Abstract
Gene expression profiling has revealed that diffuse large B cell lymphoma (DLBCL) consists of at least two distinct diseases. Patients with one DLBCL subtype, termed activated B cell–like (ABC) DLBCL, have a distinctly inferior prognosis. An untapped potential of gene expression profiling is its ability to identify pathogenic signaling pathways in cancer that are amenable to therapeutic attack. The gene expression profiles of ABC DLBCLs were notable for the high expression of target genes of the nuclear factor (NF)-κB transcription factors, raising the possibility that constitutive activity of the NF-κB pathway may contribute to the poor prognosis of these patients. Two cell line models of ABC DLBCL had high nuclear NF-κB DNA binding activity, constitutive IκB kinase (IKK) activity, and rapid IκBα degradation that was not seen in cell lines representing the other DLBCL subtype, germinal center B-like (GCB) DLBCL. Retroviral transduction of a super-repressor form of IκBα or dominant negative forms of IKKβ was toxic to ABC DLBCL cells but not GCB DLBCL cells. DNA content analysis showed that NF-κB inhibition caused both cell death and G1-phase growth arrest. These findings establish the NF-κB pathway as a new molecular target for drug development in the most clinically intractable subtype of DLBCL and demonstrate that the two DLBCL subtypes defined by gene expression profiling utilize distinct pathogenetic mechanisms.
Keywords: gene expression profiling, signal transduction, IκB kinase, microarray, apoptosis
Introduction
The Rel/nuclear factor (NF)*-κB transcription factors integrate diverse intracellular signaling pathways that are activated during normal cellular differentiation and during immune responses (1–4). NF-κB–dependent transcriptional activity is mediated by dimers of NF-κB family members (p50/105, p52/100, p65/RelA, RelB, or c-Rel), and is regulated by members of the IκB family of inhibitors, principally IκBα, which binds to NF-κB dimers and retains them in the cytoplasm. Upon phosphorylation by the IκB kinase (IKK) complex, IκBα is targeted for ubiquitination and proteasomal degradation, and released NF-κB dimers can translocate to the nucleus and activate transcription of target genes (3). NF-κB target genes encode diverse mediators of immune responses as well as regulators of cellular proliferation and apoptosis. The expression of these target genes varies, in part, with the cell type in which NF-κB is activated.
NF-κB activity is critical for normal B cell development and survival, and distinct NF-κB heterodimers participate in different stages of B cell differentiation and activation (for a review, see reference 5). NF-κB transcriptional activity in B cells primarily involves heterodimers between one of the transactivating subunits (c-Rel or RelA) and one of the nontransactivating subunits (p50, the processed form of NF-κB1 or p52, the processed form of NF-κB2). Mature murine B cell lines show constitutive low-level NF-κB activity, chiefly due to heterodimers of p50 and c-Rel (6). Targeted deletion of c-rel does not affect the number of mature B cells in mice, but such studies are complicated by functional redundancy of NF-κB family members. In mice lacking both c-rel and rela, mature B cells fail to develop due to accelerated apoptosis upon emergence of immature B cells from the bone marrow into the periphery (7). Immature B cells in such mice fail to undergo normal increases in the antiapoptotic protein, BCL-2, and its family member, A1/Bfl-1. Furthermore, transgenic overexpression of BCL-2 in c-rel and rela double mutant mice allows for partial maturation of B cells in the periphery (7). Similarly, resting B cells from mice lacking NF-κB1 show an increased rate of apoptosis, which can be prevented by enforced expression of BCL-2 (10). It is possible that the prosurvival function of basal NF-κB activity in mature B cells is the result of constitutive signaling through the B cell receptor (BCR), given that NF-κB is activated by BCR engagement and acute deletion of the BCR leads to rapid disappearance of mature B cells in mice (8).
Mature B cells also require acute increases in NF-κB activity in order to proliferate and survive in response to mitogens. Mice with inactivating mutations in c-rel and rela have normal numbers of mature B cells, but these cells are defective in their proliferative responses to BCR signaling and to lipopolysaccharide (9–11). The proliferative defect in c-Rel–deficient B cells is reflected in a block at the G1 stage of the cell cycle (10, 11). A possible mediator of NF-κB proliferative responses is IRF-4, a transcription factor that is required for proliferation of B and T lymphocytes (12) and that is induced by binding of c-Rel to NF-κB motifs in its promoter (13). However, optimal increases in cell numbers after BCR stimulation also involve inhibition of apoptosis, and NF-κB activity has again been implicated. c-Rel–deficient B cells die by apoptosis in response to BCR ligation (10, 14). The BCL-2 family member A1 is a target of NF-κB, and is induced after BCR stimulation (14). Ectopic expression of A1 in c-Rel–deficient B cells rescues the cells from apoptosis (14); BCL-2 provides similar protection from apoptosis (10, 14), but does not prevent the G1-phase block to proliferation (10). Thus, NF-κB controls both proliferation and apoptosis in B cells to maximize the response to mitogens.
NF-κB activation by mitogenic stimuli is normally self-limited, but constitutive nuclear NF-κB has been found in several types of cancers, raising the possibility that the antiapoptotic and/or pro-proliferative effects of NF-κB may contribute to malignant transformation or progression (for a review, see references 15 and 16). For example, in cell lines and some primary tumors of Hodgkin's disease, mutations of the IκBα gene result in its functional inactivation and the accumulation of p50/RelA heterodimers in the nucleus (for a review, see reference 17). In Hodgkin's disease cell lines, inhibition of this NF-κB activity by genetic means reduces cell viability (18–20). Likewise, transformation of B cells by Epstein-Barr virus induces NF-κB activity, and this activity is necessary for survival of these cells in vitro (21). In other types of lymphoid malignancies, constitutive NF-κB activity can occur occasionally due to translocations involving the NF-κB2 gene that disrupt its COOH terminus (22, 23), or by amplification of the c-rel locus (24, 25). While these various lines of investigation have led to the notion that NF-κB activation in human cancer may be of pathogenetic significance, more work is needed to demonstrate a direct link between NF-κB activity and clinical outcome.
Diffuse large B cell lymphoma (DLBCL), the most common type of nonHodgkin's lymphoma, presents an important clinical challenge given that only 40% of these patients are cured by conventional chemotherapy. The heterogeneous outcomes of these patients is due, in part, to the fact that this diagnostic category is comprised of at least two distinct disease entities that differ in their outcome after chemotherapy (26). This conclusion was drawn from an analysis of gene expression in DLBCL tumors using DNA microarrays that revealed two large DLBCL subgroups that differed in the expression of hundreds of genes. Furthermore, the gene expression profiles of these DLBCL subgroups resembled the profiles of normal B cells at different stages of differentiation, for which they were named. Germinal center B-like (GCB) DLBCLs expressed the “signature” genes of normal tonsillar germinal center B cells. Activated B cell–like (ABC) DLBCLs did not express the germinal center B cell signature genes but instead constitutively expressed genes that are normally induced in human blood B cells after BCR stimulation.
In this study, we investigated which signaling pathways might contribute to the biological and clinical differences between GCB and ABC DLBCLs. A survey of the gene expression profiles of these two DLBCL subtypes revealed that several genes expressed characteristically in ABC DLBCLs are known NF-κB target genes. We show that two DLBCL cell lines derived from ABC DLBCLs have NF-κB activation due to high constitutive IKK activity and IκBα degradation. Most importantly, abrogation of NF-κB activity was toxic to these ABC DLBCL cells and not to cell lines derived from GCB DLBCLs. This finding demonstrates that the two DLBCL subtypes have distinct pathogenetic mechanisms and establishes constitutive NF-κB activation as a target for therapeutic intervention in ABC DLBCLs.
Materials and Methods
Cell Lines.
All cell lines were maintained at 37°C in 5% carbon dioxide. ABC DLBCL lines OCI-Ly3 and OCI-Ly10, and the GCB DLBCL line OCI-Ly7, were maintained in Iscove's modified essential medium with β-mercaptoethanol (55 μM), penicillin (50 U/ml), streptomycin (50 μg/ml), and 20% heparinized normal human plasma. All other cell lines were maintained in RPMI 1640 medium with l-glutamine, Hepes, penicillin, streptomycin, and 10% FCS. As a control, SUDHL-6 cells were also cultured in the same medium as the OCI cell lines, and no increase was observed in either IκBα degradation or NF-κB DNA binding activity (data not shown).
Microarray Analysis of Gene Expression.
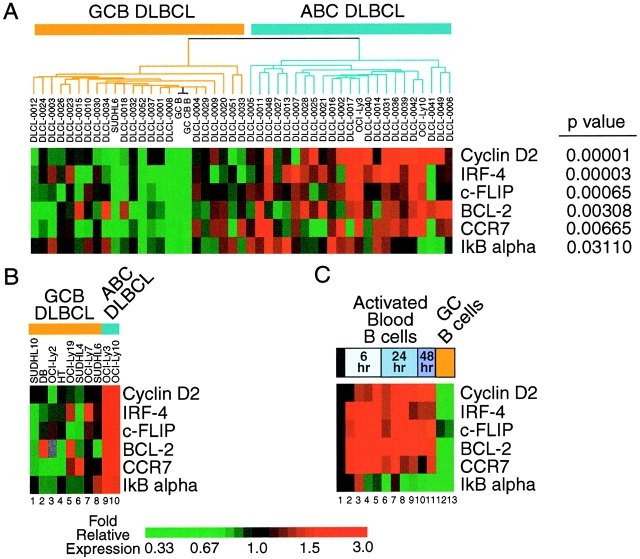
DNA microarray analysis of gene expression was performed using Lymphochip microarrays, as described previously (26). Briefly, mRNA from an experimental sample was labeled with Cy5 dUTP in a first strand cDNA reaction, and a reference cDNA probe, labeled with Cy3 dUTP, was prepared using mRNA from a pool of nine lymphoma cell lines. After hybridization of these two cDNA probes to a Lymphochip DNA microarray (27), the arrays were washed and bound fluorescent probe was detected using an Axon GenePix Scanner. The ratio of the pixel intensities for Cy5 and Cy3 fluorescence was calculated for each array element. For a given gene, these ratios were median-centered before display using a color scheme as described previously (28). Data for Figs. 1 A and C were taken from http://llmpp.nih.gov/lymphoma (26).
