Abstract
The Ste50 protein of Saccharomyces cerevisiae is a regulator of the Ste11p protein kinase. Ste11p is a member of the MAP3K (or MEKK) family, which is conserved from yeast to mammals. Ste50p is involved in all the signaling pathways that require Ste11p function, yet little is known about the regulation of Ste50p itself. Here, we show that Ste50p is phosphorylated on multiple serine/threonine residues in vivo. Threonine 42 (T42) is phosphorylated both in vivo and in vitro, and the protein kinase responsible has been identified as casein kinase I. Replacement of T42 with alanine (T42A) compromises Ste50p function. This mutation abolishes the ability of overexpressed Ste50p to suppress either the mating defect of a ste20 ste50 deletion mutant or the mating defect of a strain with a Ste11p deleted from its sterile-alpha motif domain. Replacement of T42 with a phosphorylation-mimetic aspartic acid residue (T42D) permits wild-type function in all assays of Ste50p function. These results suggest that phosphorylation of T42 of Ste50p is required for proper signaling in the mating response. However, this phosphorylation does not seem to have a detectable role in modulating the high-osmolarity glycerol synthesis pathway.
All eukaryotic cells use a highly conserved mitogen-activated protein kinase (MAPK) module as the central core of a variety of complex signal transduction pathways. These pathways respond to many external stimuli and regulate numerous cellular processes. Each MAPK cascade is typically comprised of three protein kinases: a MAP3K, or MEKK; a MAP2K, or MEK; and a MAPK. These kinases are activated by sequential phosphorylation (2, 39). Eukaryotic cells typically contain multiple MAPK modules containing unique or shared protein kinases that provide the bases for signal transduction specificity and cross talk between signaling pathways. This regulation in turn may control the pattern of signaling (i.e., transient versus sustained) and/or coordinate multiple biological processes in response to a variety of environmental cues (for reviews, see references 6, 9, 14, 27, 29, 42, and 48).
The yeast Saccharomyces cerevisiae uses five MAPK cascades to respond to different physiological stimuli. One cascade is required for a/α diploid cells undergoing meiosis, and two cascades control the developmental processes of mating and filamentation, while two other cascades control the response to environmental osmostress—the high-osmolarity glycerol synthesis (HOG) pathway regulates response to high-osmolarity stress, and the MPK pathway modulates response to low osmolarity and challenges to cell wall integrity (for reviews, see references 9, 15, 20, 23, 25, 29, and 43). Components of these MAPK cascades are shared among different modules, including those controlling mating, filamentation, and the HOG pathway (7, 17, 26, 28, 34, 35, 49). Although the extent of sharing of these components varies among different pathways, the common module among the mating, filamentation, and HOG pathways comprises at least Ste11p (MAP3K) and its two regulators—Ste20p and Ste50p.
Ste20p is the founding member of the PAK/Ste20p kinase family. The activities of these kinases are modulated by small GTPases of the Rho superfamily. Ste20p is required for the activation of Ste11p and has been shown to phosphorylate Ste11p both in vitro and in vivo (8, 18, 51). Ste50p is also implicated in the regulation of Ste11p activity. Ste50p lacks any currently identified enzymatic function but is involved in all the signaling pathways that require the function of Ste11p. The involvement of Ste50p in the modulation of Ste11p function is dependent on physical interaction of the two proteins through their respective sterile-alpha motif (SAM) domains (17, 35, 49). This interaction appears to be constitutive; however, modulation of the interaction through mutations has been shown to differentially influence Ste11p activity in the different pathways (17). The C-terminal part of Ste50p is also required for its function, but this activity requires the presence of an intact SAM domain (49). In contrast to its clear involvement in Ste11p regulation, little is known about the regulation of Ste50p itself.
Here, we show that Ste50p is a phosphoprotein that is phosphorylated at multiple serine/threonine sites in intact cells. We have identified one of these sites as amino acid residue threonine 42. Casein kinase I (CKI) is capable of phosphorylating this residue in vitro. Preventing phosphorylation of threonine 42 by its replacement at this site with alanine results in vivo in decreased signaling during the mating response but has no detectable effect on signaling in the HOG pathway. Replacement of threonine 42 with the phosphomimetic residue aspartic acid permits the wild-type function of Ste50p in both the mating and HOG pathways. It appears that phosphorylation of Ste50p by casein kinase activity in yeast modulates the function of Ste50p, and this modulation may have different effects in the different pathways that share the Ste11p-Ste50p complex.
MATERIALS AND METHODS
Materials.
