Abstract
The crystal structures of the 2C/H-2Kbm3–dEV8 allogeneic complex at 2.4 Å and H-2Kbm3–dEV8 at 2.15 Å, when compared with their syngeneic counterparts, elucidate structural changes that induce an alloresponse. The Asp77Ser mutation that imbues H-2Kbm3–dEV8 with its alloreactive properties is located beneath the peptide and does not directly contact the T cell receptor (TCR). However, the buried mutation induces local rearrangement of the peptide itself to preserve hydrogen bonding interactions between the peptide and the α1 77 residue. The COOH terminus of the peptide main chain is tugged toward the α1-helix such that its presentation to the TCR is altered. These changes increase the stability of the allogeneic peptide-major histocompatibility complex (pMHC) complex and increase complementarity in the TCR–pMHC interface, placing greater emphasis on recognition of the pMHC by the TCR β-chain, evinced by an increase in shape complementarity, buried surface area, and number of TCR–pMHC contacting residues. A nearly fourfold increase in the number of β-chain–pMHC contacts is accompanied by a concomitant 64% increase in β-chain–pMHC shape complementarity. Thus, the allogeneic mutation causes the same peptide to be presented differently, temporally and spatially, by the allogeneic and syngeneic MHCs.
Keywords: αβ T cell receptor, antigen, alloreactivity, complementarity determining regions, TCR-MHC recognition
Introduction
TCR recognition of foreign peptides bound to MHC class I molecules (pMHCs)*is the critical first step in CD8+ T lymphocyte activation. TCR pMHC ligation is followed by phosphorylation of TCR-related ζ-chains and recruitment and activation of ZAP 70. Subsequent activation of intracellular signaling cascades culminates in release of lytic granules and the induction of apoptosis in target cells. Though obligatory recognition of self-MHC molecules by TCRs is mandated by positive selection during thymic development, a significant subset of mature TCRs (1–10%; reference 1) possess the anomalous potential for alloreactive interaction and recognition of nonself MHCs.
Class I MHCs, which consist of an MHC heavy chain and a β2-microglobulin light chain, are 45-kD antigen-presenting glycoproteins expressed universally in nucleated cells. Peptides are bound to class I MHCs in an extended conformation with conserved hydrogen bonding to the peptide backbone N and C termini (2–4). The peptide side chains interact with six pockets in the peptide binding groove designated A to F (5). Frequent polymorphisms therein lead to MHC-specific peptide-binding motifs. Allelic variation in MHC I genes, arising from germline gene conversion (for a review, see reference 6), is the primary cause of graft rejection and graft-versus-host disease, the clinical manifestations of alloreactive TCR recognition.
The murine H-2Kb class I gene encodes a classical transplantation antigen within the MHC multigene cluster on chromosome 17. H-2Kbm proteins are naturally occurring mutants of the H-2Kb gene and were originally identified in skin graft experiments by correlating precise amino acid changes with altered histocompatibility (6). Most H-2Kbm molecules are characterized by clusters of amino acid substitutions, as many as five, occurring in one of either of the two amino-terminal heavy chain domains, α1 and α2 (6). H-2Kbm3 contains two mutations in the α1 domain. Substitution of serine for aspartate at position 77 determines the alloreactive phenotype, whereas replacement of alanine for lysine at position 89 determines the serologic phenotype (7). H-2Kb is responsible for positive selection of 2C TCR-bearing cytolytic T lymphocytes in transgenic mice, whereas H-2Kbm3 expression results in negative selection (8). dEV8 is a self-peptide derived from MLRQ, a component of the mitochondrial NADH ubiquinone complex, that, when bound to H-2Kbm3, is an alloligand for the 2C TCR (9).
Despite starkly contrasting biological properties, surface plasmon resonance measurements of 2C binding to H-2Kb–dEV8 and H-2Kbm3–dEV8 demonstrate that, in an isolated system, the difference in affinities and binding kinetics of 2C for these pMHCs is remarkably small (10). Although puzzling, this is not entirely unexpected. Some studies have reported good correlation between binding kinetics and the degree of T cell activation, i.e., strong agonists tend to have longer half lives than do antagonists (for a review, see reference 11), but an absolute correlation between binding kinetics and biological effect has not been experimentally observed (12, 13). However, the question remains as to how well these measurements reflect the nature of TCR–pMHC interactions in the presence of accessory molecules, such as CD8 (14–17), on the surface of the cell, and within the specialized environment of the immunological synapse (18). In addition, recent studies suggest that activated T cells have markedly increased avidity for pMHCs when compared with their naive counterparts (19).
The TCR is a cell surface glycoprotein consisting of two disulfide-linked polypeptide chains, α and β, whose binding orientation relative to the MHC-peptide binding groove was determined by crystallography to be approximately diagonal (20, 21, 23–25; for a review, see reference 22). Each chain has four possible complementarity determining regions (CDRs) with CDRs 1, 2, and 4 being germline encoded and CDR3, the most variable, being formed during thymic development by D to J and V to D gene rearrangements in the β-chain and V to J gene rearrangements in the α-chain. Generally, the α- and β-chain CDR3 loops form the majority of direct contacts to the peptide in TCR–pMHC interactions while the CDRs 1, 2, and, sometimes, 4 primarily contact the α1- and α2-helices that straddle the MHC peptide-binding groove. Thus far, four independent high resolution TCR crystal structures (≤3.2 Å) have been reported with their respective class I MHCs (20, 21, 26–28). Two independent TCRs bound to their respective class II MHCs have also been reported (29–31). In crystal structures of the A6 TCR bound to four distinct peptide–HLA-A2 complexes, no correlation was found between structure and biological activities that ranged from strong agonism to weak antagonism (32), raising the question as to how astonishingly dissimilar biological effects can be manifested by vanishingly small changes within the TCR–pMHC interface. Likewise, the structure of the 2C/H-2Kb–SIYR depicted very similar overall binding between a weak agonist and a superagonist, with functionally significant changes restricted to interaction of the P4 and P6 residues of the peptide with a “hot spot” around the TCR CDR3α and CDR3β (18). The BM3.3 TCR in complex with the H-2Kb–pBM1 MHC-peptide ligand revealed that, in the allogeneic interaction, the diagonal orientation of the TCR with respect to the antigen binding interface is preserved (28). However, comparison with the specific equivalent syngeneic complex was not possible as its structure has not yet been determined. This study enables, for the first time, a comparative analysis of the structures of a single TCR bound to both self and nonself MHC in the presence of the same bound peptide.
Materials and Methods
Affinity Measurements of Peptides Bound to H-2Kb and H-2Kbm3.
Affinity measurements for binding of peptides to soluble H-2Kb and H-2Kbm3 molecules were performed as described previously (33, 34). In brief, VSV8 peptide was labeled with 125I using the iodogen method. The 125I-VSV8 peptide was purified using a Sep-Pak column (Waters). The competition assays were performed at 23°C and 37°C as described (33) with few modifications. The binding studies were performed in 1% FCS and the free peptide was removed by gel filtration using Sephadex columns (NAP-5; Amersham Pharmacia Biotech). The dissociation constants for unlabeled peptides were determined from the molar concentrations of unlabeled peptides that gave 50% inhibition of 125I-VSV8 binding to H-2Kb or H-2Kbm3 molecules.
Protein Expression, Purification, and Crystallization.
Protein was overexpressed and purified as described previously (10). 2C and H-2Kbm3–dEV8 were mixed in 1:1 molar ratio (∼0.2 mM), and crystals of the 2C/H-2Kbm3–dEV8 complex were obtained by sitting drop vapor diffusion at 4°C with a 1:1 mixture of protein and mother liquor containing 0.1 M Tris-acetate pH 6.9, 12.5% PEG4000, and 20% glycerol.
Data Collection and Refinement Methods.
