Abstract
The PIGA gene from Toxoplasma gondii has been cloned and characterized. Like mammalian PIGA, the transmembrane and C-terminal domains are sufficient to direct localization to the parasite endoplasmic reticulum. A functional copy of PIGA is required for tachyzoite viability, demonstrating that glycosylphosphatidylinositol biosynthesis is an essential process in T. gondii.
Glycosylphosphatidylinositol (GPI)-anchored proteins dominate the surface of the Toxoplasma gondii tachyzoite (3, 19) and have been implicated in both host cell attachment and modulation of the host immune response (7, 15, 19). To further our understanding of the synthesis, trafficking, and function(s) of GPIs and GPI-anchored proteins in T. gondii, we have begun to characterize the genes involved in the parasite's GPI biosynthetic pathway.
GPI biosynthesis is a conserved pathway among eukaryotes that occurs primarily in the endoplasmic reticulum (ER) and leads to the generation of both free GPIs and GPI-anchored proteins (reviewed in references 8, 9, 13, 17, and 21). In mammalian cells, the pathway is initiated on the cytosolic face of the ER with the transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to phosphatidylinositol (PI) by the GPI-GlcNAc transferase complex (29-32). The GlcNAc transferase activity of the complex is thought to reside in the PI-glycan class A (PIGA) protein (18, 29). Loss-of-function mutations in PIGA result in a complete GPI deficiency, which consequently abolishes the surface expression of GPI-anchored proteins (reviewed in reference 2). The ability to tolerate a GPI deficiency is species specific and sometimes even life cycle stage specific (2, 10, 12, 14, 20, 22-25).
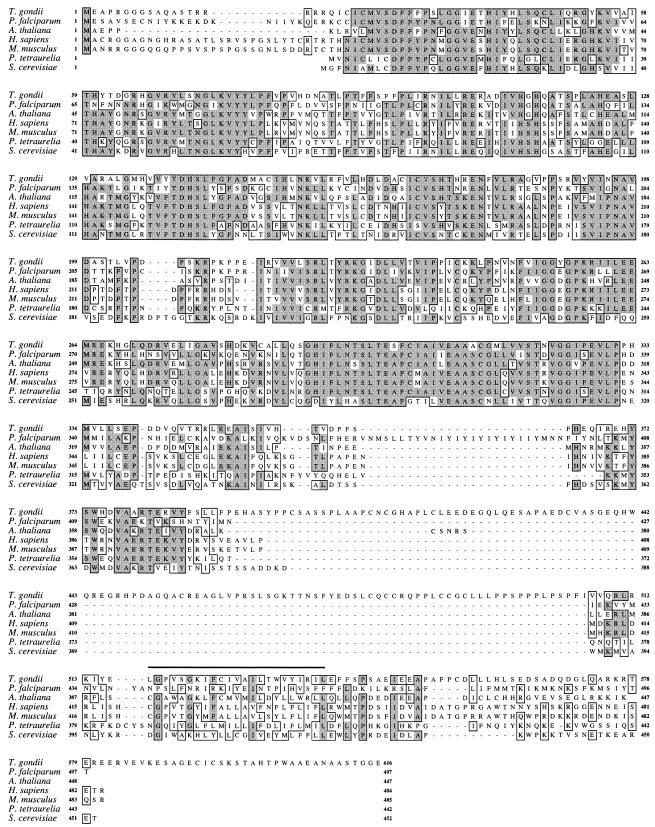
The complete PIGA cDNA of T. gondii strain RH(EP) (GenBank accession no. AY216495) was obtained and sequenced (using primers designed from an expressed sequence tag [EST1206547; http://ParaDB.cis.upenn.edu] that exhibited homology to the C-terminal region of multiple PIGA orthologues) from products of 5′ and 3′ rapid amplification of cDNA ends. Subsequent sequencing and characterization of the RH(EP) PIGA gene (GenBank accession no. AY216496) showed it to be a relatively large (∼10 kb) single-copy gene harboring 11 introns. The predicted protein sequence of T. gondii PIGA (616 amino acids) exhibits significant similarity to sequences of other PIGA orthologues (Fig. 1), particularly between residues 22 and 383 (∼50% identity and ∼70% homology), a region which contains the putative GlcNAc transferase domain. As with other PIGA proteins, T. gondii PIGA harbors a potential transmembrane domain (residues 517 to 539) followed by a stretch of mostly hydrophilic residues (residues 541 to 616) extending to the C terminus (Fig. 1). T. gondii PIGA contains an insert of ∼100 amino acids not found in other PIGA orthologues (residues 384 to 506; Fig. 1). This insert, which is also present in both genomic (http://toxodb.org/ToxoDB.shtml) and cDNA (data not shown) sequences of PIGA of T. gondii strain P(LK), exhibits no homology to any known proteins. Its functional significance, if any, is unknown.
