Abstract
Adherence of fungal cells to host substrates and each other affects their access to nutrients, sexual conjugation, and survival in hosts. Adhesins are cell surface proteins that mediate these different cell adhesion interactions. In this study, we examine the in vivo functional requirements for specific posttranslational modifications to these proteins, including glycophosphatidylinositol (GPI) anchor addition and O-linked glycosylation. The processing of some fungal GPI anchors, creating links to cell wall β-1,6 glucans, is postulated to facilitate postsecretory traffic of proteins to cell wall domains conducive to their functions. By studying the yeast sexual adhesin subunit Aga1p, we found that deletion of its signal sequence for GPI addition eliminated its activity, while deletions of different internal domains had various effects on function. Substitution of the Aga1p GPI signal domain with those of other GPI-anchored proteins, a single transmembrane domain, or a cysteine capable of forming a disulfide all produced functional adhesins. A portion of the cellular pool of Aga1p was determined to be cell wall resident. Aga1p and the α-agglutinin Agα1p were shown to be under glycosylated in cells lacking the protein mannosyltransferase genes PMT1 and PMT2, with phenotypes manifested only in MATα cells for single mutants but in both cell types when both genes are absent. We conclude that posttranslational modifications to Aga1p are necessary for its biogenesis and activity. Our studies also suggest that in addition to GPI-glucan linkages, other cell surface anchorage mechanisms, such as transmembrane domains or disulfides, may be employed by fungal species to localize adhesins.
Fungal cells modulate their adherence to one another and to environmental substrates through the activities of cell surface-localized proteins termed adhesins. Adhesins mediate these contacts during both vegetative growth and in response to differentiation cues triggering filamentous and hyphal growth forms or mating (3, 26, 40). As such, these cell-cell and cell-substrate interactions are critically important to different phases of fungal life cycles and for symbiotic or pathogenic fungi associated with algal, plant, or animal cells and are thus essential to the successful establishment and maintenance of fungal-host associations.
In the common human pathogens Candida albicans and Candida glabrata, multiple adhesins appear to participate in mediating adherence of cells to host tissues, and in many instances, their participation in virulence has been clearly demonstrated (14, 21, 31-33, 55, 60, 64, 67). Adhesins have also been found to underlie pathogenesis for other fungi, including Blastomyces dermatidis, Coccidioides immitis, Paracoccidioides brasiliensis, and a variety of other Candida species (4, 15, 25, 30, 31, 34, 38, 45, 65). Thus, adhesins are currently viewed as important virulence factors affecting fungal pathogenicity.
Studies of several adhesins from the fungal species Saccharomyces cerevisiae and C. albicans have revealed a number of general properties. The expression patterns of many of the genes encoding adhesins are regulated in response to developmental or environmental cues (19, 26, 55, 57, 59, 65). In S. cerevisiae, activation of the mating pathway induces the expression of the agglutinin genes AGA1, AGA2, and SAG1/AGα1 and the morphogenesis and agglutination-modifying gene FIG2 (19, 36, 40, 58, 76). Pseudohyphal, invasive growth differentiation and nutritional signals also serve to modulate the expression of the flocculin genes FLO1, FLO5, FLO9, FLO10, and FLO11, which is also required for biofilm formation of yeast cells (26, 42, 50, 52, 63, 73). Additionally, the C. albicans adhesin gene HWP1 was first identified as producing a differentially regulated gene product whose expression is under developmental control and is strongly induced during hyphal formation (55, 60).
Commonalities among the adhesins are also evident at the protein level. These proteins typically are relatively large and extracellular, frequently containing an extensive number of residues predicted to receive modification in the form of N- or O-linked glycosylation (39, 47). These proteins are typically maintained on the surface of the cell through a plasma membrane-resident or cell wall-linked glycophosphatidylinositol (GPI) anchor (37, 47). Finally, the majority of the adhesins also possess repeated domains that, in some cases, have been shown to be associated with adhesive activities (20, 37, 60, 65, 75).
Despite the fact that the majority of fungal adhesins studied to date receive at least one or more of these different posttranslational modifications, the significance of each of these modifications for this class of proteins remains poorly understood with respect to their function. These proteins obtain their initial cell surface location by transiting the secretory pathway; the presence of a GPI anchor is expected to aid in retention of these proteins at the cell surface following secretion. In a number of fungal species, GPI anchors of some proteins are capable of being modified by a transglycosylation reaction to become covalently associated with the β-1,6 glucan layer of the cell wall. This glucan association has been proposed to provide a means for fungal cells to more effectively expose these proteins at the surfaces of their cell walls, because residency of such proteins at the plasma membrane might be expected to keep the proteins within the interior of the cell wall given their lengths relative to the average 100- to 200-nm-thick cell wall (43, 44, 47). Similarly, the role of O-linked glycosylation of these proteins is believed to aid in their assuming extended, rodlike conformations that again aid in display of domains mediating adhesive interactions near the surface of the cell wall (14, 20, 35, 44, 75). The role of this modification has presently only been addressed by a small number of in vitro studies, despite the fact that the O-linked glycosylation pathway has been established as a general virulence determinant (6).
The components required for agglutination of S. cerevisiae cells during mating have been well described at the protein level and represent an excellent system for exploring functional requirements of adhesins. MATa cells present a bipartite agglutinin molecule that is composed of a 703-amino-acid-long, secretory pathway-processed, and GPI-anchored Aga1p bound to a 69-amino-acid polypeptide, Aga2p. This linkage occurs through a pair of disulfide bonds, and the residues involved in Aga2p are known (7). The residues within Aga1p mediating this interaction have recently been identified by us (this study) and others using different methods (56). Additionally, Aga1p contains three repeated domains: R1, R2, and a serine-threonine-rich heptapeptide repeat. Within the highly similar R1 and R2 domains, conserved subdomains can also be found, including a “WCPL” domain, whose presence has been noted in other fungal adhesins of both S. cerevisiae and C. albicans (55). The functional significance of these domains to different Aga1p functions remains to be understood.
The adhesion between the two mating cells occurs by the interaction of the Aga1p-Aga2p complex found on MATa cells with the mature 608-amino-acid GPI-anchored Sag1/Agα1 protein presented on the surface of MATα cells. Loss of AGA1, AGA2, or SAG1/AGα1 from appropriate mating cell types leads to inability of cells to adhere to mating partners in liquid medium prior to conjugation (reviewed in reference 40). The expression of the agglutinins is strongly increased by signaling during mating. In addition to being pheromone regulated, Aga2p and Agα1p are also cell type specific in their expression. The sites of Aga2p-Agα1p interaction have been mapped in these proteins, and the postsecretory trafficking (GPI anchor processing and remnant cross-linking to β-1,6 glucan) of Agα1p in the cell wall has been examined in some detail (43, 44). This system, therefore, presents an excellent model for further investigation of additional cis- and trans-acting factors that promote agglutinin function.
Because the posttranslational modifications of adhesins are postulated to enable their more effective function, this modification pathway represents a potential virulence factor whose relevance to adhesin function has not been specifically studied. In this paper, we consider the functional significance of these posttranslational modifications occurring to fungal adhesins. In particular, we have examined the relationship of glycosylation, protein size, and cell surface anchorage to the function of an endogenous yeast adhesin protein, Aga1p, whose activity can be postulated to depend on factors affecting its cell wall residency. Surprisingly, we find that GPI anchoring per se is not a requirement for normal function of the adhesin Aga1p, because we find that alternative, functional anchoring mechanisms exist for this protein. These data highlight general structural requirements for fungal adhesin activity and suggest sequence features that may characterize additional classes of adhesins, which currently remain to be identified in other fungal species.
MATERIALS AND METHODS
Growth of yeast strains.
The S. cerevisiae strains used in this study are listed in Table 1. Strains were cultured in YPAD, Sc-Leu, or Sc-Ura medium at 30°C. Cell densities were measured by A600 with a spectrophotometer (Spectronic Genesys 8). A PCR-based homologous recombination method (2) was used to delete the entire coding sequence of AGA1, replacing it with URA3. This resulted in an aga1Δ strain, as confirmed by diagnostic PCR and the absence of agglutination activity in MATa cells. A similar approach was used to construct the pmt4Δ strain. Other pmtΔ mutants were either obtained from the Yeast Genome Systematic Deletion collection or, in the case of the pmt1Δ pmt2Δ strains, were made by standard mating and sporulation procedures followed by confirmation of the genotype by diagnostic PCRs. A modified one-step transformation method (11) was used to transform yeast strains as needed.
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or referencea |
|---|---|---|
| YSE430 | MATaura3Δ leu2Δ his3Δ TRP1 met15Δ LYS2 | 74 |
| YSE431 | MATα ura3Δ leu2Δ his3Δ TRP1 MET15 lys2Δ | |
| YSE445 | MATapmt2::Kanrura3Δ leu2Δ his3Δ TRP1 met15Δ LYS2 | |
| YSE446 | MATα pmt2::Kanrura3Δ leu2Δ his3Δ TRP1 MET15 lys2Δ | |
| YSE449 | MATapmt3::Kanrura3Δ leu2Δ his3Δ TRP1 met15Δ LYS2 | |
| YSE450 | MATα pmt3::Kanrura3Δ leu2Δ his3Δ TRP1 MET15 lys2Δ | |
| YSE453 | MATapmt6::Kanrura3Δ leu2Δ his3Δ TRP1 met15Δ LYS2 | |
| YSE454 | MATα pmt6::Kanrura3Δ leu2Δ his3Δ TRP1 MET15 lys2Δ | |
| YSE619 | MATα agα1::URA3 leu2Δ his3Δ TRP1 MET15 lys2Δ | |
| YSE901 | MATapmt1::Kanrura3Δ leu2Δ his3Δ TRP1 met15Δ LYS2 | |
| YSE902 | MATα pmt1::Kanrura3Δ leu2Δ his3Δ TRP1 MET15 lys2Δ | |
| YSE903 | MATapmt1::Kanrpmt2::Kanr ura3Δ leu2Δ his3Δ TRP1 met15Δ LYS2 | This study |
| YSE904 | MATα pmt1::Kanrpmt2::Kanr ura3Δ leu2Δ his3Δ TRP1 MET15 lys2Δ | This study |
| YSE905 | MATapmt5::Kanrura3Δ leu2Δ his3Δ TRP1 met15Δ LYs2 | |
| YSE906 | MATα pmt5::Kanrura3Δ leu2Δ his3Δ TRP1 MET15 lys2Δ | |
| YSE907 | MATapmt7::Kanrura3Δ leu2Δ his3Δ TRP1 met15Δ LYS2 | |
| YSE908 | MATα pmt7::Kanrura3Δ leu2Δ his3Δ TRP1 MET15 lys2Δ | |
| YSE913 | MATaaga1::URA3 leu2Δ his3Δ TRP1 met15Δ LYS2 | This study |
| YSE995 | MATapmt4::URA3 leu2Δ his3Δ TRP1 met15Δ LYS2 | This study |
| YSE996 | MATα pmt4::URA3 leu2Δ his3Δ TRP1 MET15 lys2Δ | This study |
All strains are derived from the Yeast Genome Deletion collection (74) unless otherwise indicated.
Construction of plasmids.