Crystals were cryocooled to −170°C in a nitrogen stream. X-ray diffraction data were collected at beamline 9–1 of the Stanford Synchrotron Radiation Laboratory (SSRL) on a MAR345 image plate using a monochromatic wavelength of 1.0 Å. The 2C/H-2Kbm3–dEV8 crystals are isomorphous with the previously reported 2C/H-2Kb–SIYR crystals (18) in the orthorhombic space group P21212. The structure was determined by direct refinement using the coordinates of the 2C/H-2Kb–SIYR complex as a model (PDB code 1G6R). The structure was initially refined by rigid body methods using the seven independent TCR and MHC domains against the maximum likelihood target (35). Iterative cycles of torsion angle dynamics using a maximum likelihood target function and slow-cooling temperature protocols with the program CNS (35) were combined with manual model adjustment. Bulk solvent correction with a flat model and anisotropic correction were used throughout the refinement. The model was rebuilt in density modified, composite omit, and σA-weighted 3Fo-2Fc and Fo-Fc maps (36) using the program O (37). A final round of refinement was conducted in REFMAC5 using TLS parameters (38), yielding an Rcryst of 28.2% and an Rfree of 31.1%. Progress of the refinement was assessed by continuously monitoring continuously Rfree for cross validation (39), and avoiding divergence between Rcryst and Rfree. One of the two independent copies of the TCR in the asymmetric unit is highly disordered, as found in other 2C/pMHC crystals (18, 26). The high Wilson B factor derived from the diffraction intensities correlates with the overall average B value of the model. Analysis of the final model with PROCHECK (40) shows no outliers and 83.7% and 65.6% of residues in the most favored regions of the Ramachandran plot in the first and second complex, respectively. Shape complementarity (SC) coefficients, excluding water molecules (41), were calculated using SC as implemented in CCP4 (42) with a 1.7 Å probe. Buried surface area was calculated using the program MS with a 1.7 Å probe (43). Hydrogen bonds and TCR-pMHC contacts were identified using the programs HBPLUS (44) and CONTACSYM (45).
Crystals of H-2Kb–dEV8 and H-2Kbm3–dEV8 were grown as described previously (46) and cryocooled after soaking in mother liquor supplemented with 20% glycerol. Diffraction data for H-2Kb– dEV8 and H-2Kbm3–dEV8 were collected on the MAR345 image plate detector at beamline 9–1 at SSRL and an in-house 30 cm MAR image plate detector, respectively. All data (see Table I) were integrated and reduced with DENZO and SCALEPACK (47). The H-2Kb–dEV8 structure was determined by molecular replacement (AMORE [48]) using H-2Kb–SEV9 (PDB code 1VAB) as an overall search model. After rigid-body refinement of individual domains the H-2Kb molecules were refined using slowcool protocols as implemented in X-PLOR version 3.85 (49). Solvent mask correction was applied throughout the refinement, and Rcryst and Rfree values were used to monitor the refinement progress. The model was then subjected to several rounds of alternating slowcool/positional refinement and manual model adjusting/rebuilding using the program O (37). The structure was refined to 1.7 Å with an Rcryst and Rfree of 20.6% and 25.1%, respectively (see Table I). The structure of H-2Kbm3–dEV8 was determined by molecular replacement using H-2Kb–dEV8 as a search model and was refined to 2.1 Å with a protocol similar to that used for the refinement of H-2Kb–dEV8 to Rcryst and Rfree values of 20.6 and 25.8%, respectively (see Table I). Fig. 1 was created with Insight II (Molecular Simulations, Inc.). Figs. 2 and 3 were created with BOBSCRIPT (50) and RASTER3D (51). Fig. 4 was created with GRASP (52).
Table I.
Summary of Crystallographic Data and Refinement
| Data collection | 2C/Kbm3/dEV8 | Kb/dEV8 | Kbm3/dEV8 |
|---|---|---|---|
| Wavelength (Å) | 1.00 | 1.00 | 1.54 |
| Resolution (Å) | 2.4 (2.44–2.40)a | 1.65 (1.71–1.65) | 2.15 (2.23–2.15) |
| No. observations | 351,477 | 339,981 | 274,981 |
| No. unique reflections | 95,548 | 56,955 | 29,655 |
| Completeness (%) | 99.5 (99.3) | 86.5 (89.2) | 98.6 (99.2) |
| Rmerge b | 0.052 (0.453) | 0.058 (0.682) | 0.087 (0.472) |
| I/σ | 22.4 (2.4) | 28.6 (3.0) | 24.4 (4.0) |
| Wilson B (Å2) | 55.6 | 18.0 | 28.3 |
| Refinement statistics | |||
| Resolution (Å) | 50.0–2.4 | 10.0–1.75 | 10–2.15 |
| No. reflections | 90,694 | 47,278 | 27,550 |
| No. in test set | 4,781 | 1,878 | 1,738 |
| Rcryst c (%) | 28.2 | 20.6 | 20.6 |
| Rfree d (%) | 31.1 | 25.1 | 25.8 |
| Rmsd bonds (Å) | 0.008 | 0.010 | 0.012 |
| Rmsd angles (°) | 1.64 | 1.62 | 1.72 |
| No. waters | 726 | 401 | 277 |
| Average B (Å2) | |||
| Protein after TLS | 50 (27, 73)e | NA | NA |
| Protein before TLS | 65 (42, 88)e | 23 | 29 |
| Waters | 58 [84]f | 44 | 50 |
| Ramachandran plot (%) | |||
| Most favored | 74.7 (83.7, 65.6)e | 91.8 | 91.2 |
| Additionally allowed | 22.9 (15.5, 30.3)e | 7.3 | 8.5 |
| Generously allowed | 2.5 (0.8 4.1)e | 0.9 | 0.3 |
| Disallowed | 0.0 | 0.0 | 0.0 |
NA, not applicable.
Numbers in parentheses refer to the highest resolution shell.
Rmerge = [Σh Σi|Ii(h) − < I(h) > | / Σh Σi Ii(h)] × 100, where < I(h) > is the mean of the I(h) observation of reflection h.
Rcryst = Σhkl |Fo − Fc|/Σhkl |Fo|.
Rfree was calculated as for Rcryst, but on 4% of the data excluded from the refinement.
(complex1, complex2).
[after TLS refinement].
Figure 1.

Superposition of dEV8 peptides from the free and TCR-bound forms of pMHCs. (A) Comparison of the unbound H-2Kb–dEV8 (magenta) and unbound H-2Kbm3–dEV8 (green) complexes. (B) Comparison of the TCR-bound H-2Kb–dEV8 (PDB code 2CKB) (cyan) and TCR-bound H-2Kbm3–dEV8 (orange). (C) Comparison of the unbound H-2Kb–dEV8 (magenta) and TCR-bound H-2Kb–dEV8 (cyan) (PDB code 2CKB). (D) Comparison of the unbound H-2Kbm3–dEV8 (green) and TCR-bound H-2Kbm3–dEV8 (orange).
Figure 2.
Effect of the alloreactive mutation on peptide-MHC and peptide-MHC–TCR interactions. Hydrogen bonding networks to the peptide proximal to Asp77Ser mutation are reorganized. Hydrogen bonding around the Asp77Ser mutation is shown between the peptide and MHC (A and B) and between peptide, MHC, and TCR (C and D). (A) H-2Kb–dEV8, (B) H-2Kbm3–dEV8, (C) 2C/H-2Kb–dEV8 (PDB code 2CKB), and (D) 2C/H-2Kbm3–dEV8. Peptide in yellow, MHC in cyan, TCR in green, water molecules in red.
Figure 3.