Figure 1.
DNA microarray analysis of NF-κB target gene expression in DLBCL. Results shown are the average of two different cDNA clones used to measure expression of each NF-κB target gene. Relative expression is displayed according to the color scale shown. (A) Expression in DLBCL lymph node biopsy samples. DLBCL cases are ordered and subdivided into GCB and ABC subtypes as defined in Fig. 1 of reference 26. (B) Expression in DLBCL cell lines. (C) Expression in normal B cell differentiation and activation. Data and samples correspond to those used in Fig. 4 of reference 25. Lanes 1–11 are adult blood CD19+ B cells: lane 1, resting; lanes 2 and 6, anti-IgM; lanes 3 and 7, anti-IgM plus CD40L; lanes 4 and 8, anti-IgM plus IL-4; lanes 5 and 9, anti-IgM plus CD40L plus IL-4; lane 10, anti-IgM plus CD40L (low concentration); lane 11, anti-IgM plus CD40L (high concentration). Lanes 12 and 13 are tonsil germinal center B cells: lane 12, total B cells; lane 13, centroblasts. In a “time zero transformation,” the data have been normalized to values obtained in resting blood B cells, and therefore the first column (lane 1) is entirely black.
Western Blot Analyses.
Total cell extracts were prepared for Western blotting as described previously (29). IκBα blots were probed with antibodies to full-length hIκBα (30). Equivalent amounts of cell extract were run in each lane. IKKα blots were probed with monoclonal anti-full length IKKα (BD PharMingen). BCL–XL was detected with mAb (H-5; Santa Cruz Biotechnology, Inc.) recognizing the COOH terminus.
Electrophoretic Mobility Shift Assays.
Nuclear extracts were prepared and electrophoretic mobility shift assays (EMSAs) performed using the palindromic NF-κB binding sequence as described previously (31), along with SP1 and Oct-1 probes (Promega). Equivalent amounts of nuclear extract (10 μg) were analyzed in each assay. Antibodies for supershift EMSAs were rabbit polyclonal antibodies raised against the following peptides conjugated to Keyhole Limpet Hemocyanin: p50, NH2-terminal 12 amino acids of human p50; p65/RelA, amino acids 2–15 of human p65/RelA; c-Rel, amino acids 573–587 of human c-Rel.
IKK Assay.
Endogenous IKK complex in cell extracts was immunoprecipitated on Protein G-sepharose beads (Pharmacia γ Bind Plus) using anti-IKKγ (SC8330; Santa Cruz Biotechnology, Inc.) as primary antibody. Kinase activity of the immune complex was assayed as described (29) using Glutathione S-transferase (GST)-IκBα (amino acids 1–72) and GST-IκBα S32G/S36A (amino acids 1–72) (2 μg) as substrates (provided by H. Ellinger-Ziegelbauer, National Institutes of Health, Bethesda, MD). Assays were performed with equivalent amounts of IKK immune complex as judged by Western blots of the complexes for IKKα.
Retroviral Constructs and Transductions.
The vXY-Puro retroviral vector was prepared from pBMN-Ires-Lyt2 (provided by G. Nolan, Stanford University, Stanford, CA) by exchanging a puromycin resistance cassette from internal ribosomal entry site-Puro (pIRES; CLONTECH Laboratories, Inc.) for the coding region of mouse CD8a (Lyt-2), as reported previously (32). The enhanced green fluorescent protein (EGFP)-F vector was similarly prepared, using NcoI and XhoI to excise the coding region of farnesylated EGFP from pEGFP-F (CLONTECH Laboratories, Inc.). Human IκBα wild-type and S32G/S36A coding regions, excised from a pcDNA3.1 vector (Invitrogen) after addition of an NH2-terminal FLAG epitope by PCR by J. Mueller, were as described previously (30). NH2 terminus FLAG-tagged A1/Bfl-1 was subcloned from pcDNA3 provided by A. Karsan. BCL–XL and BCL-2 were provided in pcDNA3 by R. Youle and Y.-T. Hsu, and were subcloned after addition of an NH2 terminus hemagglutinin (HA) epitope via interim subcloning. NH2 terminus HA-tagged IKKβ was obtained by PCR from a human spleen library (CLONTECH), and K44A or S177A/S181A mutations were introduced by site-directed mutagenesis. Transductions were performed by spin infection as described previously (33).
Survival Analysis of Transduced Cells.
For enumeration of live cells after vLyt-2 retroviral transductions, measured aliquots of cultures were centrifuged and the cell pellets stained on ice for 15 min after resuspension in 10 μL of a mixture of FITC-conjugated anti–mouse CD8a (BD PharMingen; 5 μg/ml) and ethidium homodimer-1 (Molecular Probes; 2.5 μM) in FACS® buffer (5% FBS in PBS with sodium azide 0.01%). After washing with FACS® buffer, pelleted cells were resuspended in FACS® buffer containing a precise volume of a suspension of 6-μm diameter polystyrene beads (Spherotech) and analyzed on a FACSort™ flow cytometer (Becton Dickinson) with a 488-nm argon laser. CELLQuest™ software was used to identify live Lyt-2-positive cells on the basis of light scatter properties, low staining by ethidium (red fluorescence), and positive FITC fluorescence; beads were identified by their distinctive combination of low forward and high side scatter. The total number of Lyt-2-positive live cells in the original culture was then determined from the relative numbers of beads and positive cells identified in the analysis, the concentration of beads in the bead suspension, and the volumes of original cultures and aliquots. For transductions with the vEGFP-F vector, culture aliquots were simply pelleted and resuspended with beads and ethidium homodimer-1, and then similarly analyzed using EGFP fluorescence.
DNA Content Analysis after vEGFP-F Transductions.
After pelleting of culture aliquots and resuspension in 0.5 ml PBS each, tubes were vortexed during addition of 3 ml of 95% ethanol. After fixation for 15 min on ice, cells were washed once with buffer (1% FBS in PBS with sodium azide 0.01%). Cells were then incubated for 30 min at 37°C in buffer containing propidium iodide (10 μg/ml) and RNase A (0.25 mg/ml), then FACS®-analyzed without washing.
Evaluation of Anti-Ig Toxicity.
After puromycin selection, SUDHL-6 cells transduced with vXY-Puro expressing HA-tagged BCL–XL or empty vector were cultured with polyclonal goat F(ab′)2 fragments recognizing the Fc portion of human IgM (Jackson ImmunoResearch Laboratories). At various durations of culture, live cells were enumerated using reagents from the LIVE/DEAD staining kit (Molecular Probes) and compared with the number in matched untreated cultures. Measured aliquots of cultures were centrifuged and the cell pellets resuspended in PBS containing calcein AM (0.5 μM), ethidium homodimer-1 (5 μM), and a precise volume of a suspension of polystyrene beads as above. After a 10-min incubation at room temperature, the suspensions of beads and stained cells were FACS®-analyzed as above. Live cells were identified on the basis of light scatter, green calcein fluorescence, and lack of staining by ethidium.
Results
Constitutive NF-κB Activation Correlates with Gene Expression Profiles in Cell Lines.
Previous microarray studies of gene expression in primary tumors of DLBCL identified two large DLBCL subgroups that differed in expression of hundreds of genes (26). One DLBCL subgroup was termed ABC DLBCL since it was characterized by high expression of genes that are induced in blood B cells by BCR signaling. BCR stimulation activates the NF-κB pathway, and so we reexamined our published data set specifically for the expression of known NF-kB target genes. Many were found to be more highly expressed in ABC-DLBCL lines and primary tumors, and five genes were differentially expressed with a statistical significance of P < 0.05. Expression of these genes is shown in Fig. 1 A, after median centering of the raw gene expression data across the spectrum of samples illustrated. These NF-κB target genes (cyclin D2 [34], IRF-4 [13], c-FLIP [35], BCL-2 [7, 36], CCR7 [36], and IκBα [37, 38]) were highly expressed in many of the ABC DLBCLs and tended to be poorly expressed in GCB DLBCLs. Each of these NF-κB target genes were differentially expressed between the DLBCL subtypes with high statistical significance as determined by the Student's t test (Fig. 1 A). Some heterogeneity in expression of the NF-κB target genes was observed among these primary tumor specimens, which might indicate that the two major DLBCL subtypes that were defined previously might themselves include minor DLBCL subtypes that vary in the expression of NF-κB target genes.
These findings suggested that activation of the NF-κB signaling pathway is a feature common to many ABC DLBCL cases. To investigate this possibility in depth, we turned to DLBCL cell lines that are models for the two DLBCL types. Two DLBCL cell lines, OCI-Ly3 and OCI-Ly10, were shown previously to be highly related in gene expression to ABC DLBCL tumor biopsies (26). Like ABC DLBCL tumors, these cell lines failed to express germinal center B cell “signature” genes and instead expressed a set of genes that distinguished ABC DLBCL from GCB DLBCL. Conversely, the SUDHL-6 cell line strongly resembled GCB DLBCL tumors in gene expression (26). In subsequent microarray experiments, several more cell lines were found to resemble GCB DLBCLs (data not shown). In this new data set prepared from cell lines, including new microarray studies of OCI-Ly3, OCI-Ly10, and SUDHL-6, we found that many known NF-κB target genes were more highly expressed in ABC-DLBCL cell lines than in GCB DLBCL cell lines. Results for the genes used in Fig. 1 A are shown for these cell lines in Fig. 1 B, after median-centering the raw gene expression data across the new spectrum of samples. We also examined expression of these NF-κB target genes during normal B cell differentiation and activation (Fig. 1 C). As expected, these genes were upregulated during activation of blood B cells by BCR stimulation. Interestingly, normal germinal center B cells did not express these NF-κB target genes highly, which is in keeping with the lower expression of NF-κB target genes in GCB DLBCL.