The plasmids used in this study are listed in Table 2. The AGA1 gene, including 750 bases upstream of the predicted start codon and 500 bases downstream of the termination codon, was PCR amplified from genomic DNA and cloned into the yeast centromeric vector pRS315 to yield pRS315-AGA1. The primers used for this construction were P1 and P2 (see Table S1 in the supplemental material; boldface bases are the heterologous restriction sites used for cloning the fragment). The pRS315-AGA1 construct was then modified by insertion of a triple hemagglutinin (3HA) epitope at a unique SpeI site following the R2 domain of the predicted Aga1p coding sequence (amino acid 540/725), yielding pRS315-AGA1::3×HA (pSE400; pAGA1 hereafter). A 210-bp 3HA sequence was PCR amplified with primers that introduced an SpeI site at the 5′ end and an XbaI site at the 3′ end (primer pair P3-P4). The following different Aga1p GPI anchor chimeras and domain deletions were generated from this parent plasmid, pSE400. In pAGA1-GPIΔ (pSE211), the C-terminal 67 amino acids including the 38-amino-acid GPI attachment signal was deleted from pAGA1 and replaced with a unique PstI site, using a PCR-based cloning strategy (primers P1-P6 and P2-P5). The missing GPI signal was then replaced with the final 67 amino acids of Aga1p (P7-P8), Cwp2p (P9-P10), Yap3p (P11-P12), or Gas1p (P13-P14), resulting in pAGA1-(GPI-AGA1), pAGA1-(GPI-CWP2), pAGA1-(GPI-YAP3), and pAGA1-(GPI-GAS1), respectively. Similarly, the missing sequence in pAGA1-GPIΔ was replaced by the sequences encoding the transmembrane domains (TMDs) from Fus1p (amino acids 72 to 102/512) (P15-P16) and Axl2p (amino acids 499 to 541/823) (P17-P18), respectively. These clones were named pAGA1-(TMD-FUS1) and pAGA1-(TMD-AXL2). All of the clones described above were confirmed by sequencing.
TABLE 2.
Plasmids used in this study
| Plasmid | Parent vector and contents |
|---|---|
| pSE19 | pRS315 |
| pSE400 | pRS315-AGA1::3HA |
| pSE401 | pRS315-AGA1::3HA-R1Δ |
| pSE402 | pRS315-AGA1::3HA-R2Δ |
| pSE403 | pRS315-AGA1::3HA-R1ΔR2Δ |
| pSE404 | pRS315-AGA1::3HA-WCPLΔ |
| pSE405 | pRS315-AGA1::3HA-R2ΔCX4CΔ |
| pSE406 | pRS315-AGA1::3HA-R2ΔCX2CΔ |
| pSE407 | pRS315-AGA1::3HA-R2ΔWCPLΔ |
| pSE231 | pRS315-AGA1::3HA-CX2CΔ |
| pSE232 | pRS315-AGA1::3HA-CX4CΔ |
| pSE211 | pRS315-AGA1::3HA-GPIΔ |
| pSE214 | pRS315-AGA1::3HA-(GPI-AGA1) |
| pSE215 | pRS315-AGA1::3HA-(GPI-CWP2) |
| pSE217 | pRS315-AGA1::3HA-(GPI-GAS1) |
| pSE224 | pRS315-AGA1::3HA-(GPI-YAP3) |
| pSE219 | pRS315-AGA1::3HA-(TMD-FUS1) |
| pSE222 | pRS315-AGA1::3HA-(TMD-AXL2) |
| pSE225 | pRS315-AGA1::3HA-(TMD-FUS1)-61aa |
| pSE226 | pRS315-AGA1::3HA-(TMD-FUS1)-122aa |
| pSE227 | pRS315-AGA1::3HA-(TMD-FUS1)-183aa |
| pSE228 | pRS315-AGA1::3HA-(TMD-FUS1)-200aa |
| pSE204 | pRS315-AGA1::3HA-GPIΔ-Cys |
| pSE229 | pRS315-AGA1::3HA-GPIΔ-Ser |
| pSE230 | pRS315-AGA1::3HA-(TMD-FUS1)-NS |
| pSE233 | pRS315-AGA1::3HA-SD |
| pSE234 | pRS315-AGA1::3HA-(TMD-FUS1)-200aa-SD |
| pSE235 | pRS315-AGα1::3HA |
pAGA1-(TMD-FUS1) was used as the parental plasmid to construct the longer spacer-containing Aga1p chimeras. Two spacers, 1 and 2, were PCR amplified from the unique and repetitive regions between R1 and R2, with an introduced SpeI site at both ends of the fragment. Spacer 1 encodes amino acid residues 315 to 375/725 (P19-P20), and spacer 2 encodes amino acid residues 176 to 375/725 (P20-P21). Tandem dimer and trimer sequences were made from spacer 1 by limited ligation of the insert monomer sequence in the absence of vector. All of these spacers were inserted at the unique SpeI site in pAGA1, and their correct orientation was confirmed by sequencing. These chimeric proteins were named pAGA1-(TMD-FUS1)-61aa (monomer), pAGA1-(TMD-FUS1)-122aa (dimer), pAGA1-(TMD-FUS1)-183 (trimer), and pAGA1-(TMD-FUS1)-200aa (spacer 2), according to the length of the introduced spacers. To test the effect of spacer position on Aga1p function, spacer 2 was also inserted at an introduced ClaI site (33/725) in pAGA1-(TMD-FUS1), resulting in pAGA1-(TMD-FUS1)-NS (primer pair used to introduce the ClaI site, P22-P23; primer pair used to amplify spacer 2 flanked by ClaI sites, P24-P25). Spacer 2 (176 to 375/725) was also deleted from pAGA1 and pAGA1-(TMD-FUS1)-200aa, producing pAGA1-SD and pAGA1-(TMD-FUS1)-200aa-SD (P1-P26 and P2-P27). The carboxy-terminal Cys mutation resulted from the introduction of a single-base-deletion described in the text from the sequential cloning of two PCR products generated by the primer pairs P1-P40 and P2-P5. The P40 sequence matches the intended P6 primer sequence, with the exception of the inferred base deletion that gives rise to the frameshift mutation described in the text. Mutagenesis of the cysteine codon TGC to the serine codon TCT was carried out with a Stratagene QuickChange site-directed mutagenesis kit according to the manufacturer's instructions.
To create Aga1p lacking the R1 repeat (amino acid residues 32 to 149), P1 and P28 were used to generate the SalI-XmnI fragment, which was then ligated to the XmnI-XbaI fragment in pRS315. Similarly, to create Aga1p lacking the R2 repeat (amino acid residues 387 to 539), P1 and P29 were used to generate the SalI-SpeI fragment, which was ligated to the SpeI-XbaI fragment in pRS315. To delete the WCPL, CX4C, and CX2C domains from the R1 repeat, primers upstream and downstream of each region were designed, and a two-step cloning strategy was used to delete this region, which resulted in pAGA1-WCPLΔ (P1-P30 and P2-P31), pAGA1-CX4CΔ (P1-P32 and P2-P33), and pAGA1-CX2CΔ (P1-P34 and P2-P35). Deletion of R1 WCPL, CX4C, and CX2C from pAGA1-R2Δ resulted in pAGA1-R2ΔWCPLΔ, pAGA1-R2ΔCX4CΔ, and pAGA1-R2ΔCX2CΔ.
A 3HA-tagged version of AGα1 gene was introduced into the centromeric pRS315 by a strategy similar to the construction of pAGA1, and this construct was named pRS315-AGα1::3xHA (shortened to pAGα1 below) (P36-P39 and P37-38). This gene includes 1,000 bases upstream of the start codon and 500 bases downstream of the termination codon, and the 3HA (P3-P4) was inserted at an introduced XbaI site at position 573/650.
Agglutination assays.
Agglutination assays of mating yeast cells were performed as described by Zhang et al. (76). Agglutination activities were expressed as an agglutination index, AI, where AI = 1 − (2 × Aa+∝)/(Aa + A∝) (66, 76). All values presented were derived from at least three independent experiments. We observed that pmt1Δ pmt2Δ cells of both mating types displayed slowed growth and elevated levels of self-aggregation in the absence of mating partners. Statistical analyses were conducted by analysis of variance with SAS, version 8.0 (SAS Institute, Cary, N.C.).
Western blot analyses.
Yeast cells were grown to mid-log phase at 30°C in liquid medium (Sc-Leu or YPAD), diluted to an optical density at 600 nm (OD600) of 0.4 in fresh medium, and allowed to grow for 4 h before pheromone induction. Aga1p was induced by treatment with α-factor at the concentration of 2 μg/ml of cells for 2 h, while Agα1p was induced by exposing cells carrying the plasmid to 0.2× their number of MATa cells (YSE430) for 1 h. Yeast cells and medium were collected after centrifugation at 6,000 × g for 10 min at 4°C. Medium-secreted proteins, total cell lysate, Quantazyme-, dithiothreitol (DTT)-, or sodium dodecyl sulfate (SDS)-releasable proteins were collected and separated on 3 to 8% Tris-acetate gels (Invitrogen, Carlsbad, Calif.), by the methods described below. All samples were boiled at 95 to 100°C for 5 min before loading. Proteins were transferred to polyvinylidene difluoride membrane by semidry blotting and detected with anti-HA, Cdc28p, or β-1,6-glucan antibodies. For HA detection, the blotted membranes were incubated with mouse monoclonal immunoglobulin G (IgG) horseradish peroxidase (HRP)-conjugated anti-HA antibody (Santa Cruz) at a concentration of 1:5,000 in 5% nonfat milk-PBST (phosphate-buffered saline [PBS], 0.5% Tween 20) overnight. The blots were then washed three times in PBST for 30 min. Detection of the HRP activity was by chemiluminescence via a Pierce SuperSignal detection kit. Blots were imaged by X-ray film or directly detected with a Kodak/NEN 440CF imaging system. Quantification of relative Aga1p/Agα1p signals was determined with densitometry software associated with the Kodak/NEN 440CF imaging system. For Cdc28p and β-1,6-glucan detection, a similar procedure was used, except for the different antibodies. For Cdc28p, the primary antibody was anti-PSTAIR used at 1:4,000 (Sigma, St. Louis), and the secondary antibody was HRP-conjugated goat anti-mouse IgG (1:8,000) (Pierce). For β-1,6-glucan detection, the primary antibody was anti-β-1,6-glucan (1:2,000; a kind gift from Howard Bussey), and the secondary antibody was HRP-conjugated goat anti-rabbit IgG (1:5,000; Pierce).
Preparation and analysis of total cell lysates, cell wall extracts, and growth medium.
Cells were grown as described above and harvested. Secreted proteins were isolated from the growth medium by centrifugation of 10 ml of yeast cells at an OD600 of 1.3 to remove the cell pellet, followed by trichloroacetic acid (TCA) precipitation of soluble proteins from the medium (13). The precipitated proteins were dissolved or titrated in 20 μl of 0.1 M NaOH, to which 80 μl of 2× LDS sample buffer (Invitrogen) was added. To produce total cell lysates, 2 ml of yeast cells at an OD600 of 1.3 was washed twice with 50 mM potassium phosphate buffer (pH 8.0) and lysed in 100 μl of 2× LDS sample buffer with glass beads. Two to 20 μl of supernatant was loaded for each sample. Cell wall extracts were prepared by a number of different methods as follows.
(i) Quantazyme-DTT-SDS extracts of intact cells.
For Quantazyme treatment, 4 ml of cells at an OD600 of 1.3 was pelleted and washed twice in wash buffer (25 mM Tris-HCl [pH 7.5], 1 M NaCl, 0.5 mM EDTA, 1 M sorbitol with protease inhibitors). The washed cells were suspended in a mixture containing 25 mM Tris-HCl (pH 7.5), 40 mM β-mercaptoethanol, 1 mM EDTA, 1 M sorbitol with protease inhibitors, and 75 U of Quantazyme (InterSpex Products, Inc.). The mixture was incubated at 37°C for 1 h with rocking, and the supernatant was collected at 5,000 × g for 5 min at 4°C. DTT extraction of intact yeast cells was carried out as described previously (9). Briefly, 4 ml of yeast cells with an OD600 of 1.3 was pelleted, washed with water and 25 mM Tris-HCl (pH 8.5), and finally resuspended in this buffer containing 2 mM DTT. Cells were shaken vigorously for 2 h at 4°C, and the supernatant was collected by centrifugation. For SDS extraction, the same amount of cells was pelleted, washed twice with 50 mM potassium phosphate buffer (pH 8.0), and resuspended in this buffer with 1% SDS and protease inhibitors. Cells were incubated for 2 h at room temperature with rocking, and the supernatant was collected. The supernatant containing the Quantazyme-, DTT-, or SDS-releasable proteins was centrifuged at 20,000 × g for 20 min at 4°C before loading.
