Abstract
Rnt1p, the only known Saccharomyces cerevisiae RNase III endonuclease, plays important functions in the processing of precursors of rRNAs (pre-rRNAs) and of a large number of small nuclear RNAs (snRNAs) and small nucleolar RNAs (snoRNAs). While most eukaryotic RNases III, including the Schizosaccharomyces pombe enzyme Pac1p, cleave double-stranded RNA without sequence specificity, Rnt1p cleavage relies on the presence of terminal tetraloop structures that carry the consensus sequence AGNN. To search for the conservation of these processing signals, I have systematically analyzed predicted secondary structures of the 3′ external transcribed spacer (ETS) sequences of the pre-rRNAs and of flanking sequences of snRNAs and snoRNAs from sequences available in 13 other Hemiascomycetes species. In most of these species, except in Yarrowia lipolytica, double-stranded RNA regions capped by terminal AGNN tetraloops can be found in the 3′ ETS sequences of rRNA, in the 5′- or 3′-end flanking sequences of sn(o)RNAs, or in the intergenic spacers of polycistronic snoRNA transcription units. This analysis shows that RNase III processing signals and RNase III cleavage specificity are conserved in most Hemiascomycetes species but probably not in the evolutionarily more distant species Y. lipolytica.
RNases III form a family of double-stranded RNA (dsRNA) endonucleases found in both prokaryotes and eukaryotes. These enzymes are involved in the processing of a large number of stable RNAs. Prokaryotic and eukaryotic RNase III-like proteins participate in the maturation of the precursors of rRNAs (pre-rRNAs). In eukaryotes, RNases III are required for processing of the 35S pre-rRNA; in both Saccharomyces cerevisiae and Schizosaccharomyces pombe, RNases III cleave the 3′ external transcribed spacer (ETS) found in the 35S pre-rRNA (2, 3, 13, 23, 25). This processing event is one of the earliest steps in pre-rRNA processing. In addition to having a role in pre-rRNA processing, eukaryotic RNases III play important roles in the processing of several families of stable small RNAs. In S. cerevisiae, spliceosomal small nuclear RNAs (snRNAs) are processed by Rnt1p cleavage in the 3′ extension found in the precursors of U1, U2, U4, and U5 snRNAs (1, 5, 15, 21, 24). This function is conserved in fungi, at least for the U2 snRNA from S. pombe, whose 3′-end processing requires Pac1p, the S. pombe ortholog of RNase III (28). Earlier genetic data suggest that the role of Pac1p is not restricted to the U2 snRNAs but that it is also involved in the processing of other S. pombe snRNAs (20). In addition to the processing of snRNAs, Rnt1p has also been shown to have a major function in the processing of small nucleolar RNAs (snoRNAs). Some of these snoRNAs are synthesized with a 5′ extension, and Rnt1p cleavage provides an entry site for final trimming to the mature 5′ end by the 5′→3′ exonucleases Xrn1p and Rat1p (6; C. Y. Lee et al., in press). A few other snoRNAs are synthesized as polycistronic precursors, from which Rnt1p cleavage produces monocistronic intermediates that are further trimmed to the mature end by exonuclease digestion (6, 7, 17). It is not clear whether the function of RNase III in snRNA and snoRNA processing is conserved in multicellular eukaryotes. However, plant and metazoan RNases III belonging to the Dicer family are essential for processing a large number of microRNAs (10-12, 16, 18).
The identification of a large number of Rnt1p processing signals in rRNAs, snRNAs, and snoRNAs points to conserved structural features of Rnt1p substrates. Most Rnt1p dsRNA substrates are capped by tetraloop sequences carrying the weak consensus sequence AGNN (4, 6). These tetraloops are essential for cleavage activity, and the cleavage site is positioned 14 to 16 bp away from the AGNN tetraloop, suggesting that the enzyme acts as an RNA helical ruler (4). Structural analysis of these tetraloops showed that the enzyme probably recognizes a specific tetraloop conformation and that the strongest sequence requirement resides in the universal G2 position, which must be in the syn conformation (26). In contrast to Rnt1p, most other RNase III-like enzymes cleave dsRNA with only very limited or no sequence specificity. Escherichia coli RNase III specificity relies on antideterminants in the dsRNA region (27), and S. pombe Pac1p as well as human Dicer does not seem to exhibit sequence specificity. These observations raise the question of when the specificity for AGNN tetraloops arose during the evolution of RNase III enzymes.
I took advantage of the sequencing effort of the Génolevures genomic program (22), which selected a set of species representative of the Hemiascomycetes class, and performed the sequencing of a large number of random sequence tags (RSTs) to answer two questions. First, it is not known whether the function of RNase III in processing pre-rRNAs, snRNAs, and snoRNAs is conserved in Hemiascomycetes. Second, the difference in cleavage specificity between S. cerevisiae Rnt1p, which requires AGNN tetraloops, and S. pombe Pac1p, which does not seem to require this type of terminal loop, suggests that the Rnt1p specificity for AGNN tetraloops may have been acquired after the divergence of S. cerevisiae and S. pombe. The analysis of a large number of predicted RNase III substrates in Hemiascomycetes may provide an answer to these questions and help determine when this RNA structure specificity was adopted during evolution. In this study, I show that predicted Rnt1p-type cleavage sites are present in most Hemiascomycetes sequences analyzed and that most of these predicted sites exhibit terminal AGNN tetraloops that are compatible with Rnt1p cleavage specificity. These results demonstrate that RNase III processing pathways, as well as RNase III cleavage specificity, are conserved among most Hemiascomycetes, with the exception of the evolutionarily distant species Yarrowia lipolytica.
