Abstract
The pre-T cell receptor α (pTα) protein is a critical component of the pre-T cell receptor complex in early thymocytes. The expression of the pTα gene is one of the earliest markers of the T cell lineage and occurs exclusively in pre-T cells. To investigate the molecular basis of thymocyte-specific gene expression, we searched for the genomic elements regulating transcription of the mouse pTα gene. We now report that expression of the pTα gene is primarily controlled by an upstream genomic region, which can drive thymocyte-specific expression of a marker gene in transgenic mice. Within this region, we have identified two specific DNase-hypersensitive sites corresponding to a proximal promoter and an upstream transcriptional enhancer. The pTα enhancer appears to function preferentially in pre-T cell lines and binds multiple nuclear factors, including YY1. The enhancer also contains two G-rich stretches homologous to a critical region of the thymocyte-specific lck proximal promoter. Here we show that these sites bind a common nuclear factor and identify it as the zinc finger protein ZBP-89. Our data establish a novel experimental model for thymocyte-specific gene expression and suggest an important role for ZBP-89 in T cell development.
Keywords: transcription, T lymphocytes, YY1, Sp1, ZBP-89
The development of a diverse and functional lymphocyte repertoire is achieved through the precise regulation of gene expression in various lymphoid lineages and at different developmental stages (1). As a powerful approach to the study of gene regulation in the immune system, genomic elements controlling the expression of lymphocyte-specific genes have been identified and used to isolate transcription factors involved in lymphocyte development (2).
The regulatory mechanisms underlying gene expression during the specification of the T cell lineage and its subsequent effector differentiation are being rapidly uncovered. Transcriptional regulation of many T cell–specific genes, particularly the TCR genes (3), has been described in great detail. Several transcription factors, including common lymphoid factors such as PU.1 and Ikaros and T cell–specific factors, such as GATA3 and TCF1/LEF1, were found to be critical for T cell development by gene targeting techniques (4). In contrast, our understanding of stage-specific gene expression in T cell development is still limited. Although several stage-specific regulatory elements such as alternative lck promoters (5), a CD4 silencer (6, 7), and CD8 enhancers (8–10) have been identified, their mode of action is not fully understood. In particular, molecular mechanisms that bring about gene expression specifically in pre-T cells remain to be elucidated.
The pre-TCR-α (pTα)1 protein pairs with a newly synthesized TCR-β chain to form the pre-TCR, which transduces an obligatory survival signal to developing T cells at the so-called “β selection” checkpoint (11). As an essential component of the pre-TCR, pTα is critical for the efficient generation of T cells with productive TCR-β rearrangements, allelic exclusion at the TCR-β locus, and lineage commitment of α/β T cells (12). Consistent with its function at the early stages of T cell development, the pTα gene is expressed in pre-T cells in the thymus and extrathymic T cell maturation sites but not in mature thymocytes, peripheral T cells, or other cell types (13, 14). Moreover, pTα appears to be one of the earliest molecular markers of the T cell lineage, as its expression was detected in T cell progenitors in the mouse bone marrow and fetal blood (14), as well as in adult human blood (15). In view of its pre-T cell–specific expression, the pTα gene may provide an attractive model system to study the transcriptional regulation of early T cell development. To this end, we set out to characterize regulatory regions within the mouse pTα locus.
Materials and Methods
DNA Constructs.
Mouse pTα cDNA probe was amplified by reverse transcriptase (RT)-PCR as described (13) and used to screen a 129/SvJ mouse bacterial artificial chromosome genomic library (Genome Systems). A positive clone encompassing >100 kb of pTα locus was isolated and used for restriction mapping with oligonucleotide probes and for subcloning into plasmid vectors. The sequencing and mapping data were consistent with those reported previously (16).
For reporter assays, a 5′ pTα fragment spanning 1.2 kb up to the PstI site within the known 5′ untranslated region (UTR) (16) was subcloned into the HindIII site of the promoterless LacZ (Escherichia coli β-galactosidase) reporter vector pβgal-Basic (Clontech). An 8-kb XhoI-BglII 5′ fragment was subcloned into XhoI-BglII sites of this construct to create a 9-kb XhoI-PstI 5′ pTα fragment upstream of LacZ. Promoter and enhancer mapping were carried out by in-vector deletions of the above constructs or by subcloning of fragments upstream of the pTα promoter. The BstEII-MluNI enhancer fragment was subcloned into pBluescript and sequenced using T3 and T7 primers. As heterologous promoters, we used the CMV immediate early promoter and the SV40 early promoter (pβgal-Promoter vector; Clontech) subcloned upstream of LacZ in the pβgal-Basic vector. As control enhancers, we used 0.6-kb mouse CD3δ enhancer (reference 17; a gift of Dr. Cox Terhorst, Harvard Medical School) and 0.55-kb human CD2 enhancer fragment (18). To examine enhancer activity in various cell lines, 4.3-kb NheI-BglII and 0.35-kb BstEII-NarI pTα enhancer fragments were subcloned upstream of the SV40 promoter in the pβgal-Promoter vector.
For deletion analysis of the pTα enhancer, fragments were amplified by PCR using Pwo DNA polymerase (Roche Molecular Biochemicals) and subcloned into the KpnI-BglII sites of the pβgal-Promoter vector. Site-directed mutagenesis was performed using the QuikChange kit (Stratagene). All of these constructs were verified by sequencing using a primer within the SV40 promoter.
Cells.
A panel of T cell lymphomas, either derived in this laboratory from various knockout mouse strains or obtained from the American Type Culture Collection, was screened for the expression of pTα mRNA by Northern hybridization and RT-PCR (data not shown). The pTα-positive lines included LR1 (atm −/− rag-2 −/−), 642 (p53 −/−), and 799 (atm −/− p53 −/−); the pTα-negative lines included LR2 (atm −/− rag-2 −/−), EL4, and BW5147. Other cell lines included 1105, a B cell lymphoma from a c-myc– transgenic animal; MEL, a mouse erythroleukemia line; and NIH3T3 mouse fibroblasts. All lines were cultured in DMEM with 10% FCS, l-glutamine, and 2-ME.
DNase Hypersensitivity Assay.
Cells were harvested and lysed in reticulocyte standard buffer (10 mM Tris/HCl, pH 7.4, 10 mM NaCl, 5 mM MgCl2) with 0.5% NP-40. Nuclei were washed and resuspended in reticulocyte standard buffer and treated with twofold dilutions of DNase I (Roche Molecular Biochemicals) for 5 min at room temperature. The reaction was stopped with a buffer containing 1% SDS, 50 mM EDTA, and proteinase K, and DNA was extracted, digested with appropriate enzymes, and analyzed by Southern hybridization. For the comparison between different cell lines, the amount of DNase was calibrated for each line and three concentrations were chosen: (i) the minimal DNase concentration producing a visible downshift of DNA fragments after restriction digest, (ii) a twofold lower concentration, and (iii) no DNase.
The following genomic probes were used: a 0.46-kb PstI-NcoI fragment encompassing pTα exon 2 (probe 1); a 0.5-kb XhoI-HpaI fragment 10 kb 5′ of exon 1; and a 0.4-kb EcoRI-NcoI fragment 7 kb 5′ of exon 1 (probe 2).
Transgenic Mice.