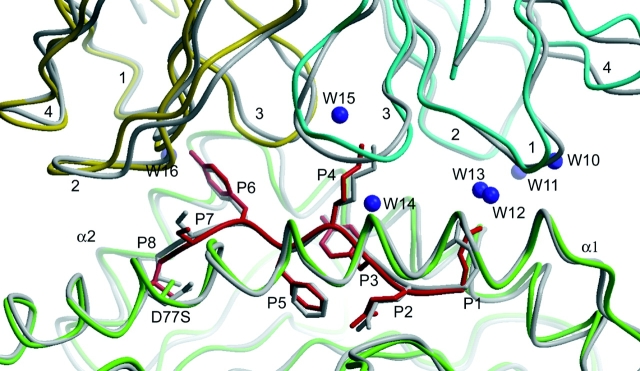
Comparison of syngeneic and allogeneic 2C TCR–pMHC complexes. The syngeneic 2C/H-2Kb–dEV8 complex is in gray; the allogeneic 2C/H-2Kbm3–dEV8 complex is colored, Vα CDRs (1–4) in cyan, Vβ CDRs (1–4) in yellow, H-2Kbm3 α1 and α2 helices in green, dEV8 (P1-P8) in red, and waters directly mediating TCR–pMHC contact in purple.
Figure 4.
A comparison of shape complementarity at the TCR–pMHC interface between the allogeneic (2C/H-2Kbm3–dEV8) and syngeneic complexes (2C/H-2Kb–dEV8). Shape complementarity is markedly increased at the TCR β-chain interface in the allogeneic complex. Shape complementarity at the TCR–pMHC interfaces was calculated using SC as implemented in CCP4 (CCP4, 1994) and mapped onto the corresponding surfaces in GRASP (reference 52). (A) 2C/H-2Kb–dEV8 (PDB code 2CKB). 2C at top and H-2Kb–dEV8 at bottom. (B) 2C/H-2Kbm3–dEV8. 2C at top and H-2Kbm3–dEV8 at bottom. The intensity of the cyan surface correlates with the magnitude of the Sc coefficient on the projected surface. Shape complementarity increases in 2C/H-2Kbm3–dEV8 at the surface projected by β-chain (area within the enclosed ovals).
Results
Structure Determination.
Alloreactivity is predicated on recognition of nonself MHCs by T cells. The question as to how single-site polymorphism in an MHC class I molecule can transform a syngeneic ligand into an allogeneic one was investigated by determining the crystal structure of the H-2Kbm3–dEV8 alloligand in both its free and TCR-bound forms. The 2.4 Å resolution of this allogeneic TCR–pMHC complex has been significantly extended over that of the original syngeneic 2C/H-2Kb–dEV8 complex at 3.2 Å. The 2C/H-2Kbm3–dEV8 structure was determined by directly refining the coordinates of the 2C/H-2Kb–SIYR complex (PDB code 1G6R), with peptide atoms removed and mutated residues truncated to alanine, against the diffraction data. The 2C/H-2Kb–SIYR crystal structure was used as a model in direct refinement because the crystals were more isomorphous with those of 2C/H-2Kbm3–dEV8 and yielded lower initial R values after rigid body refinement than did the molecular replacement solution derived after using any one of a number of different initial models. Final Rcryst and Rfree values were 28.2 and 31.1%, respectively, reflecting the extent of disorder in the second TCR–pMHC complex in the asymmetric unit, as noted in previous 2C TCR structure determinations (18, 26). However, the first complex was well-ordered and refined with relatively low B values (Table I). Both copies of the TCR–pMHC complex in the asymmetric unit include residues 1–213 of the TCR α-chain, residues 1–247 of the β-chain, residues 1–99 of the MHC β2-microglobulin light chain, residues 1–274 of the MHC heavy chain, and residues 1–8 of the peptide. The structure includes 726 water molecules, 16 carbohydrate residues, 3 glycerol molecules, and 1 acetate ion.
The resolution of the unliganded H-2Kb–dEV8 was also extended to 1.75 Å from the original 2.3 Å (26). The structure was determined by molecular replacement using wild-type H-2Kb coordinates (1VAB; reference 2) as a search model and refined to 1.75 Å with Rcryst and Rfree values of 20.6 and 25.1%, respectively. The H-2Kb–dEV8 structure includes 401 water molecules, 4 carbohydrate residues, and 2 phosphate ions. The H-2Kbm3–dEV8 structure was determined by molecular replacement using this new H-2Kb–dEV8 structure as a model and refined to 2.15 Å with Rcryst and Rfree values of 20.6 and 25.8%, respectively. The structure includes 278 water molecules, 4 carbohydrate residues, and 1 phosphate ion.
Comparison of the Unliganded Syngeneic and Allogeneic pMHCs.
The peptide-binding α1α2 domains of H-2Kb–dEV8 and H-2Kbm3–dEV8 superimpose with an r.m.s. deviation of 0.48 Å for all main-chain atoms. The peptide is bound by both MHCs in an extended conformation (Fig. 1 A) with the side chains of anchor residues Phe-P5 and Val-P8 binding in the C and F pockets, respectively. In H-2Kb–dEV8, the side-chain torsion angles at χ1 and χ2 of Tyr-P6 are –71° and –10°, respectively, while in H-2Kbm3–dEV8, a substantial rotation to angles of –175° at χ1 and –90° at χ2 results in displacement of the hydroxyl oxygen by 9.4 Å (Figs. 1 A, 2 A, and 2 B). Of interest, a previous modeling study predicted a difference in the orientation of this side chain, although in those studies the P6 side-chain rotamers in the syngeneic and allogeneic complexes were reversed (53) relative to those observed in the actual crystal structures. The torsion angle at χ1 of Ser-P7 in H-2Kb–dEV8 is –75°, compared with 57° in H-2Kbm3–dEV8 (Figs. 1 A, 2 A, and 2 B). The root mean square (r.m.s.) deviation for the peptide main-chain atoms is only 0.47 Å, but the largest deviation occurs in the four COOH-terminal residues (r.m.s. deviation 0.60 Å) and represents a shift in the peptide backbone toward the α1-helix of H-2Kbm3. Although the side chain of H-2Kbm3 Ser77 is shorter than that of H-2Kb Asp77, the hydrogen bond from the MHC side-chain hydroxyl to the main-chain nitrogen of P8 is preserved (Fig. 2 B) by a displacement of P7 and P8 backbone nitrogens by 0.9 Å and 0.5 Å, respectively, toward the α1-helix, which bears the Asp77Ser mutation.
Other structural rearrangements occur in the vicinity of Ser77 in H-2Kbm3. A different rotamer of Ser73 (χ1 = –72°) is found in the H-2Kb–dEV8 compared with H-2Kbm3–dEV8 (χ1 = –179°; Fig. 2, A and B). In H-2Kb–dEV8, the hydroxyl of Ser73 is rotated away from the Asp77 side chain, forming a hydrogen bond with the carbonyl oxygen of P5 (Fig. 2 A). In H-2Kbm3–dEV8, Ser73 Oγ hydrogen bonds to the amide nitrogen of Ser-P7 beneath which two waters (W2 and W7) are buried (Fig. 2 B), partially filling space vacated by the Asp77Ser substitution. In H-2Kb–dEV8, one water (W2) molecule is buried beneath Ser-P7 (Fig. 2 A). A water molecule (W1) coordinated by Asp77 and Thr80 side chains and the Val-P8 terminal carboxylate in most H-2Kb structures (PDB codes 1FZJ, 1FZK, 1FZO, 1KGB, 1VAC, 1VAB, 1G7Q, 1G7P), is absent in the allogeneic mutant (Fig. 2 B). In this H-2Kb–dEV8 structure, W1 is slightly beyond hydrogen bonding distance from the Asp77 carboxylate, but close enough for a polar interaction (3.8 Å; Fig. 2 A).
Comparison of Allogeneic and Syngeneic TCR–pMHC Complexes.
