
- 15.Falquet, L., M. Pagni, P. Bucher, N. Hulo, C. J. Sigrist, K. Hofmann, and A. Bairoch. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30:235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Rodriguez, L. J., J. A. Trilla, C. Castro, M. H. Valdivieso, A. Duran, and C. Roncero. 2000. Characterization of the chitin biosynthesis process as a compensatory mechanism in the fks1 mutant of Saccharomyces cerevisiae. FEBS Lett. 478:84-88. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Rodriguez, L. J., A. Duran, and C. Roncero. 2000. Calcofluor antifungal action depends on chitin and a functional high-osmolarity glycerol response (HOG) pathway: evidence for a physiological role of the Saccharomyces cerevisiae HOG pathway under noninducing conditions. J. Bacteriol. 182:2428-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gash, A. P., and M. Werner-Washburne. 2002. The genomics of yeast responses to environmental stress and starvation. Funct. Integr. Genomics 2:181-192. [DOI] [PubMed] [Google Scholar]
- 19.Ghosh, S., H. J. Blumenthal, E. Davidson, and S. Roseman. 1960. Glucosamine metabolism. V. Enzymatic synthesis of glucosamine 6-phosphate. J. Biol. Chem. 235:1265-1273. [PubMed] [Google Scholar]
- 20.Graack, H. R., U. Cinque, and H. Kress. 2001. Functional regulation of glutamine:fructose-6-phosphate aminotransferase 1 (GFAT1) of Drosophila melanogaster in a UDP-N-acetylglucosamine and cAMP-dependent manner. Biochem. J. 360:401-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henar Valdivieso, M., A. Duran, and C. Roncero. 1999. Chitin synthases in yeast and fungi. EXS 87:55-69. [DOI] [PubMed] [Google Scholar]
- 22.Holthuis, J. C., B. J. Nichols, and H. R. Pelham. 1998. The syntaxin Tlg1p mediates trafficking of chitin synthase III to polarized growth sites in yeast. Mol. Biol. Cell 9:3383-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 24.Jung, U. S., and D. E. Levin. 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signaling pathway. Mol. Microbiol. 34:1049-1057. [DOI] [PubMed] [Google Scholar]
- 25.Klis, F. M., P. Mol, K. Hellingwerf, and S. Brul. 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26:239-256. [DOI] [PubMed] [Google Scholar]
- 26.Kucharczyk, N., M. A. Denisot, F. Le Goffic, and B. Badet. 1990. Glucosamine-6-phosphate synthase from Escherichia coli: determination of the mechanism of inactivation by N3-fumaroyl-l-2, 3-diaminopropionic derivatives. Biochemistry 29:3668-3676. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, A., S. Agarwal, J. A. Heyman, S. Matson, M. Heidtman, S. Piccirillo, L. Umansky, A. Drawid, R. Jansen, Y. Liu, K. H. Cheung, P. Miller, M. Gerstein, G. S. Roeder, and M. Snyder. 2002. Subcellular localization of the yeast proteome. Genes Dev. 16:707-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lagorce, A., V. Le Berre-Anton, B. Aguilar-Uscanga, H. Martin-Yken, A. Dagkessamanskaia, and J. Francois. 2002. Involvement of GFA1, which encodes glutamine-fructose-6-phosphate amidotransferase, in the activation of the chitin synthesis pathway in response to cell-wall defects in Saccharomyces cerevisiae. Eur. J. Biochem. 269:1697-1707. [DOI] [PubMed] [Google Scholar]
- 29.Leloir, L. F., and C. E. Cardini. 1953. The biosynthesis of glucosamine. Biochim. Biophys. Acta 12:15-22. [DOI] [PubMed] [Google Scholar]
- 30.Levin, D. E., B. Bowers, C. Y. Chen, Y. Kamada, and M. Watanabe. 1994. Dissecting the protein kinase C/MAP kinase signalling pathway of Saccharomyces cerevisiae. Cell Mol. Biol. Res. 40:229-239. [PubMed] [Google Scholar]
- 31.Lohi, O., and V. P. Lehto. 1998. VHS domain marks a group of proteins involved in endocytosis and vesicular trafficking. FEBS Lett. 440:255-257. [DOI] [PubMed] [Google Scholar]
- 32.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 33.Lucero, H. A., M. J. Kuranda, and D. A. Bulik. 2002. A non-radioactive, high throughput assay for chitin synthase activity. Anal. Biochem. 305:97-105. [DOI] [PubMed] [Google Scholar]
- 34.Lussier, M., A. M. Sdicu, A. Camirand, and H. Bussey. 1996. Functional characterization of the YUR1, KTR1, and KTR2 genes as members of the yeast KRE2/MNT1 mannosyltransferase gene family. J. Biol. Chem. 271:11001-11008. [DOI] [PubMed] [Google Scholar]
- 35.Mattison, C. P., S. S. Spencer, K. A Kresge, J. Lee, and I. M. Ota. 1999. Differential regulation of the cell wall integrity mitogen-activated protein kinase pathway in budding yeast by the protein tyrosine phosphatases Ptp2 and Ptp3. Mol. Cell. Biol. 19:7651-7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milewski, S., D. Kuszczak, R. Jedrzejczak, R. J. Smith, A. J. Brown, and G. W. Gooday. 1999. Oligomeric structure and regulation of Candida albicans glucosamine-6-phosphate synthase. J. Biol. Chem. 274:4000-4008. [DOI] [PubMed] [Google Scholar]