MATERIALS AND METHODS
Sequences and secondary-structure analysis.
Sequences for rRNA 3′ ETSs and snRNAs were retrieved from the Génolevures website by using a BLAST analysis of the S. cerevisiae RSTs (http://cbi.labri.fr/Genolevures/advanced_blast.php3). For identification of the 3′ ETS sequences, the last 250 nucleotides (nt) of the 25S rRNA were used for the BLAST search, and RSTs containing more than 300 nt of sequence downstream from the 25S rRNA were selected. For sn(o)RNAs, the entire mature sn(o)RNA sequence was used. In the case of modification guide C and D box-containing (box C/D) snoRNAs, many hits were obtained; these sequences in some cases included short regions of complementarity with the rRNAs corresponding to the guide sequences. Most of the uncovered sequences whose BLAST E value was above 1.0 were discarded. For other sequences with high E values (0.1 to 0.9), the presence of the box C and box D sequences was manually verified, and sequences that did not show both of these conserved boxes were discarded. RSTs that did not contain more than 120 nt of the flanking region were also discarded. RNA secondary-structure prediction with Mfold (14) was performed by using M. Zuker's Mfold web server (29) (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/). For most sn(o)RNA sequences, only one RST was found in the Génolevures database and analyzed. For most 3′ ETS sequences, several hits were obtained for each species. In this case, two to three sequences obtained from independent RSTs were analyzed, and in all cases, the sequences and the secondary structures were found to be identical. Secondary-structure prediction was performed on 120 to 300 nt of regions flanking the 5′ end, 3′ end, or intergenic sequences. One of the 5%-suboptimal structures is shown in the figures.
Statistical evaluation of the presence of AGNN stem-loops in rRNA and sn(o)RNA flanking sequences.
To evaluate the statistical significance of the presence of predicted stem-loops capped by AGNN tetraloops, 111 sequences flanking random genes were selected from the Génolevures database. Two-hundred-nucleotide segments of these flanking sequences were folded using Mfold (14, 29) to detect the presence of predicted stem-loops of at least 4 bp capped by AGNN or XGNN (AGNN, UGNN, CGNN, or GGNN but not YNCG or GNRA) sequences. Four AGNN and 11 XGNN stem-loops were detected in these random sequences. This random sample was used for statistical evaluation with a binomial proportions test. Comparing the frequency of the presence of AGNN stem-loops in the RSTs containing flanking sequences of rRNA and sn(o)RNA to that in the random sample showed a highly significant enrichment of AGNN stem-loops in these sequences (P ≤ 1.25 × 10−22). The presence of XGNN stem-loops in the RSTs containing flanking sequences of rRNA and sn(o)RNA was also highly significant (P ≤ 9.2 × 10−21).
RESULTS
Rationale of sequence analysis.
To analyze the putative presence of RNase III processing sites in a large number of substrates and Hemiascomycetes species, I searched the Génolevures sequence database for orthologs of rRNA and sn(o)RNA sequences (see Materials and Methods). Because Rnt1p processing sites are usually present less than 300 nt from the mature sn(o)RNA sequences, I restricted the subsequent secondary-structure analysis to RSTs that contained only sufficient (>120 nt) 5′- or 3′-end-flanking sequences. I also searched the Génolevures sequence database for the presence of sequences downstream from the 25S rRNA to obtain putative 3′ ETS sequences. After these sequences were retrieved, predicted RNA secondary structures were obtained by using the Mfold algorithm (14, 29). The list of RSTs analyzed for the presence of predicted secondary structures is shown in Tables 1 and 2. Table 1 shows the RSTs used to analyze the predicted secondary structures in the 3′ ETS sequences downstream from the 25S rRNA from all 13 Hemiascomycetes species. Table 2 shows the list of RSTs used to analyze processing signals upstream (5′) or downstream (3′) from snRNAs and snoRNAs or located in the spacer of polycistronic arrays. For these sn(o)RNAs, I searched for ortholog sequences of all known Rnt1p processing substrates. Some known substrates could not be retrieved from the RSTs, or the corresponding RSTs did not contain enough flanking sequence information. Therefore, these sn(o)RNAs are not included in Table 2.
TABLE 1.
RSTs used to analyze 3′ ETS secondary structures
| RST no. | Species | Terminal loop |
|---|---|---|
| AS0AA030D08DP1 | S. bayanus | AGGA |
| AS0AA006E03TP1 | S. bayanus | AGGA |
| AV0AA009C11T1 | S. exiguus | GGGG |
| AV0AA010A07D1 | S. exiguus | GGGG |
| AT0AA005A02T1 | S. servazzii | AGAU |
| XAT0AA002F06T1 | S. servazzii | AGAU |
| AR0AA017G12CP1 | Z. rouxii | AGGA |
| AR0AA009H12TP1 | Z. rouxii | AGGA |
| AU0AA009A04D1 | S. kluyveri | AGGA |
| AU0AA010B09D1 | S. kluyveri | AGGA |
| AY0AA013E12TP1 | K. thermotolerans | AGGC |
| AY0AA014E03TP1 | K. thermotolerans | AGGC |
| AY0AA002D03TP1 | K. thermotolerans | AGGC |
| BA0AB003F06LP1 | K. lactis | AGGU |
| BA0AB031B09LP1 | K. lactis | AGGU |
| AZ0AA012F01A1 | K. marxianus | AGGU |
| AZ0AA005C01D1 | K. marxianus | AGGU |
| BB0AA027A07TP1 | P. angusta | GGUA |
| BB0AA017H04DP1 | P. angusta | GGUA |
| XBC0AA002C02D1 | D. hansenii | AGAA |
| BC0AA014H12T1 | D. hansenii | AGAA |
| AX0AA037H06TP1 | P. sorbitophila | AGGU |
| AX0AA008H11CP1 | P. sorbitophila | AGGU |
| BD0AA015E02TP1 | C. tropicalis | UAG |
| BD0AA012C11TP1 | C. tropicalis | UAG |
| BD0AA009A05DP1 | C. tropicalis | UAG |
| AW0AA031D09D1 | Y. lipolytica | GAC/GCA |
| AW0AA020C05D1 | Y. lipolytica | GAC/GCA |
| AW0AA018H09T1 | Y. lipolytica | GAC/GCA |
TABLE 2.