(ii) Quantazyme extracts of isolated cell walls.
Cell walls were obtained by breaking yeast cells (60 to 80 OD600 units) mechanically by using glass beads in 0.5 ml of 50 mM potassium phosphate buffer (pH 8.0), centrifuged at 1,000 × g for 10 min to remove unbroken cells. The glass beads were washed with 0.5 ml of breakage buffer, and the wash was combined with the original supernatant. The crude cell extract was centrifuged at 15,000 × g for 30 min at 4°C, and the pelleted cell walls were washed twice in 25 mM Tris-HCl (pH 7.5), 1 M NaCl, and 0.5 mM EDTA with protease inhibitors. The washed walls were boiled twice for 5 min in 100 μl of SDS lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 2% SDS, 10 mM DTT, 5 mM EDTA). The SDS-boiled cell walls were washed and treated with Quantazyme as described above, except that no sorbitol was added to the buffer. For experiments examining proteins releasable by Quantazyme from cell wall preparations, the supernatant was incubated with 20 μl of HA antibody-agarose conjugate (Santa Cruz) at 4°C overnight with rocking. The proteins bound to the resin were treated by the procedure of the manufacturer and detected with anti-HA or β-1,6 glucan antibodies.
RESULTS
Cell surface-anchoring requirements of a model adhesin, Aga1p.
The Aga1p:Aga2p-Agα1p adhesion system has been well dissected at the molecular level. Studies of Aga1p, however, remain incomplete with respect to the roles of GPI-glucan interactions, O-linked glycosylation, and the potential roles of conserved WCPL and CX4C domains in fungal adhesion. To address these questions, we developed strains and plasmids to enable quantitative assays of Aga1p function in agglutination (Tables 1 and 2). A deletion of the AGA1 gene, aga1Δ, was created in the BY4743 strain background and a centromeric plasmid-borne, 3HA epitope-tagged copy of AGA1 was constructed. The plasmid created, pRS315-AGA1::3xHA (pAGA1 hereafter), was then transformed into aga1Δ strains. Agglutination activity measurements showed that the epitope-tagged protein in aga1Δ strains supports an activity level equivalent to that shown by the endogenous Aga1 protein present in wild-type MATa cells. Immunoblotting of whole-cell lysates and plasma membrane or cell wall extracts indicates that the tagged form of the protein is present at induced levels in pheromone-treated or mating cells, like native Aga1p (51).
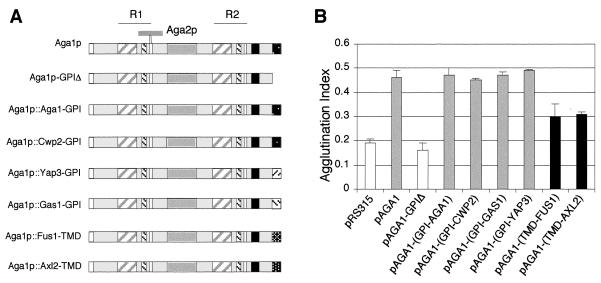
To test the relevance of the GPI processing events for Aga1p-Aga2p adhesin complex function, we used the pAGA1construct to produce tagged Aga1p derived chimeras, deletions, and insertion mutants (Materials and Methods). We first used the constructs to examine the abilities of different chimeric constructs to complement an aga1Δ strain for mating cell agglutination activity. In the chimeric Aga1 proteins shown in Fig. 1A, the native Aga1p GPI signal was deleted or exchanged for carboxy-terminal sequences specifying GPI addition and processing for three other fungal cell surface glycoproteins. A modified pAGA1 that lacks its final 67 amino acids, including the GPI anchor-specifying domain, was generated by replacing these sequences with a unique PstI site followed by a translational stop codon. Four different GPI-specifying sequences derived from the final 67 amino acids of Cwp2p, Yap3p, Gas1p, and Aga1p (as a positive control for the fusion modification) were then cloned into this construct. Cwp2p, Yap3p, and Gas1p differ in the processing of their GPI anchors. Cwp2p is efficiently processed and linked to the glucan layer (69), while Yap3p and Gas1p are expected to remain associated predominantly with the plasma membrane (1, 10, 27-29, 46, 68, 71). If the Aga1p::Cwp2-GPI fusion was active while the Aga1p::Gas1-GPI and Aga1p::Yap3-GPI fusions were not or showed reduced activity, this would support the hypothesis that glucan linkage is required for or contributes to the activity of GPI-anchored adhesins. Alternatively, should the Aga1p::Gas1-GPI and Aga1p::Yap3-GPI chimeras be fully functional, this would indicate that β-1,6 attachment of adhesins may not be a leading factor contributing to their normal activity on the fungal cell surface.
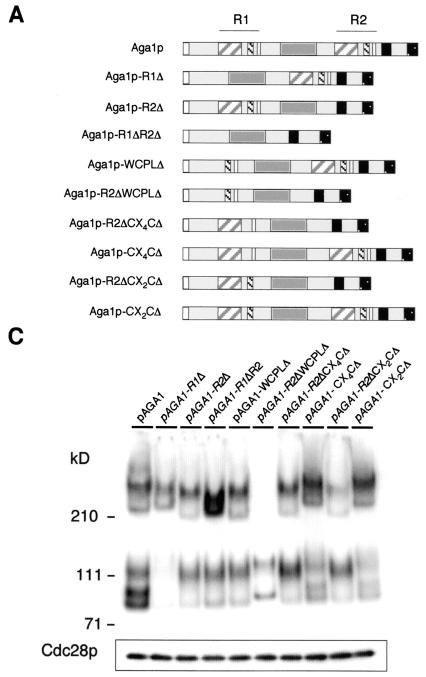
FIG. 1.
Aga1 proteins carrying heterologous GPIs or TMD anchors are functional. (A) Schematics of different chimeric Aga1 proteins. The 725-amino-acid Aga1p precursor contains a secretion signal at its N terminus and a GPI attachment signal at its C terminus. R1 and R2 repeats include amino acids 53 to 149 and 395 to 493, respectively. For each repeat, the WCPL domain (gray hatched box), CX4C motif (black hatched box), and CX2C motif (white box) are shown. A predicted interaction region between Aga1p and Aga2p is also indicated. The 3HA epitope tag inserted carboxy terminal to the R2 repeat is shown (black bar). The region between R1 and R2 contains 19 copies of an imperfect heptapeptide sequence, TSTSS/PSS (dark gray bar). In Aga1-GPIΔ, the last 67 amino acids were deleted from Aga1p and replaced with a unique PstI site in which the different GPIs and TMDs were subsequently introduced. In the different Aga1p chimeric proteins, the missing GPI signal was replaced by the final 67 amino acids that include the GPI signal of each protein indicated or by a TMD from Fus1p or Axl2p. The original GPI signal was also reintroduced as a positive control. GPI signal domains predicted to maintain residence of the protein predominantly in the plasma membrane (Yap3 and Gas1) or the cell wall (Aga1 and Cwp2) are depicted with similar fill patterns. For all of the constructs, the amino terminus is to the left; some elements are not to scale due to their small size relative to the total protein. (B) Agglutination activities of different Aga1p chimeras. MATa aga1Δ cells (YSE913) were transformed with the different constructs shown in panel A and assayed for agglutination levels in matings to wild-type MATα cells (YSE431). The data shown are the mean ± standard error of three independent experiments. Columns in the histogram are shaded according to statistical significance, with the same shading indicating no significant differences at α = the 0.5 level.
We quantified the agglutination activities supported by the different chimeras as shown in Fig. 1B. As expected, and consistent with previous studies involving deletion of the anchor-specifying sequences of Sag1/Agα1p (75), we observed that deletion of the anchor signal domain of Aga1p eliminated all activity associated with the protein. Immunoblot experiments confirmed (as seen with Sag1/Agα1p) (75) that the truncated anchorless protein is secreted through the cell wall into the culture medium (see Fig. 4C). Restoration of the Aga1p GPI signal domain to this clone restored wild-type agglutination levels. Examination of the levels of agglutination activity associated with the three Aga1::Cwp2p, Aga1::Gas1p, and Aga1::Yap3p anchor domain chimeras showed that these clones each supported wild-type agglutination activity levels. Thus, the processing and attachment of the GPI anchor to β-1,6 glucan in the cell wall may not be a prerequisite for normal levels of Aga1p activity.
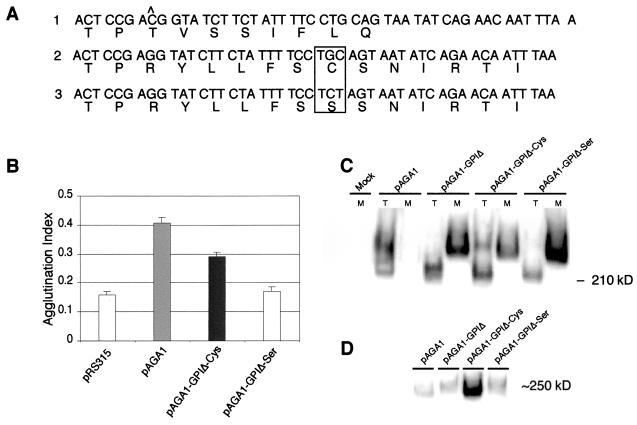
FIG. 4.
A mutant form of Aga1p anchored by a carboxy-terminal cysteine is active. (A) Predicted amino acid sequences of the carboxy termini of proteins encoded by the pAGA1-GPIΔ, pAGA1-GPIΔ-Cys, and pAGA1-GPIΔ-Ser constructs. The cytosine base in the wild-type sequence that is deleted in pAGA1-GPIΔ-Cys is indicated by ∧, and the cysteine of pAGA1-GPIΔ-Cys mutated to serine in pAGA1-GPIΔ-Ser is boxed. (B) Agglutination activities of YSE913 MATa aga1Δ strains transformed with the different plasmid constructs described in panel A. The data and representation of significance are as defined in the legend to Fig. 1B. (C) Immunoblot analysis of Aga1 proteins present in total cell lysates (T) or secreted into the culture medium (M) for the different proteins described in panels A and B. (D) Immunoblot analysis of DTT-releasable Aga1 proteins from the different constructs.
One caveat to the preceding experiment is that a recent study involving Gas1p and Yap3p has called into question the assertion that GPI-anchored proteins are sorted between either the plasma membrane or cell wall compartments due to their precursor sequences, which control modification (17). Since our results with the different GPI signals predicted to remain plasma membrane associated suggested that membrane-anchored Aga1-Aga2p complexes possessed wild-type agglutination activities, we next produced chimeras that were forced to be plasma membrane resident via the substitution of a single TMD for the GPI anchor signals. These Aga1 chimeras were constructed by using the TMDs of the type I plasma membrane proteins Fus1p and Axl2p and tested for their agglutination activities (Fig. 1A and B). As shown, these constructs both support partial agglutination activity in comparison to wild-type AGA1 and null vector control plasmids. Thus, adhesins that normally require a GPI anchor can function when retained at the cell surface plasma membrane through the presence of a transmembrane domain.