The 2C/H-2Kbm3–dEV8 complex (Fig. 3) has the same gross structural features as previously described for other class I–restricted TCR–pMHC structures with the TCR binding the pMHC in a diagonal orientation and the variable domains contacting the pMHC via their CDR loops (21, 26–28). TCR Vα-pMHC contacts are clustered in the neighborhood of the NH2-terminal half of the bound peptide, while the TCR Vβ-pMHC contacts are focused on the COOH-terminal region of the peptide. 2C binds H-2Kb–dEV8 and H-2Kbm3–dEV8 in a remarkably similar fashion (Fig. 3) with an r.m.s. deviation of 1.07 Å for all main-chain atoms of the Vα and Vβ variable domains. The pMHC-contacting CDR loops for both Vα and Vβ superimpose with an overall r.m.s. deviation for all main-chain atoms of 1.08 Å with the majority of contacts between the 2C and H-2Kb–dEV8 being conserved in the allogeneic complex. The greatest individual r.m.s. deviations are found in CDR3α and CDR3β (1.11 and 1.18 Å, respectively; Table II), CDRs that were previously identified as forming a functional hot spot for 2C TCR recognition (18). The CDR3α loop Cα atoms at Ala101 and Ala103 are displaced by 1.83 Å and 1.15 Å, respectively, in the allogeneic complex. In both complexes, the dEV8 is bound in a similar orientation (compare Fig. 1 A versus 1 B, Fig. 3) with an r.m.s. deviation for all atoms of 0.90 Å and for main-chain atoms of 0.66 Å. The three COOH-terminal residues of the peptide, proximal to the Asp77Ser mutation have an increased r.m.s deviation of 0.82 Å for backbone atoms.
Table II.
r.m.s. Deviations between the 2C TCR CDR Loops of Allogeneic H-2Kbm3 and Syngeneic H-2Kb Complexes
| CDR | r.m.s.d. (Å) |
|---|---|
| CDR1α (A:24–31) | 0.97 |
| CDR2α (A:48–55) | 0.51 |
| CDR3α (A:93–104) | 1.11 |
| CDR1β (B:26–31) | 0.74 |
| CDR2β (A:48–55) | 0.82 |
| CDR3β (B:95–107) | 1.18 |
Bound Water Molecules.
15 ordered water molecules surround the CDR1α loop, four of which mediate TCR–pMHC contact (Fig. 3). One conserved water molecule (W14; Fig. 3) bridges the hydroxyl of Tyr31 of the TCR α-chain and the amide nitrogen of Lys-P4. A second water (W10) forms hydrogen bonds with both Glu166 of the MHC heavy-chain and CDR1α Thr29 Oγ. Waters bound to the CDR1α main chain at the amide nitrogen (W12) and carbonyl oxygen of Thr29 (W13) mediate contact with the carboxylate of Glu-P1 and Oγ of Thr163, respectively (Fig. 3). An acetate ion was placed into unassigned Fo-Fc density in the solvent channel between the CDR1α loop and the peptide backbone of residues P2 and P3. The higher resolution of the allogeneic complex also reveals numerous water molecules positioned around the CDR2α loop, one of which mediates contact between the TCR and the pMHC (W11), bridging the carboxylate of MHC Glu166 and the amide nitrogen of Gly52 in the TCR α-chain.
A water molecule (W15), not previously observed, mediates contact between the Nξ of Lys-P4 and the hydroxyl of Ser93. Unambiguous density places the amide side-chain oxygen of the TCR α-chain Gln1 within hydrogen bonding distance of the backbone nitrogens of both Gly99 and Phe100 of the CDR3α. Contacts between the CDR3α loop and the pMHC are similar with the side chain of Lys-P4 protruding out from the MHC binding groove and into a pocket bounded by CDR3α and CDR3β.
For CDR1β, a water (W16) is within hydrogen bonding distance of the carbonyl oxygen of Asn28, the side-chain hydroxyl of Tyr-P6, and the carbonyl oxygen of H-2Kbm3 Lys146. As opposed to the wild-type structure, the hydrogen bonding partner of the Asn30 amide side-chain nitrogen is the backbone carbonyl oxygen of Ser-P7, not its side-chain Oγ.
The resolution (2.5 Å) of the BM3.3/H-2Kb–pBM1 structure also allowed delineation of numerous waters in the TCR–pMHC interface (28). The similar resolution of 2C/H-2Kbm3–dEV8 thus allows for comparison of water-mediated contacts in the two structures. 12 waters mediate contact between the TCR and the MHC in BM3.3/H-2Kb–pBM1, while 6 waters mediate contact in 2C/H-2Kbm3–dEV8; none are equivalent in both structures. Four waters in the peptide-binding groove are conserved between the allogeneic BM3.3/H-2Kb–pBM1 and the 2C/H-2Kbm3–dEV8 structures, but none mediates contact with the TCR in both structures.
TCR–pMHC Interface and Complementarity.
Although the Asp77Ser mutation in the H-2Kb heavy chain might have abolished the hydrogen bond between the side chain of residue 77 and the amide nitrogen of Leu-P8 (26), the hydrogen bond between these residues is preserved (compare Fig. 2, C and D). Movement of the peptide main chain toward the α1-helix maintains the Ser77 hydrogen bond with Leu-P8 and also brings Oγ of Ser-P7 within hydrogen bonding distance of heavy chain Ser73 Oγ (Fig. 2 D). The plane of the Trp147 indole ring is now rotated such that its indole nitrogen, along with the Nδ2 of Asn30 in the CDR1β loop, can make a hydrogen bond to the backbone carbonyl oxygen of Ser-P7. In the lower resolution wild-type complex, Nδ2 of CDR1β Asn30 is more than 5 Å distant from the carbonyl oxygen of Ser-P7 (Fig. 2 C). It was postulated that Nδ2 of Asn30 may hydrogen bond to the Oγ of Ser-P7 in the wild-type structure (26), but, at this resolution, the geometry and distance are not ideal for such a bond (Fig. 2 C). The novel hydrogen bond between Nδ2 of CDR1β Asn30 and the peptide in the 2C/H-2Kbm3–dEV8 complex is also absent in the 2C/H-2Kb–SIYR superagonist complex. Thus, local changes to the peptide and MHC in the vicinity of the Asp77Ser mutation actually promote a more intimate union between the TCR CDR1β loop and the pMHC.
In the 2C/H-2Kb–dEV8 structure, a total of 981 Å2 of pMHC surface area is buried by the TCR with 232 Å2 (23.6%) contributed by the peptide. In the 2C/H-2Kbm3–dEV8 structure, 2C buries 965 Å2 of pMHC surface area, approximately equal to that buried in the 2C/H-2Kb–SIYR complex, with 256 Å2 (26.5%) contributed by the peptide. Thus, even though the overall pMHC buried surface area is smaller in the mutant complex, the contribution of the peptide is slightly greater. Despite a smaller pMHC buried surface in the mutant complex, the shape complementarity (Sc) coefficient (41) is significantly higher for the mutant (0.62) than for the wild-type complex (0.41; see Fig. 5 A) and, surprisingly, even greater than that of the 2C/H-2Kb–SIYR superagonist complex (0.49).
Figure 5.

Bar graphs depicting changes in contacts, shape complementarity, and buried surface area in 2C/H-2Kbm3–dEV8 relative to 2C/H-2Kb–dEV8. Recognition by the β-chain is emphasized in the allogeneic (orange bar) vs. syngeneic (blue bar) complex. Bars at left compare pMHC interactions with the entire TCR (α, β), middle bars compare pMHC interactions with the α-chain, and bars at right compare pMHC interactions with the β-chain. (A) Number of overall contacts between TCR and pMHC and number of pMHC contacts for individual chains. (B) Sc coefficients for 2C bound to H-2Kb–dEV8 and H-2Kbm3–dEV8 and Sc coefficients for individual chains. (C) Buried pMHC surface area for 2C bound to H-2Kb–dEV8 and H-2Kbm3–dEV8, and pMHC surface area buried by individual α and β chains.
