Abstract
Sfp1, an unusual zinc finger protein, was previously identified as a gene that, when overexpressed, imparted a nuclear localization defect. sfp1 cells have a reduced size and a slow growth phenotype. In this study we show that SFP1 plays a role in ribosome biogenesis. An sfp1 strain is hypersensitive to drugs that inhibit translational machinery. sfp1 strains also have defects in global translation as well as defects in rRNA processing and 60S ribosomal subunit export. Microarray analysis has previously shown that ectopically expressed SFP1 induces the transcription of a large subset of genes involved in ribosome biogenesis. Many of these induced genes contain conserved promoter elements (RRPE and PAC). Our results show that activation of transcription from a reporter construct containing two RRPE sites flanking a single PAC element is SFP1 dependent. However, we have been unable to detect direct binding of the protein to these elements. This suggests that regulation of genes containing RRPEs is dependent upon Sfp1 but that Sfp1 may not directly bind to these conserved promoter elements; rather, activation may occur through an indirect mechanism.
The ability of a cell to properly regulate translation depends in part on the rate of ribosome biogenesis. Given the central role of translation in all aspects of cellular activity, it is not surprising that the process of ribosome biogenesis is complex, depending ultimately on a hierarchy of transcriptional, posttranscriptional, and translational regulatory mechanisms. Thus, at the transcriptional level, rRNA is transcribed from 9.1-kb ribosomal DNA (rDNA) loci, which are found in a tandem array of 100 to 200 repeats on chromosome XII (18). At any given time, approximately half of the rDNA repeats are transcriptionally silenced in a regulated manner that reflects the overall translational needs of the specific growth conditions (19). After the rRNAs are transcribed, they undergo a series of posttranscriptional processing steps by various endonucleases and exonucleases to produce the mature 18, 25, and 5.8S rRNAs (23). The 18S rRNA is incorporated, with a large set of ribosomal proteins (r-proteins), into the 40S ribosomal subunit, while the 25 and 5.8S rRNAs and r-proteins are incorporated into the 60S ribosomal subunit (23). The stoichiometry of the mature rRNAs and r-proteins and their assembly into complete ribosomal subunits is also tightly regulated. Finally, the completed subunits are exported to the cytoplasm to assemble into ribosomes (16).
Mutations that affect any steps in ribosome biogenesis will affect the ability of the cell to carry out translation at a normal level. These mutations would be expected to exhibit pleiotropic phenotypes through their general effects on a variety of cellular processes. Hence, a variety of mutations initially identified as playing a role in a specific cellular process have turned out on subsequent analysis to affect the more general process of translation. An example of this is the SFP1 gene, which encodes a protein with an unusual split zinc finger motif. SFP1 was initially identified in a screen for genes that altered import of nuclear proteins when present on high-copy-number plasmids (3). Overexpression of SFP1 was found to result in the mislocalization of several endogenous nucleolar proteins, although the null mutant did not appear to be altered in nuclear import or protein localization. These results suggested that Sfp1 played some uncharacterized role in nuclear localization.
The SFP1 gene was also identified in a differential-display screen for genes whose expression increased after DNA damage (27). Subsequent Northern blot analysis showed that the SFP1 transcript is induced sixfold after a 90-min exposure to the DNA-alkylating agent methyl methane sulfonate (MMS). Additionally, sfp1 cells were found to be more sensitive to ionizing radiation and alkylating agents than SFP1 cells, consistent with the presence of a defect in DNA repair. Finally, sfp1 mutant cells were observed to be significantly smaller than wild-type cells and showed a significant defect in their growth rate (3). Based on the precedent of wee mutants in Schizosaccharomyces pombe, we tested whether the sfp1 mutants had difficulty regulating the transition from the G2 phase of the cell cycle into mitosis. We found that the cells were in fact unable to appropriately regulate this transition, which led to the hypothesis that Sfp1 was a negative regulator of the G2/M transition after DNA damage and during the normal cell cycle. The small cell size of the sfp1 strain was also observed in a recent screen for mutations that affect critical cell size at START, which occurs late in the G1 phase (11). However, analysis in the latter work indicated that the fundamental defect in the sfp1 strain may be a defect not in regulating cell cycle progression or nuclear localization but rather in regulating ribosome biogenesis.
In this paper we further investigate the role of Sfp1 and confirm that it has an important role in ribosome biogenesis. We also present data supporting the model that Sfp1 functions as a transcriptional regulator of genes required for ribosome biogenesis.
MATERIALS AND METHODS
Strains and plasmids.
The wild-type (DN1162), sfp1 (DN1300), and kap120 (PSY1082) strains are all derivatives of strain W303 and have been described previously (21, 27). The noc4-1 strain, used in the 40S ribosomal subunit export assay, has been described previously (17). Wild-type (BY4741 MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and sfp1(5312) strains in an SC288C background were also used in this assay (4) (Research Genetics). Plasmid pIF13 (pRS424-SFP1-HAx3) was constructed by PCR amplification of the SFP1 open reading frame from a genomic subclone to include 488 bp upstream and 176 bp downstream of the open reading frame with overhanging PvuII sites. The PCR fragment was cloned into the PvuII sites of pRS424. An XmaI site was integrated immediately downstream of ATG by using a three-step PCR. Complementary oligonucleotides encoding an HAx3 epitope with XmaI-compatible ends were annealed and ligated into the pRS424-SFP1 vector containing the integrated XmaI site.
Transcription reporter constructs were constructed in the pTBA30 vector (1). Reporter constructs were made by cloning oligonucleotides with a sequence corresponding to bases −210 to −158 of the NSR1 promoter, containing two ribosomal RNA processing element (RRPE) sites flanking a single polymerase A and C (PAC) element, into the XhoI sites of pTBA30. pIF111 contains the NSR1 promoter region fragment in a single copy in pTBA30. Plasmid pIF117 contains bases −210 to −158 of the NSR1 promoter with a mutant PAC element that was made by base substitutions in the oligonucleotides cloned into the XhoI site of pTBA30 (GCGATGAGCTG to CATCGTAACTG). Plasmid pIF119 contains bases −210 to −158 of the NSR1 promoter with mutant RRPE elements that were made by base substitutions in the oligonucleotides cloned into the XhoI site of pTBA30 (GAAAAATTT to CCCCCATGG and GAAAATTTT to CCCCCTTGG). Plasmid pTBA23 was used as a control for activation (1). β-Galactosidase assays monitoring expression from these reporters were performed as previously described (12).