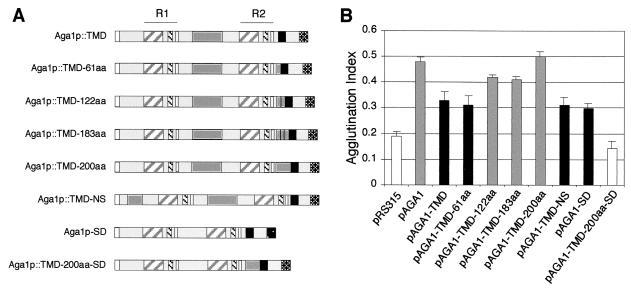
Our finding that the Aga1p::Fus1-TMD and Aga1p::Axl2-TMD chimeras are partially functional suggested—consistent with the original hypothesis that GPI-glucan transfer facilitates exposure of functional domains of adhesins to cell wall positions closer to the exterior surface of the wall—the length of the protein might similarly affect its function in the TMD-tethered context. To examine whether the size of these proteins would influence their activities, a second series of clones were constructed, inserting increasingly longer spacer domain sequences C terminal to the R1 domain of the Aga1p::Fus1-TMD chimera (Fig. 2A). The spacer domain for these clones was derived from the sequences occurring between the two major repeated domains of Aga1p, R1 and R2 (51). As Fig. 2B indicates, as the length of these membrane-resident Aga1 chimeras increases, their activity does so as well. Clones containing a spacer domain of at least 122 amino acids in length display a 30% increase in activity (86% of the activity level of wild-type Aga1p), while a chimera containing 200 inserted amino acids was found to have agglutination activity equivalent to that of the wild-type GPI-anchored Aga1p. In sum, these experiments indicate that Aga1p-Aga2p adhesin complexes can be fully functional when tethered directly to the cell surface plasma membrane, with the length of these membrane-resident adhesins being a critical modulator of their activities.
FIG. 2.
Agglutination activity of TMD-anchored Aga1p correlates with protein length. (A) Schematics of the different spacer containing constructs tested, as defined in the legend to Fig. 1A. Spacer 1 (amino acids 315 to 375/725) and spacer 2 (amino acids 176 to 375/725) were PCR amplified from coding sequences corresponding to the region between R1 and R2 and inserted into the Aga1p::Fus1-TMD construct immediately preceding the 3HA epitope tag. Tandem dimers and trimers of spacer 1 were also generated and inserted into the Aga1p::Fus1-TMD construct at the same location. For the N-terminal spacer-containing construct, spacer 2 was inserted at amino acid 33/725. Spacer deletion constructs were created by deleting the spacer 2 element (amino acids 176 to 375) from Aga1p and from Aga1p::TMD-200aa. (B) Agglutination activities of different TMD-anchored Aga1 proteins containing spacer elements of different lengths. The data and representation of significance are as defined in the legend to Fig. 1B.
The data presented above indicate that the agglutination activity increases as the spacer becomes longer when it is present carboxy terminal to the R1 domain. To test whether the position of the spacer relative to the R1 domain is important, the 200-amino-acid spacer was inserted amino terminal to the R1 domain, and the agglutination activity was measured. As shown in Fig. 2B, this agglutinin showed the same activity level as that of the pAGA1-(TMD-FUS1) construct. Thus, the positions of spacer elements relative to the R1 domain within the adhesin Aga1p are critical to the function of the protein.
To determine whether the 200-amino-acid region between the R1 and R2 domains, from which the previous spacers used in the Aga1p chimeric proteins described above were derived, plays any role in the function of the wild-type, GPI-anchored Aga1p, this domain was deleted from pAGA1. The resulting construct showed partial agglutination activity when transformed into the MATa aga1Δ strain compared with the parent pAGA1. Such a result is consistent with the notion that this domain functions as a spacer element. When the R1-R2 interdomain spacer was deleted from pAGA1-(TMD-FUS1)-200aa, the resulting construct, possessing the same length as pAGA1-(TMD-FUS1) and still containing an introduced 200-amino-acid spacer at residue 540/725, completely lost agglutination activity. These experiments indicate that the repetitive region between R1 and R2 plays an important role in the function and/or structure of the wild-type Aga1p as a spacer region that may facilitate exposure of the R1 domain. Additionally, the position of the spacer element relative to the R1 and R2 repeats within Aga1p also appears to be critical to the structure and function of the protein.
Cell wall and plasma membrane localization of wild-type Aga1p and chimeric forms.
The preceding analyses of Aga1p chimeras and domain deletion constructs further highlight the importance of determining the localization of adhesins in the plasma membrane or cell wall and whether sequences found within such proteins mediate their secretory traffic or postsecretory movement among cell surface compartments. To better characterize the trafficking requirements of Aga1p and to confirm the compartment localization of the various anchor chimeras, we carried out fractionation and immunoblotting analyses of the strains expressing these different proteins.
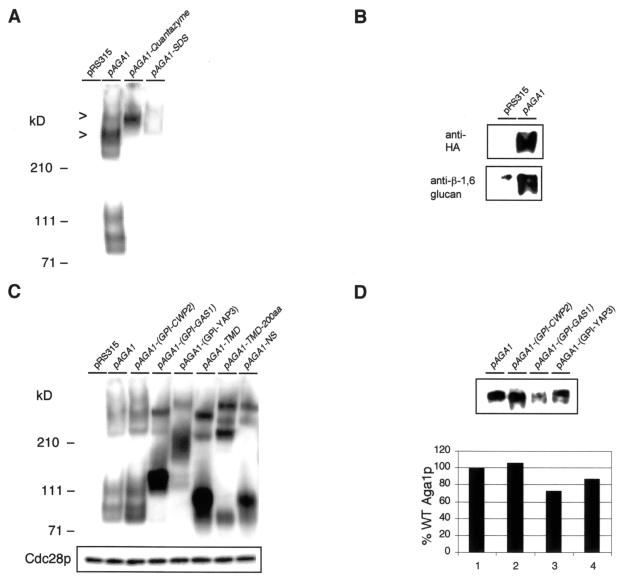
As shown in Fig. 3A, the HA epitope-tagged Aga1p is detected specifically in total cell lysates of pheromone-treated MATa cells (compare lane 1, pRS315 control, to lane 2, pAGA1). We also observed the protein to be present at relatively substantial levels in untreated cells and to increase in abundance in a time course of α-factor induction, consistent with previous studies of Aga1p (51). Three different, fully glycosylated high-molecular-mass forms of the protein (all greater than 210 kDa) were observable in the total cell lysates (Fig. 3A, lane 2, pAGA1). Of these different forms, the highest-molecular-mass form tends to be nonabundant or poorly detected in most total cell lysates. We interpret these forms shown in Fig. 3A to represent the following Aga1p species based on their sizes and extraction properties. Aga1p is initially expected to be present in the plasma membrane by virtue of its GPI lipid anchor following delivery via secretion. If Aga1p undergoes processing in the manner of Sag1/Agα1p and other cell wall-linked proteins, a processed, smaller, periplasmic form of the protein will be generated by cleavage of the GPI anchor. Following cleavage, this form of the protein would be available for interaction with the cell wall and covalent linkage of the GPI anchor remnant to β-1, 6 glucan. Thus, we interpret the three high-molecular-mass forms of Aga1p observable in total cell lysates to be a plasma membrane-anchored form (middle species), a processed periplasmic form (lower-molecular-mass form), and a cell wall-linked form (highest-molecular-mass form).
FIG. 3.
Aga1p is a GPI-anchored plasma membrane and cell wall-linked protein. (A) Immunoblot detection of Aga1p in total cell lysates and material releasable from intact cells by Quantazyme or SDS extraction. Total cell lysates of aga1Δ cells carrying either an empty vector (pRS315) or the 3HA-tagged Aga1p-encoding plasmid (pAGA1) were obtained and immunoblotted as described in Materials and Methods. Quantazyme and SDS treatment of intact aga1Δ cells carrying pAGA1 released the highest- and middle-molecular-mass species of Aga1p (marked by >). (B) Immunoblot of Quantazyme-releasable Aga1p from purified cell wall materials. The Quantazyme-releasable proteins were immunoprecipitated with HA agarose beads and detected with either anti-HA or anti-β-1,6 glucan antibodies. (C) Immunoblot of total cell lysates derived from strains expressing the chimeric Aga1 proteins shown in Fig. 1A and 2A. Twenty-microliter samples were loaded for lanes 1, 2, 3, and 8, and 2-μl samples were loaded for lanes 4 to 7. A separate blot of the same lysates (2 μl) was probed with anti-PSTAIR antibodies and detected for the levels of Cdc28p as a control for equivalence of total protein in lysates. (D) A pool of Aga1 proteins cross-reacts with antibody specific to the cell wall β-1,6 glucan in all of the GPI-anchored Aga1p chimeras. Samples were treated similarly, as indicated in panel B. The upper panel shows a representative image of the immunoblot analysis. Lanes are loaded with equivalent amounts of immunoprecipitated material, except lane 3, the loading of which is ∼0.8× that of the other samples due to a correspondingly reduced starting amount of cells used for extraction. The lower panel shows the amount of Quantazyme-releasable Aga1p in the chimeras relative to the wild-type (WT) Aga1p. The data shown are the average from two independent experiments; levels of the Aga1p::Gas1-GPI are normalized to the other samples. Histogram columns 1 to 4 represent pAGA1, pAGA1-(GPI-CWP2), pAGA1-(GPI-GAS1), and pAGA1-(GPI-YAP3), respectively.
To confirm the nature of the three different high-molecular-mass forms of Aga1p, we used extraction and fractionation methods to examine the solubility of the three Aga1p species. Intact cells were harvested and treated with SDS or Quantazyme, a β-1, 3 glucanase, as indicated in Materials and Methods. When intact cells were treated with Quantazyme, the highest-molecular-mass species of Aga1p shown above was released, which suggests that it is a cell wall-resident form (Fig. 3A, lane 3). As predicted by the processing pathway and size, the middle species (lower band in Fig. 3A, lane 4) is preferentially extracted by the detergent SDS, suggesting that it is a plasma membrane-anchored form (Fig. 3A, lane 4). A higher-molecular-mass species was also released by SDS; however, it is not clear whether this species represents a noncovalently bound cell wall form or a distinct, higher-molecular-mass plasma membrane-resident form. We also found that the middle- and highest-molecular-mass species can be partially released from intact cells by the reducing agent DTT (data not shown). Both SDS treatment and reducing conditions have previously been shown to extract different classes of cell wall-resident proteins (37). When purified cell wall fractions were prepared by sequential extraction with boiling SDS-containing buffer followed by treatment with Quantazyme, a fourth, still higher-molecular-mass species (>300 kDa) of Aga1p was released. It is predominantly, if not only, this form of Aga1p that cross-reacts with antibodies specific to β-1, 6 glucan (Fig. 3B). Thus, our data indicate that at least four forms of Aga1p are present at the surface of pheromone-treated cells. These forms are present in the plasma membrane and cell wall compartments, with a β-1, 6 glucan-linked form detectable only following Quantazyme treatment of purified cell wall material previously subjected to SDS extraction and reducing conditions. These properties are similar to those reported for other covalently linked fungal cell wall proteins carrying processed GPI anchor remnants (20, 28, 29, 37, 44, 65, 69).