FIG. 1.
ClustalW sequence alignment of PIGA orthologues from T. gondii, Plasmodium falciparum (gi23495183), Arabidopsis thaliana (gi18407913), Homo sapiens (gi219994), Mus musculus (gi1402592), Paramecium tetraurelia (gi8571458), and Saccharomyces cerevisiae (gi9755344). Regions of homology are boxed, with identity denoted by dark shading and conserved amino acid changes denoted by light shading. The predicted T. gondii PIGA transmembrane domain (amino acids 517 to 539) is denoted by a thick horizontal line.
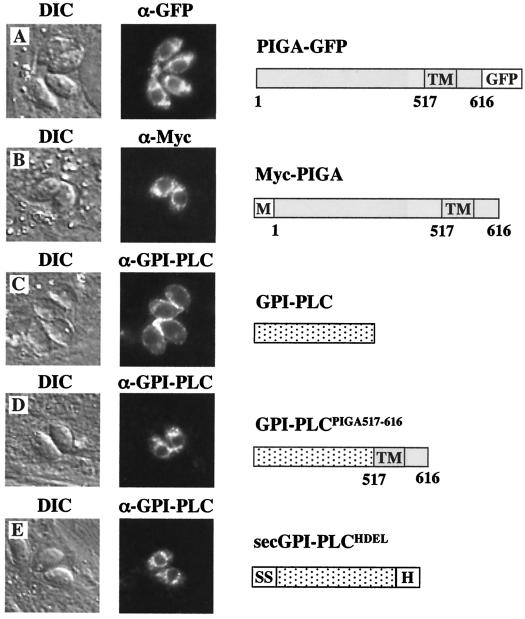
Stably expressed recombinant versions of T. gondii PIGA, containing either an N-terminal c-Myc epitope tag or a C-terminal green fluorescent protein (GFP) fusion, localized predominantly to ER-like structures encircling the nucleus of the tachyzoite (Fig. 2A and B). When transiently expressed, an engineered ER marker (Trypanosoma brucei GPI-phospholipase C [GPI-PLC] containing a secretory signal sequence and ER retention signal [secGPI-PLCHDEL]) localized to a perinuclear compartment (Fig. 2E) indistinguishable from that of PIGA.
FIG. 2.
Subcellular localization of T. gondii PIGA. PIGA with a C-terminal fusion to GFP (A) or an N-terminal c-Myc epitope tag (B) localized to ER-like structures encircling the nucleus of the parasite. As a marker for ER localization, a recombinant version of T. brucei GPI-PLC, which normally localized to the periphery of the tachyzoite (C), was engineered to localize to the ER (secGPI-PLCHDEL) by the addition of an N-terminal secretory signal sequence and the C-terminal ER-retention motif, HDEL (E). When fused to the C terminus of GPI-PLC (GPI-PLCPIGA517-616), the putative transmembrane and ER-lumenal domains (amino acids 517 to 616) of T. gondii PIGA were sufficient to alter the localization of GPI-PLC to an ER-like localization (D) indistinguishable from that of PIGA. Bar, 5 μm. M, c=Myc; SS, signal sequence; H, HDEL; TM, transmembrane; DIC, differential interference contrast.