Translation drug sensitivity.
Yeast strains were grown in a rich medium (YPD, containing 2% [wt/vol] peptone, 1% [wt/vol] yeast extract, and 2% [wt/vol] dextrose). For determination of sensitivity to translation drugs, cultures were grown to mid-log phase (optical density at 600 nm [OD600], 0.5) and plated for confluent growth on YPD plates. Filter disks were placed on the plates, and 10 μl of 1 M paromomycin, 25 mM hygromycin, or 5 mM cycloheximide was spotted onto each disk. Plates were incubated at 30°C for 48 h (wild type) or 72 h (sfp1). Sensitivity is expressed as the average radius of inhibition of growth.
MMS sensitivity.
Yeast strains were grown in a rich medium (YPD). Cultures were grown to mid-log phase (OD600, 0.5), serially diluted 10-fold, and spotted onto YPD plates containing 0.009% MMS. Plates were incubated at 30°C for 72 h (sfp1) and photographed.
Determination of total translation.
Wild-type (DN1162) and sfp1 (DN1300) cultures were grown in a synthetic medium (SD, containing 0.67% [wt/vol] yeast nitrogen base supplemented with amino acids and 2% [wt/vol] dextrose) lacking methionine (SD−Met) at 30°C to an OD600 of 0.5. At time zero, cold methionine was added to a final concentration of 0.5 μM and [35S]methionine was added to a final concentration of 1 μCi/ml [EXPRE35S35S (35S)Protein Labeling Mix, containing 7.9 mCi of [35S]methionine/ml; New England Nuclear Life Sciences]. At given times, 1-ml aliquots were taken in duplicate from each culture and the OD600 was measured. Trichloroacetic acid (TCA) was added to a final concentration of 2.5%, and aliquots were placed on ice for 10 min, followed by incubation at 70°C for 20 min. The total lysate was then filtered through a 25-mm-pore-size Whatman GF/C filter. The filters were washed twice with 5 ml of 5% TCA and twice with 5 ml of 95% ethanol; then they were allowed to air dry for 2 h. Filters were counted by scintillation, and the values for duplicate samples were averaged.
Polyribosome profile analysis.
Polyribosome preparation was performed based on a procedure described previously (2). One hundred milliliters of wild-type (DN1162) and sfp1 (DN1300) cultures were grown to an OD600 of 0.5, and cycloheximide was added to a final concentration of 0.1 mg/ml. Ice was added directly to the cultures, and the cells were pelleted by centrifugation at 12,000 × g for 5 min. The cell pellets were washed once in 10 ml of ice-cold lysis buffer (10 mM Tris-HCl [pH 7.5], 0.1 M NaCl, 30 mM MgCl2, 50 μg of cycloheximide/ml, 200 μg of heparin/ml, 0.2% diethyl pyrocarbonate) and resuspended in 0.5 ml of cold lysis buffer in 1.5-ml microcentrifuge tubes. A 250-μl volume of glass beads was added, and the cells were lysed by vortexing eight times for 30 s each time. The lysates were centrifuged for 5 min at 12,000 × g and 4°C, and the supernatants were transferred to clean tubes. The OD260 was measured, and 5 to 10 OD260 units was layered onto a 7-ml linear sucrose gradient (7 to 49% [wt/vol] sucrose in gradient buffer, consisting of 50 mM Tris acetate [pH 7.0], 50 mM NH4Cl, 12 mM MgCl2, 1 mM dithiothreitol, and 0.1% diethyl pyrocarbonate). The gradients were centrifuged in a Beckman SW28 rotor at 27,000 rpm for 4 h and fractionated, and the sucrose gradient profiles were recorded by absorption at A254.
r-protein localization assays.
The Rpl11b-green fluorescent protein (GFP) fusion plasmid (21) was transformed into wild-type (DN1162), sfp1 (DN1300), and kap120 (PSY1082) strains. Cells were grown in selective medium at 30°C to mid-log phase (OD600, 0.5) and shifted to 37°C for 1 h. Three milliliters of culture was then removed and fixed by adding formaldehyde to a final concentration of 3.7% and agitating at 30°C for 30 min. Cells were harvested and washed twice with 1 ml of solution P (0.1 M KPi [pH 6.5]-1.2 M sorbitol) and placed at 4°C. The remaining culture was shifted back to 30°C, and 3-ml aliquots were removed at 30 and 60 min and fixed as described above. Cells were permeabilized by adding Triton X-100 to a final concentration of 0.5% and incubating at room temperature for 10 min. 4′,6′-Diamidino-2-phenylindole (DAPI) was added to fixed cells to a final concentration of 1 μg/ml and incubated for 3 min at room temperature. Cells were washed twice in 1 ml of 1× phosphate-buffered saline and stored at 4°C in 100 μl of 1× phosphate-buffered saline. Rpl11b-GFP was visualized by fluorescence microscopy using a Zeiss microscope. Rps2-GFP was localized as described previously (17). The Rps2-GFP fusion plasmid (17) was transformed into wild-type, sfp1, and noc4-1 strains, which were all in a S288C strain background (4) (Research Genetics). Fixing and DAPI staining were performed as described above.
Pulse-chase labeling of rRNA.
rRNA processing was assayed in the wild-type and sfp1 mutant strains based on a procedure described previously (21). One-hundred-milliliter cultures were grown in SD−Met to a final OD600 of 0.5. Cells were harvested and resuspended in 4 ml of SD−Met, and 250 μCi of l-[methyl-3H]methionine (1.0 mCi/ml; New England Nuclear Life Sciences) was added. Cells were incubated for 5 min at 25°C. After 3 min, cultures were chased with cold methioine in −Met medium to a final concentration of 5.1 mM. Aliquots (1 ml) were removed at various time points and pelleted, and the supernatant was removed before the pellets were placed on dry ice. Total RNA was prepared from labeled cells by extraction with hot acid-phenol (6). Ten thousand counts per minute of each sample was resolved on a 1.2% formaldehyde gel. RNA was transferred to a nylon membrane (Qiabrane; Qiagen) by capillary transfer. The membrane was air dried and sprayed with En3Hance (New England Nuclear Life Sciences), according to the manufacturer's protocol. Blots were exposed to Kodak X-Omat/AR film for 3 days at −80°C and quantitated using Alphaease FC software (Alpha Innotech).
rRNA levels.