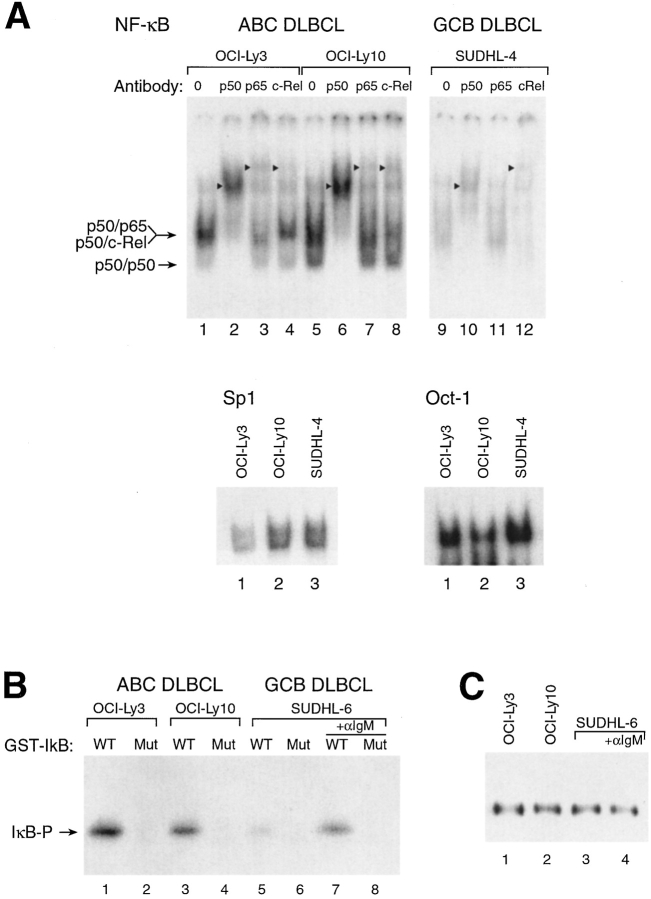
EMSAs confirmed constitutive NF-κB activation in ABC DLBCL cell lines (Fig. 2 A). Nuclear extracts of the ABC DLBCL cell lines contained high levels of NF-κB DNA binding activity whereas two GCB DLBCL cell lines had much lower levels; this difference was not seen in control EMSAs using Oct-1 and Sp1 probes. Addition of antibodies specific for p50 to the EMSAs revealed that most of the NF-κB DNA binding activity involved heterodimers containing the p50 NF-κB1 polypeptide. Although less apparent, supershifted complexes were also produced by antibodies to both the p65 and c-Rel subunits in the EMSAs of ABC DLBCL cell lines, demonstrating that these cell lines had nuclear p50/RelA and p50/c-Rel heterodimers. In contrast, the nuclear NF-κB DNA binding activity in the GCB cell line SUDHL-4 was much lower, and was largely mediated by p50/c-Rel heterodimers.
Figure 2.
NF-κB and IKK kinase are constitutively active in ABC DLBCL but not GCB DLBCL cell lines. (A) EMSAs of nuclear extracts from DLBCL cell lines for NF-κB DNA-binding activity. Supershifting antibodies are indicated, as is the mobility of NF-κB heterodimers. Arrowheads indicate the position of supershifted bands. The identity of the p50/p50 homodimer species was established by comparison with extracts of cells transfected with (and overexpressing) p50 (data not shown). EMSAs using Oct-1 and Sp1 probes showed no increased binding activity in ABC DLBCL lines relative to GCB DLBCL lines. Equivalent amounts of protein (10 μg) were used for all reactions. Comparable results were obtained with the GCB DLBCL line SUDHL-6 (data not shown). (B) IKK kinase assay of anti-IKKγ immune complexes from DLBCL cell lines. GST-IκBα (amino acids 1–72) was substrate as follows: WT, wild-type IκBα amino acids 1–72; Mut, IκBα amino acids 1–72 with phosphoacceptor serine residues 32 and 36 substituted by glycine and alanine, respectively. Where indicated, SUDHL-6 cells were activated with anti-IgM (Jackson ImmunoResearch Laboratories) for 10 min before extraction. (C) Western blot analysis of immunoprecipitated IKKγ complexes used in B. Western blots were developed with an antibody to IKKα.
Constitutive IKK Activity and IκB Degradation in ABC DLBCL Cell Lines.
In normal cells, two important regulators of NF-κB activity are IκBα and its kinase IKK. We used an in vitro kinase assay to measure the activity of IKK in the DLBCL cell lines, using equivalent amounts of immunoprecipitated IKK protein (Fig. 2, B and C). IKK from unstimulated ABC DLBCL cell lines produced significantly greater phosphorylation of a glutathione S-transferase-IκBα fusion protein than did IKK from the GCB DLBCL cell line SUDHL-6. After activation of SUDHL-6 by cross-linking its BCR, however, IKK activity rose to levels comparable to those observed in unstimulated ABC DLBCL cell lines. The specificity of the kinase activity detected was confirmed by the lack of phosphorylation of a mutant IκB substrate in which the serines that are phosphorylated by IKK were substituted with glycine and alanine residues. Therefore, in ABC DLBCL cell lines, the greater NF-κB DNA binding activity and the expression of the NF-κB target genes are likely due to constitutive activity of IKK.
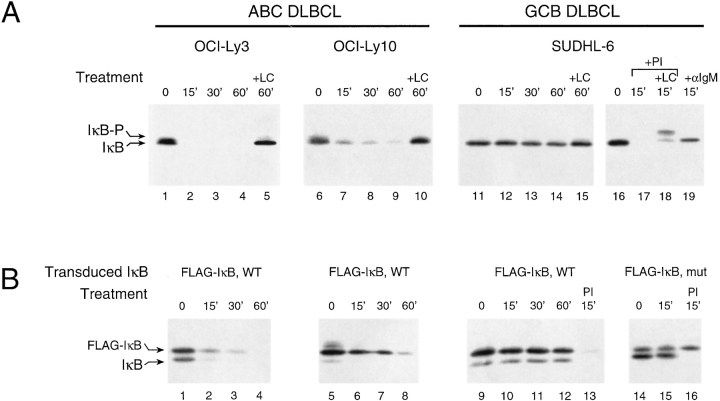
This conclusion was supported by studies of IκBα protein turnover in the DLBCL cell lines. Because IκBα transcription is itself positively regulated by NF-κB activity (37, 38), the steady-state level of IκBα protein does not necessarily reflect the rate of phosphorylation by IKK and subsequent degradation. Indeed, full-length IκBα protein is found at comparable levels in our untreated cell lines (Fig. 3 A). However, when ABC DLBCL cell lines were treated with cycloheximide (CHX) to block new protein synthesis, the level of IκBα declined rapidly through phosphorylation and degradation (Fig. 3 A). By contrast, the IκBα level in SUDHL-6 was significantly more stable upon CHX treatment unless the cells were also stimulated by BCR cross-linking or by PMA and ionomycin treatment (Fig. 3 A). Lactacystin, a drug that inhibits degradation of ubiquitinated IκBα by proteasomes, prevented the CHX-induced loss of IκBα in resting ABC DLBCL cell lines and in stimulated SUDHL-6 cells (Fig. 3 A). In related studies, FLAG-tagged wild-type IκBα was expressed in the DLBCL cell lines using a retroviral vector (vXY-Puro) which also translates a puromycin resistance gene located 3′ of an internal ribosomal entry site element. After retroviral transduction and puromycin selection, polyclonal populations of infected cells were studied for expression and metabolism of FLAG-tagged IκBα. Wild-type FLAG-tagged IκBα was metabolized in the same manner as the native protein in the three cell lines, with rapid degradation observed in the ABC DLBCL cell lines (Fig. 3 B). This confirms the differences in IκBα degradation rates between the cell lines shown in Fig. 3 A and shows that these differences are not due to possible alterations in the endogenous IκBα genes in these cell lines. Furthermore, these findings show that the machinery for constitutive IκBα degradation in ABC-DLBCL lines is robust and is not affected by the expression of additional exogenous wild-type IkBα.
Figure 3.
(A) Rapid, constitutive, and proteasome-dependent degradation of IκBα in ABC DLBCL cell lines. Western blot analysis of IκBα in extracts of CHX-treated cells (5 × 105 cells per lane, 20 μg/ml CHX). SUDHL-6 showed stable IκBα unless cells were activated for the times indicated by PMA (Sigma-Aldrich; 40 ng/ml plus ionomycin) (Calbiochem; 2 μM) (PI) or anti-IgM (50 μg/ml). The more slowly migrating form of IκBα (IκB-P) seen in cells treated with clastolactacystin β−lactone (LC; Calbiochem, 80 μM), the active metabolite of lactacystin, most likely represents phosphorylated IκB. (B) Ectopically expressed wild-type IκBα was degraded similarly to endogenous IκBα in ABC and GCB DLBCL cell lines. Western blot analysis of IκBα in extracts of CHX-treated cells stably transduced with retroviruses expressing FLAG epitope-tagged wild-type IκBα or super-repressor IκBα (S32G/S36A) and treated with CHX with and without PI for the times indicated. In SUDHL-6, the super-repressor form of FLAG-tagged IκBα did not affect the degradation of endogenous IκBα induced by PI but was itself resistant to degradation.
Inhibition of Constitutive NF-κB Activity Is Toxic to ABC DLBCL Cell Lines.