Previous studies have shown that the transmembrane domain of mammalian PIGA, together with the 23 residues immediately C terminal to this domain, is sufficient for ER targeting and/or retention (31). Like its mammalian counterpart, T. gondii PIGA lacks an obvious N-terminal signal sequence but appears to localize to the ER. To determine whether the transmembrane and C-terminal domains of T. gondii PIGA direct localization, residues 517 to 616 were fused to the C terminus of GPI-PLC (GPI-PLCPIGA517-616). In transient-expression studies, GPI-PLCPIGA517-616 exhibited a localization pattern similar to that of PIGA (Fig. 2D) and strikingly different from the peripheral localization of GPI-PLC lacking this domain (Fig. 2C), indicating that the signals required for ER targeting of T. gondii PIGA are also located within the transmembrane and C-terminal domains. Heterologously expressed GPI-PLC can induce a GPI deficiency in other parasites through the cleavage of GPI intermediates (10, 22). Stable expression of wild-type GPI-PLC exhibited little effect on tachyzoites (data not shown); however, it was not possible to generate clones stably expressing GPI-PLC targeted to the ER, suggesting that GPI-PLC expression at the site of GPI biosynthesis was lethal.
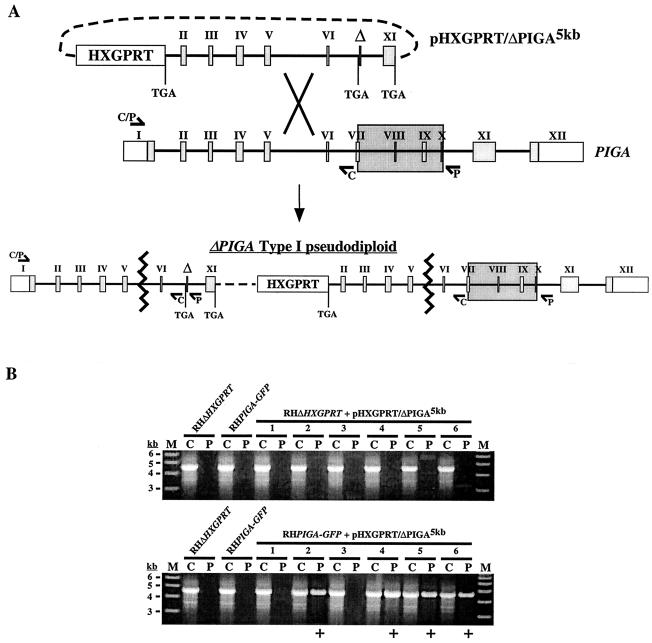
When chemical mutagenesis (27), insertional mutagenesis (6), and targeted gene disruption of PIGA, each coupled with selection (using Clostridium septicum alpha toxin) (11, 33) for a GPI-anchored protein deficiency, were used to attempt to generate GPI-deficient parasites, the results were uniformly unsuccessful, suggesting that GPI biosynthesis is required for tachyzoite viability. To show this conclusively, we attempted to disrupt PIGA in the presence of a second copy of the gene. A knockout vector (pHXGPRT/ΔPIGA5kb) was designed such that integration within the region located upstream of the internal deletion (type I) would result in a pseudodiploid (5, 26, 28) harboring two nonfunctional PIGA alleles (Fig. 3A).
FIG. 3.
Targeted disruption of PIGA. (A) Schematic of the gene disruption strategy. The pHXGPRT/ΔPIGA5kb knockout vector was constructed such that single-crossover homologous recombination into the PIGA locus upstream of the deletion would generate a type I pseudodiploid with two nonfunctional PIGA alleles. To generate ΔPIGA5kb, the 5′ end of PIGA was truncated within exon 1 (removing the 5′ untranslated region and coding sequence corresponding to residues 1 to 47) and the 3′ end was truncated into exon 11 (removing the coding sequence for the putative transmembrane and ER-lumenal domains [residues 519 to 616]). An internal sequence corresponding to a highly conserved region of PIGA (amino acids 303 to 368) was also deleted, and stop codons were engineered into all cloning junctions within the ΔPIGA5kb allele. Type I integration resulted in the harboring by the pairs upstream allele of the ∼1.8-kb deletion (marked by Δ; large shaded box indicates the corresponding sequence in the wild-type allele), which could be detected by PCR (using primer set P) as a product of ∼4.6 kb. The P primer set was found to be incapable of amplifying the predicted product (∼6.5 kb) from wild-type genomic DNA. (B) Six independent stable transgenic populations (lane pairs 1 to 6) derived from either parental parasites (RHΔHXGPRT) or parasites harboring a stable second copy of PIGA (RHPIGA-GFP) were screened for integration of the plasmid into PIGA (see text and elsewhere in this legend). ΔPIGA type I pseudodiploids were not detected in any of the six populations derived from parental parasites, while four out of six populations from RHPIGA-GFP parasites yielded a positive PCR product result (+). Lanes C, control primers; lanes P, pseudodiploid-specific primers; lanes M, molecular mass markers.