Five-milliliter cultures were grown in rich medium to an OD600 of 0.5. Cells were pelleted, and total RNA was isolated by using hot acid-phenol extraction (6). Equal concentrations of total RNA were loaded onto a 1.2% formaldehyde agarose gel. The RNA was then transferred to a nitrocellulose filter, and the rRNA gel was probed by Northern blot analysis for 25 and 18S rRNAs as well as ACT1 message (5). Quantitation to determine the ratios of 25 and 18S rRNAs to ACT1 was performed using IPLabgel (Molecular Dynamics).
Chromatin immunoprecipitation.
Plasmids pIF13 (pRS424-SFP1-HAx3) and pIF12 (pRS424-SFP1) were transformed into DN1300 (sfp1). Chromatin immunoprecipitations were performed as described previously (13). Oligonucleotide primers were chosen to amplify approximately 300- and 200-bp fragments from the promoters of Sfp1 target genes and control genes, respectively. The following primers were used to amplify the promoters of putative Sfp1 target genes: for NSR1, 5′ GCCTGATGTATGGGTCCCCATG 3′ and 5′ CCTGCCTGGGTTGAGTGATCCG 3′; for RPA190, 5′ GCCCGCTATCGGAGGGTCTCAG 3′ and 5′ CACTGTATAGTTAATGGATACCTAAG 3′; for SIK1, 5′ GAAGATAATACTGTATACTAGTG 3′ and 5′ GCCATCTTTCACTAAATACTGTTC 3′; for NOP5, 5′ CAAGATCCTGAACCAGCGTTC 3′ and 5′ GAGCATAACCAGCTGAAGTTTC 3′; for SEH1, 5′ CAAACCTTCACCTGGTGGTGCG 3′ and 5′ CATCATGAACTAAATCATCATGCCCAC; and for YGR250C, 5′ GTCGGAGATTCCCTATTGGGCGG 3′ and 5′ CATCCTCGAAACTTCTGATGCGG 3′. One microliter of product from total chromatin and immunoprecipitation reactions was used as a template in the PCRs.
RESULTS
sfp1 cells are sensitive to drugs that inhibit translation.
Small cell size in yeast often correlates with defects in the process of translation (8). To determine if the small cell size of an sfp1 mutant is consistent with a defect in translation, sfp1 cells were examined for sensitivity to drugs that interfere with translation. Lawns of wild-type and sfp1 cells were grown on plates that contained filter disks with the antibiotics paromomycin, cycloheximide, and hygromycin. Cells that are sensitive to the antibiotic show a zone of inhibited growth around the filter disk. The size of this zone of inhibition is relative to the sensitivity of the cells to the particular antibiotic. The zone of inhibited growth for each drug was significantly larger for the sfp1 mutant strain than for the wild-type strain, indicating that the sfp1 mutant was particularly sensitive to these translation drugs (Table 1). In contrast, sfp1 cells showed no sensitivity to drugs that do not have an effect on translation, such as caffeine, hydrogen peroxide, or calcofluor white (data not shown).
TABLE 1.
Translation drug sensitivity
| Drug | Zone of inhibition (mm)
|
|
|---|---|---|
| Wild type | sfp1 mutant | |
| Cycloheximide | 11.5 ± 0.9 | 16.5 ± 1.5 |
| Paromomycin | 1.6 ± 0.2 | 7.5 ± 1.5 |
| Hygromycin | 3.7 ± 0.7 | 12.8 ± 1.3 |
sfp1 cells exhibit a reduced translation rate.
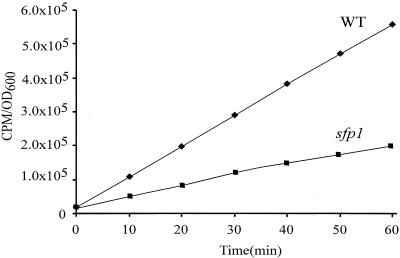
To determine if the reduced cell size and sensitivity to translation drugs of the sfp1 mutant were the results of a global defect in translation, the total translation of sfp1 cells was compared with that of wild-type cells in the same background. The rate at which [35S]methionine was incorporated into newly synthesized proteins by each strain was measured over time. sfp1 cells exhibited at least a 2.5-fold defect in the rate of incorporation of the label (Fig. 1).
FIG. 1.
The rate of translation is reduced in sfp1 cells. [35S]methionine was added to wild-type and sfp1 cultures grown in SD−Met medium, and total incorporation of the label was measured at the indicated time points by TCA precipitation of the aliquots and counting of the radioactivity in the pellet. The sfp1 mutant shows a ∼2.5-fold decrease in the rate of methionine incorporation.
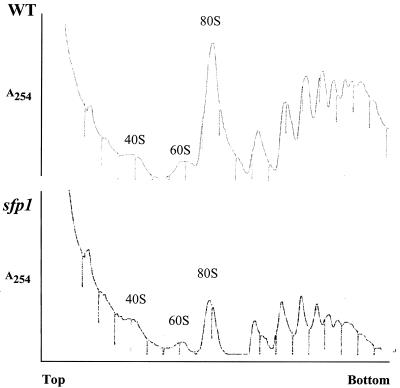
sfp1 cells show reduced levels of 80S ribosomes and higher-order polyribosomes.