Restriction endonucleases and DNA-modifying enzymes were obtained from New England Biolabs and Amersham Biosciences. High-fidelity Taq thermostable DNA polymerase and tablet protease inhibitors were purchased from Roche Molecular Biochemicals. Acid-washed glass beads (450 to 600 μm in diameter), synthetic α mating factor, protease inhibitors, and bovine serum albumin were purchased from Sigma. α mating factor was dissolved in 90% methanol at a concentration of 1.0 mg/ml and stored at −20°C. Plasmids pGEX-4T-3 and pGEX-2TK, glutathione-Sepharose beads, glutathione, and protein A/G-Sepharose beads were obtained from Amersham Biosciences. The antibody against glutathione S-transferase (GST) was described previously (50). The anti-Ste50p antibodies were raised in rabbits against bacterially expressed, purified GST-Ste50p. GST-Ste50 protein fusions were detected using rabbit polyclonal anti-GST antibodies (49). Rabbit polyclonal antibodies directed against Ste5p were described previously (22). Anti-Fus3 goat polyclonal antibodies were from Santa Cruz Biotechnology. Nitrocellulose membranes were from Bio-Rad. Horseradish peroxidase-conjugated secondary antibodies were from Santa Cruz Biotechnology. The enhanced chemiluminescence assay system was purchased from Amersham Biosciences.
Yeast strains and manipulations.
Yeast media, culture conditions, and manipulations of yeast strains were as described previoulsy (40). Yeast transformations with circular or linearized plasmid DNA were carried out after treatment of yeast cells with lithium acetate (40). The yeast strains used in this study are listed in Table 1.
TABLE 1.
Yeast strains used in this study
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| W303-1A | MATaade2 ura3 his3 leu2 trp1 can1 | R. Rothstein |
| W303-1B | MATα ade2 ura3 his3 leu2 trp1 can1 | R. Rothstein |
| YCW241 | W303-1A FUS1::LacZ::LEU2 | 49 |
| YCW315 | W303-1A Δste50::TRP1 | 49 |
| YCW316 | W303-1B Δste50::TRP1 | 49 |
| YCW350 | W303-1A Δste20::TRP1 Δste50::KanR | This study |
| YCW360 | MATα ura3 leu2 his3 Δssk2::LEU2 Δssk22::LEU2 Δste50::TRP1 | 49 |
| YCW365 | MATaura3 leu2 his3 Δssk2::LEU2 Δssk22::LEU2 Δste50::TRP1 | 49 |
| YCW393 | MATahis3 leu2 trp1 ura3 Δste50::TRP1 sst1::hisG FUS1::LacZ::LEU2 | This study |
| YCW466 | MATaura3 leu2 his3 ssk2Δ::LEU2 ssk22Δ::LEU2 STE11ΔSAM | This study |
| YCW1151 | MATα ura3 leu2 his3 Δssk1::KanRyck1 yck2ts STE11ΔSAM | This study |
| DC17 | MATα his1 | J. Hicks |
| DC16 | MATa his1 | J. Hicks |
Plasmid constructions.
Yeast expression constructs carrying different fragments of STE50 were generated by PCR (41) using appropriate primers and cloned into pYEX-4T2. The starting plasmid for constructing the STE50 mutants was pVL57 (49). To make Ste50p (1-67), pVL57 was digested with EcoRI and BglII, blunt ended with T4 DNA polymerase, and religated to create pCW250. To make Ste50p(99-346), the fragment of STE50 was PCR amplified with primers OCW78 (5′-CGGGATCCATGAGAGACAGCAAGTTG-3′ [BamHI site is underlined]) and OCW18 (5′-GGGAATTCTTAGAGTCTTCCACCGGG-3′ [EcoRI site is underlined]), digested with BamHI and EcoRI, and cloned into pYEX-4T2 to give rise to pCW206. To make the serine/threonine (S/T)-to-alanine (A) mutations in Ste50p, site-directed mutagenesis by PCR was used with specific mutagenic primer pairs and flanking primers of OCW18 and OCW104: 5′-CGGAATTCATGTCCCCTATACTAGG-3′, which annealed to the GST sequence within pVL57. The PCR products were digested and cloned as BamHI-EcoRI fragments into pYEX-4T2, and mutations were confirmed by DNA sequencing. pCW259 contains the serine 12-to-alanine mutation (S12A), pCW260 contains S16A, pCW261 contains T24A, pCW262 contains S33A, pCW263 contains S36A, pCW264 contains T42A, pCW265 contains S46A/T47A, and pCW266 contains T53A. To make centromere-containing plasmid versions of the STE50 constructs that express STE50 under its own promoter, the 5′-flanking sequences of STE50 were amplified by PCR from genomic DNA with primers OCW87 (5′-TCCCCGCGGGTCCGGTGAAAAGTAT-3′ [SpeI site is underlined]) and OCW88 (5′-CGGGATCCTGATTTGCTATCTCTGCTAG-3′ [EcoRI site is underlined]). These PCR products were digested with BamHI and SpeI and cloned along with the BamHI-EcoRI fragment of STE50 from pVL57 into the SpeI and EcoRI sites of pRS316. The resulting construct, pCW267, contained an 800-bp 5′ flanking sequence of STE50 with an added BamHI site upstream of the STE50 coding sequence. The mutant alleles of STE50 were then cloned by exchanging the BamHI-EcoRI fragments from pCW259, pCW260, pCW261, pCW262, pCW263, pCW264, pCW265, and pCW266 with that of pCW267 to generate pCW276, pCW277, pCW278, pCW279, pCW280, pCW281, pCW282, and pCW283. Plasmid pCW379 expressing Ste50p with a substitution of aspartic acid for T42 was created in a similar manner. Multiple S/T-to-A mutations were created by multiple-round site-directed PCR mutagenesis. Several yeast constructs expressing Ste50p bearing multiple S/T-to-A substitutions as GST fusions were created: pCW372, with the N-terminal six-S/T substitution by A (6A); pCW373, with the N-terminal eight-S/T substitution by A, except T42 (7A/T42); and pCW401, with the N-terminal eight-S/T substitution by A (8A).