Graphical mapping of the TCR–pMHC complementarity onto the individual surfaces of the TCR and pMHC (Fig. 4) demonstrates that the increase in shape complementarity is most pronounced for the CDR loops of the β-chain and pMHC residues proximal to, and including, the COOH-terminal residues of the peptide. Sc coefficients calculated independently for the TCR α- and β-chains confirm this observation; 2C bound to H-2Kb has Sc coefficients of 0.48 and 0.43 for the α and β chains, respectively (Fig. 5 A). When bound to H-2Kbm3, the 2C α- and β-chains have Sc coefficients of 0.59 and 0.72, respectively. The summation of protein–protein contacts also indicates a shift in emphasis toward recognition of the pMHC by the 2C β-chain. Of the 121 contacts tabulated between 2C and H-2Kbm3–dEV8, 66 are established by the β-chain and 55 by the α chain (Fig. 5 B). In contrast, the 2C/H-2Kb–dEV8 interaction is dominated by contacts with the α-chain, 71 contact versus only 18 for the β-chain (26). The vast majority of the additional β-chain contacts in the mutant complex arise from van der Waals interactions; therefore, the changes at the interface are less well reflected by changes in the buried surface area calculations using a 1.7 Å probe. However, when the pMHC surface areas buried by the two chains of the TCR are calculated independently for the mutant and wild-type complexes, the surface area buried by the β-chain is slightly greater in the allogeneic complex, 462 Å2 versus 454 Å2, even though the total pMHC surface area buried in the syngeneic complex is greater. A concomitant slight decrease in pMHC surface area buried by the α-chain is also found in the mutant complex (466 Å2) versus the wild-type (486 Å2; Fig. 5 C). Of interest, the Sc coefficient calculated for the BM3.3/H-2Kb–pBM1 structure (PDB code 1FO0) is 0.60, and its total buried pMHC surface area was determined to be significantly smaller (1478 Å2; reference 28) than that for 2C/H-2Kb–dEV8 (2039 Å2).
Comparison of Liganded and Unliganded H-2Kbm3–dEV8.
As in the syngeneic structures, the side-chain of Tyr-P6 in H-2Kbm3–dEV8 must shift to accommodate the CDR1β loop of 2C. However, in contrast to the equivalent wild-type structures (Fig. 1 C), the χ1 and χ2 angles of P6 change little between the bound and unbound H-2Kbm3–dEV8 (Fig. 1 D), and the bulky aromatic ring of the side chain is repositioned only by a local lateral movement of the peptide main chain toward the α2-helix, which is also seen in the syngeneic complex. The Cα carbon of P6 is displaced by 1.5 Å toward the α2-helix. However, relative to the syngeneic complex, the P7 backbone nitrogen is pulled closer to the α1-helix in order to preserve the hydrogen bond from MHC residue 77 to the peptide in the alloreactive complex. Despite significant local changes in the disposition of the peptide, buried waters hydrogen bonded to the carbonyl oxygen of Phe-P5 (W6) and the amide nitrogen of Tyr-P6 (W5) are conserved (Fig. 2, B and D) with coordinates being shifted the same distance and in the same direction as the peptide main chain. The side-chain conformation of Glu152 in the α2-helix must be reorganized to avoid clashing with the main-chain shift in the peptide.
The Ser-P7 χ1 angle is rotated 172° in the unliganded H-2Kbm3–dEV8 to −48° in the liganded molecule (Fig. 1, B versus D). In the allogeneic pMHC, movement of the peptide main chain toward the α2-helix upon TCR ligation increases the distance between the hydroxyl of Ser73 and the amide nitrogen of Ser-P7 with which it forms a hydrogen bond (Fig. 2 B). This increase in the hydrogen bonding distance may weaken the bond sufficiently such that Oγ of Ser73 preferentially hydrogen bonds with Oγ of Ser-P7 (Fig. 2 D). However, what causes the dissolution of the hydrogen bond between Oγ of Ser-P7 and the terminal carboxyl of Val-P8 (compare Fig. 2, B and D) in the liganded H-2Kbm3–dEV8 is not clear.
Stability of H-2Kb and H-2Kbm3 Complexes.
In competitive binding assays, dEV8 was shown to bind H-2Kbm3 with greater affinity than H-2Kb at both 23°C (1.3 versus 11.1 nM, respectively) and 37°C (30.4 versus 90.1 nM, respectively; Table III). As a control, binding to the peptide OVA8 was measured for both H-2Kb and H-2Kbm3. The affinity of H-2Kbm3 for OVA8 was less than that for H-2Kb at both 23°C (13.3 versus 4.5 nM, respectively) and 37°C (160.2 versus 82.2 nM, respectively). This would indicate that the increase in affinity of H-2Kbm3 for dEV8 is likely not a reflection of an increase in affinity for peptides in general.
Table III.
Affinity Measurements of Peptides Binding to H-2Kb and H-2Kbm3
| 23°C KD (nM) |
37°C KD (nM) |
|
|---|---|---|
| H-2Kb | ||
| OVA8 | 4.5 | 82.2 |
| dEV8 | 11.1 | 90.1 |
| H-2Kbm3 | ||
| OVA8 | 13.3 | 160.2 |
| dEV8 | 1.3 | 30.4 |
The values are the means of three independent experiments. The standard errors of these values are between 10–15%.
Discussion
The difference in affinities of 2C for H-2Kb–dEV8 and H-2Kbm3–dEV8 are small and likely close to the margin of error for the system (surface plasmon resonance) in which binding was quantified. Nevertheless, the dissociation rate of 2C for H-2Kbm3–dEV8 (4.8 ± 0.4 s−1 × 10−2) is measurably decreased relative to that for H-2Kb–dEV8 (18.5 ± 2.0 s−1 × 10−2; reference 10); a longer half-life for this interaction would be in agreement with a generally established relationship between off-rates and degree of agonism (11). Although the mutation that confers alloreactivity to H-2Kbm3 does not directly contact the TCR, it does result in repositioning of the peptide such that a novel hydrogen bond is formed between the TCR and the peptide and the number of van der Waals contacts between the TCR and the pMHC in the neighborhood of the peptide is significantly increased. This hydrogen bond is formed between Asn30 of CDR1β and Ser-P7, which is positioned above the Asp77Ser mutation (Fig. 2 D). Loss of a single hydrogen bond in a protein–protein interaction can reduce the affinity of the interaction by as much as three orders of magnitude (54). The novel hydrogen bonding network, additional van der Waals contacts in the 2C/H-2Kbm3–dEV8 complex, and altered peptide presentation convert this interaction into an allogeneic one that results in negative selection. Another possible explanation for the alloreactive nature of H-2Kbm3–dEV8 is an increase in affinity of the allogeneic MHC for the dEV8 peptide relative to the syngeneic molecule (Table III). In a related study, the cell-surface density of the H-2Ld alloantigen appeared to dictate positive or negative selection (55). An ∼16-fold decrease in the H-2Ld cell-surface density resulted in positive rather than negative selection of 2C-bearing cytotoxic T lymphocytes. However, the differences in H-2Ld cell-surface density between negatively and positively selecting cells in this study would appear to be greater than could be explained by a threefold increase in affinity, such as that seen for H-2Kbm3–dEV8 at 37°C.
One model proposed to explain the frequency of alloreactive T cells is dependent on presentation of a unique set of self-peptides by allogeneic MHC molecules (56). The H-2Kbm3 system would appear to be in disagreement with this peptide-dependent hypothesis because presentation of the same peptide results in both positive and negative selection depending on the allelic variant of the MHC by which it is presented. However, the mutations in the MHC alter the disposition of the peptide such that the TCR may be “seeing” an altered view of the peptide when presented by the mutant MHC. Thus, although the peptide is chemically identical in the syngeneic and allogeneic pMHC molecules, from the perspective of the TCR, it may be unique when considered in the context of the syngeneic and allogeneic pMHCs. The structural data presented here, therefore, can be reconciled with this model. A second hypothesis, based on allogeneic TCRs directly discriminating between the polymorphic changes in the α-helices of the allogeneic MHC (57), does not fit in this case, as the definitive alloreactive mutation in H-2Kbm3 is buried in the peptide-binding groove and does not directly contact the TCR.