Because we found that each of the different Aga1 chimeras carrying the GPI anchors of Cwp2p, Gas1p, or Yap3p and a Fus1p-TMD-200aa-anchored form confer full Aga1p-Aga2p complex activity in agglutination, we also examined the levels and cell wall localization of these different forms. Immunoblot analyses of the constructs tested in the experiments shown in Fig. 1 and 2 demonstrated that all of the chimeric constructs were expressed and present in cells at levels equivalent to, or in the case of TMD and membrane GPI-containing chimeras, ∼10- to 20-fold greater than that of Aga1p in wild-type cells. In these experiments, we consistently observed that TMD-anchored Aga1 chimeras and those containing GPI anchors predicted to remain predominantly in the plasma membrane (Gas1p, Yap3p) were present in greater abundance than the wild-type Aga1p (Fig. 3C). At present, we do not know whether this apparent difference is due to differential stability of the chimeras in question or because they may transit secretory compartments more rapidly or efficiently than the wild-type Aga1p. We examined whether such cells were similar or less active in agglutination under diluted-cell conditions in which wild-type Aga1p-bearing cells display ∼50% of the agglutination activity seen under the standard, higher-cell-density assay conditions we employed for our studies. No apparent differences were seen among any of these strains and wild-type strains: all demonstrated a reduction in activity approximately similar to that of the wild-type strain (data not shown). Thus, despite their relatively greater abundance at the cell surface, at present we have no evidence to suggest that any of the GPI- or spacer-containing TMD-anchored Aga1p chimeras are intrinsically less active than the wild-type Aga1p.
When the different GPI-anchored chimeras were examined for their levels of β-1,3 glucanase-releasable Aga1p, similar amounts of each cross-reacted with antibody specific to β-1, 6 glucan, indicating linkage to the cell wall (Fig. 3D). This suggests that postsecretory processing of GPI anchors occurs in both the cell wall and predicted plasma membrane GPI-anchored Aga1 chimeras constructed in this study. Since >10-fold more Aga1p was detected in total cell lysates derived from the plasma membrane GPI-anchored chimeras, the efficiency of this postsecretory processing is apparently much lower for proteins containing plasma membrane GPI anchors. The TMD-anchored chimera was observed to completely lack a pool of Aga1p cross-linked to β-1, 6 glucan (data not shown), consistent with its absence of a suitable GPI remnant on this protein to function as an acceptor for covalent linkage to the cell wall. These results lend support to the notion that the localization efficiencies of GPI-anchored proteins in the cell wall and plasma membrane represent a continuum ranging from predominantly cell wall resident to predominantly membrane resident. Thus, our studies are consistent with a cell wall active form of the native Aga1p being present on pheromone-treated cells, which arises through postsecretory processing and traffic events.
A disulfide-dependent linkage mechanism also supports Aga1p agglutination functions.
In the process of preparing the GPIΔ construct described in the preceding section, a construct encoding an Aga1 protein with a truncated and slightly altered carboxy-terminal domain was serendipitously produced. This mutant, hereafter denoted pAGA1-GPIΔ-Cys, was originally designed to truncate the final 67 amino acids from Aga1p, as does the pAGA1-GPIΔ construct. Initial agglutination activity tests of the pAGA1-GPIΔ-Cys mutant indicated that the encoded protein possessed moderate levels of agglutination activity (described below). We determined the DNA sequence of this clone and discovered a frameshift mutation that was inadvertently encoded by an oligonucleotide primer (presumably a failure sequence from the original synthesis) used to generate the clone. The frameshift produced by the pAGA1-GPIΔ-Cys sequence alters the AGA1 open reading frame in a manner that rapidly leads to the truncation of the reading frame, not, however, before encoding a single cysteine residue near the predicted carboxy terminus of the protein, as shown in Fig. 4A. Because initial tests of this construct indicated that it possessed partial agglutinin activity, and because initial studies of Aga1p cell wall residency indicated that a fraction of the protein was releasable from intact cells by treatment with reducing agents, we proceeded to mutate the cysteine residue by creating a cysteine-to-serine substitution as shown in Fig. 4A. These constructs were tested to determine the agglutination activities they were able to support as depicted in Fig. 4B. The results demonstrated that the carboxy-terminal cysteine-containing clone, pAGA1-GPIΔ-Cys, possessed 54% of the level of wild-type pAGA1 activity. In contrast, the Aga1 protein containing the Cys-to-Ser substitution as encoded by pAGA1-GPIΔ-Ser had a null level of activity indistinguishable from that of pAGA1-GPIΔ or vector-only-containing cells. Thus, the Aga1p-Aga2p adhesin complex can be functionally anchored to the cell surface by a single disulfide linkage near the carboxy terminus of Aga1p.
To investigate the localization of the GPIΔ, -Cys, and -Ser frameshifted Aga1 proteins, we performed immunoblots of the culture medium and total cell lysate fractions from cells carrying these constructs. In the strain containing the wild-type pAGA1, very little Aga1p is secreted into the growth medium. In contrast, essentially all of the cell wall form of Aga1p is secreted into the medium in the strains containing pAGA1-GPIΔ or pAGA1-GPIΔ-Ser (Fig. 4C). The strain containing pAGA1-GPIΔ-Cys retains at the cell surface about one-third of the amount of the cell wall form observed for the wild-type Aga1p, consistent with the partial agglutination activity conferred by this construct. This anchorage occurs through the introduced Cys residue at the C terminus of the truncated Aga1 protein, because 10-fold more Aga1p is released by DTT from pAGA1-GPIΔ-Cys than from the wild-type pAGA1 and pAGA1-GPIΔ-Ser (Fig. 4D).
Domains of Aga1p required for function in agglutination.
Fungal adhesins are frequently characterized by the presence of repeated domains that are often rich in serine and threonine residues that represent potential sites of O-linked glycosylation. The structural significance of most of these domains for the function of the proteins that contain them remains to be determined. Roy et al. (51) noted the repeated structure of the agglutinin Aga1p in their cloning and characterization of the gene. Aga1p contains two ∼100-amino-acid-long repeated regions designated R1 and R2 separated by a central region containing ∼19 copies of a serine-threonine-rich heptapeptide sequence, TSTSS/PSS (Fig. 1A). Sharkey et al. (55) reported the sequence of the C. albicans HWP1 gene and predicted protein and noted the presence of a well-conserved WCPL domain present in a number of other adhesins, including Aga1p. This 21- to 49-residue-long domain is characterized by the presence of several highly conserved threonine residues spread throughout the domain, and the tetrapeptide sequence W/YCPL at its carboxy terminus. In further examining the sequence elements of this domain, we found some of these domains contain a conserved pair of cysteine residues separated by 4 amino acids (CX4C) and located in the amino-terminal portion of the domain. The sequences of the adhesins Aga1p and Fig2p also contain the CX4C motif; however, it is present in these proteins at positions separate from and carboxy terminal to the WCPL domains; in Aga1p, it is found as part of the R1 and R2 domain elements. This suggests that the CX4C and WCPL domain elements, although they may perform a common or related function in these proteins, appear to be structurally independent elements in these proteins. In addition to these elements, in Aga1p, the R1 and R2 domains also contain a second CX2C cysteine pair.
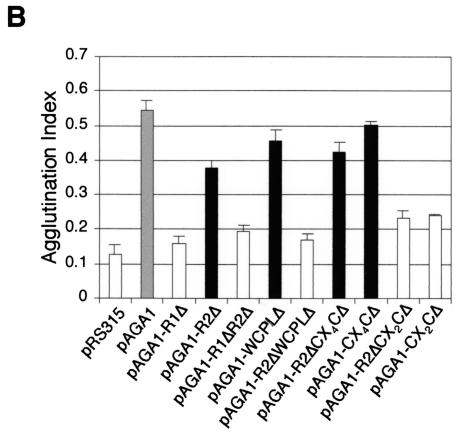
To better understand the functional significance of these different domains in Aga1p to its activities in agglutination, we constructed several deletion derivatives that lacked these different elements (Fig. 5A). We assayed the agglutination activities associated with each of these constructs as shown in Fig. 5B. Our results showed that deletion of the R1 domain or the CX2C element within this domain eliminated all agglutinin activity. Our results also show that the R2 domain is only partially dispensable for agglutination activities, because we found that elimination of the R2 domain led to an ∼30% reduction in agglutination activity of Aga1p. These results would be most consistent with the R2 domain facilitating extension of the molecule and therefore the exposure of the R1 domain in more peripheral cell wall domains closer to the surface of the wall.
FIG. 5.
Functional domains of Aga1p R1 and R2 as revealed by deletion analysis. (A) Schematics of deletion derivatives lacking different elements, as defined in the legend to Fig. 1A. (B) Agglutination activities of the different Aga1p mutants shown in panel A. White columns indicate <25% of wild-type Aga1p-mediated agglutination activity, black columns indicate >50% of wild-type activity, and the gray column represents wild-type activity. (C) Immunoblot of total cell lysates derived from the deletion mutants shown in panels A and B. A separate blot of the same lysates was probed with anti-PSTAIR antibodies and detected for the levels of Cdc28p as a control for equivalence of total protein in lysates.
To assess the roles of individual domain elements within the R1 repeat, we constructed deletions of the R1 WCPL, CX4C, and CX2C domains individually and in combination with the deletion of the entire R2 domain. The second set of constructs provides the context to assess the roles of these domains in the absence of potential redundant contributions from cognate elements within the R2 domain. Figure 5B shows that deletion of the R1 WCPL domain alone produces a small reduction in adhesin activity. Similarly, significant effects were not associated with removing the CX4C element, either alone or in combination with the R2 domain. When the R1 WCPLΔ and R2Δ mutations are combined, Aga1p adhesin function is completely eliminated. This suggests that either a specific redundant and essential function exists for the two WCPL domains, or the combined deletion of both these domains alters the structure of Aga1p such that it is either too short to function at the cell surface in displaying Aga2p, or it is no longer properly trafficked by the secretory pathway.
Next, we used immunoblotting to check whether partial reductions in agglutinin function associated with loss of the WCPL- or cysteine-containing elements or the entire repeated domains R1 or R2 are the result of defects in biogenesis or trafficking of Aga1p. Analysis of the truncated proteins present in total cell lysates (Fig. 5A) indicated that Aga1p with deletion of both the R1 WCPL and R2 domains was retained in the endoplasmic reticulum and Golgi, because no high-molecular-mass form of this protein was detectable in cell lysates (Fig. 5C). This suggests that deletion of both these domains shortens or dramatically alters the structure of Aga1p such that it is no longer properly trafficked by the secretory pathway. None of the other truncated proteins, including that encoded by the R1ΔR2Δ construct, displayed substantial trafficking problems, manifested as the similar amounts of cell wall and plasma membrane forms of these different mutant Aga1 proteins compared to the amount of the wild-type Aga1p. In sum, these data indicate that the R1 CX2C and WCPL domains are necessary for complete activity of the Aga1 adhesin, with the former playing a critical role in its function.
Cell type-dependent and -independent agglutinin requirements for O-linked modifications.
The Aga1p-Aga2p a-agglutinin complex is composed of proteins that have been shown to undergo modification in the form of O-linked glycan addition to the many serine and threonine residues found in these proteins. This is particularly the case for Aga1p, which is composed of ∼50% Ser/Thr residues (51). Because the conserved WCPL domain and frequently one of the residues internal to the CX4C cysteine pair, as well as the 19 heptapeptide repeats found between the R1 and R2 repeats, are all characterized by the presence of these hydroxyl-containing amino acids, we sought to determine whether this posttranslational modification was important to the function of the Aga1p-Aga2p a-agglutinin complex. The S. cerevisiae genome encodes seven open reading frames similar to those coding for protein mannosyltransferases (PMTs), represented by the genes PMT1 to PMT7 (61). These enzymes are responsible for addition of the first mannose of an O-linked chain to the hydroxy amino acid side chain, and their substrate specificities, while still relatively poorly understood, have recently begun to be characterized (23, 24, 61). These previous studies indicated that among the PMT1, PMT2, PMT3, and PMT4 deletion mutants tested, only PMT1 and PMT2 appeared to play roles in the glycosylation of Aga2p.