RSTs used to analyze processing signals in sn(o) RNAsa
| RNA(s) | RNA type | Pr | RST no. | Species | Terminal loop |
|---|---|---|---|---|---|
| snR7 (U5) | snRNA | 3′ | AR0AA020B08CP1 | Z. rouxii | AGUU |
| snR7 (U5) | snRNA | 3′ | AZ0AA010E03T1 | K. marxianus | Not found |
| snR7 (U5) | snRNA | 3′ | BA0AB004C05LP1 | K. lactis | Not found |
| snR14 (U4) | snRNA | 3′ | AU0AA006A07T1 | S. kluyveri | AGUA |
| snR14 (U4) | snRNA | 3′ | AS0AA004D02DP1 | S. bayanus | Not found |
| snR14 (U4) | snRNA | 3′ | AW0AA001A07D1 | Y. lipolytica | Not found |
| snR19 (U1) | snRNA | 3′ | AV0AA015A01T1 | S. exiguus | AGGA |
| snR20 (U2) | snRNA | 3′ | AU0AA010F08T1 | S. kluyveri | AGUU |
| snR17 (U3) | C/D | 3′ | BA0AB014F05SP1 | K. lactis | AGGU |
| snR17 (U3) | C/D | 3′ | BA0AB015G04SP1 | K. lactis | AGGU |
| snR17 (U3) | C/D | 3′ | AR0AA032D06TP1 | Z. rouxii | AGGU |
| snR17 (U3) | C/D | 3′ | XBB0AA001B07T1 | P. angusta | Not found |
| snR36 | H/ACA | 5′ | AV0AA014A06T1 | S. exiguus | AGGU |
| snR40 | C/D | 5′ | BA0AB006D02LP1 | K. lactis | GGGC/AGUA |
| snR40 | C/D | 5′ | AR0AA017D12CP1 | Z. rouxii | AGUC |
| snR40 | C/D | 5′ | BC0AA012E09T2 | D. hansenii | Not foundb |
| snR40 | C/D | 5′ | AY0AA002D05DP1 | K. thermotolerans | UGCG/AGCG |
| snR46 | H/ACA | 5′ | AZ0AA006A01T1 | K. marxianus | AGGA |
| snR47 | C/D | 5′ | AS0AA02F07DP1 | S. bayanus | AGAU |
| snR47 | C/D | 5′ | AZ0AA001C06T1 | K. marxianus | AGAA |
| snR47 | C/D | 5′ | BD0AA013A04DP1 | C. tropicalis | AGAA |
| snR52 | C/D | 5′ | XAS0AA001E11DP1 | S. bayanus | Not found |
| snR52 | C/D | 5′ | AR0AA030G04CP1 | Z. rouxii | AGGC |
| snR52 | C/D | 5′ | BA0AB025A11SP1 | K. lactis | AGGC |
| snR52 | C/D | 5′ | AZ0AA004A04T1 | K. marxianus | AGGC |
| snR52 | C/D | 5′ | AY0AA013B09DP1 | K. thermotolerans | AGUU |
| snR56 | C/D | 5′ | AS0AA018H07TP1 | S. bayanus | UGGU |
| snR56 | C/D | 5′ | XAY0AA001E01DP1 | K. thermotolerans | AGUU |
| snR57-55 | C/D | P | AZ0AA001F03D1 | K. marxianus | AGAG |
| snR57-55 | C/D | P | AR0AA011A04CP1 | Z. rouxii | AGUU |
| snR57-55 | C/D | P | AX0AA034B01CP1 | P. sorbitophilia | AGAA |
| snR55-61 | C/D | P | AZ0AA001F03D1 | K. marxianus | AGUU |
| snR55-61 | C/D | P | BA0AB011B10SP1 | K. lactis | Not found |
| snR60 | C/D | 5′ | BA0AB005H01SP1 | K. lactis | AGGA |
| snR62 | C/D | 5′ | XAV0AA001B11T1 | S. exiguus | AGGU |
| snR62 | C/D | 5′ | AZ0AA009E05D1 | K. marxianus | AGGC |
| snR64 | C/D | 5′ | BA0AB036B05SP1 | K. lactis | AGGA |
| snR68 | C/D | 5′ | AS0AA001F09DP1 | S. bayanus | AGGA |
| snR68 | C/D | 5′ | BA0AB018E07LP1 | K. lactis | Not found |
| snR68 | C/D | 5′ | AY0AA009A03TP1 | K. thermotolerans | AGCU |
| snR71 | C/D | 5′ | AR0AA013G05CP1 | Z. rouxii | AGUU |
| snR70-51 | C/D | P | BA0AB036A02SP1 | K. lactis | GGCU |
| snR70-51 | C/D | P | BA0AB031B05LP1 | K. lactis | GGCU |
| snR70-51 | C/D | P | AZ0AA014D07T1 | K. marxianus | GGGC |
| snR78-77 | C/D | P | BA0AB037H07SP1 | K. lactis | AGUU |
| snR77-76 | C/D | P | + | K. lactis | AGAC |
| snR75-74 | C/D | P | BA0AB027E08SP1 | K. lactis | AGUU |
| snR77-76 | C/D | P | AS0AA022E11TP1 | S. bayanus | AGUU |
| snR75-74 | C/D | P | AS0AA022E11TP1 | S. bayanus | AGUU |
| snR79 (Z9) | C/D | 5′ | AS0AA023F01TP1 | S. bayanus | AGGA |
| snR79 (Z9) | C/D | 5′ | AT0AA011G09T1 | S. servazzii | AGUU |
| snR79 (Z9) | C/D | 5′ | AR0AA022C07CP1 | Z. rouxii | AGAU |
sn(o)RNA type is given according to S. cerevisiae nomenclature. In the case of the snR78-72 operon, a contig was obtained from two K. lactis RSTs (BA0AB037H07SP1 and BA0AB027E08SP1) (+). Pr, processing signal; P, polycistronic.