To determine if Sfp1 is directly involved in the process of translation or is required for the biosynthesis of translation components, we compared the sucrose gradient profiles of the ribosomal subunits and polyribosomes in wild-type and sfp1 cells (2). The polyribosome profiles from wild-type and sfp1 cells were significantly different (Fig. 2). The level of 40S subunits in the sfp1 strain appeared to be largely unaffected. However, sfp1 cells showed slightly reduced levels of 60S ribosomal subunits. Additionally, the first peak in the polyribosome fraction from the sfp1 cells appeared to be a doublet. This type of profile has been previously observed in ribosome biogenesis mutants (7, 24). If the proportion of the 60S subunit is decreased relative to the 40S subunit, this doublet may be caused by the 40S subunits loading onto the mRNA but being unable to form active ribosomes. The defect in the ribosome profile in the sfp1 mutants therefore appears to be slightly greater for the 60S subunits than for the 40S subunits.
FIG. 2.
Polyribosome profiles show that sfp1 cells have reduced levels of 80S ribosomes and higher-order polyribosomes. Extracts from wild-type and sfp1 cells were resolved on 7 to 47% sucrose gradients. The OD254 was monitored for each sample. Positions of the 40, 60, and 80S ribosome peaks are shown.
The levels of higher-order polysomes were also reduced in the sfp1 strain (Fig. 2). The polysome peaks represent multiple ribosomes per transcript. In the wild-type strain, a large proportion of the ribosomes were associated in higher-order polysomes. However, in sfp1 cells, the majority of the ribosomes were associated with lower-order polysomes.
The 60 and 40S ribosomal subunits are not exported in an sfp1 strain.
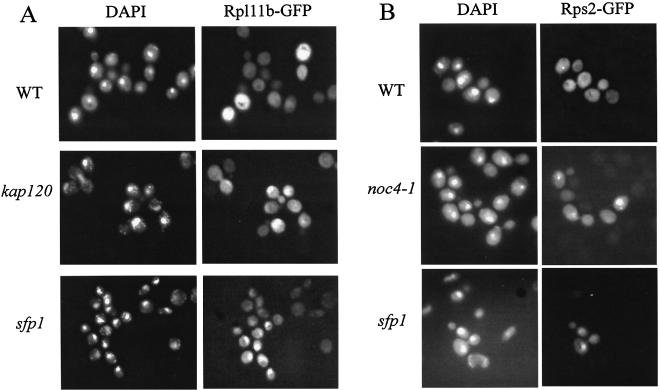
To assemble the ribosomal subunits, r-proteins must be imported into the nucleus. The assembled ribosomes are then exported from the nucleus. Since SFP1 was originally identified based on a phenotype that altered import or export from the nucleus (3), it was therefore possible that the defects in translation and 60S ribosomal subunit levels in the sfp1 mutant could be caused by improper localization of ribosomal components. To determine if the sfp1 mutant causes defects in any of these steps in ribosome biogenesis, we used an assay to monitor the presence and export of the 60S subunit (21). Rpl11b is an r-protein that is incorporated into the 60S ribosomal subunit during the later stages of ribosome biogenesis. In wild-type cells the Rpl11b-GFP fusion protein was evenly distributed in the cytoplasm (Fig. 3A) (21). In contrast, it has been shown previously that the Rpl11b-GFP fusion protein accumulates in the nucleus in mutants, such as kap120 mutants, which have defects in rRNA processing and r-protein import or export (Fig. 3A) (21). We observed a similar accumulation of Rpl11b-GFP in the nuclei of sfp1 cells (Fig. 3A).
FIG. 3.
sfp1 cells are defective in nuclear export of the 60 and 40S ribosomal subunits. (A) DAPI staining and localization of the RPL11b-GFP fusion protein (60S component) are shown for wild-type, kap120, and sfp1 cells. (B) DAPI staining and localization of the Rps2-GFP fusion protein (40S component) are shown for wild-type, noc4-1, and sfp1 cells.
We also assayed the ability of an sfp1 cell to export the 40S ribosomal subunit by using a Rps2-GFP fusion protein (17). Rps2 is incorporated into the small subunit, and in a wild-type cell, Rps2-GFP is evenly distributed in the cytoplasm. It has been shown that in mutants involved in nuclear export of the 40S subunit, such as noc4-1 mutants, there is an accumulation of Rps2-GFP in the nucleus (17). In the noc4-1 control strain, we observed nuclear accumulation of Rps2-GFP, but to a lesser extent than we saw for Rpl11b-GFP in the kap120 strain (Fig. 3B). In the sfp1 strain, we observed nuclear accumulation of Rps2-GFP in a low percentage of cells (Fig. 3B).
sfp1 cells demonstrate a delay in rRNA processing.
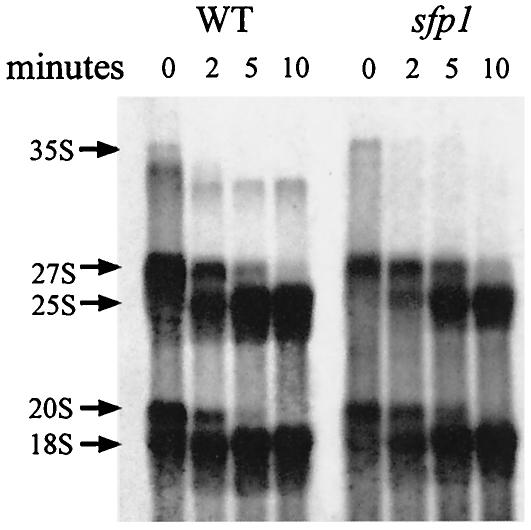
The nuclear accumulation of Rpl11b-GFP can be a result of a defect in rRNA processing (21). To examine if Sfp1 was involved in rRNA processing, an rRNA pulse-chase experiment was performed to determine if specific steps in processing were blocked. rRNA was radiolabeled in vivo with l-[methyl-3H]methionine, and aliquots were removed at time points after the chase to monitor the processing of the 3H-labeled transcript into mature 18 and 25S rRNAs. In a wild-type cell, the majority of the 3H-labeled pre-rRNA was processed to mature 25 and 18S rRNAs by the 5-min time point (Fig. 4). However, in the sfp1 cells, there was a significant delay in processing of the 27S pre-rRNA into the mature 25S rRNA. Quantification of the gel showed that 84% of the 27S pre-rRNA was processed to 25S rRNA at 5 min in a wild-type strain, while only 60% of the 27S precursor rRNA was processed into mature 25S rRNA in the sfp1 strain. Since the 25S rRNA is incorporated into the 60S ribosomal subunit, the delay in processing of the 27S pre-rRNA may explain the reduced levels of 60S ribosomal subunits and the export defect. There is also a slight delay in processing of the 20S pre-rRNA into mature 18S rRNA in the sfp1 strain. In wild-type cells, 90% of the 20S pre-rRNA was processed to the mature 18S rRNA at the 5-min time point, compared to 64% in the sfp1 strain.