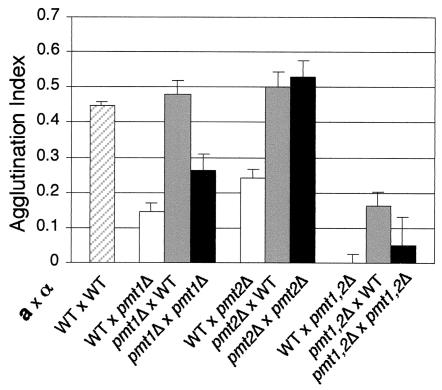
To investigate the effects caused by the absence of different protein mannosyltransferases genes on a- and α-agglutinin activities and glycosylation of Aga1p and Sag1/Agα1p, we tested the agglutination abilities of mutants in all seven of the PMT genes both in unilateral (a pmtΔ × α WT [wild type] and a WT × α pmtΔ) and bilateral (a pmtΔ × α pmtΔ) agglutination reactions. Similar to other studies failing to find vegetative growth phenotypes associated with the loss of the PMT5, PMT6, or PMT7 genes (61), we found no significant differences relative to wild-type strains in agglutination levels in either unilateral or bilateral contexts for strains missing these genes or PMT3 or PMT4 (data not shown). In examining the effects of the loss of the PMT1 and PMT2 genes, we were surprised to find no effects on agglutination levels due to the absence of these genes individually from MATa cells, but there was a strong effect caused by the loss of each of these genes from MATα cells (Fig. 6). Analysis of cells carrying both mutations (pmt1Δ pmt2Δ double mutants) demonstrated a collective and apparently redundant requirement for the PMT1 and PMT2 genes in both MATa and MATα cells, as evidenced by the effects seen in both unilateral and bilateral crosses involving these strains. These data strongly suggest that these two gene products act redundantly on the a- and α-agglutinin components and that a greater functional requirement exists in MATα cells for proper glycosylation of the α-agglutinin.
FIG. 6.
Mating type-dependent and -independent agglutination defects of strains mutated in either or both PMT1 and PMT2. Agglutination indices are shown for both bilateral and reciprocal unilateral matings of wild-type (WT), pmt1Δ, pmt2Δ, and pmt1Δ pmt2Δ mutants. For each mating, the first partner listed is the MATa strain and the second strain is the MATα strain, as indicated to the lower left of the histogram.
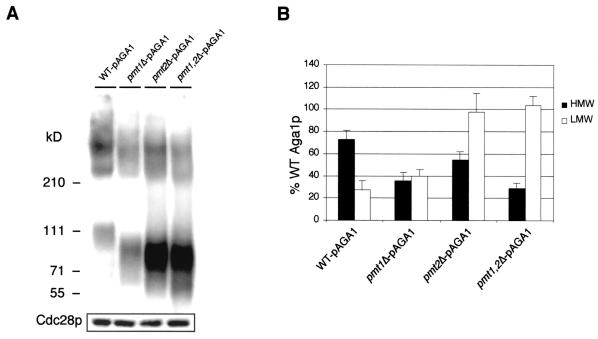
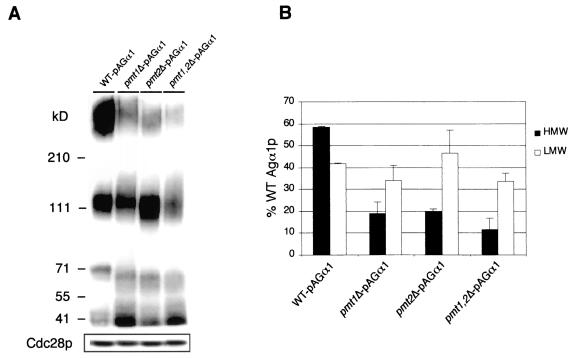
To confirm that the PMT1 and PMT2 genes play a role in glycosylation of Aga1p, we transformed MATa wild-type, pmt1Δ, pmt2Δ, and pmt1Δ pmt2Δ strains with the pAGA1 plasmid and immunoblotted SDS-polyacrylamide gel electrophoresis-separated total cell lysates as described in Materials and Methods. The results shown in Fig. 7A confirmed that both Pmt1p and Pmt2p are involved in the O-glycosylation of Aga1p, as manifested by the marked decrease in high-molecular-mass Aga1p and increase in low-molecular-mass, immature forms of Aga1p in the pmt mutants. Quantification of the high- and low-molecular-mass Aga1p in different pmt mutants revealed that 1.5- to 4-fold more low-molecular-mass Aga1p exists in the pmt mutants (Fig. 7B).
FIG. 7.
Aga1p is a substrate of Pmt1p and Pmt2p. (A) Immunoblot of total cell lysates from wild-type (WT) and pmt mutant strains transformed with pAGA1. A separate blot of the same lysates was probed with anti-PSTAIR antibodies and detected for the levels of Cdc28p as a control for equivalence of total protein in lysates. (B) Quantification of Aga1p from the wild type and pmt mutants. The data shown are based on three independent immunoblots, and bars indicate standard errors. The total cellular pool of Aga1p, including high-molecular-mass (HMW; >210 kDa) and low-molecular-mass (LMW; 70 to 120 kDa) forms, from the wild-type strain was normalized to 1.0, and the amount of each form in the pmt mutant cells was expressed as a percentage of this value.
To test whether Pmt1p and Pmt2p are, as would be expected from the preceding agglutination activity data, involved in O-glycosylation of Agα1p, a 3HA-tagged Agα1p was constructed in the yeast centromeric vector pRS315. This plasmid, pRS315-AGα1::3HA (pAGα1 hereafter), restored full agglutination activity to a MATα agα1Δ strain (data not shown). pAGα1 was transformed into MATα wild-type, pmt1Δ, pmt2Δ, and pmt1Δ pmt2Δ strains, and immunoblots were used to detect, in total cell lysates, whether Pmt1p and Pmt2p are responsible for O-glycosylation of Agα1p. As shown in Fig. 8A and B, at least twofold less high-molecular-mass Agα1p was found in pmt1Δ, pmt2Δ, and pmt1Δ pmt2Δ strains, confirming that Pmt1p and Pmt2p O-glycosylate Agα1p.
FIG. 8.
Sag1/Agα1p is a substrate of Pmt1p and Pmt2p. (A) Immunoblot of total cell lysates from wild-type (WT) and pmt mutant strains transformed with pSAG1/AGα1. A separate blot of the same lysates was probed with anti-PSTAIR antibodies and detected for the levels of Cdc28p as a control for equivalence of total protein in lysates. (B) Quantification of Sag1/Agα1p from the wild type and pmt mutants. The data shown are based on three independent immunoblots; bars indicate standard errors. The total cellular pool of Sag1/Agα1p, including high molecular mass (HMW; >250 kDa) and low molecular mass (LMW; 100 to 150 kDa) forms, from the wild-type strain was normalized to 1.0, and the amount of each form in the pmt mutant cells was expressed as a percentage of this value.
DISCUSSION
Anchorage mechanisms of fungal adhesins.
The overwhelming majority of fungal adhesins studied to date localize to the cell surface through the presence of GPI anchors predicted to be processed and subsequently cross-linked to β-1,6 glucan. This mechanism has apparently been successfully adopted by a wide variety of adhesins that bind to proteins, extracellular matrix carbohydrates, or present substrate domains for covalent transglutamylating reactions (14, 32, 33, 40, 62, 63, 65). In this study, we show that fungal adhesins can be partially or fully functional when cell surface anchored by at least three “alternative” mechanisms. Chimeric and fully functional Aga1 proteins were constructed that bear GPI anchors predicted to be plasma membrane rather than cell wall resident. Aga1 proteins anchored in the plasma membrane due to the introduction of single-pass, type I transmembrane protein domains were also found to be active. Partially functional Aga1p mutants were generated that contained a single carboxy-terminal cysteine residue capable of forming a disulfide with cell wall-resident components.
The alternative anchoring mechanisms for adhesins we document rely on both noncovalent and covalent linking mechanisms, whose roles have been well described in localizing fungal proteins in the cell wall (37, 39, 47). In addition, secreted proteins containing TMDs will, depending on their topology, project any number of extracellular domains above the plasma membrane. Depending upon signal peptide processing events and the relative locations of subsequent TMDs, such extracellular domains may be quite extensive in size, an example of which are the cell wall-resident, extracellular domains of the Wsc-related family of sensor proteins for cell integrity signaling pathways in budding yeasts (70). Our results with different Aga1p::TMD chimeric proteins indicate that such a cell surface mechanism is also feasible for adhesins.
Our finding that TMD and disulfide-anchored adhesins are capable of partial-to-normal levels of adhesin function suggests that, given the appropriate protein structure, such an anchoring mechanism can be utilized by natural adhesins of some fungal species. Thus, alternative adhesin anchoring mechanisms could underlie to a potentially significant extent some of the activities observed in other fungal cell adhesion contexts, both in budding yeast and in other species, that are yet to be characterized at the molecular level. For example, a wide variety of adhesins and adhesin-related proteins have been described in the human commensal pathogen C. albicans and its related species. Where the contributions of individual adhesin genes to fungal cell-host cell adherence have been quantified, residual adherence interactions in strains deleted of individual components are often observed. Thus, these redundancies indicate the potential for a variety of proteins, both known and unknown, to participate in producing adhesion levels characterizing wild-type strains.
The role of GPI anchor-β-1,6 glucan interactions in promoting Aga1p agglutinin activity.
The rationale for studying the anchorage of certain glycoproteins to the glucan layer of the fungal cell wall began with the hypothesis that fungal adhesion-mediating proteins are likely to require this modification to be effectively displayed on the surface of the cell wall (43). If these proteins remained anchored on the plasma membrane, even if their polypeptide backbone were fully extended, they might be unable to project beyond the glucan layer to mediate adhesion (43). Thus, an effort was made to identify glucan-linked forms of these GPI-anchored proteins. Studies by P. Lipke and colleagues (43, 44) first documented the existence of forms of the MATα cell agglutinin Sag1/Agα1p that are part of a postsecretory pathway for the maturation of this protein leading to its localization in the β-1,6 glucan of the cell wall. More recent studies have determined the cis elements of the GPI anchor addition signal sequences preferred by the processing pathway components for designating a protein for anchor modification and the efficiency with which different anchors may be processed (27-29, 69, 71). Taken together with the observation that many adhesins in budding yeast may be predicted to undergo addition of GPI anchors that are processed in this manner (10, 27), these observations suggest an underlying importance for this processing event for the efficient display of cell surface adhesins. While this argument appears attractive, a direct test of the requirement of this processing event for the normal activity of an adhesin had never been made, and the necessity for such a pathway for Sag1/Agα1p has been questioned (7). The GPI anchor addition signal had only been shown to be essential for retention of Agα1p in the MATα cell wall, instead of secretion of the agglutinin to the medium as occurs in the complete absence of this signal (44, 75).
Our studies both support some elements of this model and call into question other of its components. Critically, when the GPI addition signal region of Aga1p was substituted with those of the cell wall-resident Cwp2 protein or the predicted plasma membrane-resident proteins Gas1 and Yap3, all produced fully functional agglutinins. Fractionation and extraction experiments support the expectation that a much greater fraction of the Gas1-GPI or Yap3-GPI-anchored proteins are resident in the plasma membrane than the cell wall, in contrast to the situation observed for the wild-type Aga1p and -Cwp2p GPI-anchored forms. Because there is a greater steady-state plasma membrane-resident pool of the Gas1p- and Yap3p-anchored Aga1 chimeras, our studies are consistent with these anchors undergoing processing and cell wall attachment less efficiently than those of Cwp2p and Aga1p. Nonetheless, a fraction of the Aga1 chimeras with Gas1- and Yap3-derived anchors do become processed and linked to the cell wall, consistent with the observations of others (17). It is possible that the abilities of the Gas1p and Yap3p GPI anchors to produce sufficient quantities of cell wall glucan-linked forms of the Aga1p-Aga2p adhesin to mediate agglutinin function might be to some extent dependent on the significantly induced expression levels of Aga1p in response to mating pheromone signaling. Regardless of their abundance, such constructs possessed full agglutination activity, while TMD-anchored constructs of equivalent size possessed only partial activity. Thus, we conclude that GPI context and cell wall residency contribute to, but are not essential for all of, the activity level of Aga1p observed in wild-type cells.