The construct containing a 9-kb pTα 5′ fragment upstream of LacZ and SV40 intron/Poly(A) sequences in the pβgal-Basic vector was digested with XhoI/SalI and gel-purified to remove the vector backbone. DNA was microinjected into fertilized oocytes from FVB mice, and transgenic founders were bred with wild-type FVB mice. A 0.25-kb AspI-PstI probe from the 3′ end of the 9-kb pTα fragment hybridized with 5- and 1.6-kb EcoRV fragments from the endogenous pTα gene and the transgene, respectively, and was used for transgene copy number determination by PhosphorImager (Molecular Dynamics) quantitation. Total RNA was extracted from various organs of 4-wk-old F1-transgenic mice, and Poly(A) RNA was prepared from 250 μg total RNA using biotin-oligo(dT)/streptavidin magnetic beads (Roche Molecular Biochemicals). Transgene expression was analyzed by Northern hybridization with a 5′ 1.3-kb fragment of LacZ gene.
RNase Protection Assay.
An NcoI fragment spanning 1.6 kb 5′ of the pTα first exon was subcloned in the reverse orientation into the NcoI site of pSL301 vector and linearized with EcoRV. A 0.4-kb α-[32P]dUTP–labeled riboprobe was synthesized using T3 RNA polymerase and hybridized with 20 μg total RNA from the indicated cell lines. After digestion with RNase T1 and A, the protected fragments were resolved on a 6% denaturing sequencing gel in parallel with a DNA sequencing ladder as a marker.
Transfections.
Cells were transferred into 6-well plates and transfected with 1–2 μg DNA/well in duplicate, using lipid reagents according to the manufacturers' instructions. Cell lines LR1, BW5147, MEL, and NIH3T3 were transfected using Fugene 6 reagent (Roche Molecular Biochemicals); cells from cell lines LR2 and 642 were transfected using Superfect reagent (Qiagen). LacZ reporter vectors containing no promoter (pβgal-Basic) or SV40 promoter (pβgal-Promoter) were used as controls in all experiments. After 24 h, cells were harvested and β-galactosidase activity was measured using a chemiluminescent assay (Clontech or Tropix) in the scintillation counter. The results represent mean cpm ± range of duplicate transfections. Note that these are arbitrary units depending on the assay conditions; in particular, the assay from Tropix was found to be 10–100 times more sensitive than the assay from Clontech.
Electrophoretic Mobility Shift Assay.
For crude nuclear extract preparation, cell nuclei were isolated by hypotonic lysis, and proteins were extracted with a buffer containing 0.4 M NaCl. Each binding reaction (15 μl) contained 10 fmol (105 cpm) γ-[32P] dATP–labeled double-stranded oligonucleotide probe, 3–5 μg nuclear protein extract, 0.5 μg poly(dI-dC), and, where indicated, a 100-fold excess of unlabeled oligonucleotide or 2 μg Ab. Binding was performed for 20 min at room temperature in a final concentration of 50 mM NaCl; 20 mM Tris/HCl, pH 7.5; 1 mM EDTA; 1 mM dithiothreitol; and 10% glycerol. Protein–DNA complexes were separated on 4% nondenaturing polyacrylamide gels in 0.5× Tris/borate/EDTA buffer and visualized by autoradiography.
Oligonucleotide probes 1–6 spanning the core enhancer are depicted (see Fig. 6). Other probes included consensus oligonucleotides for YY1 (Santa Cruz Biotechnology), Sp1 (Promega Corp.), and Ikaros (IK-BS2; reference 19), and an lck proximal promoter −365/−328 probe (lckB; reference 20). Anti-YY1 mAb and anti-Sp1 and -Sp3 polyclonal Ab were purchased from Santa Cruz Biotechnology. Anti-ZBP89 rabbit antiserum was provided by Drs. Juanita Merchant and David Law (University of Michigan, Ann Arbor, MI) and was used as protein A–purified IgG fraction. Purified anti-CD3 mAb 145-2C11 was used as a negative control.
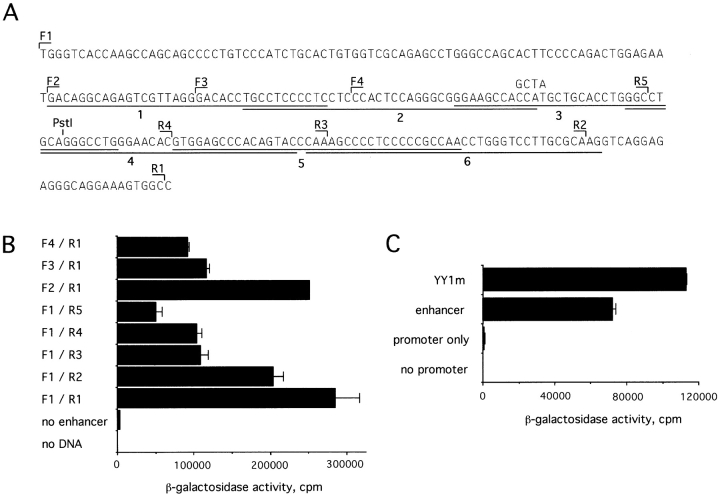
Figure 6.
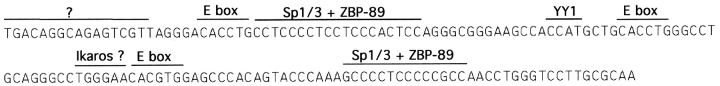
Fine mapping of the pTα enhancer. (A) Sequence of the 0.25-kb BstEII-MluNI enhancer fragment, showing forward (F) and reverse (R) primers used for amplification of the deletion fragments. The oligonucleotide probes 1–6 used for EMSA are underlined. The PstI site and the mutation introduced at the putative YY1 binding site are indicated. The sequence is available from EMBL/GenBank/ DDBJ under accession no. AF132612. In B and C, the enhancer fragments were subcloned upstream of the SV40 promoter/ LacZ reporter gene and assayed by transient transfection of LR1 cells. (B) Nested enhancer deletions were amplified by PCR with the indicated primer pairs. (C) The YY1 site mutation shown above (YY1m) was introduced into the 0.25-kb enhancer fragment by site-directed mutagenesis.
Potential transcription factor binding sites were analyzed using the TRANSFAC database and software (http://transfac.gbf.de; reference 21).
Results
Specific DNase-hypersensitive Sites Upstream of the pTα Gene.