- 36a.Mishra, C., C. E. Semino, K. J. McCreath, H. de la Vega, B. J. Jones, C. A. Specht, and P. W. Robbins. 1997. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast 13:327-336. [DOI] [PubMed] [Google Scholar]
- 37.Misra, S., B. M. Beach, and J. H. Hurley. 2000. Structure of the VHS domain of human Tom1 (target of myb 1): insights into interactions with proteins and membranes. Biochemistry 39:11282-11290. [DOI] [PubMed] [Google Scholar]
- 38.Ono, N., T. Yabe, M. Sudoh, T. Nakajima, T. Yamada-Okabe, M. Arisawa, and H. Yamada-Okabe. 2000. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology 146:385-391. [DOI] [PubMed] [Google Scholar]
- 39.Orlean, P., E. Arnold, and W. Tanner. 1985. Apparent inhibition of glycoprotein synthesis by S. cerevisiae mating pheromones. FEBS Lett. 184:313-317. [DOI] [PubMed] [Google Scholar]
- 40.Osmond, B. C., C. A. Specht, and P. W. Robbins. 1999. Chitin synthase III: synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc. Natl. Acad. Sci. USA 96:11206-11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popolo, L., D. Gilardelli, P. Bonfante, and M. Vai. 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J. Bacteriol. 179:463-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ram, A. F., J. C. Kapteyn, R. C. Montijn, L. H. Caro, J. E. Douwes, W. Baginsky, P. Mazur, H. van den Ende, and F. M. Klis. 1998. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 180:1418-1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts, C. J., B. Nelson, M. J. Marton, R. Stoughton, M. R. Meyer, H. A. Bennett, Y. D. He, H. Dai, W. L. Walker, T. R. Hughes, M. Tyers, C. Boone, and S. H. Friend. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287:873-880. [DOI] [PubMed] [Google Scholar]
- 44.Roncero, C. 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41:67-78. [DOI] [PubMed] [Google Scholar]
- 45.Santos, B., and M. Snyder. 1997. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 136:95-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schekman, R., and V. Brawley. 1979. Localized deposition of chitin on the yeast cell surface in response to mating pheromone. Proc. Natl. Acad. Sci. USA 76:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shimizu, J., Y. Okumura, K. Yoda, and M. Yamasaki. 1997. A glutamine synthetase mutant of Saccharomyces cerevisiae shows defect in cell wall. J. Gen. Appl. Microbiol. 43:157-162. [DOI] [PubMed] [Google Scholar]
- 49.Spellman, P. T., G. Sherlock, M. Q. Zhang, V. R. Iyer, K. Anders, M. B. Eisen, P. O. Brown, D. Botstein, and B. Futcher. 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9:3273-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terashima, H., N. Yabuki, M. Arisawa, K. Hamada, and K. Kitada. 2000. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264:64-74. [DOI] [PubMed] [Google Scholar]
- 51.Trilla, J. A., A. Duran, and C. Roncero. 1999. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 145:1153-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Valdivia, R. H., D. Baggott, J. S. Chuang, and R. W. Schekman. 2002. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev. Cell 2:283-294. [DOI] [PubMed] [Google Scholar]
- 53.Valdivieso, M. H., L. Ferrario, M. Vai, A. Duran, and L. Popolo. 2000. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 182:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watzele, G., and W. Tanner. 1989. Cloning of the glutamine:fructose-6-phosphate amidotransferase gene from yeast. Pheromonal regulation of its transcription. J. Biol. Chem. 264:8753-8758. [PubMed] [Google Scholar]
- 55.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 56.Yanagisawa, K., D. Resnick, C. Abeijon, P. W. Robbins, and C. B. Hirschberg. 1990. A guanosine diphosphatase enriched in Golgi vesicles of Saccharomyces cerevisiae. Purification and characterization. J. Biol. Chem. 265:19351-19355. [PubMed] [Google Scholar]
- 57.Zheng, J., M. Khalil, and J. F. Cannon. 2000. Glc7p protein phosphatase inhibits expression of glutamine-fructose-6-phosphate transaminase from GFA1. J. Biol. Chem. 275:18070-18078. [DOI] [PubMed] [Google Scholar]
- 58.Zhu, H., M. Bilgin, R. Bangham, D. Hall, A. Casamayor, P. Bertone, N. Lan, R. Jansen, S. Bidlingmaier, T. Houfek, T. Mitchell, P. Miller, R. A. Dean, M. Gerstein, and M. Snyder. 2001. Global analysis of protein activities using proteome chips. Science 293:2101-2105. [DOI] [PubMed] [Google Scholar]
- 59.Ziman, M., J. S. Chuang, and R. W. Schekman. 1996. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell 7:1909-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ziman, M., J. S. Chuang, M. Tsung, S. Hamamoto, and R. Schekman. 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 9:1565-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]