The above studies suggested that constitutive NF-κB activation in ABC DLBCL cell lines proceeds through a classical pathway of IKK-mediated IκBα degradation. To inhibit NF-κB activation in these cell lines, we used a “super-repressor” IκBα that cannot be phosphorylated by IKK and, as a result, can inhibit acute induction of NF-κB activity (30). In multiple attempts to introduce this mutant IκBα into ABC DLBCL lines by retroviral transduction, we consistently observed that there were few or no surviving cells at 2 d after addition of puromycin. In contrast, large numbers of surviving ABC DLBCL cells were obtained with the control empty retrovirus and with a wild-type IκBα retrovirus. The toxicity of the super-repressor IκBα for ABC DLBCL cell lines was selective since transduction of GCB DLBCL cell lines with this virus and with control virus yielded comparably high numbers of surviving cells. In stably transduced SUDHL-6 cells, wild-type IκBα was readily degraded upon PI treatment, but super-repressor IκBα was resistant to this treatment, as expected (Fig. 3 B).
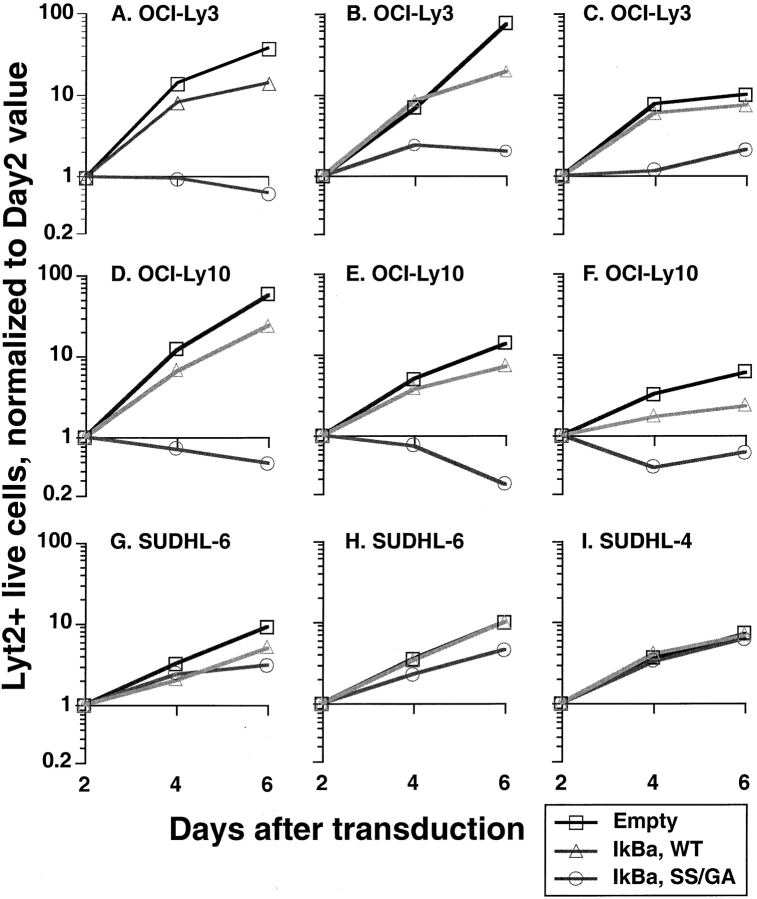
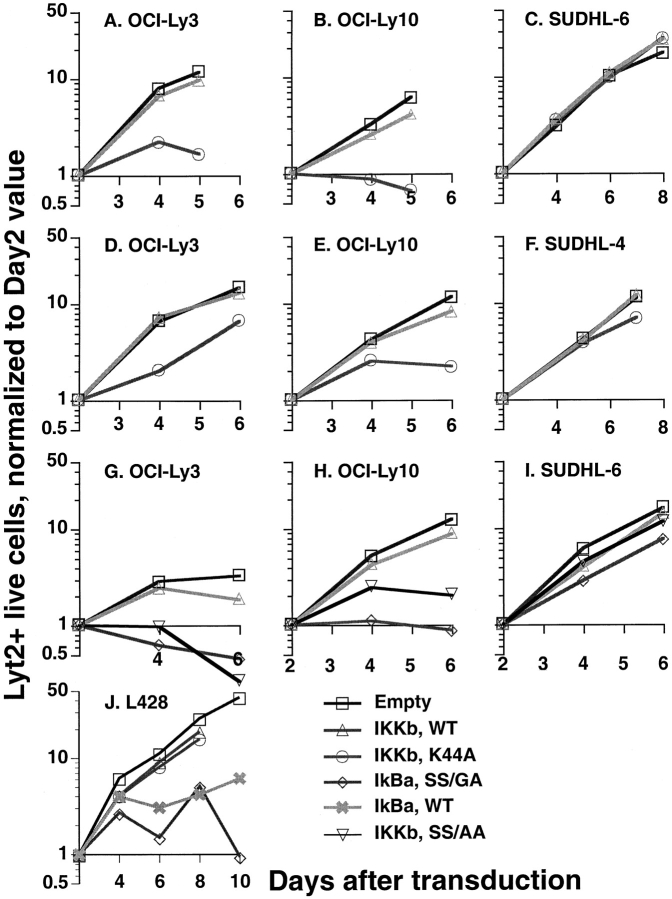
To confirm and quantitate the toxicity of super-repressor IκBα for ABC DLBCL cells, we expressed super-repressor IκBα using a retroviral vector (vLyt-2) that also expressed mouse Lyt-2, a surface marker not found in human cells. After infection by vLyt-2 retroviruses, the survival and growth of transduced cells was directly measured by flow cytometry for Lyt-2. ABC and GCB DLBCL cell lines were transduced with vLyt2 retroviruses expressing super-repressor or wild-type IκBα, or with control virus. 2 d after infection, the three retroviruses each yielded Lyt-2-positive cells in all cell lines with comparable efficiency (data not shown). After this time, however, the ABC DLBCL cells transduced with the super-repressor IκBα retrovirus either declined in number or accumulated at a slower rate than cells transduced with the wild-type IκBα virus or with control virus (Fig.4 A–F). On the other hand, GCB DLBCL cell lines were unaffected by transduction with super-repressor IκBα retrovirus, and increased in number exponentially over time (Fig. 4 G-I). These results establish that ABC and GCB DLBCL cell lines differ in their dependence on NF-κB activity for survival.
Figure 4.
Responses of DLBCL cell lines to transduction with super-repressor IκBα. The results of individual experiments are shown; those shown for the same cell line were obtained from separate transductions performed at different times. For each experiment, equal numbers of cells were transduced with vLyt-2 retrovirus containing wild-type IκBα, SS mutant (S32G/S36A) IκBα, or insert-empty vector. For each vector, equal volumes of the same viral supernatants were applied to the different lines. On the days shown after transduction, live transduced cells were quantified by flow cytometry as described in Materials and Methods, using FITC-conjugated anti–mouse CD8a (Lyt-2), ethidium, and beads. Results from each cell line, vector, and transduction were normalized to the corresponding day 2 value.
Constitutive NF-κB Activation in ABC Lines Proceeds through IKK.
These results suggested that constitutive IKK activity was necessary for survival of ABC DLBCL cells since the super-repressor IκBα that we used cannot be phosphorylated by IKK. To test this hypothesis further, we transduced the DLBCL cell lines with a catalytically inactive form of IKKβ that was previously shown to function as a dominant negative inhibitor of IKK activity (39). Retroviral transduction of this mutant IKKβ was toxic to ABC DLBCL cell lines (Fig. 5 A, B, D, and E) but not to GCB DLBCL cell lines (Fig. 5 C and F). In contrast, ABC DLBCL cell lines infected with a retrovirus expressing wild-type IKKβ or with a control retrovirus proliferated exponentially. Activation of the IKK complex requires phosphorylation of serine residues 177 and 181 in an “activation loop” of IKKβ and substitution of alanine for those serines results in a dominant negative form of IKKβ (40). Retroviral transduction of this dominant negative form of IKKβ was toxic to OCI-Ly3 and OCI-Ly10 cells but not SUDHL-6 cells (Fig. 5 G–I). In control experiments, we examined the effect of the various NF-κB inhibitors on the growth of a Hodgkin's lymphoma cell line, L428, which has constitutive nuclear NF-κB that does not require IKK activity. L428 has NF-κB activity due to an inactivating mutation in the IκBα gene (20) and therefore, as would be expected, growth of L428 was inhibited by both wild-type and super-repressor IκBα but not by the catalytically inactive dominant negative form of IKKβ (Fig. 5 H). Taken together, these results demonstrate that the viability of ABC DLBCL cell lines depends critically upon constitutive IKK activity leading to nuclear NF-κB activation of target genes.
Figure 5.
Responses of DLBCL cell lines to transduction with mutant IKKβ. The results of individual experiments are shown; those shown for the same cell line were obtained from separate transductions performed at different times. For each cell line, equal numbers of cells were transduced with wild-type IKKβ, kinase-inactive mutant (K44A) IKKβ, activation-loop mutant (S177A/S181/A) IKKβ, or insert-empty vLyt-2 retrovirus. Hodgkin's lymphoma cell line L428 was also transduced with vLyt-2 retrovirus containing forms of IκBα or IKKβ. For each vector, equal amounts of the same viral supernatants were applied to the different lines. Live transduced cells were enumerated by flow cytometry on days shown, as in Fig. 4. Results from each cell line, vector, and transduction were normalized to the corresponding day 2 value.
NF-κB Inhibition Causes Apoptosis and Cell-Cycle Arrest in ABC DLBCL Lines.