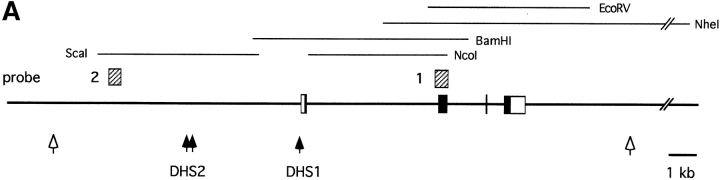
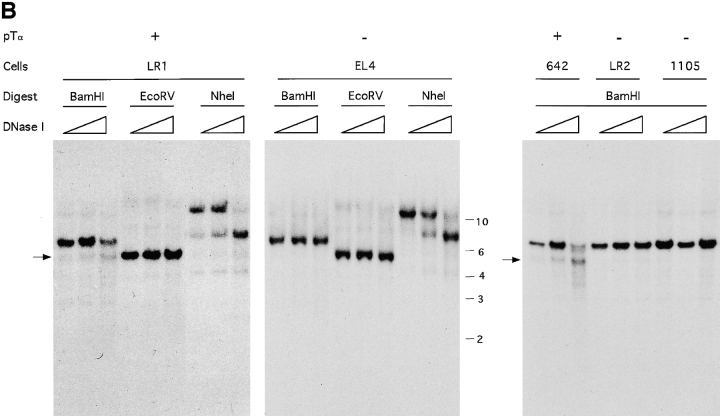
Because regulatory genomic regions often display increased sensitivity to DNase I treatment, we searched for such DNase-hypersensitive sites (DHS) in the pTα locus of T cell lymphomas manifesting or lacking pTα expression. We first used a probe corresponding to pTα exon 2 in conjunction with several restriction digests shown in Fig. 1 A. No strong DHS correlating with pTα expression were detected within or downstream of the pTα gene (EcoRV and NheI digests in Fig. 1 B and data not shown). A nonspecific DHS was detected 4 kb downstream of the last exon in all cell lines tested, confirming the validity of the analysis (NheI digest, Fig. 1 B). In contrast, a specific site (DHS 1) was found immediately upstream of the first exon in pTα-positive but not pTα-negative T cell lines (BamHI digest, Fig. 1 B).
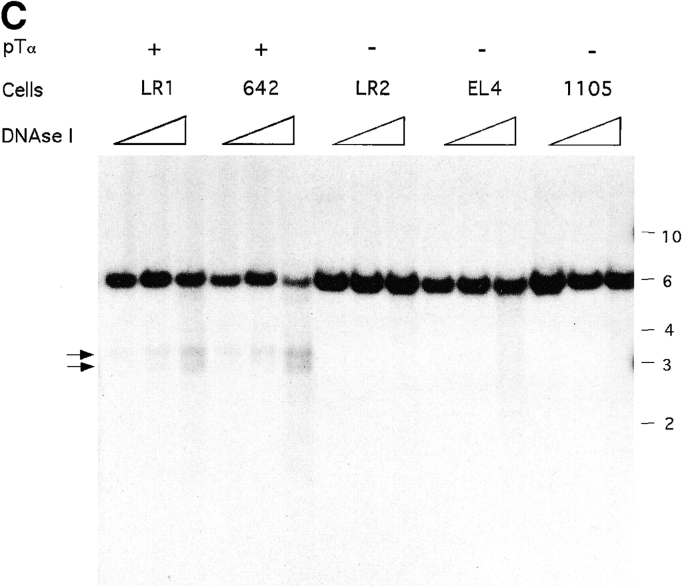
Figure 1.
DNase hypersensitivity analysis of the mouse pTα locus. (A) Map of the pTα gene showing restriction fragments and genomic probes used. The exons and UTR (16) are shown as filled and empty boxes, respectively. The approximate positions of nonspecific (empty arrowheads) and specific (filled arrowheads) DHS are indicated. In B, genomic DNA from pTα-positive and -negative cell lines was treated with increasing concentrations of DNase I, digested with indicated enzymes, and hybridized with probe 1. The band corresponding to DHS 1 is indicated (arrow). The positions of DNA size markers (kb) are shown. In C, the same DNA samples were digested with ScaI and hybridized with probe 2. The bands corresponding to DHS 2 are indicated (arrows).
To examine the regions further upstream of the pTα gene, we initially used a probe detecting a 10-kb BglII fragment 5′ of the gene (data not shown). This preliminary analysis suggested the presence of a specific DHS within the region; however, a nonspecific DHS immediately 3′ to the probe precluded more precise mapping. To better localize the potential second DHS, we used a more downstream fragment as a probe (probe 2, Fig. 1 A). As shown in Fig. 1 C, these experiments revealed the presence of two closely located DHS, collectively referred to as DHS 2, ∼4–4.5 kb upstream of the first pTα exon specifically in pTα-positive cell lines. Thus, the genomic region 5′ of pTα harbors at least two specific DHS and is likely to play a major role in the regulation of pTα expression.
The Region Upstream of pTα Can Drive Thymocyte-specific Transgene Expression.
To verify that putative regulatory regions upstream of the pTα gene were sufficient for pre-T cell–specific gene expression, we created transgenic mice carrying the marker gene LacZ under the control of a pTα 5′ fragment. The transgene contained 9 kb of pTα 5′ region, including both DHS and a part of the known 5′ UTR, upstream of LacZ and SV40 intron and Poly(A) signal. The heterozygous progeny of transgenic founders were analyzed for transgene expression by Northern hybridization with a LacZ probe.
Of six transgenic lines analyzed, two lines manifested detectable LacZ expression in the thymus. Both lines expressed LacZ in the thymus but not in the spleen; the line carrying fewer copies of the transgene (four copies) was analyzed in more detail. As shown in Fig. 2, the marker gene was expressed exclusively in the thymus but not in the spleen, lymph nodes, or other organs, with a pattern and abundance comparable to that of the endogenous pTα gene. Thus, a 9-kb 5′ genomic fragment of the pTα gene supported thymus-specific transgene expression in two transgenic mouse lines. We therefore conclude that this fragment contains all information necessary for pTα expression in thymocytes.
Figure 2.
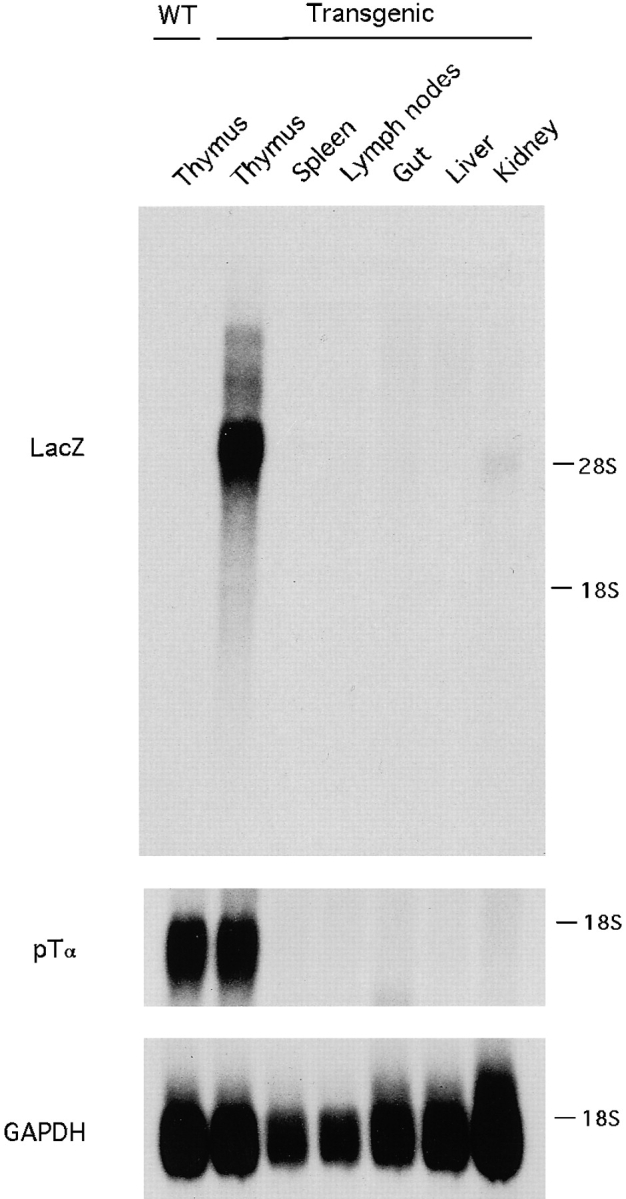
Expression of a LacZ marker gene driven by pTα upstream fragment in transgenic mice. Poly(A) RNA from the indicated organs of transgenic or wild-type (WT) littermates was analyzed by Northern hybridization with a LacZ probe, and then the blot was stripped and rehybridized with the pTα cDNA probe and GAPDH cDNA probe. The blot was exposed for 4 h for LacZ and pTα probes and 0.5 h for GAPDH probe. The positions of 28S and 18S rRNA are indicated.