These results help confirm that postsecretory processing of the GPI anchor for any given protein, while influenced by sequence context in the region of the GPI signal (28, 29, 69, 71), occurs with different efficiencies. This leads to a range of residencies for GPI proteins from primarily cell wall localized to primarily plasma membrane retained, rather than a binary choice between these compartments, and is therefore consistent with studies documenting residency in both the cell wall and the plasma membrane for Gas1p and Yap3p (1, 17, 46, 68).
Certain properties pertaining to the differentiated states of fungal cells may also promote or permit the activities of plasma membrane-resident adhesins. Our results suggest plasma membrane-resident GPI-anchored adhesins may be active in contributing to adhesion interactions immediately after secretion and before postsecretory traffic to the cell wall. In the case of the Aga1p-Aga2p adhesin, the context of mating cell differentiation and the abundantly induced expression of these proteins in response to mating cell signaling may further play a role in enabling activity of membrane-resident forms. Haploid mating cells alter gene expression in response to the presence of mating pheromones, and the cell wall is specifically altered in a number of ways in these cells, principally as a thinned cell wall glucan layer in the region of the mating projection (22, 41). Such cells also possess altered chitin and glucan compositions in response to stimulation of chitin synthase expression and β-1,3 glucan synthesis by complexes containing effectors of Rho1 signaling (5, 16, 18, 49, 54, 58, 72). Because these cells are cell cycle arrested at the G1/S transition, a variety of cell wall mannoproteins also change in their abundance relative to cells progressing without arrest through the cell cycle (37, 57). As a group, these changes to the cell wall produce a specialized environment within which mating adhesins normally function.
Domain organization constraints of fungal adhesins.
Most fungal adhesins possess repeated domain structures, and this is true of both of the agglutinins Aga1p and Sag1/Agα1p. Here we have analyzed the roles of different repeated subdomains found within Aga1p. One simple type of repeat function in such proteins is illustrated by the ∼19 imperfect TSTSS/PSS heptapeptide repeats located between the R1 and R2 domains of Aga1p. This domain is predicted to be an O-glycosylation substrate based on its high number of serine and threonine residues and the presence of proline residues that may contribute to the use of such sites (51, 61). We found that deleting this domain from the native Aga1p reduced its activity, while introducing dimers and trimers of this sequence domain into carboxy-terminal regions of TMD-anchored Aga1 proteins increased their activities progressively with increasing length of the introduced domain. Introduction of this domain at a site amino terminal to the R1 domain of Aga1p failed to increase activity of the TMD-anchored agglutinin. These observations are consistent with this simple sequence performing a spacing function that aids in projecting other subdomains of Aga1p possessing adhesion activities to positions closer to the exterior surface of the cell wall. Such a spacer- or display-facilitating function has recently been demonstrated for a similarly serine-threonine-rich region found in the C. glabrata adhesin Epa1p (20). An alternative possible function for this domain that is not necessarily mutually exclusive with the previous function is that this interdomain spacer might be required to keep R1 and R2 domains physically separated to confer full Aga1p-Aga2p-mediated agglutination activity. Such a separated organization of domains might be important if the R2 domain is capable of binding Aga2p as discussed below.
We also investigated the domain requirements for the R1 and R2 repeated regions and their subdomains. A recent study examined the ability of different portions of these domains to bind and restore active structure to a purified fragment of Aga2p and found the CX2C motif within the R1 domain to play a critical role in mediating this activity in this biochemical assay (56). Our studies agree with these findings in most respects and confirm their relevance in vivo. We also found the R1 CX2C motif to play an essential role in mediating the function of Aga1p in vivo. By analyzing the activity of Aga1p with the R2 domain deleted, we found that the R2 domain is required for the full level of agglutination activity mediated by the wild-type Aga1p. These data are consistent with the R2 domain acting as a spacer to further extend the R1 domain of the molecule into peripheral regions of the cell wall near its surface. Whether Aga2p is also capable of associating with the R2 domain remains an open question, and our data do not exclude the possibility that the R2 domain functions in a weakly redundant manner to the R1 domain. Such an interaction would create cooperative binding interactions between Aga1p-displayed Aga2p molecules and Sag1/Agα1 proteins presented on MATα cells. The structure of Sag1/Agα1p also contains three domains that share sequence similarity and which have been previously proposed to mediate such cooperative interactions (12, 75). If such an activity does exist for the R2 domain, our data indicate that it must be obligate to a primary function of the R1 domain in organizing the Aga2p molecules that initiate interactions with Sag1/Agα1p. Indeed, in the case of the Aga1p-Aga2p adhesin complex, the actual in vivo stoichiometry of Aga2p to Aga1p polypeptides remains to be demonstrated.
O-linked glycosylation is required for proper adhesin activity in vivo.
The functional importance of PMT-mediated modification and subsequent trafficking of cell surface-resident proteins has been previously demonstrated for the cell surface proteins Mid2p and Axl2/Bud10p (48, 53). Our studies revealed a MATα cell-specific requirement for the activities, alone and in combination, of PMT1 and PMT2 for normal levels of adhesin function. Previous studies have shown that Aga2p is a substrate of the enzymes encoded by these genes in vivo and that purified Aga2p displays reduced activity when chemically deglycosylated in vitro (8, 24). The conditions required for substantial β elimination of glycans from proteins are relatively harsh, however, and are a caveat to these results. Here we find that MATa cells carrying deletions of either PMT1 or PMT2 alone show no reduction in agglutination activities, despite such strains underglycosylating both Aga1p and Aga2p (24). We show that only MATa pmt1Δpmt2Δ cells display significant agglutination defects and by immunoblot analysis demonstrate substantially reduced levels of high-molecular-mass Aga1p found at the cell surface. Currently we cannot conclude whether the agglutination defect seen in pmt1Δ pmt2Δ MATa cells is due to underglycosylation solely of Aga1p or Aga2p or both components of the complex. In contrast, MATα cells showed defects in both single and double mutants of PMT1 and PMT2, and Sag1/Agα1p was correspondingly underglycosylated in such cells. The nature of this cell type-specific requirement for PMT activity in agglutinin function remains to be completely understood.
Our data underscore the roles of Pmt1p and Pmt2p in the proper O-glycosylation and function of Sag1/Agα1p. The O-glycosylation of this protein has been described by Chen et al. (12); interestingly, the majority of the sites modified by addition of O-linked glycan were found to lie in the carboxy half of the protein, despite many potential sites existing in the amino-terminal half of the polypeptide. One unusual result we obtained was the apparent rescue of normal agglutination activities of MATα pmt1Δ and, to a much greater extent, pmt2Δ cells when mated in a bilateral context (e.g., pmt2Δ × pmt2Δ), an effect we found reproducible based on multiple experiments involving several independently derived strains. It is difficult to speculate as to the basis of such an effect. Loss of the adhesin Fig2p appears to similarly produce bilateral and partially asymmetric, α-cell-type-specific effects on agglutination (36, 76). Perhaps the adhesins represent substrates of the PMT enzymes that are in competition in some cell types and whose deletion alters the proportion and perhaps identities of residues posttranslationally modified in such contexts. It is possible that such alternatively modified forms of the same proteins may interact in complementary or nonproductive ways to affect adhesin complex function differently in unilateral or bilateral contexts. Alternatively, differences in cell wall structure caused by the altered glycosylation of Pmt1p and Pmt2p substrate cell wall mannoproteins might lead to complementary effects on wall composition and structure that rescue or facilitate agglutination. The observations of such cell type-, substrate-, and pattern-specific occurrences of posttranslational modification sites in adhesins highlight the many aspects of the pathways leading to these modifications that are still yet to be fully understood.
Supplementary Material
Acknowledgments
We are grateful to Paula Sundstrom and Janet Staab for technical suggestions regarding immunoblot analysis of cell wall mannoproteins; Peter Orlean for discussions regarding GPI anchoring signals; and John Belote, Melissa Pepling, and members of our laboratory for helpful discussions and comments on the manuscript. We also thank Timothy Phoenix for assistance in constructing and characterizing deletions of AGA1.
This work was supported by a graduate training fellowship from the Consortium for Plant Biotechnology Research (GO12026-153) to G.H. and a grant from NIH/NIDCR DE14443-01 to S.E.
Footnotes
The supplemental material for this article may be found at http://ec.asm.org/
REFERENCES
- 1.Ash, J., M. Dominguez, J. J. M. Bergeron, D. Y. Thomas, and Y. Bourbonnais. 1995. The yeast proprotein convertase encoded by YAP3 is a glycophosphatidylinositol-anchored protein that localizes to the plasma membrane. J. Biol. Chem. 270:20847-20854. [DOI] [PubMed] [Google Scholar]
- 2.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman, J., and P. E. Sudbery. 2002. Candida albicans: a molecular revolution built on lessons from budding yeast. Nat. Rev. Genet. 3:918-930. [DOI] [PubMed] [Google Scholar]
- 4.Brandhorst, T. T., P. J. Rooney, T. D. Sullivan, and B. S. Klein. 2002. Using new genetic tools to study the pathogenesis of Blastomyces dermatidis. Trends Microbiol. 10:25-30. [DOI] [PubMed] [Google Scholar]
- 5.Buehrer, B. M., and B. Errede. 1997. Coordination of the mating and cell integrity mitogen-activated protein kinase pathways in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6517-6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buurman, E. T., C. Westwater, B. Hube, A. J. Brown, F. C. Odds, and N. A. Gow. 1998. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 95:7670-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cappellaro, C., C. Baldermann, R. Rachel, and W. Tanner. 1994. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and α-agglutinin. EMBO J. 13:4737-4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappellaro, C., K. Hauser, V. Mrsa, M. Watzele, G. Watzele, C. Gruber, and W. Tanner. 1991. Saccharomyces cerevisiae a- and α-agglutinin: characterization of their molecular interaction. EMBO J. 10:4081-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cappellaro, C., V. Mrsa, and W. Tanner. 1998. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 180:5030-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caro, L. H., H. Tettelin, J. H. Vossen, A. F. Ram, H. van den Ende, and F. M. Klis. 1997. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477-1489. [DOI] [PubMed] [Google Scholar]
- 11.Chen, D., B. Yang, and T. Kuo. 1992. One-step transformation of yeast in stationary phase. Curr. Genet. 21:83-84. [DOI] [PubMed] [Google Scholar]
- 12.Chen, M. H., Z. M. Shen, H. Zhao, P. C. Kahn, and P. N. Lipke. 1995. Structure of Saccharomyces cerevisiae alpha-agglutinin. Evidence for a yeast cell wall protein with multiple immunoglobulin-like domains with atypical disulfides. J. Biol. Chem. 270:26168-26177. [DOI] [PubMed] [Google Scholar]
- 13.Coligan, J. E., B. M. Dunn, D. W. Speicher, and P. T. Wingfield (ed.). 1999. Current protocols in protein science, vol. 1, p. 5.2.12. Wiley Interscience, New York, N.Y.