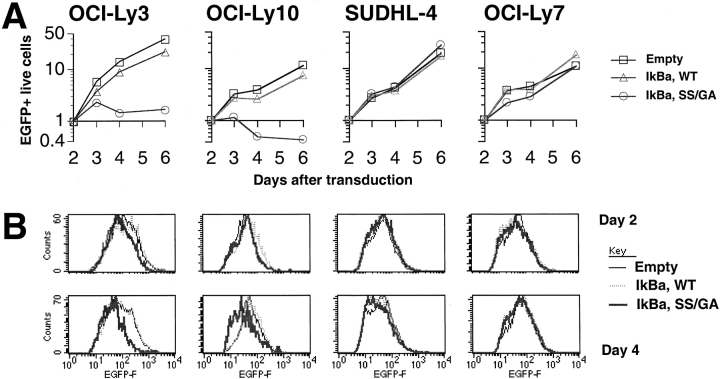
In many of the experiments presented above, blockade of NF-κB signaling in ABC DLBCL cell lines resulted in a net decrease in cell numbers, providing indirect evidence that NF-κB prevents cell death in these cells. Nevertheless, it was also possible that a cell cycle arrest might contribute to the decreased cell numbers after NF-κB inhibition. To test this possibility, and to provide direct evidence for cell death after NF-κB inhibition, we turned to a different retroviral vector that had two advantages over the ones used previously. This bicistronic vector expresses a form of EGFP modified to contain a farnesylation signal, which we thought might facilitate the detection of dying cells given the known stability of the EGFP protein and the membrane insertion of the farnesylated domain. Furthermore, this retroviral construct had higher transduction efficiencies, thus permitting the recovery of sufficient transduced cells to perform an analysis of the cell cycle. Transduction of super-repressor IκBα into ABC DLBCL cell lines using this new retroviral vector reduced the number of live cells relative to control cells transduced with either a wild-type IκBα retrovirus or with an empty retrovirus (Fig. 6 A). Again, super-repressor IκBα was not toxic to GCB DLBCL cell lines.
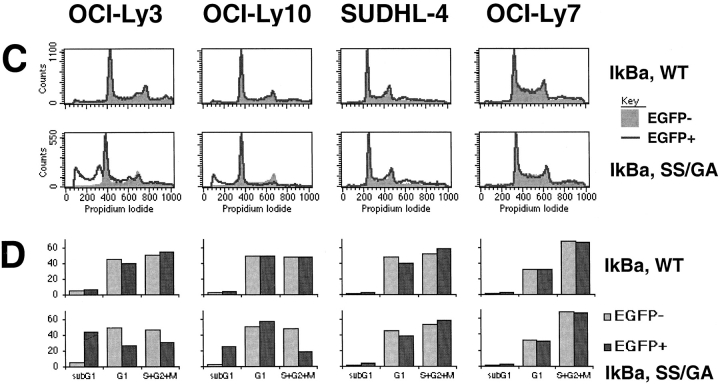
Figure 6.
Toxicity of super-repressor IκBα in ABC DLBCL lines. (A) Response of ABC DLBCL lines OCI-Ly3 and OCI-Ly10, and GCB DLBCL lines SUDHL-4 and OCI-Ly7, to transduction with bicistronic retroviruses expressing EGFP as well as wild-type IκBα, super-repressor SS/GA mutant (S32G/S36A) IκBα, or no insert. Absolute numbers of live EGFP+ cells were enumerated by flow cytometry on days shown, and normalized to the corresponding day 2 value. (B) Histograms of EGFP fluorescence in live transduced cells. For each cell line and day after transduction, histograms for live EGFP+ cells from the three retroviruses were normalized based on the maximum count value for each. (C) Histograms of DNA content, based on propidium iodide staining of all cells in cultures 4 d after transduction. For each cell line and retrovirus, histograms for EGFP− and EGFP+ cells were normalized based on the maximum count value for each. The histograms for EGFP− cells are shown shaded and filled, while EGFP+ cells are shown as a line. Results for empty retrovirus (data not shown) were similar to those for wild-type IκBα. (D) Relative proportions of sub-G1, G1, and G2+M phase cells 4 d after transduction. Values are shown for the comparable EGFP− and EGFP+ cells.
Interestingly, the histogram of EGFP fluorescence in living transduced ABC DLBCL cells showed a shift of the cell population over time toward lower values with the super-repressor IκBα (Fig. 6 B). This effect was not seen with ABC DLBCL cells transduced with wild-type IκBα or empty retroviruses nor was it seen in GCB DLBCL cells transduced with super-repressor IκBα retrovirus. Since the retroviruses used are bicistronic, it is likely that the decrease in EGFP fluorescence reflects a parallel change in the histogram of super-repressor IκBα expression. This shift became even more pronounced with time, and was also present in earlier experiments using the vLyt2 retrovirus (data not shown). Given that the overall numbers of ABC DLBCL cells transduced with the super-repressor IκBα retrovirus decreased or stayed constant over time, these results suggested that cells expressing high levels of EGFP and super-repressor IκBα were negatively selected from the population due to cell death.
To provide further evidence for cell death in super-repressor IκBα transduced cells, and to evaluate cell cycle progression, we measured DNA content by flow cytometry. Changes due to super-repressor IκBα in ABC DLBCL lines were most readily apparent at 4 d after transduction (Fig. 6 C and D). Both ABC DLBCL cell lines showed a decreased proportion of EGFP+ cells corresponding to the S and G2+M cell cycle phases, indicating a partial G1-phase cell cycle arrest. Furthermore the cell lines showed an increase in cells with less than 2N DNA content, a finding associated with apoptotic cell death. These changes were not seen in ABC DLBCL cells transduced with wild-type IκBα or empty retroviruses, nor were they seen in GCB DLBCL lines expressing super-repressor IκBα. Thus, inhibition of NF-κB had two effects on ABC DLBCLs: cell cycle arrest and cell death.
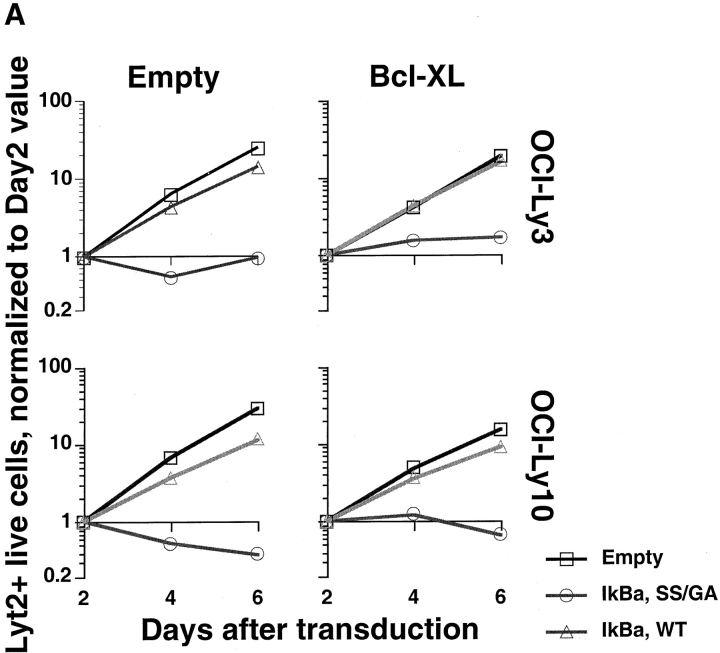
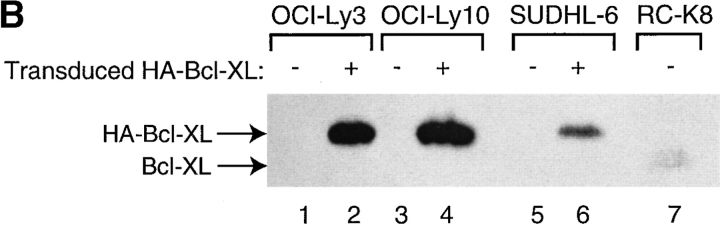
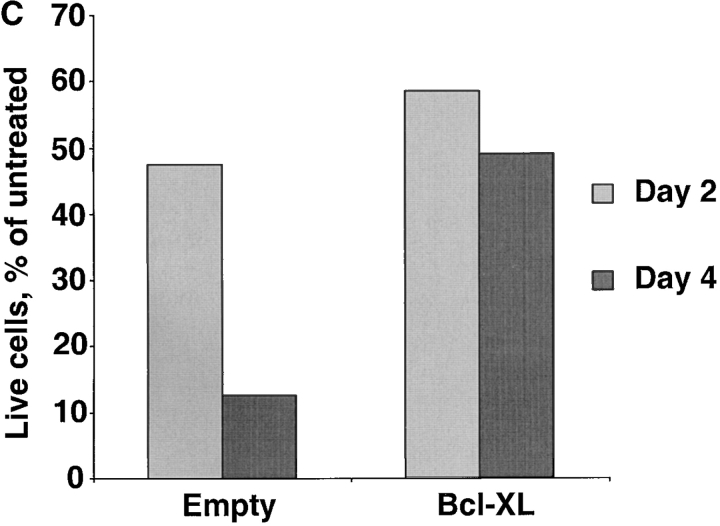
Several known NF-κB target genes are antiapoptotic members of the BCL-2 family, including BCL-2 itself, A1 and BCL–XL (14, 41–46). Therefore, we tested whether the antiapoptotic activity of BCL-2 family members could protect ABC DLBCL cells against the toxicity caused by inactivating NF-κB. OCI-Ly3 and OCI-Ly10 cells were first transduced with control or BCL–XL-expressing vXY-Puro retroviruses; after puromycin selection, resistant cells were superinfected with vLyt-2 retroviruses expressing IκBα (wild-type or super-repressor) or with control vLyt-2 retrovirus. BCL–XL afforded no protection against the toxic effects of super-repressor IκBα (Fig. 7 A), despite the fact that the transduced OCI-Ly3 and OCI-Ly10 cells expressed high levels of BCL–XL protein (Fig. 7 B). Likewise, transduction of OCI-Ly3 cells with retroviruses expressing A1 and BCL-2 did not block the effects of super-repressor IκBα (data not shown). In control experiments, we found that retrovirally mediated expression of BCL–XL was able to protect SUDHL-6 cells against apoptosis caused by cross-linking of the surface Ig receptor, demonstrating that this retrovirus can express functionally meaningful levels of BCL–XL (Fig. 7 B and C). These results demonstrate that the prosurvival effect of NF-κB in ABC DLBCL cell lines is likely due to the activity of multiple NF-κB target genes, and cannot be simply mimicked by expression of a single antiapoptotic BCL-2 family member.