Numerous reports have identified water molecules as compensatory elements in protein–protein interactions, occupying positions vacated by mutated side chains. In keeping with this observation, an additional water molecule is found in the cavity partially vacated by the aspartate to serine mutation at position 77 in the heavy chain in H-2Kbm3–dEV8. In the H-2Kbm1–VSV8 complex, a water molecule reestablishes hydrogen bonding to the peptide where it has been abolished by the Glu152Ala mutation (58). In the complexes of the HyHEL-5 (54) and D1.3 (59) antibodies bound to lysozyme, buried water molecules occupy cavities formed by Arg68Lys and Asp18Ala mutations, respectively. In the Phe78Ser mutant of Rac (60), a member of the RHO GTPase family, and in the Leu41Ala mutant of ROP (61), an RNA-binding protein involved in plasmid replication, waters occupy analogous positions. In H-2Kb–dEV8, the water molecule bound to the carbonyl oxygen of Tyr6 in the peptide is coordinated by Oδ2 of heavy chain Asp77. In H-2Kbm3–dEV8, the conserved water bound to the Tyr6 carbonyl is coordinated by a second water which occupies the cavity expanded by the Asp77Ser mutation. The novel cavity-filling water in turn bridges the hydroxyl oxygens of MHC heavy chain Ser73 and Ser77. Thus, the H-2Kbm3–dEV8 complex is another example of the important accessory role of solvent in stabilizing protein-ligand interfaces, as well as playing a direct role in the ligand receptor recognition.
Thus far, only limited data are available delineating the role of bound water in the TCR–pMHC interface. The BM3.3/H-2Kb–pBM1 structure was the first to reveal an extensive network of bound waters in a TCR–pMHC interface (28), although waters were originally identified in the 2C/H-2Kb–dEV8 complex (26). The unusual conformation and size of the CDR3α loop in BM3.3/H-2Kb–pBM1 complex creates a cavity which is filled by ∼30 water molecules. The structure of 2C/H-2Kbm3–dEV8 complex also highlights numerous waters in the TCR–pMHC interface. As more TCR–pMHC structures are determined to higher resolution, the role of waters in TCR–pMHC binding will be elucidated, much as it was in the case of antibody–antigen recognition. Higher resolution structures of antibody–antigen complexes have demonstrated that bound waters participate extensively in shaping the complementary surfaces of the antibody and antigen (62–66).
The BM3.3/H-2Kb–pBM1 structure formally established that, in an allogeneic interaction, the TCR–pMHC binding orientation is still diagonal (28); however, as the structure for the corresponding TCR/self-ligand complex has not yet been determined, the degree of similarity with the syngeneic complex cannot be ascertained. The 2C/H-2Kbm3–dEV8 complex confirms the observation that allogeneic TCR–pMHC interactions are oriented diagonally and, furthermore, demonstrates that the structural differences between an allogeneic and syngeneic interaction can be exceedingly subtle. In the BM3.3/H-2Kb–pBM1 and 2C/H-2Kbm3–dEV8 interactions, β-chain–pMHC contacts are increased and become emphasized. However, with only two allogeneic structures available, whether this correlates with alloreactivity itself remains conjecture. Of note, the H-2Ld–QL9 alloligand has been predicted to increase substantially the interactions with the 2C β-chain through a COOH-terminal bulge in the peptide formed to accommodate a prominent ridge in the floor of the H-2Ld binding groove (67). However, it is apparent that relatively innocuous, even buried, polymorphisms can alter the presentation of the peptide itself, and, hence, lead to alloreactive TCR specificity and biological outcome.
Although the overall change in individual structures, as measured by r.m.s. deviations, appears minimal, shape complementarity coefficients and buried surface area calculations, as well as the distribution of TCR–pMHC contacts, indicate that the summation of changes amounts to a subtle, but global shift, in the nature of TCR binding such that β-chain–pMHC interactions become a more dominant feature. The 2C/H-2Kbm3–dEV8 is the first structure for which an analysis of this type has been possible, pointing to the importance of accumulating more structural information that addresses the question as to how TCRs distinguish between syngeneic and allogeneic ligands. Such information will provide invaluable insights into immunological phenomena that directly affect human health and disease.
Acknowledgments
The authors gratefully acknowledge helpful advice and assistance from Robyn Stanfield, Jeffrey Speir, and Samantha Greasley, and excellent technical support from Randy Stefanko and Ahteri Forcada-Lowrie. We thank the staff of Stanford Synchrotron Radiation Laboratory (SSRL) beamline 9-1 for data collection support.
This work was supported by National Institutes of Health grant AI-42266 (I.A. Wilson), National Institutes of Health grants CA-58896 (I.A. Wilson) and AI-42267, (L. Teyton), National Institutes of Health training fellowship AI-07244 (J.G. Luz), a post-doctoral fellowship from the German Academic Exchange Service and Skaggs Institute (M.G. Rudolph), and a CJ Martin Fellowship (987071) from the National Health and Medical Research Council of Australia and The Austin Research Institute. This is manuscript number 14170-MB from the Scripps Research Institute. Coordinates of 2C/H-2Kbm3–dEV8, H-2Kbm3–dEV8, and H-2Kb–dEV8 have been deposited in the PDB with accession codes 1JTR, 1LEK, and 1LEG, respectively.
M. Huang's present address is Division for Hemostasis and Thrombosis, Beth Israel Deaconess Medical Center, Mail Drop RE 319, 330 Brookline Ave., Boston, MA 02215.
K.C. Garcia's present address is Department of Microbiology and Department of Immunology, Stanford University School of Medicine, Fairchild D319, 299 Campus Dr., Stanford, CA 94305-5124.
V. Apostolopoulos's present address is The Austin Research Institute, Immunology and Vaccine Laboratory, Studley Rd., Heidelberg, Victoria 3084, Australia.
Footnotes
Abbreviations used in this paper: pMHC, peptide-MHC; r.m.s., root mean square; SC, shape complementarity.