Characterization of a Proximal pTα Promoter.
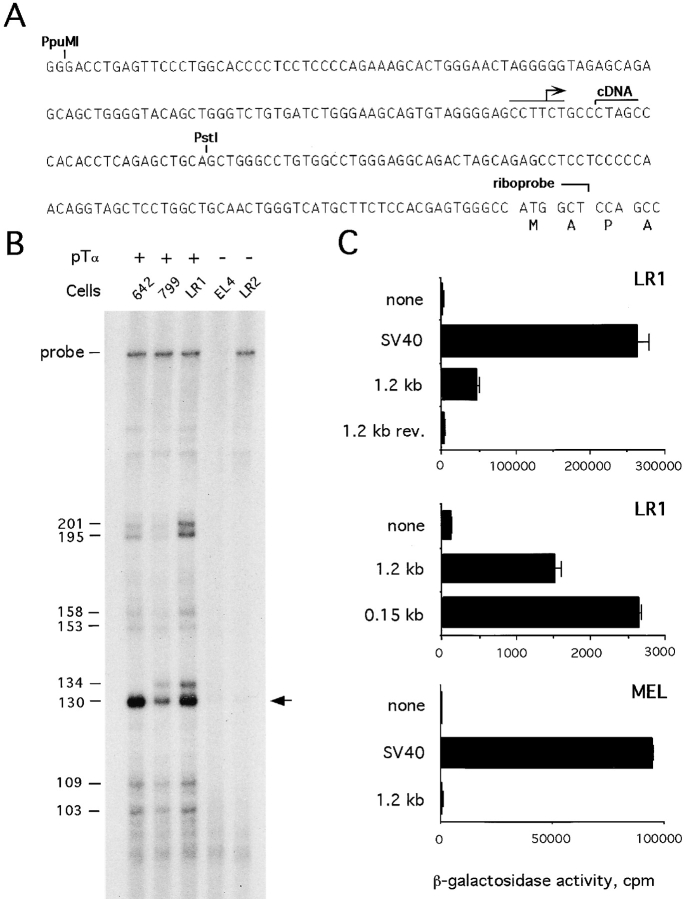
Because of its position 5′ of the first exon, the first DHS (DHS 1) was likely to reflect the activity of a proximal promoter. To confirm this notion, we analyzed the 5′ transcriptional start of the pTα gene by RNase protection assay, using a probe spanning 0.4 kb immediately upstream of the translation start site (Fig. 3 A). Fig. 3 B shows the presence in T cell lymphomas of heterogeneous pTα transcripts initiated within 100–200 bp of ATG, with the most abundant transcript corresponding to a short 5′ UTR of ∼124 bp. This position is only 5 bp 5′ of the longest pTα cDNA clone (16) and might represent a major transcription start site.
Figure 3.
Characterization of the pTα proximal promoter. (A) The sequence immediately upstream of the translation start site (16). A putative transcription start site, the 5′ end of the longest cDNA clone (16), and the 3′ end of a riboprobe used for RNase protection are shown. The PstI site represents the 3′ end of the cloned pTα promoter fragments. The PpuMI-PstI fragment corresponds to a 0.15-kb core pTα promoter. (B) RNase protection was performed using RNA from the indicated cell lines and the 414-bp riboprobe shown above. The positions of a free probe and of the sequencing ladder fragments (bp) corresponding to the bands are indicated. A major protected fragment corresponding to the putative transcription start site is marked (arrow). (C) LacZ reporter constructs were transiently transfected into LR1 or MEL cells, and β-galactosidase activity was determined with a chemiluminescent assay. The promoters included SV40 promoter, 1.2-kb pTα promoter in direct or reverse (rev.) orientation, and 0.15-kb pTα promoter.
To examine the promoter function of the sequences adjacent to the identified transcription start site, a 1.3-kb fragment spanning this region was subcloned upstream of a LacZ (β-galactosidase) reporter gene and transfected into the T cell line LR1. This early passage cell line, derived from an atm −/− rag-2 −/− thymoma, has an immature T cell phenotype (Thy-1hiCD4loCD8loCD2loCD25−CD44−TCR-β−pTα+) and can be transfected using lipid reagents. As shown in Fig. 3 C, the pTα upstream fragment manifested a relatively weak orientation-dependent promoter activity. Using 5′ deletions of the fragment, the promoter function was localized to a 0.15-kb PpuMI-PstI fragment containing 115 bp 5′ of the putative transcription start site. Although the low activity of pTα promoter precluded the analysis of its function in other T cell lines, the promoter was inactive in an erythroleukemia cell line, MEL (Fig. 3 C) and in NIH3T3 fibroblasts (not shown). Thus, DHS 1 apparently corresponds to a short proximal promoter that might be specific at least for lymphoid cells.
Characterization of an Upstream pTα Enhancer.
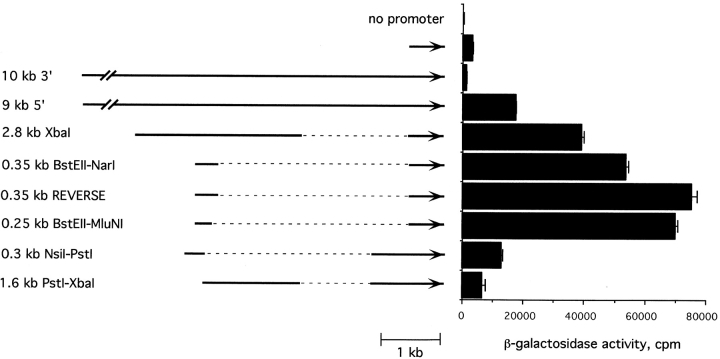
To search for the possible distal enhancers in the pTα locus, we examined the effect of larger pTα genomic fragments in a transient transfection assay with LR1 T cells. As shown in Fig. 4, a 9-kb 5′ pTα fragment, used in the transgene as described above, was more active than a 0.5-kb 5′ fragment containing the core promoter. Using in-vector deletions of the 9-kb region, we first localized the enhancer activity to a 2.8-kb XbaI fragment. By creating a series of nested 5′ and 3′ deletions of this fragment (data not shown), this activity was further mapped to a 0.35-kb region between BstEII and NarI sites, 4 kb upstream of the pTα promoter. Fig. 4 demonstrates that this fragment and a smaller 0.25-kb BstEII-MluNI fragment were fully active as transcriptional enhancers. In contrast, 5′ or 3′ truncations at a PstI site within this region significantly reduced the enhancer function. Importantly, these mapping data are consistent with the location of DHS 2, suggesting that the described pTα enhancer is at least one regulatory region corresponding to DHS 2.
Figure 4.