Only 120 nt of flanking sequence was available.
Conservation of RNase III processing sites in the 3′ ETS sequences of the pre-rRNAs.
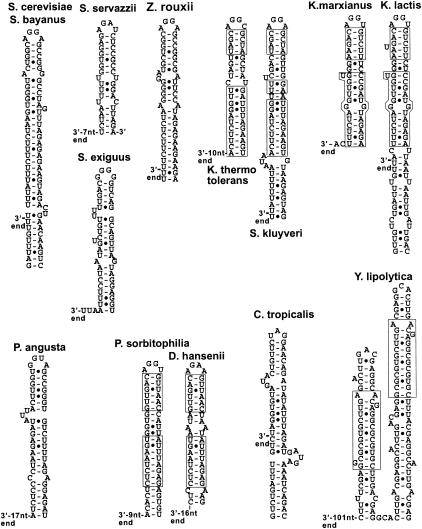
Ribosomal DNA (rDNA) is present in a large number of copies per haploid genome for all the Hemiascomycetes species analyzed (22). Because the Génolevures sequencing program was based on the random sequencing of genomic fragments, rDNA sequences are represented in multiple copies in the RSTs of the Génolevures database for all of these species. This feature allowed the retrieval of several copies of 3′ ETS sequences for all 13 Hemiascomycetes species (Table 1). For all of these species, extensive secondary structures could be predicted immediately downstream from the mature 3′ end of the 25S rRNA (Fig. 1). For most Hemiascomycetes species, the predicted secondary structures are capped by terminal AGNN tetraloops (Table 1; Fig. 1). The occurrence of the stems capped by AGNN tetraloops is highly significant compared to their occurrence in a random sequence population extracted from the Génolevure database (P ≤ 1.25 × 10−22) (see Materials and Methods). The predicted structures suggest that the 3′ ETS sequence is cleaved by an RNase III-like activity in these species and that the Rnt1p orthologs have retained the specificity for AGNN tetraloops. Closely related species have conserved several base pairs in their 3′ ETS secondary structures (compare Saccharomyces kluyveri and Kluveromyces thermotolerans, Kluveromyces marxianus and Kluveromyces lactis, and Pichia sorbitophilia and Debaryomyces hansenii), and the sequences and therefore the secondary structures are identical in S. cerevisiae and Saccharomyces bayanus. For Saccharomyces exiguus and Pichia angusta, the terminal loops exhibit a G at the first position. However, it is known that Rnt1p can tolerate a G at the first position of the tetraloop, as long as the tetraloop does not adopt a GNRA fold (8). The other exceptions were found in Candida tropicalis and in Y. lipolytica, for which the 3′ ETS sequences did not exhibit this type of terminal tetraloop structure in any of the RST sequences analyzed. Interestingly, two predicted stem-loops structures were found in the 3′ ETS of Y. lipolytica. These two stem-loops may correspond to genuine processing signals, as a particular sequence has been conserved in both of these stems (Fig. 1). Thus, these two stems may correspond to duplicated processing signals. The stems are not capped by AGNN tetraloops but rather by trinucleotide loops. Overall, analysis of the 3′ ETS sequences suggests that RNase III processing in the 3′ ETS occurs in most Hemiascomycetes species and that the RNase III activities involved have a specificity for dsRNA capped by AGNN tetraloops, with the exception of Y. lipolytica and possibly C. tropicalis (see below). Not surprisingly, Y. lipolytica is the most distant species from S. cerevisiae on the evolutionary scale (22).
FIG. 1.
Predicted secondary structures in the 3′ ETS sequences of the pre-rRNAs in various Hemiascomycetes. Distances in nucleotides to the 3′ ends of the 25S rRNAs are indicated. Distances are only approximate, since the exact 3′ ends have not been experimentally mapped but are estimated from the S. cerevisiae sequence. The sequences of the 3′ ETSs are identical in S. cerevisiae and S. bayanus. Base pairs that have been conserved in closely related species (S. kluyveri and K. thermotolerans, K. marxianus and K. lactis, and P. sorbitophilia and D. hansenii) are boxed. The boxed sequence shown for Y. lipolytica corresponds to a sequence conserved in both stems.