Figure 7.
BCL-2 family members fail to block the toxicity of NF-κB inhibition. (A) The indicated ABC DLBCL cell lines were transduced with vXY-Puro retrovirus expressing BCL–XL or control retrovirus and puromycin-selected, then transduced with vLyt-2 retrovirus expressing forms of IκBα or control retrovirus. Live vLyt2-transduced cells were quantitated as indicated in the legend to Fig. 4. Values represent the average of two experiments with BCL–XL-transduced lines and three with control lines. (B) Western blot analysis for expression of endogenous and transduced (HA-tagged) BCL–XL, probed with antibody to BCL–XL COOH terminus. Lines transduced with vXY-Puro retrovirus expressing BCL–XL were puromycin-selected. Other lines shown were untransduced, including RC-K8, a DLBCL line with high expression of endogenous BCL–XL mRNA and protein. (C) Effect of BCL–XL on survival of SUDHL-6 after BCR cross-linking. In cells transduced with BCL–XL or control retrovirus, the number of live cells in cultures treated with anti-IgM was compared with that in matched untreated cultures.
Discussion
Recently, gene expression profiling was able to stratify DLBCL patients into two subgroups that had markedly divergent clinical outcomes after multiagent chemotherapy (26). However, given the extensive differences in gene expression between these two DLBCL subtypes, it was not clear which individual genes or signaling pathways contributed critically to this biological and clinical dichotomy. ABC DLBCL tumors expressed genes that are characteristically activated in blood B cells by signaling through the BCR. One of the signaling pathways prominently engaged after BCR stimulation is the NF-κB pathway, and ABC DLBCLs frequently expressed NF-κB target genes highly. Conversely, GCB DLBCLs generally had low expression of NF-κB target genes, as did normal germinal center B cells, from which this DLBCL subtype is putatively derived. Two DLBCL cell lines were indistinguishable in gene expression from ABC DLBCL tumors overall, and consequently expressed NF-κB targets highly. We found that these cell lines had constitutive nuclear NF-κB, IκBα degradation, and IKK activity, whereas cell lines derived from GCB DLBCL lacked this evidence of NF-κB pathway activation. Most importantly, dominant interference with the NF-κB pathway was toxic to ABC DLBCL cells, but not to GCB DLBCL cells, thus validating NF-κB and its upstream activating pathways as molecular targets in ABC DLBCL.
There are two general models to explain the constitutive NF-kB activation in ABC DLBCL. The first is that ABC DLBCLs may be derived from a B cell differentiation stage at which NF-κB is normally activated. Although the cell of origin of ABC DLBCL has not been clearly defined, a recently described minor germinal center B cell subtype is an intriguing candidate. Approximately 5% of germinal center B cells show decreased expression of BCL-6, increased expression of Blimp-1 and expression of IRF-4 (47, 48), all features of ABC DLBCL. These cells also have a plasmacytic morphology suggesting that they may be intermediates on the path to terminal plasmacytic differentiation. If ABC DLBCLs are derived from this cell type, then the activity of NF-κB in these tumors may reflect the physiological use of this pathway at this differentiation stage.
In an alternative model, the activation of NF-κB is an oncogenic event that occurs in ABC DLBCLs and is unrelated to the physiological role of NF-κB in B cell differentiation. Indeed, the avian retrovirus v-rel gene encodes a potent oncogene that causes lymphoid tumors in chickens (49), and recently it has been shown that human c-Rel can also transform primary chicken spleen cells (50). The genomic locus containing the human c-rel gene is amplified and overexpressed in 23% of DLBCLs (51), especially in those arising in extranodal sites, but the mechanistic significance of this is unknown. Another relatively rare genomic event in B cell malignancies is alteration of the NF-κB2 gene by translocation and deletion of its 3′ end (22, 23, 52). Removal of the 3′ end of the NF-κB2 gene increases the NF-κB2 mRNA expression and deletes the COOH-terminal transrepression domain of NF-κB2, creating a constitutive activator of transcription (53, 54) that can transform 3T3 fibroblasts (55). These relatively infrequent genomic modifications of the c-rel and NF-κB2 loci are unlikely to account for the activation of the NF-κB pathway in ABC DLBCL since this DLBCL subtype accounts for about half of all DLBCLs (26), and since no overexpression of the c-rel or NF-κB2 genes is evident in ABC DLBCLs (data not shown).
NF-κB family members can, in some cases, function as transforming oncogenes, but their primary role in malignant transformation may be to prevent apoptosis that can be caused by various other oncogenes. For example, transformation by activated ras, BCR-ABL and by the HTLV-I tax protein requires activation of the NF-κB pathway (56–59). Each of these oncogenes activates signaling pathways that can trigger apoptosis, but they also activate NF-κB, directly or indirectly, thereby preventing apoptosis and promoting cellular transformation. Other oncogenes such as c-myc cause apoptosis and not cellular transformation under conditions of growth factor deprivation (60), and NF-κB activation by serum-derived PDGF can protect cells against this apoptosis (61). These observations suggest an intriguing hybrid model to explain NF-kB activation in ABC DLBCL: physiological NF-κB activation at a particular stage of B cell differentiation might permit certain oncogenic events to occur that would otherwise cause apoptosis in the absence of NF-kB. From this perspective, the normal induction of NF-κB during immune responses might be viewed as a predisposing event to lymphoma formation.
In keeping with the established ability of the NF-κB pathway to prevent cell death (14), inhibition of this pathway in ABC DLBCL cell lines decreased cell numbers and caused the appearance of cells with sub-genomic DNA content. However, the mechanisms underlying this pro-survival effect of NF-κB are likely to be multifactorial. In various cell types, NF-κB can transcriptionally induce one or more antiapoptotic BCL-2 family members, including A1, BCL-2, and BCL–XL, and ABC DLBCL cell lines express BCL-2 (Fig. 1 B) and A1 (data not shown). Forced expression of A1 or BCL–XL alone have been reported to protect multiple cell types against apoptosis after NF-κB inhibition (14, 45, 46, 62). Given these considerations, we were surprised that forced expression of BCL-2, A1, or BCL-XL did not affect the toxicity of NF-κB inhibition to ABC DLBCL cell lines. However, it has been shown that in certain contexts, the simultaneous expression of multiple antiapoptotic NF-κB target genes is required to protect against apoptosis induced after NF-κB inhibition, and some of these (TRAF1, TRAF2, c-IAP1, and c-IAP2) are not BCL-2 family members (63). Furthermore, it is possible that primary activation of the NF-κB pathway in ABC DLBCLs initiates an elaborate regulatory cascade that establishes a new signaling homeostasis within the cell, including pathways other than NF-κB.
In addition to its survival effects, activity of the NF-κB pathway was necessary for cell cycle progression in ABC DLBCLs. NF-κB inhibition resulted in a G1-phase cell cycle arrest of these cells, implying that primary or secondary targets of NF-κB are required for their cell cycle progression. The NF-κB target gene cyclin D2 is a potential mediator of these effects, given that D-type cyclins are required for progression beyond G1 phase, and that cyclin D2 expression is highly characteristic of ABC DLBCL cell lines and primary tumors. Furthermore, induction of cyclin D2 expression is required for the proliferative response of B cells to BCR activation (64), which is especially significant given that the gene expression profile of ABC DLBCL resembles that of BCR-activated B cells. Regardless of the specific cause, cell-cycle arrest itself causes apoptosis in many types of tumor cells, and the loss of NF-κB–driven proliferative factors (such as cyclin D2) may therefore contribute to the cytoxicity of NF-κB inhibitors in ABC DLBCL lines.
The nuclear NF-κB activity in ABC DLBCL cell lines results from constitutive IKK activity, but the upstream signaling pathways activating IKK in these cells are not yet clear. Most known NF-κB activating stimuli lead to phosphorylation of IKKβ in a regulatory loop of the kinase domain (3, 40, 65). This mechanism is also likely to account for IKK activity in ABC DLBCL since a dominant negative version of IKKβ with mutations in this loop was toxic to these cells. A multitude of signaling pathways lead to activation of IKK (2, 4). Signaling through the BCR activates phospholipase Cγ2, BLNK, and Btk which appear to act in concert to activate IKK (66–69). ABC DLBCLs express many genes in common with BCR-stimulated blood B cells, beyond just the NF-κB target genes (26). Thus, constitutive activation of NF-κB in ABC DLBCL may be due to a signaling cascade that mimics BCR stimulation and leads to IKK activation. A future understanding of the upstream signaling pathway that leads to IKK activation in ABC DLBCL may provide new molecular targets for pharmacological intervention in this disease.
Finally, our results demonstrate the power of gene expression profiling to reveal the activity of signaling pathways in cancer that are potentially therapeutic targets. The present observation that ABC DBLCL, but not GCB DLBCL, often engages the NF-κB pathway, validates the view that these lymphoma subtypes are distinct disease entities that have disparate pathogenetic mechanisms. We observed previously that these subtypes were also quite different in the overall clinical course of patients, but it is important to emphasize that this clinical difference occurred in the context of multiagent, anthracycline-based chemotherapy (26). In this regard, the ability of NF-κB to inhibit responses to cancer therapeutic agents (15) may contribute to the refractory clinical behavior of ABC DLBCL. Furthermore, inhibition of NF-κB can synergize with chemotherapy to kill tumor cells (15). Thus, we propose that clinical trials should be initiated in DLBCL in which chemotherapy is combined with pharmacological inhibition of the NF-κB pathway (70, 71), and that these trials should include gene expression profiling of the tumor cells to correlate the response of the patients with the molecular phenotype of their cancers. In this fashion, pretreatment gene expression profiling in DLBCL may ultimately be used to identify which pathogenic signaling pathways are active in a particular case, allowing the appropriate choice of specific therapy to be made.