Mapping of the pTα upstream enhancer. The reporter constructs were transiently transfected into LR1 cells, and β-galactosidase activity was determined 24 h later. The indicated upstream genomic pTα fragments were subcloned 5′ of the pTα promoter fragments (shown as arrows) in the LacZ reporter vector. A 10-kb downstream pTα genomic fragment was used as a control. The BstEII-NarI fragment was used either in forward or reverse orientation.
Fig. 4 also demonstrates that pTα enhancer increased transcription when placed in either orientation and at a variable distance from the promoter. To examine its activity on heterologous promoters, the larger 4.3-kb and the smaller 0.35-kb fragments containing the pTα enhancer were subcloned upstream of the SV40 early promoter. As shown in Fig. 5 A, both pTα enhancer fragments significantly increased the activity of the SV40 promoter in LR1 T cells. The activity of pTα enhancer in LR1 cells was stronger than that of previously described T cell–specific enhancers of CD3δ (Fig. 5 A) and CD2 (not shown) genes and was also observed with another strong heterologous promoter, the CMV immediate early promoter (data not shown). Thus, the identified upstream enhancer of the pTα gene appears as a powerful, bona fide transcriptional enhancer, functioning irrespective of distance, orientation, or the corresponding promoter (22).
Figure 5.
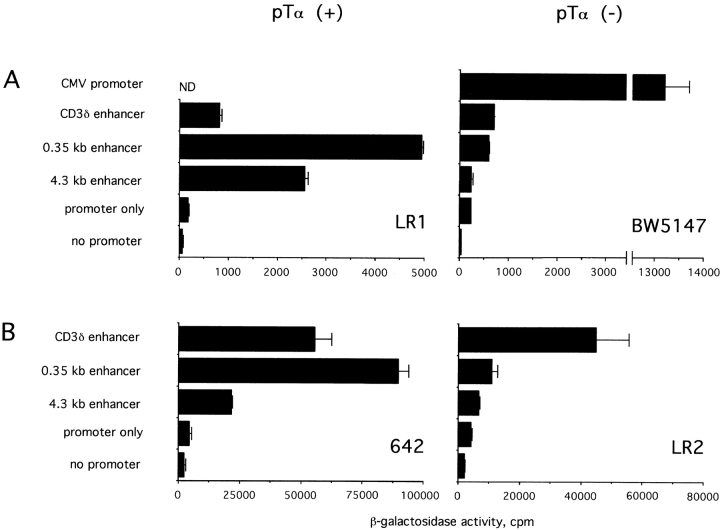
Activity of the pTα enhancer in various T cell lines. The 4.3- and 0.35-kb pTα enhancer fragments and the control CD3δ enhancer were subcloned upstream of the SV40 promoter/LacZ reporter gene. (A) The cells were transfected using Fugene 6 reagent. A CMV promoter/LacZ construct was used as a positive control for transfection of BW5147 cells. In B, the cells were transfected using Superfect reagent.
To examine the cell and stage specificity of the pTα enhancer, the enhancer/SV40 promoter constructs were introduced into a pTα-negative T cell line, BW5147. As shown in Fig. 5 A, the large enhancer fragment was completely inactive, whereas the small, 0.35-kb fragment produced only a minor increase similar to the CD3δ enhancer. Similar results were obtained with smaller enhancer fragments (not shown). This was not due to a limiting transfection efficiency, as the CMV promoter produced a strong reporter activity in these cells. A similarly low activity of the pTα enhancer was observed in nonlymphoid MEL and NIH3T3 cells (data not shown). In another series of experiments, the same reporter constructs were introduced into pTα-positive (642) or -negative (LR2) T cell lines. These early passage T cell lymphomas could be transfected with a comparable low efficiency by the same protocol. Fig. 5 B demonstrates that the pTα enhancer increased the promoter activity in 642 cells but was scarcely functional in LR2 cells compared with the control CD3δ enhancer. Altogether, these data suggest, but do not prove, that the pTα enhancer is preferentially active in pre-T cells as compared with mature T cells or nonlymphoid cells.
Nuclear Factors Binding to the Core pTα Enhancer.
The sequence of the 0.25-kb BstEII-MluNI enhancer fragment was determined and is shown in Fig. 6 A. Nested 5′ and 3′ deletions of this region were produced by PCR using the indicated primers, subcloned into the SV40 promoter/ LacZ reporter vector, and assessed for their activity in LR1 cells (Fig. 6 B). This analysis revealed a core enhancer of 149 bp, defined by primers F2 and R2. Further deletions in this region significantly decreased the enhancer activity; the regions between F2/F3, R2/R3, and R4/R5 appeared particularly important. These data are consistent with the deleterious effect of truncations at the PstI site within the core enhancer region (Fig. 4).
Next, we examined nuclear proteins binding to the pTα enhancer by electrophoretic mobility shift assay (EMSA) using nuclear extracts from several pTα-positive or -negative T cell lines, a B cell line, and an erythroleukemia cell line. As probes, we used six double-stranded oligonucleotides spanning the core enhancer (Fig. 6 A) or larger promoter and enhancer DNA fragments. This analysis revealed multiple distinct nuclear factors interacting with the core enhancer; however, we were unable to detect any DNA– protein complexes appearing specifically in pTα-expressing T cell lines.
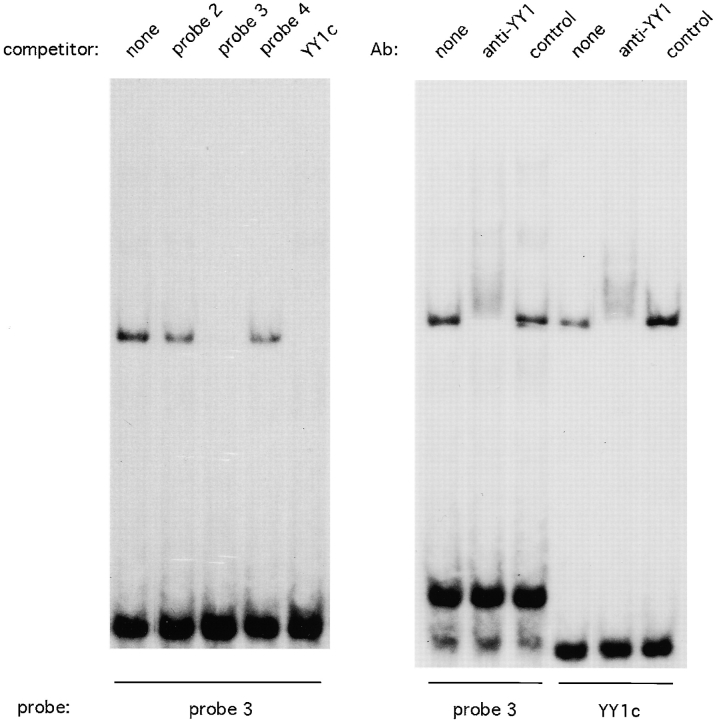
The core enhancer sequence contained a potential binding site (CCAT; reference 23) for the transcription factor YY1. Indeed, probe 3, containing the putative YY1 site, formed a complex, found in all cells examined, that could be specifically competed by YY1 consensus oligonucleotide (Fig. 7). Furthermore, Fig. 7 shows that the factor binding to both probes could be supershifted by anti-YY1 Ab, thus confirming its identity as YY1. To explore the functional role of this interaction, the YY1 binding site was mutated so that the mutant sequence was unable to compete with YY1 binding (not shown). As shown in Fig. 6 C, the YY1 site mutation resulted in a minor, but consistent, increase in enhancer activity. Thus, the core pTα enhancer appears to interact with YY1 transcription factor, which might contribute to the repression of its activity.