FIG. 4.
sfp1 cells exhibit a delay in 27 and 20S pre-rRNA processing. rRNAs from wild-type and sfp1 cells were pulse-labeled in vivo by using l-[methyl-3H]methionine. Samples were harvested and processed at the indicated time points, electrophoresed on a formaldehyde gel, transferred to a membrane, and visualized by exposure to film after being treated with a fluorescence enhancer. The positions of the 35, 27, 25, 20, and 18S rRNAs are shown.
sfp1 cells have reduced levels of rRNA.
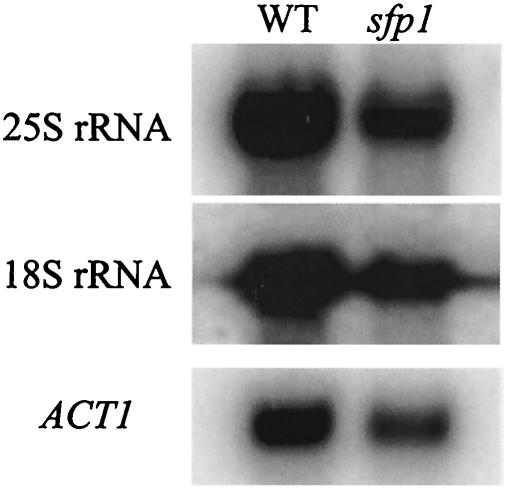
Defects in transcription of rDNA loci also cause a defect in ribosome biogenesis (19). To examine if Sfp1 has a role in the expression of the rDNA loci, rRNA levels in the sfp1 cells were tested. Total RNA was purified from both wild-type and sfp1 cultures, and equal concentrations of total RNA were resolved on a formaldehyde agarose gel. To confirm that this defect was specific to rRNA, Northern blotting was performed using probes specific to the 25 and 18S rRNAs. As a loading control, the blot was also probed with an ACT1-specific probe. The ratio of both 25 and 18S rRNAs to ACT1 message was 2.7 to 1 in the wild-type strain. In contrast, the level of rRNA expression was slightly decreased in the sfp1 strain, with a ratio of 2.1 to 1 (Fig. 5).
FIG. 5.
sfp1 cells have reduced levels of rRNA. Equal concentrations of total RNA purified from a wild-type strain and an sfp1 strain were electrophoresed on a formaldehyde agarose gel. The RNA was then transferred to a nylon membrane and probed by a Northern blot procedure with labeled probes specific for 25S rRNA (top), 18S rRNA (center), and ACT1 (bottom) The levels of hybridization of the 25S rRNA, 18S rRNA, and ACT1 probe to the wild-type and sfp1 RNA samples were measured on a phosphorimager.
Activation of transcription from RRPE elements is SFP1 dependent.
Recent microarray data have implicated Sfp1 as being a transcriptional activator of genes involved in ribosome biogenesis (11). Many of these genes contain conserved promoter elements called RRPE and PAC (10, 25). It is possible that Sfp1 functions through these elements to activate transcription. To test if this activation is Sfp1 dependent, a reporter construct was made that contains the RRPE and PAC elements from the NSR1 promoter (bases −110 to −158) regulating expression of a CYC1-lacZ fusion. While the construct lacking the RRPE and PAC sites showed background levels of expression in a wild-type strain (Table 2, pTBA30), a construct containing the sites (pIF111) showed almost a fourfold-higher level of expression, indicating that these elements were functioning as weak activator sites. The level of expression was decreased threefold, to background levels, in the sfp1 strain. In contrast, overexpression of SFP1 caused a twofold increase in expression from this construct compared to that in the wild type. These data suggest that transcription from these reporter constructs containing RRPE and PAC elements is SFP1 dependent. There was no significant difference between the wild-type and sfp1 strains' abilities to activate transcription of a lacZ reporter containing an activation sequence from the CYC1 promoter but lacking the RRPE and PAC elements (data not shown). The reduced growth rate and small size of sfp1 cells therefore have no effect on their ability to activate transcription from a promoter that is independent of the RRPE and PAC elements.
TABLE 2.
Sfp1-mediated regulation through RRPE and PAC elements
| Plasmid | β-Galactosidase activitya in the following strain:
|
||
|---|---|---|---|
| Wild type | sfp1 | SFP1 (2μ) | |
| pTBA30 (blank) | 2.2 ± 0.1 | 2.5 ± 0.7 | 3.1 ± 0.6 |
| pIF111 (RRPE + PAC) | 7.9 ± 0.6 | 2.3 ± 0.3 | 17.5 ± 1.6 |
| pIF117 (RRPE + pacb) | 21.5 ± 0.3 | 3.3 ± 0.4 | 111.1 ± 11.6 |
| pIF119 (rrpeb + PAC) | 2.5 ± 0.1 | 2.3 ± 0.7 | 3.0 ± 0.3 |
Measured in Miller units.
Mutant site.
We next constructed mutations in both the PAC and RRPE elements and assayed the abilities of wild-type and sfp1 strains to activate transcription from these reporter constructs. In a wild-type strain background, mutation of the PAC element caused almost a threefold increase in activation over that with the wild-type reporter construct (Table 2, pIF117 versus pIF111). Additionally, in a strain overexpressing SFP1, mutation of the PAC element caused a sixfold increase in activation (Table 2, pIF117 versus pIF111). These data suggest that the PAC element serves as a repressor element in the context of this reporter. There was no activation from this construct in the sfp1 mutant strain. Mutation of the RRPE elements caused complete loss of activation from the reporter construct in a wild-type strain (Table 2, pIF119). Furthermore, in a strain overexpressing SFP1, there was still a failure to activate transcription from this construct. This would imply that the RRPE elements function as activating elements in the context of this promoter and that this activation is SFP1 dependent.