Acknowledgments
The authors wish to thank Andreas Rosenwald and Elaine Hurt for contributing microarray data to Fig. 1 B; the National Institutes of Health Donor Center for providing normal human plasma; Art Shaffer for preparation of the EGFP-F retrovirus; and all those who provided reagents and cell lines.
Footnotes
Abbreviations used in this paper: ABC, activated B cell–like; BCR, B cell receptor; CHX, cycloheximide; DLBCL, diffuse large B cell lymphoma; EGFP, enhanced green fluorescent protein; EMSA, electrophoretic mobility shift assay; GCB, germinal center B-like; HA, hemagglutinin; IKK, IκB kinase; NF, nuclear factor; PI, PMA and ionomycin.
References
- 1.Baldwin, A.S., Jr. 1996. The NF-κB and I κB proteins: new discoveries and insights. Annu. Rev. Immunol. 14:649–683. [DOI] [PubMed] [Google Scholar]
- 2.Ghosh, S., M.J. May, and E.B. Kopp. 1998. NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225–260. [DOI] [PubMed] [Google Scholar]
- 3.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621–663. [DOI] [PubMed] [Google Scholar]
- 4.Siebenlist, U., G. Franzoso, and K. Brown. 1994. Structure, regulation and function of NF-κB. Annu. Rev. Cell Biol. 10:405–455. [DOI] [PubMed] [Google Scholar]
- 5.Gugasyan, R., R. Grumont, M. Grossmann, Y. Nakamura, T. Pohl, D. Nesic, and S. Gerondakis. 2000. Rel/NF-κB transcription factors: key mediators of B-cell activation. Immunol. Rev. 176:134–140. [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto, S., M.J. Schmitt, and I.M. Verma. 1994. Qualitative changes in the subunit composition of κB-binding complexes during murine B-cell differentiation. Proc. Natl. Acad. Sci. USA. 91:5056–5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grossmann, M., L.A. O'Reilly, R. Gugasyan, A. Strasser, J.M. Adams, and S. Gerondakis. 2000. The anti-apoptotic activities of Rel and RelA required during B-cell maturation involve the regulation of Bcl-2 expression. EMBO J. 19:6351–6360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam, K.P., R. Kuhn, and K. Rajewsky. 1997. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 90:1073–1083. [DOI] [PubMed] [Google Scholar]
- 9.Doi, T.S., T. Takahashi, O. Taguchi, T. Azuma, and Y. Obata. 1997. NF-κB RelA-deficient lymphocytes: normal development of T cells and B cells, impaired production of IgA and IgG1 and reduced proliferative responses. J. Exp. Med. 185:953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grumont, R.J., I.J. Rourke, L.A. O'Reilly, A. Strasser, K. Miyake, W. Sha, and S. Gerondakis. 1998. B lymphocytes differentially use the Rel and nuclear factor κB1 (NF-κB1) transcription factors to regulate cell cycle progression and apoptosis in quiescent and mitogen-activated cells. J. Exp. Med. 187:663–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kontgen, F., R.J. Grumont, A. Strasser, D. Metcalf, R. Li, D. Tarlinton, and S. Gerondakis. 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes Dev. 9:1965–1977. [DOI] [PubMed] [Google Scholar]
- 12.Mittrucker, H.W., T. Matsuyama, A. Grossman, T.M. Kundig, J. Potter, A. Shahinian, A. Wakeham, B. Patterson, P.S. Ohashi, and T.W. Mak. 1997. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 275:540–543. [DOI] [PubMed] [Google Scholar]
- 13.Grumont, R.J., and S. Gerondakis. 2000. Rel induces interferon regulatory factor 4 (IRF-4) expression in lymphocytes: modulation of interferon-regulated gene expression by rel/nuclear factor κB. J. Exp. Med. 191:1281–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grumont, R.J., I.J. Rourke, and S. Gerondakis. 1999. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Genes Dev. 13:400–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baldwin, A.S. 2001. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-κB. J. Clin. Invest. 107:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rayet, B., and C. Gelinas. 1999. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 18:6938–6947. [DOI] [PubMed] [Google Scholar]
- 17.Staudt, L.M. 2000. The molecular and cellular origins of Hodgkin's disease. J. Exp. Med. 191:207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bargou, R.C., F. Emmerich, D. Krappmann, K. Bommert, M.Y. Mapara, W. Arnold, H.D. Royer, E. Grinstein, A. Greiner, C. Scheidereit, and B. Dorken. 1997. Constitutive nuclear factor-κB-RelA activation is required for proliferation and survival of Hodgkin's disease tumor cells. J. Clin. Invest. 100:2961–2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bargou, R.C., C. Leng, D. Krappmann, F. Emmerich, M.Y. Mapara, K. Bommert, H.D. Royer, C. Scheidereit, and B. Dorken. 1996. High-level nuclear NF-κB and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood. 87:4340–4347. [PubMed] [Google Scholar]
- 20.Krappmann, D., F. Emmerich, U. Kordes, E. Scharschmidt, B. Dorken, and C. Scheidereit. 1999. Molecular mechanisms of constitutive NF-κB/Rel activation in Hodgkin/Reed-Sternberg cells. Oncogene. 18:943–953. [DOI] [PubMed] [Google Scholar]
- 21.Cahir-McFarland, E.D., D.M. Davidson, S.L. Schauer, J. Duong, and E. Kieff. 2000. NF-κB inhibition causes spontaneous apoptosis in Epstein-Barr virus-transformed lymphoblastoid cells. Proc. Natl. Acad. Sci. USA. 97:6055–6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fracchiolla, N.S., L. Lombardi, M. Salina, A. Migliazza, L. Baldini, E. Berti, L. Cro, E. Polli, A.T. Maiolo, and A. Neri. 1993. Structural alterations of the NF-κB transcription factor lyt-10 in lymphoid malignancies. Oncogene. 8:2839–2845. [PubMed] [Google Scholar]
- 23.Neri, A., C.C. Chang, L. Lombardi, M. Salina, P. Corradini, A.T. Maiolo, R.S. Chaganti, and R. Dalla-Favera. 1991. B cell lymphoma-associated chromosomal translocation involves candidate oncogene lyt-10, homologous to NF-κB p50. Cell. 67:1075–1087. [DOI] [PubMed] [Google Scholar]
- 24.Joos, S., M.I. Otano-Joos, S. Ziegler, S. Bruderlein, S. du Manoir, M. Bentz, P. Moller, and P. Lichter. 1996. Primary mediastinal (thymic) B-cell lymphoma is characterized by gains of chromosomal material including 9p and amplification of the REL gene. Blood. 87:1571–1578. [PubMed] [Google Scholar]
- 25.Lu, D., J.D. Thompson, G.K. Gorski, N.R. Rice, M.G. Mayer, and J.J. Yunis. 1991. Alterations at the rel locus in human lymphoma. Oncogene. 6:1235–1241. [PubMed] [Google Scholar]
- 26.Alizadeh, A.A., M.B. Eisen, R.E. Davis, C. Ma, I.S. Lossos, A. Rosenwald, J.C. Boldrick, H. Sabet, T. Tran, X. Yu, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 403:503–511. [DOI] [PubMed] [Google Scholar]
- 27.Alizadeh, A., M. Eisen, R.E. Davis, C. Ma, H. Sabet, T. Tran, J. Powell, L. Yang, G. Marti, T. Moore, et al. 1999. The lymphochip: a specialized cDNA microarray for the genomic-scale analysis of gene expression in normal and malignant lymphocytes. Cold Spring Harbor Symp. Quant. Biol. 64:71–78. [DOI] [PubMed] [Google Scholar]
- 28.Eisen, M.B., P.T. Spellman, P.O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA. 95:14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ellinger-Ziegelbauer, H., K. Brown, K. Kelly, and U. Siebenlist. 1997. Direct activation of the stress-activated protein kinase (SAPK) and extracellular signal-regulated protein kinase (ERK) pathways by an inducible mitogen-activated protein kinase/ERK kinase kinase 3 (MEKK) derivative. J. Biol. Chem. 272:2668–2674. [DOI] [PubMed] [Google Scholar]
- 30.Brown, K., S. Gerstberger, L. Carlson, G. Franzoso, and U. Siebenlist. 1995. Control of IκB-α proteolysis by site-specific, signal-induced phosphorylation. Science. 267:1485–1488. [DOI] [PubMed] [Google Scholar]
- 31.Bours, V., P.R. Burd, K. Brown, J. Villalobos, S. Park, R.P. Ryseck, R. Bravo, K. Kelly, and U. Siebenlist. 1992. A novel mitogen-inducible gene product related to p50/p105-NF-κB participates in transactivation through a κB site. Mol. Cell. Biol. 12:685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaffer, A.L., X. Yu, Y. He, J. Boldrick, E.P. Chan, and L.M. Staudt. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 13:199–212. [DOI] [PubMed] [Google Scholar]
- 33.Quong, M.W., D.P. Harris, S.L. Swain, and C. Murre. 1999. E2A activity is induced during B-cell activation to promote immunoglobulin class switch recombination. EMBO J. 18:6307–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hinz, M., P. Loser, S. Mathas, D. Krappmann, B. Dorken, and C. Scheidereit. 2001. Constitutive NF-κB maintains high expression of a characteristic gene network, including CD40, CD86, and a set of antiapoptotic genes in Hodgkin/Reed-Sternberg cells. Blood. 97:2798–2807. [DOI] [PubMed] [Google Scholar]