Conservation of 5′-end processing signals.
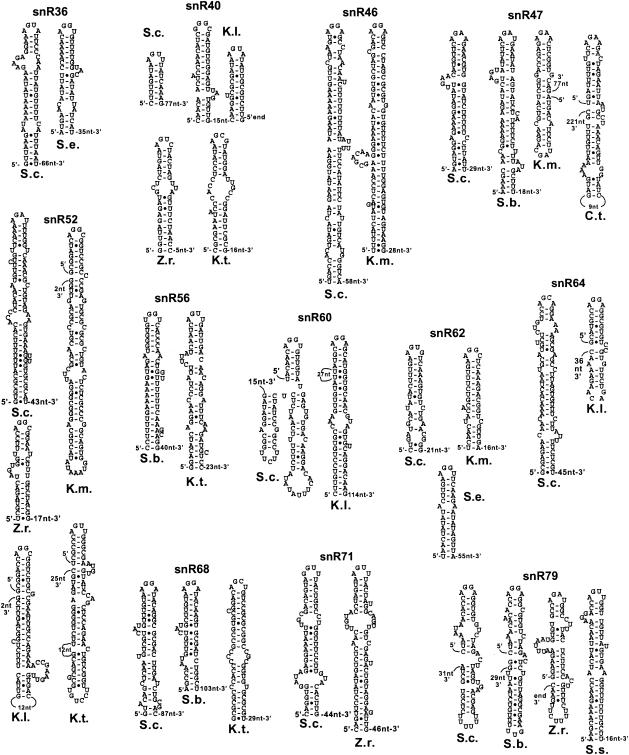
A large number of independently transcribed box H/ACA and box C/D snoRNAs are processed by RNase III cleavage in the 5′ extension of the precursor, followed by exonucleolytic digestion (6; Lee et al., in press). I searched for orthologs of these snoRNAs and analyzed the 5′-end-flanking sequences for the presence of predicted secondary structures. A total of 27 orthologous sequences flanked by sufficient 5′-end-flanking sequences were found for a total number of 12 different snoRNAs (two box H/ACA snoRNAs, snR36, and snR46, and 10 box C/D snoRNAs) (Table 2; Fig. 2). In most cases, extensive secondary structures could be predicted at a short distance upstream from the mature snoRNA sequences, and these stems were capped by AGNN tetraloops. Interestingly, the distances between the last nucleotide of the stem and the 5′ end of the snoRNA are often comparable for S. cerevisiae and the other species. In some cases, the predicted processing signals seem to have diverged from a single stem to two stems which potentially stack onto each other to reconstitute a processing signal, as described previously for the snR40 5′-end processing signal in S. cerevisiae (6). This is the case for snR47 in K. marxianus and for snR52 in all Kluyveromyces species. For some very close species, for example, S. cerevisiae and S. bayanus, the sequence of the stems has diverged, but compensatory mutations have maintained the overall architecture of the stem-loop (e.g., in snR47). In some other cases, the upper part of the stem near the tetraloop has conserved some sequence similarities between S. cerevisiae and other species (e.g., in snR47, snR62, snR71, and snR79).
FIG. 2.
Predicted secondary structures in the 5′-end processing signals of Hemiascomycetes snoRNAs. The distances in nucleotides are from the last nucleotides of the stems to the predicted 5′ ends of the mature snoRNAs. Distances are only approximate, since the exact 5′ ends have not been experimentally mapped but are estimated from the S. cerevisiae sequence. S.c., S. cerevisiae; S.b., S. bayanus; S.e., S. exiguus; S.s., Saccharomyces servazzii; K.l., K. lactis; K.t., K. thermotolerans; K.m., K. marxianus; Z.r., Z. rouxii.
This analysis did not reveal any ortholog of snoRNA sequences from the Pichia genus or from D. hansenii and Y. lipolytica. Since these organisms are the most evolutionarily distant from S. cerevisiae, the sequences of these snoRNAs may have diverged enough so that orthologous sequences can hardly be found with standard BLAST procedures. Several matches of very short sequences with high P values were obtained for some snoRNAs in some of these species, but they were not analyzed further. However, some of the matches may represent true orthologous sequences. Interestingly, a true ortholog of the snR47 snoRNA was identified in C. tropicalis, and a predicted AGAA stem-loop was found in the 5′-end-flanking sequence of this snoRNA. This finding suggests that even though AGNN stem-loops are absent from the 3′ ETS of the rDNA, an RNase III activity that recognizes AGNN tetraloops is present in C. tropicalis. It is unknown whether the same activity processes the rRNA on the 3′ ETS or whether a duplicated RNase III activity specialized in processing the rRNA 3′ ETS exists without specificity for dsRNA capped by AGNN tetraloops.
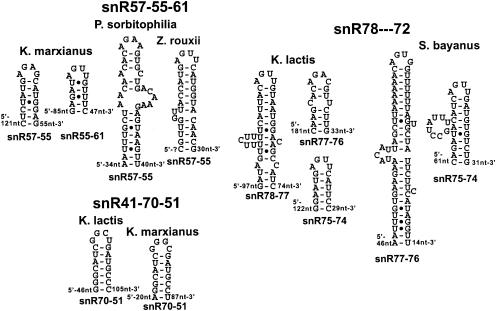
Conservation of polycistronic box C/D snoRNA processing signals.