- 14.Cormack, B. P., N. Ghori, and S. Falkow. 1999. An adhesin of the yeast pathogen Candida glabrata mediating adherence to human epithelial cells. Science 285:578-582. [DOI] [PubMed] [Google Scholar]
- 15.Cutler, J. E. 1991. Putative virulence factors of Candida albicans. Annu. Rev. Microbiol. 45:187-218. [DOI] [PubMed] [Google Scholar]
- 16.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance and thermotolerance. Microbiology 146:2121-2132. [DOI] [PubMed] [Google Scholar]
- 17.de Sampaio, G., J. P. Bourdineaud, and G. J. M. Lauquin. 1999. A constitutive role for GPI anchors in Saccharomyces cerevisiae: cell wall targeting. Mol. Microbiol. 34:247-256. [DOI] [PubMed] [Google Scholar]
- 18.Dijkgraaf, G. J., M. Abe, Y. Ohya, and H. Bussey. 2002. Mutations in Fks1p affect the cell wall content of beta-1, 3- and beta-1, 6-glucan in Saccharomyces cerevisiae. Yeast 19:671-690. [DOI] [PubMed] [Google Scholar]
- 19.Erdman, S. E., L. Lin, M. Malczynski, and M. Snyder. 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140:461-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frieman, M. B., J. M. McCafferey, and B. P. Cormack. 2002. Modular domain structure in the Candida glabrata adhesin Epa1p, a β1, 6 glucan-cross-linked cell wall protein. Mol. Microbiol. 2:479-492. [DOI] [PubMed] [Google Scholar]
- 21.Fu, Y., G. Rieg, W. A. Fonzi, P. H. Belanger, J. E. Edwards, Jr., and S. G. Filler. 1998. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect. Immun. 66:1783-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gammie, A. E., V. Brizzio, and M. D. Rose. 1998. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell 9:1395-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gentzsch, M., and W. Tanner. 1996. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15:5752-5759. [PMC free article] [PubMed] [Google Scholar]
- 24.Gentzsch, M., and W. Tanner. 1997. Protein-O-glycosylation in yeast: protein-specific mannosyltransferases. Glycobiology 7:481-486. [DOI] [PubMed] [Google Scholar]
- 25.Gow, N. A. R. 1994. Growth and guidance of the fungal hypha. Microbiology 140:3193-3205. [DOI] [PubMed] [Google Scholar]
- 26.Guo, B., C. A. Styles, Q. Feng, and G. R. Fink. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97:12158-12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamada, K., S. Fukuchi, M. Arisawa, M. Baba, and K. Kitada. 1998. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:53-59. [DOI] [PubMed] [Google Scholar]
- 28.Hamada, K., H. Terashima, M. Arisawa, and K. Kitada. 1998. Amino acid sequence requirement for efficient incorporation of glycosylphosphatidylinositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 273:26946-26953. [DOI] [PubMed] [Google Scholar]
- 29.Hamada, K., H. Terashima, M. Arisawa, N. Yabuki, and K. Kitada. 1999. Amino acid residues in the ω-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J. Bacteriol. 181:3886-3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanna, S. A., J. L. Monteiro da Silva, and M. J. S. M. Giannini. 2000. Adherence and intracellular parasitism of Paracoccidioides brasiliensis in Vero cells. Microbes Infection 2:877-884. [DOI] [PubMed] [Google Scholar]
- 31.Hostetter, M. K. 1994. Adhesins and ligands involved in the interaction of Candida spp. with epithelial and endothelial surfaces. Clin. Microbiol. Rev. 7:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoyer, L., R. Fundyga, J. Hecht, J. Kapteyn, F. Klis, and J. Arnold. 2001. Characterization of agglutinin-like sequence genes from non-albicans Candida and phylogenetic analysis of the ALS family. Genetics 157:1555-1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoyer, L. L., S. Scherer, A. R. Shatzman, and G. P. Livi. 1995. Candida albicans ALS1: domains related to a Saccharomyces cerevisiae sexual agglutinin separated by a repeating motif. Mol. Microbiol. 15:39-54. [DOI] [PubMed] [Google Scholar]
- 34.Hung, C.-Y., J.-J. Yu, K. R. Seshan, U. Reichard, and G. T. Cole. 2002. A parasitic phase-specific adhesin of Coccidioides immitis contributes to the virulence of this respiratory fungal pathogen. Infect. Immun. 70:3443-3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jentoft, N. 1990. Why are proteins O-glycosylated? Trends Biochem. Sci. 15:291-294. [DOI] [PubMed] [Google Scholar]
- 36.Jue, C. K., and P. N. Lipke. 2002. Role of Fig2p in agglutination in Saccharomyces cerevisiae. Eukaryot. Cell 1:843-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapteyn, J. C., H. van Den Ende, and F. M. Klis. 1999. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426:373-383. [DOI] [PubMed] [Google Scholar]
- 38.Klein, B. S. 2000. Molecular basis of pathogenicity in Blastomyces dermitidis: the importance of adhesion. Curr. Opin. Microbiol. 3:339-343. [DOI] [PubMed] [Google Scholar]
- 39.Klis, F. M. 1994. Cell wall assembly in yeast. Yeast 10:851-869. [DOI] [PubMed] [Google Scholar]
- 40.Lipke, P. N., and J. Kurjan. 1992. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 56:180-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipke, P. N., A. Taylor, and C. E. Ballou. 1976. Morphogenic effects of α-factor on Saccharomyces cerevisiae a cells. J. Bacteriol. 127:610-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu, C.-F., J. Kurjan, and P. N. Lipke. 1994. A pathway for cell wall anchorage of Saccharomyces cerevisiae α-agglutinin. Mol. Cell. Biol. 14:4825-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu, C. F., R. C. Montijn, J. L. Brown, F. Klis, J. Kurjan, H. Bussey, and P. N. Lipke. 1995. Glycosyl phosphatidylinositol-dependent cross-linking of alpha-agglutinin and beta 1, 6-glucan in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 128:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matthews, R. C. 1994. Pathogenicity determinants of Candida albicans: potential targets for immunotherapy? Microbiology 140:1505-1511. [DOI] [PubMed] [Google Scholar]
- 46.Nuoffer, C., P. Jenö, A. Conzelmann, and H. Riezman. 1991. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 11:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orlean, P. 1997. Biogenesis of yeast wall and surface components, p. 229-362. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular biology of the yeast Saccharomyces, vol. III. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 48.Philip, B., and D. E. Levin. 2001. Wsc1 and Mid2 are cell surface sensors for cell wall integrity signaling that act through Rom2, a guanine nucleotide exchange factor for Rho1. Mol. Cell. Biol. 21:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qadota, H., I. Ishii, A. Fujiyama, Y. Ohya, and Y. Anraku. 1992. RHO gene products, putative small GTP-binding proteins, are important for activation of the CAL1/CDC43 gene product, a protein geranylgeranyl-transferase in Saccharomyces cerevisiae. Yeast 8:735-741. [DOI] [PubMed] [Google Scholar]
- 50.Reynolds, T. B., and G. R. Fink. 2001. Bakers' yeast, a model for fungal biofilm formation. Science 291:878-881. [DOI] [PubMed] [Google Scholar]
- 51.Roy, A., C. F. Lu, D. L. Marykwas, P. N. Lipke, and J. Kurjan. 1991. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol. Cell. Biol. 11:4196-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rupp, S., E. Summers, H. Lo, H. Madhani, and G. Fink. 1999. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 18:1257-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanders, S., M. Gentzsch, W. Tanner, and I. Herskowitz. 1999. O-glycosylation of Axl2/Bud10p by Pmt4p is required for its stability, localization and function in daughter cells. J. Cell Biol. 145:1177-1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheckman, R., and V. Brawley. 1979. Localized deposition of chitin on the yeast cell surface in response to mating pheromone. Proc. Natl. Acad. Sci. USA 76:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharkey, L. L., M. D. McNemar, S. M. Saporito-Irwin, P. S. Sypherd, and W. A. Fonzi. 1999. HWP1 functions in the morphological development of Candida albicans downstream of EFG1, TUP1, and RBF1. J. Bacteriol. 181:5273-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen, Z. M., L. Wang, J. Pike, C. K. Jue, H. Zhao, H. de Nobel, J. Kurjan, and P. N. Lipke. 2001. Delineation of functional regions within the subunits of the Saccharomyces cerevisiae cell adhesion molecule a-agglutinin. J. Biol. Chem. 276:15768-15775. [DOI] [PubMed] [Google Scholar]
- 57.Smits, G., H. van den Ende, and F. Klis. 2001. Differential regulation of cell wall biogenesis during growth and development in yeast. Microbiology 147:781-794. [DOI] [PubMed] [Google Scholar]
- 58.Sprague, G. F., and J. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 657-744. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular biology of the yeast Saccharomyces, vol. II. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 59.Staab, J., S. Bradway, P. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]
- 60.Staab, J. F., C. A. Ferrer, and P. Sundstrom. 1996. Developmental expression of a tandemly repeated, proline- and glutamine-rich amino acid motif on hyphal surfaces on Candida albicans. J. Biol. Chem. 271:6298-6305. [DOI] [PubMed] [Google Scholar]
- 61.Strahl-Bolsinger, S., M. Gentzsch, and W. Tanner. 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426:297-307. [DOI] [PubMed] [Google Scholar]
- 62.Stratford, M. 1992. Yeast flocculation: receptor definition by mnn mutants and concanavalin A. Yeast 8:635-645. [DOI] [PubMed] [Google Scholar]
- 63.Stratford, M. 1992. Yeast flocculation: reconciliation of physiological and genetic viewpoints. Yeast 8:25-38. [DOI] [PubMed] [Google Scholar]
- 64.Sundstrom, P. 1999. Adhesins in Candida albicans. Curr. Opin. Microbiol. 2:353-357. [DOI] [PubMed] [Google Scholar]
- 65.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 66.Terrance, K., and P. N. Lipke. 1981. Sexual agglutination in Saccharomyces cerevisiae. J. Bacteriol. 148:889-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsuchimori, N., L. L. Sharkey, W. A. Fonzi, S. W. French, J. E. Edwards, Jr., and S. G. Filler. 2000. Reduced virulence of HWP1-deficient mutants of Candida albicans and their interactions with host cells. Infect. Immun. 68:1997-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vai, M., E. Gatti, E. Lacana, L. Popolo, and L. Alberghina. 1991. Isolation and deduced amino acid sequence of the gene encoding gp115, a yeast glycophospholipid-anchored protein containing a serine-rich region. J. Biol. Chem. 266:12242-12248. [PubMed] [Google Scholar]
- 69.Van der Vaart, J. M., R. te Biesebeke, J. W. Chapman, H. Y. Toschka, F. M. Klis, and C. T. Verrips. 1997. Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl. Environ. Microbiol. 63:615-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Verna, J., A. Lodder, K. Lee, A. Vagts, and R. Ballester. 1997. A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 94:13804-13809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vossen, J. H., W. H. Muller, P. N. Lipke, and F. M. Klis. 1997. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 179:2202-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe, D., M. Abe, and Y. Ohya. 2001. Yeast Lrg1p acts as a specialized RhoGAP regulating 1, 3-beta-glucan synthesis. Yeast 18:943-951. [DOI] [PubMed] [Google Scholar]
- 73.Watari, J., Y. Takata, M. Ogawa, H. Sahara, S. Koshino, M. L. Onnela, U. Airaksinen, R. Jaatinen, M. Penttila, and S. Keranen. 1994. Molecular cloning and analysis of the yeast flocculation gene FLO1. Yeast 10:211-225. [DOI] [PubMed] [Google Scholar]
- 74.Winzeler, E., D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. Boeke, H. Bussey, A. Chu, C. Connelly, K. Davis, F. Dietrich, S. Dow, M. El Bakkoury, F. Foury, S. Friend, E. Gentalen, G. Giaever, J. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 75.Wojciechowicz, D., C. F. Lu, J. Kurjan, and P. N. Lipke. 1993. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein α-agglutinin, a member of the immunoglobulin superfamily. Mol. Cell. Biol. 13:2554-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, M., D. Bennett, and S. E. Erdman. 2002. Maintenance of mating cell integrity requires the adhesin Fig2p. Eukaryot. Cell 1:811-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.