Figure 7.
The pTα enhancer binds transcription factor YY1. EMSA was performed with LR1 nuclear extract and labeled probe 3 from pTα enhancer or YY1 consensus probe (YY1c). In the left panel, the reactions included a 100-fold excess of unlabeled enhancer probes (Fig. 6) or YY1 consensus probe. In the right panel, anti-YY1 mAb or a control mAb were added to the reactions.
C-rich Sites in the Core Enhancer Bind ZBP-89.
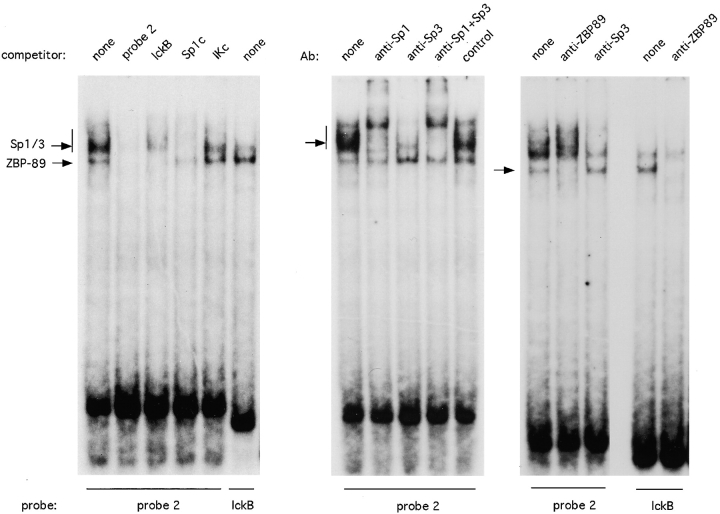
It was reported previously that the thymocyte-specific lck proximal promoter contains a G-rich site that appears critical for the promoter function and binds a T cell nuclear factor (factor B) (20). Because the pTα enhancer sequence features two similar C-rich stretches, we examined the factors binding to these sites and their possible relation to factor B of the lck promoter. Enhancer probes 2, 5, and 6 produced a similar pattern of nuclear complexes and effectively competed with each other for binding (not shown); we therefore concluded that the 5′ and 3′ C-rich sites bind the same nuclear factors. Fig. 8 demonstrates that enhancer probe 2, encompassing the 5′ C-rich site, formed two distinct complexes in T cell nuclear extracts. The upper complex was specifically competed out by an Sp1 consensus probe, whereas the lower complex was competed out by the G-rich B site of lck promoter (lckB); moreover, the labeled lckB probe formed a single complex of similar size. Antibody supershift experiments confirmed that the upper complex consisted of Sp1 and Sp3 transcription factors (Fig. 8, center panel). Thus, the C-rich enhancer regions bind Sp1 family proteins and another protein identical to the B complex of lck promoter.
Figure 8.
The pTα enhancer binds Sp family transcription factors and ZBP-89. EMSA was performed with EL4 nuclear extract and labeled enhancer probe 2 or lckB probe. Cold competitor oligonucleotides included enhancer probe 2, lckB probe, Sp1 consensus probe (Sp1c), or a control Ikaros consensus probe (IKc). Antibodies to transcription factors were added as shown. The complexes containing Sp1/Sp3 or ZBP-89 are indicated (arrows).
A recently cloned zinc finger transcription factor, ZBP-89 (BFCOL1, BERF-1), was shown to bind long, G-rich stretches in several promoters (24–26) and, therefore, represented a good candidate for the observed binding activity. We tested this possibility and found that anti–ZBP-89 Ab specifically blocked the formation of the lower complex with probe 2 and of the major complex with lckB probe (Fig. 8, right panel). Thus, ZBP-89 appears to interact with two sites within the pTα enhancer core and with a critical site of the lck proximal promoter. These observations suggest a possible role for ZBP-89 in thymocyte-specific gene expression.
Discussion
This study was aimed at establishing a model to study the regulation of pre-T cell–specific gene transcription. The regulation of genes expressed at the early stages of B cell development has been extensively studied, and stage-specific transcription factors such as EBF (early B cell factor) have been identified (27). In contrast, the mechanisms of stage-specific gene expression in T cells are less well understood. Certain transcription factors appear to regulate specific stages of T cell development, as illustrated by the role of LKLF (lung Kruppel-like factor) in the maintenance of mature T cell quiescence (28). A clear example of reciprocal transcriptional regulation in early versus mature T cells is the alternative promoter usage at the lck tyrosine kinase gene (5). In particular, the lck proximal promoter was proven to function specifically in immature thymocytes (20) and has been extensively used to target transgene expression to early T cells. In another case, a thymocyte-specific enhancer was found in the third intron of Thy-1 gene (29). Furthermore, stage-specific silencers and enhancers were described in the CD4 (6, 7) and CD8 (8–10) loci, respectively. Despite this progress, the molecular basis of an apparently common pattern of pre-T cell– specific transcription is obscure.
The expression of the pTα gene has been examined in great detail and was shown to occur specifically in pre-T cells (13, 14). Recently, the expression of an alternatively spliced pTα isoform (lacking the extracellular Ig domain) was described (30, 31) and proposed to occur in mature T cells as well as in pre-T cells (30). This isoform was also observed in the original analysis of pTα expression but was not detected in mature T cells (14). Similarly, we could not detect its expression in the spleen or in pTα-negative T cell lines (Reizis, B., unpublished results). Therefore, the expression of a shorter pTα isoform in mature T cells is likely to occur, if at all, only in a specialized minor population of T cells or at extremely low levels. Thus, by and large, the pTα gene appears to be expressed in early T cells and as such represents a valuable model for stage-specific T cell gene expression.
Although the mouse and human pTα genes have been extensively mapped and sequenced (16, 31), the regulation of pTα gene expression has not been studied. We now report that mouse pTα gene transcription is regulated primarily by an upstream genomic region. It is possible, however, that additional genomic elements within, downstream of, or farther upstream of the gene might contribute to its regulation. In particular, possible locus control regions conferring position-independent transgene expression remain to be identified in the pTα locus; the nonspecific DHS 5′ and 3′ of the gene are candidates for such elements. Another likely regulatory region is the recently described sequence in the first pTα intron, which is conserved between mouse and human genes (31). We found that a fragment containing this sequence lacked any detectable enhancer activity in a transient transfection assay (data not shown); thus, it might function as a silencer or an element regulating chromatin accessibility. Further studies are required to delineate the complete hierarchy of pTα transcriptional regulation. In any case, our data indicate that the cell and stage specificity of pTα expression are fully determined by upstream elements.
Within the pTα upstream region, we have identified a proximal promoter and an enhancer located 4 kb 5′ of the promoter. As in many T cell–specific genes, the promoter appeared relatively weak and is most likely insufficient for pTα expression. The pTα enhancer, on the other hand, manifested high activity in transient transfection assays and appeared to function preferentially in pTα-positive pre-T cell lines. It is possible, however, that additional elements in the vicinity of the enhancer contribute to its specificity. In this regard, it is noteworthy that DHS 2 actually consists of two sites separated by ∼0.3–0.4 kb; the precise nature of these sites is currently under investigation. In addition, the enhancer may require its cognate pTα promoter to achieve full specificity.