In S. cerevisiae, several box C/D snoRNAs are expressed as polycistronic transcription units and processed by Rnt1p cleavage followed by exonuclease processing (6, 7, 17). In these cases, the Rnt1p cleavage sites are often made of short stems capped by AGNN tetraloops, which stack onto longer stems where cleavage occurs (17). Partial orthologous sequences were found for the operon expressing seven snoRNAs (snR78 to -72 [snR78-72]) in S. bayanus, K. lactis, and P. sorbitophilia (Table 2; Fig. 3). The S. bayanus RST contained all snoRNAs but snR78, the K. lactis sequences contained snR78-75, and P. sorbitophilia contained snR75-73. Since the distances between the mature snoRNAs in these RSTs are usually 120 nt or less, these species have probably conserved the polycistronic mode of expression of these snoRNAs. For S. bayanus and K. lactis, RNase III processing signals similar to those observed in S. cerevisiae could be predicted (Fig. 3). For simplicity, the longer stems onto which the AGNN short stem-loops potentially stack were not represented in Fig. 3. No secondary structures capped by AGNN tetraloops could be identified for the P. sorbitophilia sequences.
FIG. 3.
Predicted secondary structures in intergenic spacers of polycistronic precursors of box C/D snoRNAs. The names of the polycistronic transcription units are indicated in large print above the names of the species. The locations of the stem-loops between snoRNAs as well as the distances in nucleotides to each of the neighboring snoRNAs are indicated.
Orthologous sequences for the tricistronic snR57-snR55-snR61 snoRNA transcription unit were identified in K. marxianus, Zygosaccharomyces rouxii, and P. sorbitophilia. Although no AGNN stem-loop can be predicted from the S. cerevisiae sequence (unpublished data), polycistronic processing sites could be predicted in all of these three species (Fig. 3), either between snR57 and snR55 or between snR55 and snR61, strongly suggesting that these three snoRNAs are also expressed from a polycistronic precursor. Orthologs of the snR41-snR70-snR51 tricistronic expression unit were identified in K. lactis and K. marxianus (Table 2; Fig. 3), with short GGNN stem-loops between each species. While this sequence diverges from the AGNN consensus, it is known that Rnt1p can tolerate a G at the first position (8). Overall, these results suggest that the polycistronic mode of expression of some of the box C/D snoRNAs has been conserved in Hemiascomycetes and that the corresponding orthologs of RNase III play a role in the processing of these species. For some of the snoRNAs, no processing signal could be identified (e.g., between snR55 and snR61 for K. lactis and P. sorbitophilia). Despite the absence of processing signals between snR55 and snR61, it is possible that a single processing signal between snR57 and snR55 is sufficient to separate the three snoRNAs; that is, if the AGNN stem-loop located between snR57 and snR55 coaxially stacks onto a downstream stem that loops out the second snoRNA (snR55), dsRNA cleavage on both sides of snR55 may be sufficient to separate the three snoRNAs from each other, as was observed for snR75 (17). For the other snoRNA species, the absence of the predicted AGNN tetraloop does not necessarily mean that these structures do not exist; previous computer searches for secondary structures in the polycistronic arrays revealed that the processing signals are usually harder to predict than are rRNA or 5′-end processing signals, even if they have been identified in vivo (4). Therefore, the absence of predicted sites does not necessarily mean that they do not exist in vivo.
Conservation of 3′-end processing signals.
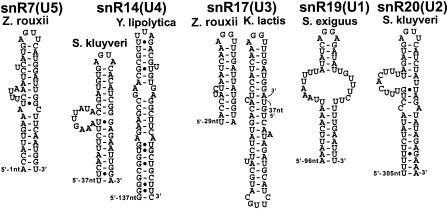
The 3′-end processing of snRNAs and snoRNAs by RNase III is clearly a dispensable processing pathway. This conclusion is suggested by the fact that the 3′ ends of most sn(o)RNAs that are processed by Rnt1p can be generated by exonucleases or by the polyadenylation machinery in the absence of Rnt1p (1, 5, 15, 21, 24). For example, the U5 snRNA is present in S. cerevisiae with two forms; the longer one is processed through a Rnt1p-dependent pathway, while the shorter form is processed through a Rnt1p-independent pathway (5). S. exiguus also exhibits these two forms, while S. kluyveri, K. lactis, and Y. lipolytica exhibit only one form (19). These observations suggest that for these three species, the dependence on RNase III for processing has been lost. Eleven RSTs were obtained with sufficient 3′-end-flanking sequences for spliceosomal snRNAs or the U3 snoRNA. The conservation of stem-loop structures capped by AGNN tetraloops was not as strong for these 3′-end processing signals as those described previously for 5′-end processing signals (Table 2; Fig. 4). Only one AGUU stem-loop was found for Z. rouxii U5 snRNA, while no AGNN stem-loop could be predicted in RSTs from K. marxianus and K. lactis. This negative result is consistent with the observation that only one form of U5 is present in K. lactis, probably processed through an RNase III-independent pathway. Similarly, only one species out of three showed an AGNN stem-loop for the U4 snRNA (Table 2; Fig. 4). The conservation of the stem-loop structure seems stronger for the U3 snoRNA, for which both K. lactis and Z. rouxii exhibited AGNN stem-loops (Fig. 4). Overall, these phylogenetic observations strengthen previous biochemical data suggesting that 3′-end processing of snRNAs by RNase III is a redundant pathway that may have been lost during the recent evolution of yeast species.
FIG. 4.
Predicted secondary structures in the 3′ extensions of snRNAs and of the U3 snoRNA. The estimated distances in nucleotides to the mature 3′ ends of the sn(o)RNAs are indicated.