Parental [RH(EP)ΔHXGPRT) (26, 28)] and RH(EP)ΔHXGPRT/PIGA-GFP (referred to hereafter as RHPIGA-GFP) parasites were each transfected (16, 28) with undigested pHXGPRT/ΔPIGA5kb in six independent experiments. Stable expression of PIGA-GFP was confirmed by both immunofluorescence (Fig. 2A) and Western blot (data not shown) experiments. A forward primer directed against the 5′ untranslated region of PIGA (which was not present in ΔPIGA5kb) and a reverse primer located directly downstream of the deleted region (primer set P) (Fig. 3A) were used for PCR to screen stable transgenic populations for the presence of type I ΔPIGA pseudodiploids. These primers yield a product of ∼4.6 kb that is pseudodiploid specific, since amplification through the deleted region was not possible using genomic DNA as a template (Fig. 3B). As a control, the same upstream primer and a reverse primer located immediately upstream of the deleted region in the ΔPIGA5kb allele (primer set C) were used to amplify a product of ∼4.6 kb from both the wild-type and pseudodiploid PIGA alleles (Fig. 3A). Type I ΔPIGA pseudodiploids were not detected in any of the six independent populations from the parental parasites. However, they were present in four of the six populations derived from RHPIGA-GFP parasites (Fig. 3B), indicating that a second copy of PIGA was sufficient to rescue the parasite upon disruption of the wild-type gene. Integration into the genomic locus was confirmed both by Southern blot analysis of independent clones and by the absence of a reverse transcription-PCR product corresponding to the PIGA transcript in these clones (data not shown).
These results demonstrate that GPI biosynthesis is essential for viability in T. gondii, and they identify the GPI biosynthetic pathway as a potential target for the development of new chemotherapeutics against this parasite. It is not clear whether the lethal consequences of PIGA disruption in T. gondii result from a deficiency in GPI-anchored proteins, free GPIs, or both. Future experiments aimed at disrupting components of the T. gondii GPI transamidase complex (1, 4, 12, 20, 34) should resolve this question and reveal the relative importance of GPI-anchored proteins in the T. gondii life cycle.
Acknowledgments
We thank Kojo Mensa-Wilmot for helpful advice and for generously providing GPI-PLC constructs and antibodies. We also thank Dominique Soldati, Con Beckers, David Sibley, and Michael White for providing constructs, David Roos for helpful advice, and Mary Tierney, Doug Johnson, and members of the laboratory for critical reading of the manuscript.
This work was supported by PHS grants AI42355 (G.E.W.) and CA22435 (Vermont Cancer Center) and through the Vermont EPSCoR program under NSF grant EPS-9874685.
REFERENCES
- 1.Benghezal, M., A. Benachour, S. Rusconi, M. Aebi, and A. Conzelmann. 1996. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 15:6575-6583. [PMC free article] [PubMed] [Google Scholar]
- 2.Bessler, M., A. Schaefer, and P. Keller. 2001. Paroxysmal nocturnal hemoglobinuria: insights from recent advances in molecular biology. Transfus. Med. Rev. 15:255-267. [DOI] [PubMed] [Google Scholar]
- 3.Black, M. W., and J. C. Boothroyd. 2000. Lytic cycle of Toxoplasma gondii. Microbiol. Mol. Biol. Rev. 64:607-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, R., S. Udenfriend, G. M. Prince, S. E. Maxwell, S. Ramalingam, L. D. Gerber, J. Knez, and M. E. Medof. 1996. A defect in glycosylphosphatidylinositol (GPI) transamidase activity in mutant K cells is responsible for their inability to display GPI surface proteins. Proc. Natl. Acad. Sci. USA 93:2280-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donald, R. G., and D. S. Roos. 1998. Gene knock-outs and allelic replacements in Toxoplasma gondii: HXGPRT as a selectable marker for hit-and-run mutagenesis. Mol. Biochem. Parasitol. 91:295-305. [DOI] [PubMed] [Google Scholar]