Our analysis of the nuclear factors interacting with the core pTα enhancer (Figs. 7 and 8, and Reizis, B., unpublished data) suggests a preliminary model of its architecture (Fig. 9). We could detect at least two distinct factors binding to the 5′ end of the sequence, and these interactions appear important for the enhancer function, as evidenced by the F2/F3 deletion (Fig. 6). The two 5′ E boxes appear to have formed identical complexes, whereas the 3′ E box represents a consensus binding site for bHLH-ZIP transcription factors and might bind a distinct set of proteins. The enhancer features a perfect Ikaros binding site, and probe 4, spanning this site, formed a nuclear complex that was specifically inhibited by Ikaros consensus probe IK-BS2 (19) and vice versa; however, we were unable to confirm the identity of this factor using anti-Ikaros antiserum. Nevertheless, Ikaros protein isoforms or Ikaros-related factors might interact with this site in vivo and contribute to the repression of the enhancer by recruiting it to centromeric heterochromatin in pTα-negative cells (32, 33). In addition, transcription factor YY1 binds to the middle portion of the enhancer and appears to repress its activity. YY1 is a multifunctional factor implicated in, among other things, the repression of tissue-specific genes such as Ig and globin genes, possibly due to the recruitment of corepressors such as histone deacetylases (34). Thus, the pTα enhancer may be subject to the complex regulation at the level of chromatin modification.
Figure 9.
The proposed architecture of the pTα enhancer. Putative binding sites for nuclear factors are marked.
The pTα enhancer contains two C-rich sites, and deletion of the 3′ site compromised the enhancer function (Fig. 6). We found that these sites bind Sp1 and Sp3, the related ubiquitous transcriptional activator and repressor proteins, respectively (35). The true role of Sp proteins in the regulation of the pTα enhancer might be difficult to establish; indeed, the expression of many genes thought to be regulated by Sp1 was not affected in its absence (35). It should be noted, however, that Sp1 is abundantly expressed in the thymus (36) and therefore might play a specific role in thymocyte gene expression.
In addition, these C-rich sites, as well as an important G-rich site in the proximal lck promoter, bind a common nuclear factor identified here as ZBP-89. The truncated clone of ZBP-89, htβ, was originally cloned as a zinc finger protein binding to the CACCC box in the human TCR-α promoter (37). Recently, the full length protein ZBP-89/ BFCOL1/BERF-1 was cloned as a protein binding to long, G-rich stretches in various promoters (24–26, 38, 39). This protein appears to be capable of both transcriptional activation (25, 26, 37) and repression (24, 26, 38), possibly depending on the DNA context and cell type; the precise function of ZBP-89 on the pTα enhancer remains to be established. Despite its apparently ubiquitous expression, we have found that ZBP-89 mRNA is expressed in the thymus at significantly higher levels than in any other organ tested, including the spleen (Reizis, B., unpublished results). Indeed, six out of eight cDNA clones corresponding to ZBP-89 in the mouse expressed sequence tag database are derived from the thymus library. In addition, the ZBP-89 complex was easily detectable by EMSA in T cell lines but not in nonlymphoid cells such as MEL. Together with the observed binding of ZBP-89 to the regulatory elements active specifically in early T cells, these observations suggest an important role of ZBP-89 in T cell development.
A proposed model of the pTα core enhancer differs significantly from the core enhancers of TCR genes (3); in particular, it lacks obvious binding sites for T cell–specific transcription factors such as GATA3 or TCF1/LEF1. Furthermore, we were unable to detect enhancer-binding nuclear factors directly correlating with pTα expression in T cell lines. This may be due to the technical limitations of our approach, or it might reflect the requirement for additional regulatory sites as discussed above. Another possibility, however, is the existence of protein cofactors interacting with the enhancer-binding proteins and conferring cell and stage specificity on the enhancer. Indeed, the bHLH proteins, YY1 and Sp1, are known to be involved in complex interactions with other proteins, which are essential for their function on a particular regulatory region. Similarly, ZBP-89 is likely to undergo cell type–specific regulation by other proteins. Future studies using this and other models should delineate the precise mechanism of stage-specific gene expression in T cells.
Acknowledgments
We thank Anne Harrington for oocyte injections, Christoph Westphal and Cathy O'Hara for cell lines, and Juanita Merchant for anti-ZBP89 antiserum. We are grateful to Mark Bedford for his crucial advice and support and to Robert Weiss, Jennifer Michaelson, Kevin Fitzgerald, and Yasumasa Ishida for many helpful discussions.
Abbreviations used in this paper
- DHS
DNase-hypersensitive site
- EMSA
electrophoretic mobility shift assay
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- pTα
pre-TCR-α
- UTR
untranslated region
Footnotes
B. Reizis is supported by the Cancer Research Institute Postdoctoral Fellowship.
References
- 1.Glimcher LH, Singh H. Transcription factors in lymphocyte development—T and B cells get together. Cell. 1999;96:13–23. doi: 10.1016/s0092-8674(00)80955-1. [DOI] [PubMed] [Google Scholar]
- 2.Clevers HC, Grosschedl R. Transcriptional control of lymphoid development: lessons from gene targeting. Immunol Today. 1996;17:336–343. doi: 10.1016/0167-5699(96)10019-0. [DOI] [PubMed] [Google Scholar]
- 3.Leiden JM. Transcriptional regulation of T cell receptor genes. Annu Rev Immunol. 1993;11:539–570. doi: 10.1146/annurev.iy.11.040193.002543. [DOI] [PubMed] [Google Scholar]
- 4.Clevers H, Ferrier P. Transcriptional control during T cell development. Curr Opin Immunol. 1998;10:166–171. doi: 10.1016/s0952-7915(98)80245-8. [DOI] [PubMed] [Google Scholar]
- 5.Wildin RS, Garvin AM, Pawar S, Lewis DB, Abraham KM, Forbush KA, Ziegler SF, Allen JM, Perlmutter RM. Developmental regulation of lck gene expression in T lymphocytes. J Exp Med. 1991;173:383–393. doi: 10.1084/jem.173.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 7.Siu G, Wurster AL, Duncan DD, Soliman TM, Hedrick SM. A transcriptional silencer controls the developmental expression of the CD4 gene. EMBO (Eur Mol Biol Organ) J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellmeier W, Sunshine MJ, Losos K, Littman DR. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- 9.Hostert A, Garefalaki A, Mavria G, Tolaini M, Roderick K, Norton T, Mee PJ, Tybulewicz VL, Coles M, Kioussis D. Hierarchical interactions of control elements determine CD8α gene expression in subsets of thymocytes and peripheral T cells. Immunity. 1998;9:497–508. doi: 10.1016/s1074-7613(00)80633-0. [DOI] [PubMed] [Google Scholar]