Mutational analysis of Sfp1 shows that the two C-terminal zinc fingers are essential for Sfp1 function.
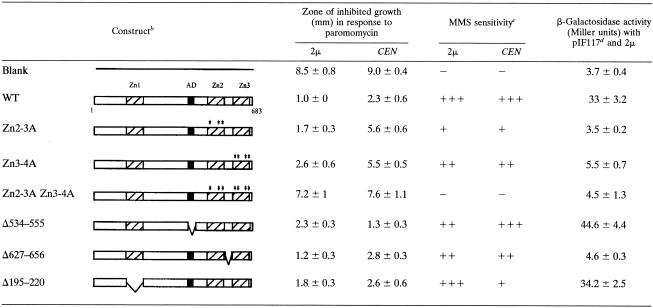
Examination of the Sfp1 sequence reveals several zinc finger motifs and an acidic domain that may be required for its function. To investigate if these domains are required for Sfp1 function, we constructed a series of deletions and point mutations in the protein and examined their phenotypes by assaying for the abilities of the mutants to complement the increased sensitivity to translation drugs of an sfp1 mutant strain. The two C-terminal fingers exhibit extensive homology with each other. Deletion of both C-terminal zinc fingers (Δ606-682) conferred sensitivity to translation drugs comparable to that of the sfp1 strain. However, deletion of each of the C-terminal fingers individually (Δ606-620 or Δ663-682) partially complemented the mutant phenotype (data not shown). These results suggest that they may perform redundant functions, possibly binding similar DNA sequences.
It is possible that deleting one individual zinc finger may alter the folding of the protein and the relative position of the second zinc finger. To address this concern, the conserved cysteine and histidine residues that typically chelate the zinc atom to form a zinc finger were converted to alanine. Disruption of both C-terminal zinc fingers (Znf2-3A Znf3-4A) (C605A, H618A, H623A, C661A, C664A, H667A, and H680A) in either low or high copy numbers caused a phenotype that was identical to that of an sfp1 strain (Table 3). However, disruption of the individual C-terminal zinc fingers (Znf2-3A or Znf3-4A) resulted in only a very slight sensitivity in this assay when they were expressed in high copy numbers (Table 3). However, when these disruptions were expressed in a single copy, the sensitivity to translation drugs was much more dramatic. This suggests that the proteins are partially functional and that high-copy expression of these mutants can compensate for a partial loss of function. In contrast to the effects of the C-terminal zinc fingers, deletion of the N-terminal zinc finger (amino acids 195 to 220), caused no substantial phenotype, indicating that this single zinc finger is not as important in Sfp1 function as the C-terminal fingers.
TABLE 3.
Domain analysis of Sfp1a
A summary of the deletion and point mutational analysis is shown.
Diagrams show the deleted or mutated regions of each construct. Zn1, Zn2, and Zn3 indicate the three zinc finger domains. AD indicates the position of the acidic domain. WT, wild type.
Sensitivity to MMS was measured qualitatively. A score of +++ indicate the least sensitivity, and a score of − indicates the most sensitivity.
For plasmid pIF117, see Table 2.
Many zinc finger transcription factors contain multiple zinc finger domains that are conservatively spaced with respect to each other, usually 6 to 8 residues apart (26). Sfp1, on the other hand, is unusual because 39 amino acids separate the two C-terminal zinc fingers. To determine if the spacing between the fingers is critical for the function of the protein, the linker between the two fingers was deleted (Δ627-656). This spacing mutant fully complemented the sfp1-null mutant when assayed for drug sensitivity and MMS (Table 3).
Sfp1 also contains an acidic domain. In other proteins, acidic domains are often implicated in transcriptional activation and are thought to interact with histones or other trans-acting proteins to activate transcription (9, 15, 22). DNA microarray analysis and our transcription reporter assays indicate that Sfp1, when overexpressed, induces a large number of genes involved in ribosome biogenesis, suggesting that it is functioning as an activator protein. We therefore deleted the acidic domain of Sfp1 to determine if it was required for function in vivo. However, this mutant complemented the sfp1 mutant phenotype for translation drug and MMS sensitivity, indicating that, if Sfp1 is functioning as a transcriptional activator, it does not require the acidic domain. All of the mutant constructs were also tested for their abilities to activate transcription from the reporter with a mutated PAC element (pIF117) at a high copy number (Table 3). Most of the mutations had the same effect in the transcription reporter assay as they did in the translation drug assay. Mutations in either of the two C-terminal zinc fingers resulted in a complete loss of activation from this reporter, while deletion of the N-terminal zinc finger or the acidic domain resulted in levels of activation comparable to that of the wild type. Interestingly, while altering the spacing between the two C-terminal zinc fingers had no effect on translation drug sensitivity, this deletion resulted in a loss of transcriptional activation from the reporter. It is quite possible that the transcription assay is a much more sensitive assay than the translation drug disk diffusion assay.
DISCUSSION
Sfp1 is required for ribosome biogenesis.
The role of Sfp1 in the cell has been a puzzle since it was first identified in a screen for factors involved in nuclear import (3). At high copy numbers, SFP1 blocked nuclear targeting of nucleolar proteins, yet the null mutant did not appear to have a significant nuclear import defect. SFP1 was also identified through its induction in the presence of DNA-damaging agents, such as MMS and ionizing radiation (27). It was shown that sfp1 mutants fail to arrest at the G2/M stage of the cell cycle after DNA damage, suggesting that Sfp1 has an important role in the DNA damage checkpoint. However, the most noticeable phenotypes of the sfp1 mutants are their small cell size and slow growth (3, 11, 27). The small size of the sfp1 mutant is similar to that caused by the “wee” mutation in S. pombe, which affects the G2/M transition and allows progression into the cell cycle before the cell has reached a particular size (20). This array of phenotypes has made the functional role of Sfp1 in the cells somewhat of a puzzle.