References
- 1.Sherman, L.A., and S. Chattopadhyay. 1993. The molecular basis of allorecognition. Annu. Rev. Immunol. 11:385–402. [DOI] [PubMed] [Google Scholar]
- 2.Fremont, D.H., M. Matsumura, E.A. Stura, P.A. Peterson, and I.A. Wilson. 1992. Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science. 257:919–927. [DOI] [PubMed] [Google Scholar]
- 3.Matsumura, M., D.H. Fremont, P.A. Peterson, and I.A. Wilson. 1992. Emerging principles for the recognition of peptide antigens by MHC class I molecules. Science. 257:927–934. [DOI] [PubMed] [Google Scholar]
- 4.Madden, D.R., J.C. Gorga, J.L. Strominger, and D.C. Wiley. 1991. The structure of HLA-B27 reveals nonamer self-peptides bound in an extended conformation. Nature. 353:321–325. [DOI] [PubMed] [Google Scholar]
- 5.Saper, M.A., P.J. Bjorkman, and D.C. Wiley. 1991. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 Å resolution. J. Mol. Biol. 219:277–319. [DOI] [PubMed] [Google Scholar]
- 6.Nathenson, S.G., J. Geliebter, G.M. Pfaffenbach, and R.A. Zeff. 1986. Murine major histocompatibility complex class-I mutants: molecular analysis and structure-function implications. Annu. Rev. Immunol. 4:471–502. [DOI] [PubMed] [Google Scholar]
- 7.Pullen, J.K., H.D. Hunt, R.M. Horton, and L.R. Pease. 1989. The functional significance of two amino acid polymorphisms in the antigen-presenting domain of class I MHC molecules. Molecular dissection of Kbm3. J. Immunol. 143:1674–1679. [PubMed] [Google Scholar]
- 8.Sha, W.C., C.A. Nelson, R.D. Newberry, J.K. Pullen, L.R. Pease, J.H. Russell, and D.Y. Loh. 1990. Positive selection of transgenic receptor-bearing thymocytes by Kb antigen is altered by Kb mutations that involve peptide binding. Proc. Natl. Acad. Sci. USA. 87:6186–6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tallquist, M., T. Yun, and L. Pease. 1996. A single T cell receptor recognizes structurally distinct MHC/peptide complexes with high specificity. J. Exp. Med. 184:1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia, K.C., M.D. Tallquist, L.R. Pease, A. Brunmark, C.A. Scott, M. Degano, E.A. Stura, P.A. Peterson, I.A. Wilson, and L. Teyton. 1997. αβ T-cell receptor interactions with syngeneic and allogeneic ligands: affinity measurements and crystallization. Proc. Natl. Acad. Sci. USA. 94:13838–13843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, M., J. Boniface, Z. Reich, D. Lyons, J. Hampl, B. Arden, and Y. Chien. 1998. Ligand recognition by αβ T cell receptors. Annu. Rev. Immunol. 16:523–544. [DOI] [PubMed] [Google Scholar]
- 12.Kersh, G.J., E.N. Kersh, D.H. Fremont, and P.M. Allen. 1998. High- and low-potency ligands with similar affinities for the TCR: the importance of kinetics in TCR signaling. Immunity. 9:817–826. [DOI] [PubMed] [Google Scholar]
- 13.Baker, B.M., S.J. Gagnon, W.E. Biddison, and D.C. Wiley. 2000. Conversion of a T cell antagonist into an agonist by repairing a defect in the TCR/peptide/MHC interface: implications for TCR signaling. Immunity. 13:475–484. [DOI] [PubMed] [Google Scholar]
- 14.Garcia, K.C., C.A. Scott, A. Brunmark, F.R. Carbone, P.A. Peterson, I.A. Wilson, and L. Teyton. 1996. CD8 enhances formation of stable T-cell receptor/MHC class I molecule complexes. Nature. 384:577–581. [DOI] [PubMed] [Google Scholar]
- 15.Sykulev, Y., R.J. Cohen, and H.N. Eisen. 1995. The law of mass action governs antigen-stimulated cytolytic activity of CD8+ cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA. 92:11990–11992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho, B.K., K.C. Lian, P. Lee, A. Brunmark, C. McKinley, J. Chen, D.M. Kranz, and H.N. Eisen. 2001. Differences in antigen recognition and cytolytic activity of CD8(+) and CD8(−) T cells that express the same antigen-specific receptor. Proc. Natl. Acad. Sci. USA. 98:1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniels, M.A., and S.C. Jameson. 2000. Critical role for CD8 in T cell receptor binding and activation by peptide/major histocompatibility complex multimers. J. Exp. Med. 191:335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degano, M., K.C. Garcia, V. Apostolopoulos, M.G. Rudolph, L. Teyton, and I.A. Wilson. 2000. A functional hot spot for antigen recognition in a superagonist TCR/MHC complex. Immunity. 12:251–261. [DOI] [PubMed] [Google Scholar]
- 19.Fanmy, T.M. 2001. Increased TCR avidity after T cell activation: a mechanism for sensing low-density antigen. Immunity. 14:135–143. [PubMed] [Google Scholar]
- 20.Garcia, K.C., M. Degano, R.L. Stanfield, A. Brunmark, M.R. Jackson, P.A. Peterson, L. Teyton, and I.A. Wilson. 1996. An αβ T cell receptor structure at 2.5Å and its orientation in the TCR-MHC complex. Science. 274:209–219. [DOI] [PubMed] [Google Scholar]
- 21.Garboczi, D.N., P. Ghosh, U. Utz, Q.R. Fan, W.E. Biddison, and D.C. Wiley. 1996. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 384:134–141. [DOI] [PubMed] [Google Scholar]
- 22.Garcia, K.C., L. Teyton, and I.A. Wilson. 1999. Structural basis of T cell recognition. Annu. Rev. Immunol. 17:369–397. [DOI] [PubMed] [Google Scholar]
- 23.Wilson, I.A. 1999. Perspectives: protein structure. Class-conscious TCR? Science. 286:1867–1868. [DOI] [PubMed] [Google Scholar]
- 24.Hennecke, J., and D.C. Wiley. 2001. T cell receptor-MHC interactions up close. Cell. 104:1–4. [DOI] [PubMed] [Google Scholar]
- 25.Teng, M.K., A. Smolyar, A.G.D. Tse, J.H. Liu, J. Kiu, R.E. Hussey, S.G. Nathenson, H.C. Chang, E.L. Reinherz, and J.H. Wang. 1998. Identification of a common docking topology with substantial variation among different TCR-peptide-MHC complexes. Curr. Biol. 8:409–412. [DOI] [PubMed] [Google Scholar]
- 26.Garcia, K.C., M. Degano, L.R. Pease, M. Huang, P.A. Peterson, L. Teyton, and I.A. Wilson. 1998. Structural basis of plasticity in T cell receptor recognition of a self peptide-MHC antigen. Science. 279:1166–1172. [DOI] [PubMed] [Google Scholar]
- 27.Ding, Y.H., K.J. Smith, D.N. Garboczi, U. Utz, W.E. Biddison, and D.C. Wiley. 1998. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 8:403–411. [DOI] [PubMed] [Google Scholar]
- 28.Reiser, J.B., C. Darnault, A. Guimezanes, C. Gregoire, T. Mosser, A.-M. Schmitt-Verhulst, J.C. Fontecilla-Camps, B. Malissen, D. Housset, and G. Mazza. 2000. Crystal structure of a T cell receptor bound to an allogeneic MHC molecule. Nat. Immunol. 1:291–297. [DOI] [PubMed] [Google Scholar]
- 29.Reinherz, E.L., K. Tan, L. Tang, P. Kern, J. Liu, Y. Xiong, R.E. Hussey, A. Smolyar, B. Hare, R. Zhang, et al. 1999. The crystal structure of a T cell receptor in complex with peptide and MHC class II. Science. 286:1913–1921. [DOI] [PubMed] [Google Scholar]
- 30.Hennecke, J., A. Carfi, and D.C. Wiley. 2000. Structure of a covalently stabilized complex of a human αβ T-cell receptor, influenza HA peptide and MHC class II molecule, HLA-DR1. EMBO J. 19:5611–5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hennecke, J., C.D. Wiley. 2002. Structure of a complex of the human α/β T cell receptor (TCR) HA1.7, influenza hemagglutinin peptide, and major histocompatibility complex class II molecule, HLA-DR4 (DRA0101 and DRB10401): insight into TCR cross-restriction and alloreactivity. J. Exp. Med. 195:571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding, Y.H., B.M. Baker, D.N. Garboczi, W.E. Biddison, and D.C. Wiley. 1999. Four A6-TCR/peptide/HLA-A2 structures that generate very different T cell signals are nearly identical. Immunity. 11:45–56. [DOI] [PubMed] [Google Scholar]