- 35.Micheau, O., S. Lens, O. Gaide, K. Alevizopoulos, and J. Tschopp. 2001. NF-κB signals induce the expression of c-FLIP. Mol. Cell. Biol. 21:5299–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, J., G.W. Peet, D. Balzarano, X. Li, P. Massa, R.W. Barton, and K.B. Marcu. 2001. Novel NEMO/IKKγ and NF-κB target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276:18579–18590. [DOI] [PubMed] [Google Scholar]
- 37.Chiao, P.J., S. Miyamoto, and I.M. Verma. 1994. Autoregulation of IκB α activity. Proc. Natl. Acad. Sci. USA. 91:28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scott, M.L., T. Fujita, H.C. Liou, G.P. Nolan, and D. Baltimore. 1993. The p65 subunit of NF-κB regulates IκB by two distinct mechanisms. Genes Dev. 7:1266–1276. [DOI] [PubMed] [Google Scholar]
- 39.Zandi, E., D.M. Rothwarf, M. Delhase, M. Hayakawa, and M. Karin. 1997. The IκB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 91:243–252. [DOI] [PubMed] [Google Scholar]
- 40.Mercurio, F., H. Zhu, B.W. Murray, A. Shevchenko, B.L. Bennett, J. Li, D.B. Young, M. Barbosa, M. Mann, A. Manning, and A. Rao. 1997. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science. 278:860–866. [DOI] [PubMed] [Google Scholar]
- 41.Chen, C., L.C. Edelstein, and C. Gelinas. 2000. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-x(L). Mol. Cell. Biol. 20:2687–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheng, Q., H.H. Lee, Y. Li, T.P. Parks, and G. Cheng. 2000. Upregulation of Bcl-x and Bfl-1 as a potential mechanism of chemoresistance, which can be overcome by NF-κB inhibition. Oncogene. 19:4936–4940. [DOI] [PubMed] [Google Scholar]
- 43.D'Souza, B., M. Rowe, and D. Walls. 2000. The bfl-1 gene is transcriptionally upregulated by the Epstein-Barr virus LMP1, and its expression promotes the survival of a Burkitt's lymphoma cell line. J. Virol. 74:6652–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, H.H., H. Dadgostar, Q. Cheng, J. Shu, and G. Cheng. 1999. NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc. Natl. Acad. Sci. USA. 96:9136–9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang, C.Y., D.C. Guttridge, M.W. Mayo, and A.S. Baldwin. 1999. NF-κB induces expression of the Bcl-2 homologue A1/Bfl-1 to preferentially suppress chemotherapy-induced apoptosis. Mol. Cell. Biol. 19:5923–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zong, W.X., L.C. Edelstein, C. Chen, J. Bash, and C. Gelinas. 1999. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNFα-induced apoptosis. Genes Dev. 13:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Angelin-Duclos, C., G. Cattoretti, K.I. Lin, and K. Calame. 2000. Commitment of B lymphocytes to a plasma cell fate is associated with Blimp-1 expression in vivo. J. Immunol. 165:5462–5471. [DOI] [PubMed] [Google Scholar]
- 48.Falini, B., M. Fizzotti, A. Pucciarini, B. Bigerna, T. Marafioti, M. Gambacorta, R. Pacini, C. Alunni, L. Natali-Tanci, B. Ugolini, et al. 2000. A monoclonal antibody (MUM1p) detects expression of the MUM1/IRF4 protein in a subset of germinal center B cells, plasma cells, and activated T cells. Blood. 95:2084–2092. [PubMed] [Google Scholar]
- 49.Gilmore, T.D. 1999. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene. 18:6925–6937. [DOI] [PubMed] [Google Scholar]
- 50.Gilmore, T.D., C. Cormier, J.-J. Jims, and M.-E. Gapuzan. 2001. Malignant transformation of primary chicken spleen cells by human transcription factor c-Rel. Oncogene. 20:7098–7103. [DOI] [PubMed] [Google Scholar]
- 51.Houldsworth, J., S. Mathew, P.H. Rao, K. Dyomina, D.C. Louie, N. Parsa, K. Offit, and R.S. Chaganti. 1996. REL proto-oncogene is frequently amplified in extranodal diffuse large cell lymphoma. Blood. 87:25–29. [PubMed] [Google Scholar]
- 52.Neri, A., N.S. Fracchiolla, E. Roscetti, S. Garatti, D. Trecca, A. Boletini, L. Perletti, L. Baldini, A.T. Maiolo, and E. Berti. 1995. Molecular analysis of cutaneous B- and T-cell lymphomas. Blood. 86:3160–3172. [PubMed] [Google Scholar]
- 53.Chang, C.C., J. Zhang, L. Lombardi, A. Neri, and R. Dalla-Favera. 1995. Rearranged NFKB-2 genes in lymphoid neoplasms code for constitutively active nuclear transactivators. Mol. Cell. Biol. 15:5180–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim, K.E., C. Gu, S. Thakur, E. Vieira, J.C. Lin, and A.B. Rabson. 2000. Transcriptional regulatory effects of lymphoma-associated NFKB2/lyt10 protooncogenes. Oncogene. 19:1334–1345. [DOI] [PubMed] [Google Scholar]
- 55.Ciana, P., A. Neri, C. Cappellini, F. Cavallo, M. Pomati, C.C. Chang, A.T. Maiolo, and L. Lombardi. 1997. Constitutive expression of lymphoma-associated NFKB-2/Lyt-10 proteins is tumorigenic in murine fibroblasts. Oncogene. 14:1805–1810. [DOI] [PubMed] [Google Scholar]
- 56.Yamaoka, S., H. Inoue, M. Sakurai, T. Sugiyama, M. Hazama, T. Yamada, and M. Hatanaka. 1996. Constitutive activation of NF-κB is essential for transformation of rat fibroblasts by the human T-cell leukemia virus type I Tax protein. EMBO J. 15:873–887. [PMC free article] [PubMed] [Google Scholar]
- 57.Finco, T.S., J.K. Westwick, J.L. Norris, A.A. Beg, C.J. Der, and A.S. Baldwin, Jr. 1997. Oncogenic Ha-Ras-induced signaling activates NF-κB transcriptional activity, which is required for cellular transformation. J. Biol. Chem. 272:24113–24116. [DOI] [PubMed] [Google Scholar]
- 58.Arsura, M., F. Mercurio, A.L. Oliver, S.S. Thorgeirsson, and G.E. Sonenshein. 2000. Role of the IκB kinase complex in oncogenic Ras- and Raf-mediated transformation of rat liver epithelial cells. Mol. Cell. Biol. 20:5381–5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reuther, J.Y., G.W. Reuther, D. Cortez, A.M. Pendergast, and A.S. Baldwin, Jr. 1998. A requirement for NF-κB activation in Bcr-Abl-mediated transformation. Genes Dev. 12:968–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Evan, G.I., A.H. Wyllie, C.S. Gilbert, T.D. Littlewood, H. Land, M. Brooks, C.M. Waters, L.Z. Penn, and D.C. Hancock. 1992. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 69:119–128. [DOI] [PubMed] [Google Scholar]
- 61.Romashkova, J.A., and S.S. Makarov. 1999. NF-κB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 401:86–90. [DOI] [PubMed] [Google Scholar]
- 62.Karsan, A., E. Yee, and J.M. Harlan. 1996. Endothelial cell death induced by tumor necrosis factor-α is inhibited by the Bcl-2 family member, A1. J. Biol. Chem. 271:27201–27204. [DOI] [PubMed] [Google Scholar]
- 63.Wang, C.Y., M.W. Mayo, R.G. Korneluk, D.V. Goeddel, and A.S. Baldwin. 1998. NF-κB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science. 281:1680–1683. [DOI] [PubMed] [Google Scholar]
- 64.Lam, E.W., J. Glassford, L. Banerji, N.S. Thomas, P. Sicinski, and G.G. Klaus. 2000. Cyclin D3 compensates for loss of cyclin D2 in mouse B-lymphocytes activated via the antigen receptor and CD40. J. Biol. Chem. 275:3479–3484. [DOI] [PubMed] [Google Scholar]
- 65.Delhase, M., M. Hayakawa, Y. Chen, and M. Karin. 1999. Positive and negative regulation of IκB kinase activity through IKKβ subunit phosphorylation. Science. 284:309–313. [DOI] [PubMed] [Google Scholar]
- 66.Tan, J.E., S.C. Wong, S.K. Gan, S. Xu, and K.P. Lam. 2001. The adaptor protein BLNK is required for B cell antigen receptor-induced activation of NFκ-B and cell cycle entry and Survival of B lymphocytes. J. Biol. Chem. 276:20055–20063. [DOI] [PubMed] [Google Scholar]
- 67.Petro, J.B., and W.N. Khan. 2001. Phospholipase C-γ2 couples Bruton's tyrosine kinase to the NF-κB signaling pathway in B lymphocytes. J. Biol. Chem. 276:1715–1719. [DOI] [PubMed] [Google Scholar]
- 68.Petro, J.B., S.M. Rahman, D.W. Ballard, and W.N. Khan. 2000. Bruton's tyrosine kinase is required for activation of IκB kinase and nuclear factor κB in response to B cell receptor engagement. J. Exp. Med. 191:1745–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bajpai, U.D., K. Zhang, M. Teutsch, R. Sen, and H.H. Wortis. 2000. Bruton's tyrosine kinase links the B cell receptor to nuclear factor κB activation. J. Exp. Med. 191:1735–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Adams, J., M. Behnke, S. Chen, A.A. Cruickshank, L.R. Dick, L. Grenier, J.M. Klunder, Y.T. Ma, L. Plamondon, and R.L. Stein. 1998. Potent and selective inhibitors of the proteasome: dipeptidyl boronic acids. Bioorg. Med. Chem. Lett. 8:333–338. [DOI] [PubMed] [Google Scholar]
- 71.Umezawa, K., A. Ariga, and N. Matsumoto. 2000. Naturally occurring and synthetic inhibitors of NF-κB functions. Anticancer Drug Des. 15:239–244. [PubMed] [Google Scholar]