DISCUSSION
In this study, I show that RNA processing signals obeying S. cerevisiae RNase III specificity rules are present in most Hemiascomycetes species analyzed in the Génolevures program. Predicted RNase III cleavage sites are present in all four categories of processing signals described so far: 3′ ETS, 5′ end, 3′ end, and polycistronic (Fig. 5). The most complete phylogenetic picture was obtained for the rRNA 3′ ETS sequences, since they were found in abundance in the Génolevures RSTs and since they are easily identified with a standard BLAST analysis due to the strong conservation of the 25S rRNA sequence. Strikingly, Rnt1p-type stem-loops were present in all species but C. tropicalis and Y. lipolytica (Table 1; Fig. 1). In the case of snoRNAs, orthologous sequences were harder to obtain for evolutionarily distant species such as Pichia spp., D. hansenii, C. tropicalis, and Y. lipolytica, because these noncoding RNAs are small and because their sequences may have diverged significantly. Therefore, standard BLAST analysis revealed only a limited number of substrates for these species. Nevertheless, the purpose of this study was not to identify the full set of snoRNAs orthologs for all these species but rather to identify examples that would reveal the conservation of Rnt1p-type processing sites. Data obtained on the 3′ ETS sequences of D. hansenii and P. sorbitophilia, on the polycistronic signal between snR57 and snR55 in P. sorbitophilia, and on snR47 in C. tropicalis, however, suggest that an Rnt1p-like activity may exist in Pichia, D. hansenii, and C. tropicalis, since some of the predicted RNA processing signals identified from these species show AGNN-type secondary structures. In C. tropicalis, only one snoRNA substrate could be found, and it obeys the AGNN rule (snR47). The other predicted substrate, the 3′ ETS, does not show any AGNN tetraloop. One possible explanation is that the same activity processes the snoRNAs and the rRNA on the 3′ ETS sequences but that the 3′ ETS activity does not require AGNN tetraloops. This activity has been shown for S. cerevisiae's U18 snoRNA (9), where Rnt1p cleavage occurs without the presence of AGNN tetraloops. Alternatively, it is possible that a duplicated RNase III activity that is specialized for the processing of the rRNA 3′ ETS exists but has lost the specificity for dsRNA capped by AGNN tetraloops.
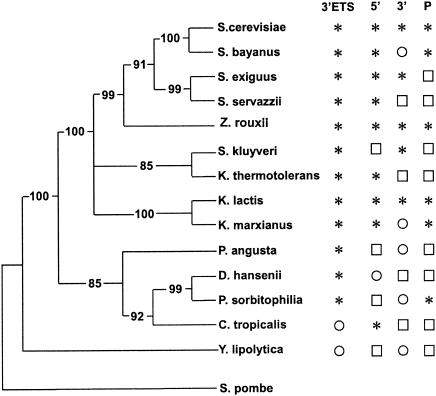
FIG. 5.
Phylogenetic distribution of AGNN stem-loop processing signals in Hemiascomycetes. The cladogram of Hemiascomycetes was constructed from rDNA sequences of the 25S rRNAs by using phylogenetic distances obtained from reference 22 and with S. pombe as an outgroup. Asterisks indicate the presence of at least one predicted AGNN stem-loop processing signal. Open squares indicate that no RST was found for this category of processing signal in the particular species. Open circles indicate that while at least one RST was available, no AGNN stem-loop could be detected.
The presence of the processing signals suggests that most Hemiascomycetes species have conserved the mode of expression and processing of these noncoding RNAs. While the conservation of RNase III cleavage in the 3′ ETS sequence was expected, given the functional conservation described for S. pombe (23), the conservation of RNase III processing sites upstream from mature snoRNA sequences and in intergenic snoRNA sequences suggests that these snoRNAs have conserved their mode of expression and processing from monocistronic or polycistronic precursors. In most cases, the sequences of the flanking sequences have more or less strongly diverged, but the secondary structures have been conserved due to the selection pressure to maintain the RNase III cleavage sites. While this prediction has not been tested experimentally, the phylogenetic argument is probably strong enough to support the prediction that the RNase III orthologs cleave the 5′ extensions of the precursors, or the intergenic spacers in the polycistronic precursors, and that cleavage is followed by exonucleolytic processing.
Overall, the sum of data obtained from this phylogenetic analysis suggests that Rnt1p-type processing exists, at least for one category of the substrates for each Hemiascomycetes species analyzed by the Génolevures program, with the exception of Y. lipolytica (Fig. 5). Interestingly, this species is the most evolutionarily distant species from S. cerevisiae when a cladogram is established by using rDNA sequences (Fig. 5) and seems to have diverged early from other Hemiascomycetes species. Based on these observations, two scenarios are possible. It is possible that the specificity of RNase III for dsRNA capped by AGNN tetraloops was present in the common ancestor of all these Hemiascomycetes but that the Y. lipolytica RNase III somehow evolved and lost the specificity for the tetraloops even though this specificity was conserved in all other species. Alternatively, it is possible that the specificity for AGNN tetraloops was acquired after the divergence between Y. lipolytica and all other Hemiascomycetes species analyzed. In both cases, the events require a rapid coevolution of the enzyme specificity and the substrate sequences. While the second scenario seems more parsimonious since S. pombe Pac1p does not rely on AGNN tetraloop recognition, further analysis of RNase III processing signals in more distant Ascomycetes species will possibly reveal which is the more likely evolutionary scenario.