- 33.Matsumura, M., Y. Saito, M.R. Jackson, E.S. Song, and P.A. Peterson. 1992. In vitro peptide binding to soluble empty class I major histocompatibility complex molecules isolated from transfected Drosophila melanogaster cells. J. Biol. Chem. 267:23589–23595. [PubMed] [Google Scholar]
- 34.Saito, Y., P.A. Peterson, and M. Matsumura. 1993. Quantitation of peptide anchor residue contributions to class I major histocompatibility complex molecule binding. J. Biol. Chem. 268:21309–21317. [PubMed] [Google Scholar]
- 35.Brünger, A.T., P.D. Adams, G.M. Clore, W.L. DeLano, G.P., R.W. Grosse-Kunstleve, J.-S. Jiang, J. Kuszewski, N. Nilges, N.S. Pannu, R.J. Read, et al. 1998. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Crystallogr. D54:905–921. [DOI] [PubMed] [Google Scholar]
- 36.Read, R.J. 1986. Improved fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A42:140–149. [Google Scholar]
- 37.Jones, T.A., S. Cowan, J.Y. Zou, and M. Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47:110–119. [DOI] [PubMed] [Google Scholar]
- 38.Winn, M.D., M.N. Isupov, and G.N. Murshudov. 2001. Use of TLS parameters to model anisotropic displacements in macromolecular refinement. Acta Crystallogr. D57:122–133. [DOI] [PubMed] [Google Scholar]
- 39.Brünger, A.T. 1992. Free R-value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 355:472–475. [DOI] [PubMed] [Google Scholar]
- 40.Laskowski, R.A., M.W. MacArthur, D.S. Moss, and J.M. Thornton. 1993. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26:283–291. [Google Scholar]
- 41.Lawrence, M.C., and P.M. Colman. 1993. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234:946–950. [DOI] [PubMed] [Google Scholar]
- 42.CCP4. 1994. The Collaborative Computational Project Number 4, suite programs for protein crystallography. Acta Crystallogr. D50:760–763. [DOI] [PubMed] [Google Scholar]
- 43.Connolly, M.L. 1983. Analytical molecular surface calculation. J. Appl. Crystallogr. 16:548–558. [Google Scholar]
- 44.McDonald, I.K., and J.M. Thornton. 1994. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238:777–793. [DOI] [PubMed] [Google Scholar]
- 45.Sheriff, S., W.A. Hendrickson, and J.L. Smith. 1987. Structure of myohemerythrin in the azidomet state at 1.7/1.3 Å resolution. J. Mol. Biol. 197:273–296. [DOI] [PubMed] [Google Scholar]
- 46.Stura, E.A., M. Matsumura, D.H. Fremont, Y. Saito, P.A. Peterson, and I.A. Wilson. 1992. Crystallization of murine major histocompatibility complex class I H-2Kb with single peptides. J. Mol. Biol. 228:975–982. [DOI] [PubMed] [Google Scholar]
- 47.Otwinowski, Z., and W. Minor. 1997. Processing of x-ray diffraction data collected in oscillation mode. Meth. Enzymol. 276:307–326. [DOI] [PubMed] [Google Scholar]
- 48.Navaza, J. 1994. AMoRe: an automated package for molecular replacement. Acta Crystallogr. A50:157–163. [Google Scholar]
- 49.Brünger, A.T. 1992. X-PLOR,Version 3.1. X-PLOR, Version 3.1. Yale University Press, New Haven, CT.
- 50.Esnouf, R.M. 1997. An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15:132–134. [DOI] [PubMed] [Google Scholar]
- 51.Merritt, E.A., and M.E.P. Murphy. 1994. Raster3D Version 2.0 - A program for photorealistic molecular graphics. Acta Crystallogr. D50:869–873. [DOI] [PubMed] [Google Scholar]
- 52.Nicholls, A., K.A. Sharp, and B. Honig. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 11:281–296. [DOI] [PubMed] [Google Scholar]
- 53.Tallquist, M.D., A.J. Weaver, and L.R. Pease. 1998. Degenerative recognition of alloantigenic peptides on a positively-selecting class I molecule. J. Immunol. 160:802–809. [PubMed] [Google Scholar]
- 54.Chacko, S., E. Silverton, L. Kam-Morgan, S. Smith-Gill, G. Cohen, and D. Davies. 1995. Structure of an antibody-lysozyme complex unexpected effect of conservative mutation. J. Mol. Biol. 245:261–274. [DOI] [PubMed] [Google Scholar]
- 55.Cook, J.R., E.M. Wormstall, T. Hornell, J. Russell, J.M. Connolly, and T.H. Hansen. 1997. Quantitation of the cell surface level of Ld resulting in positive versus negative selection of the 2C transgenic T cell receptor in vivo. Immunity. 7:233–241. [DOI] [PubMed] [Google Scholar]
- 56.Matzinger, P., and M.J. Bevan. 1977. Hypothesis: why do so many lymphocytes respond to major histocompatibility antigens? Cell. Immunol. 29:1–5. [DOI] [PubMed] [Google Scholar]
- 57.Bevan, M.J. 1984. High determinant density may explain the phenomenon of alloreactivity. Immunol. Today. 5:128–130. [DOI] [PubMed] [Google Scholar]
- 58.Wolan, D.W., L. Teyton, M.G. Rudolph, B. Villmow, S. Bauer, D.H. Busch, and I.A. Wilson. 2001. Crystal structure of the murine NK cell-activating receptor NKG2D at 1.95 A. Nat. Immunol. 2:248–254. [DOI] [PubMed] [Google Scholar]
- 59.Dall'Acqua, W., E.R. Goldman, W. Lin, C. Teng, D. Tsuchiya, H. Li, X. Ysern, B.C. Braden, Y. Li, S.J. Smith-Gill, and R.A. Mariuzza. 1998. A mutational analysis of binding interactions in an antigen-antibody protein-protein complex. Biochemistry. 37:7981–7991. [DOI] [PubMed] [Google Scholar]
- 60.Hirshberg, M., R.W. Stockley, G. Dodson, and M.R. Webb. 1997. The crystal structure of human rac1, a member of the rho-family complexed with a GTP analogue. Nat. Struct. Biol. 4:147–152. [DOI] [PubMed] [Google Scholar]
- 61.Vlassi, M., G. Cesareni, and M. Kokkinidis. 1999. A correlation between the loss of hydrophobic core packing interactions and protein stability. J. Mol. Biol. 285:817–827. [DOI] [PubMed] [Google Scholar]
- 62.Bhat, T.N., G.A. Bentley, G. Boulot, M.I. Greene, D. Tello, W. Dall'Acqua, H. Souchon, F.P. Schwarz, R.A. Mariuzza, and R.J. Poljak. 1994. Bound water molecules and conformational stabilization help mediate an antigen-antibody association. Proc. Natl. Acad. Sci. USA. 91:1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kondo, H., M. Shiroishi, M. Matsushima, K. Tsumoto, and I. Kumagai. 1999. Crystal structure of anti-Hen egg white lysozyme antibody (HyHEL-10) Fv-antigen complex. Local structural changes in the protein antigen and water-mediated interactions of Fv-antigen and light chain-heavy chain interfaces. J. Biol. Chem. 274:27623–27631. [DOI] [PubMed] [Google Scholar]
- 64.Li, Y., H. Li, S.J. Smith-Gill, and R.A. Mariuzza. 2000. Three-dimensional structures of the free and antigen-bound Fab from monoclonal antilysozyme antibody HyHEL-63. Biochemistry. 39:6296–6309. [DOI] [PubMed] [Google Scholar]
- 65.Mylvaganam, S.E., Y. Paterson, and E.D. Getzoff. 1998. Structural basis for the binding of an anti-cytochrome c antibody to its antigen: crystal structures of FabE8-cytochrome c complex to 1.8 A resolution and FabE8 to 2.26 A resolution. J. Mol. Biol. 281:301–322. [DOI] [PubMed] [Google Scholar]
- 66.Ochoa, W.F., S.G. Kalko, M.G. Mateu, P. Gomes, D. Andreu, E. Domingo, I. Fita, and N. Verdaguer. 2000. A multiply substituted G-H loop from foot-and-mouth disease virus in complex with a neutralizing antibody: a role for water molecules. J. Gen. Virol. 81:1495–1505. [DOI] [PubMed] [Google Scholar]
- 67.Speir, J.A., K.C. Garcia, A. Brunmark, M. Degano, P.A. Peterson, L. Teyton, and I.A. Wilson. 1998. Structural basis of 2C TCR allorecognition of H-2Ld peptide complexes. Immunity. 8:553–562. [DOI] [PubMed] [Google Scholar]