- 6.Donald, R. G., and D. S. Roos. 1995. Insertional mutagenesis and marker rescue in a protozoan parasite: cloning of the uracil phosphoribosyltransferase locus from Toxoplasma gondii. Proc. Natl. Acad. Sci. USA 92:5749-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dzierszinski, F., M. Mortuaire, M. F. Cesbron-Delauw, and S. Tomavo. 2000. Targeted disruption of the glycosylphosphatidylinositol-anchored surface antigen SAG3 gene in Toxoplasma gondii decreases host cell adhesion and drastically reduces virulence in mice. Mol. Microbiol. 37:574-582. [DOI] [PubMed] [Google Scholar]
- 8.Ferguson, M. A. 1999. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112:2799-2809. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson, M. A., J. S. Brimacombe, S. Cottaz, R. A. Field, L. S. Guther, S. W. Homans, M. J. McConville, A. Mehlert, K. G. Milne, J. E. Ralton, et al. 1994. Glycosyl-phosphatidylinositol molecules of the parasite and the host. Parasitology 108:S45-S54. [DOI] [PubMed] [Google Scholar]
- 10.Garg, N., M. Postan, K. Mensa-Wilmot, and R. L. Tarleton. 1997. Glycosylphosphatidylinositols are required for the development of Trypanosoma cruzi amastigotes. Infect. Immun. 65:4055-4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon, V. M., K. L. Nelson, J. T. Buckley, V. L. Stevens, R. K. Tweten, P. C. Elwood, and S. H. Leppla. 1999. Clostridium septicum alpha toxin uses glycosylphosphatidylinositol-anchored protein receptors. J. Biol. Chem. 274:27274-27280. [DOI] [PubMed] [Google Scholar]
- 12.Hilley, J. D., J. L. Zawadzki, M. J. McConville, G. H. Coombs, and J. C. Mottram. 2000. Leishmania mexicana mutants lacking glycosylphosphatidylinositol (GPI):protein transamidase provide insights into the biosynthesis and functions of GPI-anchored proteins. Mol. Biol. Cell 11:1183-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikezawa, H. 2002. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 25:409-417. [DOI] [PubMed] [Google Scholar]
- 14.Ilgoutz, S. C., J. L. Zawadzki, J. E. Ralton, and M. J. McConville. 1999. Evidence that free GPI glycolipids are essential for growth of Leishmania mexicana. EMBO J. 18:2746-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacquet, A., L. Coulon, J. De Neve, V. Daminet, M. Haumont, L. Garcia, A. Bollen, M. Jurado, and R. Biemans. 2001. The surface antigen SAG3 mediates the attachment of Toxoplasma gondii to cell-surface proteoglycans. Mol. Biochem. Parasitol. 116:35-44. [DOI] [PubMed] [Google Scholar]
- 16.Karsten, V., H. Qi, C. J. Beckers, and K. A. Joiner. 1997. Targeting the secretory pathway of Toxoplasma gondii. Methods 13:103-111. [DOI] [PubMed] [Google Scholar]
- 17.Kinoshita, T., K. Ohishi, and J. Takeda. 1997. GPI-anchor synthesis in mammalian cells: genes, their products, and a deficiency. J. Biochem. (Tokyo) 122:251-257. [DOI] [PubMed] [Google Scholar]
- 18.Kostova, Z., D. M. Rancour, A. K. Menon, and P. Orlean. 2000. Photoaffinity labelling with P3-(4-azidoanilido)uridine 5′-triphosphate identifies gpi3p as the UDP-GlcNAc-binding subunit of the enzyme that catalyses formation of GlcNAc-phosphatidylinositol, the first glycolipid intermediate in glycosylphosphatidylinositol synthesis. Biochem. J. 350:815-822. [PMC free article] [PubMed] [Google Scholar]
- 19.Lekutis, C., D. J. Ferguson, M. E. Grigg, M. Camps, and J. C. Boothroyd. 2001. Surface antigens of Toxoplasma gondii: variations on a theme. Int. J. Parasitol. 31:1285-1292. [DOI] [PubMed] [Google Scholar]