Acknowledgments
I thank the Génolevures program for their sequencing effort and C. Coffinier for critical reading of the manuscript.
This work was supported by NIGMS grant R01 GM-61518 from the NIH.
REFERENCES
- 1.Abou Elela, S., and M. Ares, Jr. 1998. Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J. 17:3738-3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abou Elela, S., H. Igel, and M. Ares, Jr. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85:115-124. [DOI] [PubMed] [Google Scholar]
- 3.Allmang, C., and D. Tollervey. 1998. The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol. 278:67-78. [DOI] [PubMed] [Google Scholar]
- 4.Chanfreau, G., M. Buckle, and A. Jacquier. 2000. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl. Acad. Sci. USA 97:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chanfreau, G., S. A. Elela, M. Ares, Jr., and C. Guthrie. 1997. Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev. 11:2741-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chanfreau, G., P. Legrain, and A. Jacquier. 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284:975-988. [DOI] [PubMed] [Google Scholar]
- 7.Chanfreau, G., G. Rotondo, P. Legrain, and A. Jacquier. 1998. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17:3726-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danin-Kreiselman, M., C. Y. Lee, and G. Chanfreau. 2003. RNase III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11:1279-1289. [DOI] [PubMed] [Google Scholar]
- 9.Giorgi, C., A. Fatica, R. Nagel, and I. Bozzoni. 2001. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 20:6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grishok, A., A. E. Pasquinelli, D. Conte, N. Li, S. Parrish, I. Ha, D. L. Baillie, A. Fire, G. Ruvkun, and C. C. Mello. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106:23-34. [DOI] [PubMed] [Google Scholar]
- 11.Hutvagner, G., J. McLachlan, A. E. Pasquinelli, E. Balint, T. Tuschl, and P. D. Zamore. 2001. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293:834-838. [DOI] [PubMed] [Google Scholar]
- 12.Ketting, R. F., S. E. Fischer, E. Bernstein, T. Sijen, G. J. Hannon, and R. H. Plasterk. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kufel, J., B. Dichtl, and D. Tollervey. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5:909-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 15.Morlando, M., P. Greco, B. Dichtl, A. Fatica, W. Keller, and I. Bozzoni. 2002. Functional analysis of yeast snoRNA and snRNA 3′-end formation mediated by uncoupling of cleavage and polyadenylation. Mol. Cell. Biol. 22:1379-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park, W., J. Li, R. Song, J. Messing, and X. Chen. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12:1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qu, L.-H., A. Henras, Y.-J. Lu, H. Zhou, W.-X. Zhou, Y.-Q. Zhu, J. Zhao, Y. Henry, M. Caizergues-Ferrer, and J.-P. Bachellerie. 1999. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 19:1144-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reinhart, B. J., E. G. Weinstein, M. W. Rhoades, B. Bartel, and D. P. Bartel. 2002. MicroRNAs in plants. Genes Dev. 16:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roiha, H., E. O. Shuster, D. A. Brow, and C. Guthrie. 1989. Small nuclear RNAs from budding yeasts: phylogenetic comparisons reveal extensive size variation. Gene 82:137-144. [DOI] [PubMed] [Google Scholar]
- 20.Rotondo, G., M. Gillespie, and D. Frendewey. 1995. Rescue of the fission yeast snRNA synthesis mutant snm1 by overexpression of the double-strand-specific PacI ribonuclease. Mol. Gen. Genet. 247:698-708. [DOI] [PubMed] [Google Scholar]
- 21.Seipelt, R. L., B. Zheng, A. Asuru, and B. C. Rymond. 1999. U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res. 27:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Souciet, J.-L., M. Aigle, F. Artiguenave, G. Blandin, M. Bolotin-Fukuhara, E. Bon, P. Brottier, S. Casaregola, J. de Montigny, B. Dujon, P. Durrens, C. Gaillardin, A. Lépingle, B. Llorente, A. Malpertuy, C. Neuvéglise, O. Ozier-Kalogéropoulos, S. Potier, W. Saurin, F. Tekaia, C. Toffano-Nioche, M. Wésolowski-Louvel, P. Wincker, and J. Weissenbach. 2000. Genomic exploration of the hemiascomycetous yeasts: 1. A set of yeast species for molecular evolution studies. FEBS Lett. 487:3-12. [DOI] [PubMed] [Google Scholar]
- 23.Spasov, K., L. I. Perdomo, E. Evakine, and R. N. Nazar. 2002. RAC protein directs the complete removal of the 3′ external transcribed spacer by the PacI nuclease. Mol. Cell 9:433-437. [DOI] [PubMed] [Google Scholar]
- 24.van Hoof, A., P. Lennertz, and R. Parker. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol. 20:441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, H., H. Xu, L. J. Miraglia, and S. T. Crooke. 2000. Human RNase III is a 160-kDa protein involved in preribosomal RNA processing. J. Biol. Chem. 275:36957-36965. [DOI] [PubMed] [Google Scholar]
- 26.Wu, H., P. K. Yang, S. E. Butcher, S. Kang, G. Chanfreau, and J. Feigon. 2001. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J. 20:7240-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, K., and A. W. Nicholson. 1997. Regulation of ribonuclease III processing by double-helical sequence antideterminants. Proc. Natl. Acad. Sci. USA 94:13437-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou, D., D. Frendewey, and S. M. Lobo Ruppert. 1999. Pac1p, an RNase III homolog, is required for formation of the 3′ end of U2 snRNA in Schizosaccharomyces pombe. RNA 5:1083-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]