- 10.Zhang X, Seong R, Piracha R, Larijani M, Heeney M, Parnes JR, Chamberlain JW. Distinct stage-specific cis-active transcriptional mechanisms control expression of T cell coreceptor CD8α at double- and single-positive stages of thymic development. J Immunol. 1998;161:2254–2266. [PubMed] [Google Scholar]
- 11.von Boehmer H, Fehling HJ. Structure and function of the pre-T cell receptor. Annu Rev Immunol. 1997;15:433–452. doi: 10.1146/annurev.immunol.15.1.433. [DOI] [PubMed] [Google Scholar]
- 12.von Boehmer H, Aifantis I, Azogui O, Feinberg J, Saint-Ruf C, Zober C, Garcia C, Buer J. Crucial function of the pre-T-cell receptor (TCR) in TCRβ selection, TCRβ allelic exclusion and αβ versus γδ lineage commitment. Immunol Rev. 1998;165:111–119. doi: 10.1111/j.1600-065x.1998.tb01234.x. [DOI] [PubMed] [Google Scholar]
- 13.Saint-Ruf C, Ungewiss K, Groettrup M, Bruno L, Fehling HJ, von Boehmer H. Analysis and expression of a cloned pre-T cell receptor gene. Science. 1994;266:1208–1212. [PubMed] [Google Scholar]
- 14.Bruno L, Rocha B, Rolink A, von Boehmer H, Rodewald H-R. Intra- and extra-thymic expression of the pre-T cell receptor alpha gene. Eur J Immunol. 1995;25:1877–1882. doi: 10.1002/eji.1830250713. [DOI] [PubMed] [Google Scholar]
- 15.Bruno L, Res P, Dessing M, Cella M, Spits H. Identification of a committed T cell precursor population in adult human peripheral blood. J Exp Med. 1997;185:875–884. doi: 10.1084/jem.185.5.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehling HJ, Laplace C, Mattei MG, Saint-Ruf C, von Boehmer H. Genomic structure and chromosomal location of the mouse pre-T cell receptor alpha gene. Immunogenetics. 1995;42:275–281. doi: 10.1007/BF00176445. [DOI] [PubMed] [Google Scholar]
- 17.Georgopoulos K, van der Elsen P, Bier E, Maxam A, Terhorst C. A T cell specific enhancer is located in a DNAse I-hypersensitive area at the 3′ end of the CD3-δ gene. EMBO (Eur Mol Biol Organ) J. 1988;7:2401–2407. doi: 10.1002/j.1460-2075.1988.tb03085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lake RA, Wotton D, Owen MJ. A 3′ transcriptional enhancer regulates tissue-specific expression of the human CD2 gene. EMBO (Eur Mol Biol Organ) J. 1990;9:3129–3136. doi: 10.1002/j.1460-2075.1990.tb07510.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molnar A, Georgopoulos K. The Ikaros gene encodes a family of functionally diverse zinc finger DNA-binding proteins. Mol Cell Biol. 1994;14:8292–8303. doi: 10.1128/mcb.14.12.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen JM, Forbush KA, Perlmutter RM. Functional dissection of the lck proximal promoter. Mol Cell Biol. 1992;12:2758–2768. doi: 10.1128/mcb.12.6.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heinemeyer T, Chen X, Karas H, Kel AE, Kel OV, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackwood EM, Kadonaga JT. Going the distance: a current view of enhancer action. Science. 1998;281:60–63. doi: 10.1126/science.281.5373.60. [DOI] [PubMed] [Google Scholar]
- 23.Srivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang 1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Merchant JL, Iyer GR, Taylor BR, Kitchen JR, Mortensen ER, Wang Z, Flintoft RJ, Michel JB, Bassel-Duby R. ZBP-89, a Kruppel-like zinc finger protein, inhibits epidermal growth factor induction of the gastrin promotor. Mol Cell Biol. 1996;16:6644–6653. doi: 10.1128/mcb.16.12.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hasegawa T, Takeuchi A, Miyaishi O, Isobe K, de Crombrugghe B. Cloning and characterization of a transcription factor that binds to the proximal promoters of the two mouse type I collagen genes. J Biol Chem. 1997;272:4915–4923. doi: 10.1074/jbc.272.8.4915. [DOI] [PubMed] [Google Scholar]
- 26.Passantino R, Antona V, Barbieri G, Rubino P, Melchionna R, Cossu G, Feo S, Giallongo A. Negative regulation of β enolase gene transcription in embryonic muscle is dependent upon a zinc finger factor that binds to the G-rich box within the muscle-specific enhancer. J Biol Chem. 1998;273:484–494. doi: 10.1074/jbc.273.1.484. [DOI] [PubMed] [Google Scholar]
- 27.Reya T, Grosschedl R. Transcriptional regulation of B-cell differentiation. Curr Opin Immunol. 1998;10:158–165. doi: 10.1016/s0952-7915(98)80244-6. [DOI] [PubMed] [Google Scholar]
- 28.Kuo CT, Veselits ML, Leiden JM. LKLF: a transcriptional regulator of single-positive T cell quiescence and survival. Science. 1997;277:1986–1990. doi: 10.1126/science.277.5334.1986. [DOI] [PubMed] [Google Scholar]
- 29.Vidal M, Morris R, Grosveld F, Spanopoulou E. Tissue-specific control elements of the Thy-1 gene. EMBO (Eur Mol Biol Organ) J. 1990;9:833–840. doi: 10.1002/j.1460-2075.1990.tb08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barber DF, Passoni L, Wen L, Geng L, Hayday AC. The expression in vivo of a second isoform of pTα: implications for the mechanism of pTα action. J Immunol. 1998;161:11–16. [PubMed] [Google Scholar]
- 31.Saint-Ruf C, Lechner O, Feinberg J, von Boehmer H. Genomic structure of the human pre-T cell receptor α chain and expression of two mRNA isoforms. Eur J Immunol. 1998;28:3824–3831. doi: 10.1002/(SICI)1521-4141(199811)28:11<3824::AID-IMMU3824>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 32.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 33.Klug CA, Morrison SJ, Masek M, Hahm K, Smale ST, Weissman IL. Hematopoietic stem cells and lymphoid progenitors express different Ikaros isoforms, and Ikaros is localized to heterochromatin in immature lymphocytes. Proc Natl Acad Sci USA. 1998;95:657–662. doi: 10.1073/pnas.95.2.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Y, Lee J, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 35.Lania L, Majello B, de Luca P. Transcriptional regulation by the Sp family proteins. Int J Biochem Cell Biol. 1997;29:1313–1323. doi: 10.1016/s1357-2725(97)00094-0. [DOI] [PubMed] [Google Scholar]
- 36.Saffer JD, Jackson SP, Annarella MB. Developmental expression of Sp1 in the mouse. Mol Cell Biol. 1991;11:2189–2199. doi: 10.1128/mcb.11.4.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y, Kobori JA, Hood L. The htβ gene encodes a novel CACCC box-binding protein that regulates T-cell receptor gene expression. Mol Cell Biol. 1993;13:5691–5701. doi: 10.1128/mcb.13.9.5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law GL, Itoh H, Law DJ, Mize GJ, Merchant JL, Morris DR. Transcription factor ZBP-89 regulates the activity of the ornithine decarboxylase promotor. J Biol Chem. 1998;273:19955–19964. doi: 10.1074/jbc.273.32.19955. [DOI] [PubMed] [Google Scholar]
- 39.Law DJ, Tarle SA, Merchant JL. The human ZBP-89 homolog, located at chromosome 3q21, represses gastrin gene expression. Mamm Genome. 1998;9:165–167. doi: 10.1007/s003359900711. [DOI] [PubMed] [Google Scholar]