- 20.Lillico, S., M. C. Field, P. Blundell, G. H. Coombs, and J. C. Mottram. 2003. Essential roles for GPI-anchored proteins in African trypanosomes revealed using mutants deficient in GPI8. Mol. Biol. Cell 14:1182-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McConville, M. J., and A. K. Menon. 2000. Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids (review). Mol. Membr. Biol. 17:1-16. [DOI] [PubMed] [Google Scholar]
- 22.Mensa-Wilmot, K., J. H. LeBowitz, K. P. Chang, A. al-Qahtani, B. S. McGwire, S. Tucker, and J. C. Morris. 1994. A glycosylphosphatidylinositol (GPI)-negative phenotype produced in Leishmania major by GPI phospholipase C from Trypanosoma brucei: topography of two GPI pathways. J. Cell Biol. 124:935-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagamune, K., T. Nozaki, Y. Maeda, K. Ohishi, T. Fukuma, T. Hara, R. T. Schwarz, C. Sutterlin, R. Brun, H. Riezman, and T. Kinoshita. 2000. Critical roles of glycosylphosphatidylinositol for Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 97:10336-10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naik, R. S., E. A. Davidson, and D. C. Gowda. 2000. Developmental stage-specific biosynthesis of glycosylphosphatidylinositol anchors in intraerythrocytic Plasmodium falciparum and its inhibition in a novel manner by mannosamine. J. Biol. Chem. 275:24506-24511. [DOI] [PubMed] [Google Scholar]
- 25.Naik, R. S., G. Krishnegowda, and D. C. Gowda. 2003. Glucosamine inhibits inositol acylation of the glycosylphosphatidylinositol anchors in intraerythrocytic Plasmodium falciparum. J. Biol. Chem. 278:2036-2042. [DOI] [PubMed] [Google Scholar]
- 26.Pfefferkorn, E. R., and S. E. Borotz. 1994. Toxoplasma gondii: characterization of a mutant resistant to 6-thioxanthine. Exp. Parasitol. 79:374-382. [DOI] [PubMed] [Google Scholar]
- 27.Pfefferkorn, E. R., and L. C. Pfefferkorn. 1979. Quantitative studies of the mutagenesis of Toxoplasma gondii. J. Parasitol. 65:364-370. [PubMed] [Google Scholar]
- 28.Roos, D. S., R. G. Donald, N. S. Morrissette, and A. L. Moulton. 1994. Molecular tools for genetic dissection of the protozoan parasite Toxoplasma gondii. Methods Cell Biol. 45:27-63. [DOI] [PubMed] [Google Scholar]
- 29.Tiede, A., C. Nischan, J. Schubert, and R. E. Schmidt. 2000. Characterisation of the enzymatic complex for the first step in glycosylphosphatidylinositol biosynthesis. Int. J. Biochem. Cell Biol. 32:339-350. [DOI] [PubMed] [Google Scholar]
- 30.Watanabe, R., N. Inoue, B. Westfall, C. H. Taron, P. Orlean, J. Takeda, and T. Kinoshita. 1998. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 17:877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watanabe, R., T. Kinoshita, R. Masaki, A. Yamamoto, J. Takeda, and N. Inoue. 1996. PIG-A and PIG-H, which participate in glycosylphosphatidylinositol anchor biosynthesis, form a protein complex in the endoplasmic reticulum. J. Biol. Chem. 271:26868-26875. [DOI] [PubMed] [Google Scholar]
- 32.Watanabe, R., Y. Murakami, M. D. Marmor, N. Inoue, Y. Maeda, J. Hino, K. Kangawa, M. Julius, and T. Kinoshita. 2000. Initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-P and is regulated by DPM2. EMBO J. 19:4402-4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wichroski, M. J., J. A. Melton, C. G. Donahue, R. K. Tweten, and G. E. Ward. 2002. Clostridium septicum alpha-toxin is active against the parasitic protozoan Toxoplasma gondii and targets members of the SAG family of glycosylphosphatidylinositol-anchored surface proteins. Infect. Immun. 70:4353-4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, J., S. Nagarajan, J. J. Knez, S. Udenfriend, R. Chen, and M. E. Medof. 1997. The affected gene underlying the class K glycosylphosphatidylinositol (GPI) surface protein defect codes for the GPI transamidase. Proc. Natl. Acad. Sci. USA 94:12580-12585. [DOI] [PMC free article] [PubMed] [Google Scholar]