Defects in translation are also frequent causes of small cell size (8). We reasoned that it was therefore possible that Sfp1 is involved in the process of translation as well. This model has been supported by the finding that Sfp1 is required for wild-type levels of expression of the genes involved in ribosome biogenesis (11). We have shown that expression and processing of rRNA are significantly delayed in a sfp1 mutant and that r-proteins are not properly localized in the cell. The defects in expression, processing, and localization of ribosomal components are the likely cause for the slight reduction in the number of 60S subunits, and therefore the reduced numbers of higher-order polysomes, that we observed in the gradient profiles. The reduced number of polysomes is likely the cause of the decrease in total translation and the sensitivity to translation inhibitors that we observed in the sfp1 mutant.
The role of Sfp1 as a transcriptional activator of genes involved in ribosome biogenesis can be used to explain most of the phenotypes previously identified. It was shown that SFP1 overexpression interfered with proper nuclear localization of a mitochondrial protein containing a nuclear localization sequence (3). The higher levels of Sfp1 in the cell likely stimulated ribosome biogenesis. Since ribosomal subunits and r-proteins are continuously shuttled in and out of the nucleus, abnormally high levels of ribosomal subunit trafficking may interfere with the localization of other nuclear proteins or prevent complete assembly of ribosomal subunits. This model is supported by the finding that the sfp1 mutant showed no defect in localization of nuclear proteins (3).
The role of Sfp1 in transcriptional regulation of ribosome biogenesis genes may also explain the DNA damage and checkpoint phenotypes previously observed in an sfp1 mutant strain (27). The inability of sfp1 cells to arrest at the G2/M checkpoint may be the result of the cell being unable to properly synthesize the proteins required to respond to DNA damage. Furthermore, the induction of SFP1 transcription in response to treatment with MMS may be likened to the induction of stress response proteins. Since a large amount of proteins must be present to respond to DNA damage, Sfp1 may be stimulating ribosome biogenesis in order for the cell to synthesize the proteins needed to respond to these stresses. Alternatively, it is possible that this could be the result of an adaptation response. SFP1 expression was shown to be induced after prolonged exposure to MMS (27). Since many cell stresses cause ribosome biogenesis to be reduced, over time this biogenesis is likely to be restored as the cells adapt to these stresses. However, it remains intriguing that G2/M progression in the normal cell cycle, i.e., in the absence of stress, is also affected in the sfp1 mutant (27).
Role of Sfp1 as a transcription factor.
Recent microarray experiments have shown that Sfp1 is required for the expression of a large number of genes involved in ribosome biogenesis (11). Given that Sfp1 contains DNA-binding and activator motifs that are common to many transcription factors, it is possible that Sfp1 is functioning as a transcriptional regulatory protein of these genes. Many of the genes induced by ectopic expression of Sfp1 have two conserved promoter elements, termed RRPE and PAC (10, 25), that are often variably spaced with respect to each other. We have shown that transcription activated by the RRPE promoter elements is Sfp1 dependent in vivo. β-Galactosidase reporter assays show that in the absence of Sfp1, transcription from a reporter construct containing RRPE and PAC elements is reduced threefold in comparison to the expression in the wild-type strain (Table 2, pIF111). In contrast, high-copy expression of Sfp1 caused more than a twofold increase in the level of expression of the reporter. Thus, it is possible that Sfp1 may directly activate transcription through these elements. Mutational analysis of RRPE and PAC suggests that the PAC element serves as a repressing element and the RRPE elements function to activate transcription from these promoters in an Sfp1-dependent manner.
If Sfp1 was directly binding to these promoters, one attractive model is that individual zinc fingers in Sfp1 recognize each of these elements to regulate transcription at these promoters. The two C-terminal zinc fingers of Sfp1 are highly conserved and likely bind to very similar DNA sequences. In support of this model, we have shown that mutations in both C-terminal zinc fingers completely eliminate the function of the protein. Mutations in one finger or the other give rise to a partially defective phenotype with respect to translation drug sensitivity, suggesting that they make similar contributions to the function of the protein. The loss of one of the zinc finger motifs likely reduces the binding affinity of the protein for DNA or protein cofactors, resulting in partial activity.
Sfp1 was shown to bind to promoters of a subset of genes involved in ribosome biogenesis, as well as genes involved in nuclear import, in a genomewide chromatin immunoprecipitation screen (14). However, most of these promoters lacked RRPE and PAC elements, and none of these genes were upregulated when Sfp1 was overexpressed, or repressed in the absence of Sfp1 (11). Additionally, none of the genes identified in the Sfp1 microarray study were bound by Sfp1 in the genomic chromatin immunoprecipitation analysis. This would suggest that if Sfp1 is regulating genes containing RRPE and PAC elements, this regulation is likely occurring through an indirect mechanism. In support of this model, using chromatin immunoprecipitation assays with an epitope-tagged Sfp1 that complements the sfp1 mutant phenotype, we have been unable to show that Sfp1 is specifically associated with several different promoters of genes containing RRPE and PAC elements that are involved in ribosome biogenesis (data not shown). Additionally, we were unable to show that Sfp1 was bound to the promoters of several of the genes identified by the global chromatin immunoprecipitation experiment (data not shown). We have also been unable to detect Sfp1-dependent binding to fragments containing the RRPE and PAC elements by electrophoretic mobility shift assays using crude yeast extracts (data not shown). It is possible that Sfp1 does not directly bind to DNA at these promoters and is therefore unable to cross-link with high efficiency. Alternatively, Sfp1 may be acting indirectly, possibly by activating an activator protein that functions at these promoters. In the absence of Sfp1, the activator may not be expressed or may fail to bind the RRPE elements, preventing transcription from these constructs. This hypothesis is supported by data showing that, in the absence of Sfp1, activation from a construct containing RRPE and PAC elements is reduced to background levels compared to activation in a wild-type strain. It is also supported by data showing that mutation of the RRPE elements causes a complete loss of activation from these constructs. Mutation of the PAC element causes an increase in activation from this construct (Table 2, pIF117), suggesting that it binds by a repressor. This activation is induced sixfold when SFP1 is overexpressed, and activation is lost in an sfp1 strain. These results suggest that Sfp1 is acting indirectly as a transcription factor at these promoters, most likely by activating an activator protein. This model is also supported by the fact that levels of Sfp1 in a wild-type cell are very low (27), calling into question if there is enough protein in the cell to bind to the large number of promoters that were identified in the microarray experiments (11).
If Sfp1 is functioning as a transcription factor that specifically binds to the promoters of a subset of ribosomal biogenesis genes, or serves as a potential upstream regulator, it is not surprising that deletion of Sfp1 imparts the translation-specific defects we have observed. Failure to activate transcription of the genes containing RRPE and PAC elements would have drastic implications within the cell. The failure to fully activate many of these genes would significantly reduce the cell's ability to synthesize ribosomes, resulting in a global reduction of cellular translation, which in turn would result in defects in many essential cellular functions.
Acknowledgments
We are deeply indebted to Terri Goss Kinzy for advice and protocols. We thank Pam Silver for the Rpl11b-GFP plasmid and kap120 strain. We also thank Ed Hurt and Herbert Tschochner for the noc4-1 strain and Rps2-GFP plasmid.
This work is supported by a grant from the National Institutes of Health (GM57058) to D.N. and A.K.V.
REFERENCES
- 1.Acton, T. B., H. Zhong, and A. K. Vershon. 1997. DNA-binding specificity of Mcm1: operator mutations that alter DNA-bending and transcriptional activities by a MADS box protein. Mol. Cell. Biol. 17:1881-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baim, S. B., D. F. Pietras, D. C. Eustice, and F. Sherman. 1985. A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol. Cell. Biol. 5:1839-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blumberg, H., and P. Silver. 1991. A split zinc-finger protein is required for normal yeast growth. Gene 107:101-110. [DOI] [PubMed] [Google Scholar]
- 4.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 5.Brown, T., and K. Mackey. 1997. Analysis of RNA by Northern and slot blot hybridization, p. 4.9.1-4.9.6. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley and Sons, Inc., New York, N.Y.
- 6.Collart, M. A., and S. Oliviero. 1997. Preparation of yeast RNA, p. 13.12.1-13.12.5. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley and Sons, Inc., New York, N.Y. [DOI] [PubMed]
- 7.Deshmukh, M., Y. F. Tsay, A. G. Paulovich, and J. L. Woolford, Jr. 1993. Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol. 13:2835-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartwell, L. H., and M. W. Unger. 1977. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J. Cell Biol. 75:422-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hope, I. A., and K. Struhl. 1986. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell 46:885-894. [DOI] [PubMed] [Google Scholar]
- 10.Hughes, J. D., P. W. Estep, S. Tavazoie, and G. M. Church. 2000. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 296:1205-1214. [DOI] [PubMed] [Google Scholar]
- 11.Jorgensen, P., J. L. Nishikawa, B. J. Breitkreutz, and M. Tyers. 2002. Systematic identification of pathways that couple cell growth and division in yeast. Science 297:395-400. [DOI] [PubMed] [Google Scholar]
- 12.Keleher, C. A., C. Goutte, and A. D. Johnson. 1988. The yeast cell-type-specific repressor alpha 2 acts cooperatively with a non-cell-type-specific protein. Cell 53:927-936. [DOI] [PubMed] [Google Scholar]
- 13.Kuo, M. H., and C. D. Allis. 1999. In vivo cross-linking and immunoprecipitation for studying dynamic protein:DNA associations in a chromatin environment. Methods 19:425-433. [DOI] [PubMed] [Google Scholar]
- 14.Lee, T. I., N. J. Rinaldi, F. Robert, D. T. Odom, Z. Bar-Joseph, G. K. Gerber, N. M. Hannett, C. T. Harbison, C. M. Thompson, I. Simon, J. Zeitlinger, E. G. Jennings, H. L. Murray, D. B. Gordon, B. Ren, J. J. Wyrick, J. B. Tagne, T. L. Volkert, E. Fraenkel, D. K. Gifford, and R. A. Young. 2002. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science 298:799-804. [DOI] [PubMed] [Google Scholar]
- 15.Ma, J., and M. Ptashne. 1987. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell 48:847-853. [DOI] [PubMed] [Google Scholar]
- 16.Mager, W. H., and R. J. Planta. 1991. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol. Cell. Biochem. 104:181-187. [DOI] [PubMed] [Google Scholar]
- 17.Milkereit, P., D. Strauss, J. Bassler, O. Gadal, H. Kuhn, S. Schutz, N. Gas, J. Lechner, E. Hurt, and H. Tschochner. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40S subunits. J. Biol. Chem. 278:4072-4081. [DOI] [PubMed] [Google Scholar]
- 18.Petes, T. D. 1979. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. USA 76:410-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Planta, R. J. 1997. Regulation of ribosome synthesis in yeast. Yeast 13:1505-1518. [DOI] [PubMed] [Google Scholar]
- 20.Russell, P., and P. Nurse. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast. Cell 45:145-153. [DOI] [PubMed] [Google Scholar]
- 21.Stage-Zimmermann, T., U. Schmidt, and P. A. Silver. 2000. Factors affecting nuclear export of the 60S ribosomal subunit in vivo. Mol. Biol. Cell 11:3777-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Struhl, K. 1989. Helix-turn-helix, zinc-finger, and leucine-zipper motifs for eukaryotic transcriptional regulatory proteins. Trends Biochem. Sci. 14:137-140. [DOI] [PubMed] [Google Scholar]
- 23.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33:261-311. [DOI] [PubMed] [Google Scholar]
- 24.Vilardell, J., and J. R. Warner. 1997. Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol. 17:1959-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wade, C., K. A. Shea, R. V. Jensen, and M. A. McAlear. 2001. EBP2 is a member of the yeast RRB regulon, a transcriptionally coregulated set of genes that are required for ribosome and rRNA biosynthesis. Mol. Cell. Biol. 21:8638-8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfe, A., L. Nekludova, and C. Pabo. 1999. DNA recognition by Cys2 His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 3:183-212. [DOI] [PubMed] [Google Scholar]
- 27.Xu, Z., and D. Norris. 1998. The SFP1 gene product of Saccharomyces cerevisiae regulates G2/M transitions during the mitotic cell cycle and DNA-damage response. Genetics 150:1419-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]