For bacterial expression constructs, pVL56 expressing the full-length Ste50p in pGEX-2TK was used as the starting construct. Plasmid pCW249 was created to express the N-terminal amino acids (aa) 1 to 67 of Ste50p by digesting pVL56 with EcoRI and BglII, blunt ending it with T4 DNA polymerase, and religation. Plasmids similar to pCW249, expressing the N-terminal 67 aa of Ste50p, were created for all the multiple S/T-to-A mutants. Plasmid pCW360 had the substitution of alanine for threonine 42 (T42A), pCW358 had the N-terminal five S/T residues replaced with alanine (5A), pCW380 had the N-terminal six S/T residues replaced with alanine (6A), pCW359 had the N-terminal eight S/T residues replaced with alanine (8A), and pCW381 had the N-terminal eight S/T residues replaced with alanine, except T42 (7A/T42).
Preparation of GST fusion proteins from Escherichia coli and yeast.
The GST fusion proteins were expressed in E. coli strain UT5600 (New England Biolabs), extracted, bound to glutathione-Sepharose beads, and eluted with glutathione as previously described (51). The eluted proteins were then concentrated and washed with storage buffer (50 mM Tris-HCl, pH 7.5, 200 mM KCl, 1 mM dithiothreitol, and 10% glycerol) by centrifugation using the Centricon-30 system (Amicon Inc.) and stored at −80°C. For yeast GST fusion expression constructs, including the yeast ORF protein kinase library, cells were induced with galactose for 5 h with GAL promoter-driven GST fusion vectors (55) or with the addition of 0.5 mM CuSO4 for 2 h with CUP1 promoter-driven GST fusion vectors. Total cell extracts were prepared as described previously (49). Purification of yeast GST fusion proteins were performed with yeast extracts and glutathione-Sepharose beads according to the procedure described previously for purification of E. coli GST fusion proteins (51). The eluted proteins were washed with storage buffer, concentrated with the Centricon 30 system, and stored at −80°C.
Protein kinase assays,32P metabolic labeling, and phosphopeptide mapping.
In vitro kinase assays were performed essentially as described previously (51). The reactions were carried out in 30 μl of kinase buffer containing 2 μM [γ-32P]ATP (∼2 × 104 Ci/mol), and the reaction mixtures were incubated at 30°C for 30 min. The reaction mixture was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the labeled products were visualized by either autoradiography or phosphorimaging (Molecular Dynamics).
The in vivo metabolic labeling with [32P]orthophosphate was performed with yeast cells (ssk2 ssk22 Δste50) carrying different alleles of the STE50 gene as GST fusions under the control of the CUP1 promoter. Exponentially growing cells from selective medium were inoculated into phosphate-free YPD medium (54) at an optical density at 600 nm (OD600) of ∼0.2 for 3 h, and copper sulfate was then added to the medium to a final concentration of 0.5 mM and incubated for 2 h. Approximately 10 OD600 units of cells were then harvested, resuspended in 5 ml of fresh phosphate-free YPD medium containing 1.5 mCi of [32P]orthophosphate (Amersham Biosciences), and incubated for 1 h at 30°C. Yeast total cell extracts were prepared, and labeled Ste50p was purified with glutathione-Sepharose beads as described for the purification of GST fusion protein from yeast. Two-dimensional (2-D) tryptic phosphopeptide mapping analysis was performed essentially as described previously (3, 12).
Subcellular fractionation of Ste50p-containing protein complexes.
Cells expressing GST-Ste50p or GST-Ste50-T42Ap were grown in liquid Ura-dropout medium to late exponential phase. The cultures were divided into two aliquots, and one aliquot of each was treated with α-factor at a final concentration of 3 μM for 90 min at 30°C. Equivalents of 250 OD600 units of cells were harvested by centrifugation; washed with ice-cold extraction buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, 1 mM dithiothreitol, 2 mM EDTA, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and 1 mM Na3VO4 and supplemented with protease inhibitor cocktail tablets (Roche Molecular Biochemicals); and disrupted using a French press. The homogenates were centrifuged at 6,000 × g for 5 min at 4°C to remove cell debris. The supernatant aliquots, corresponding to ∼50 OD280 units, were loaded on an eight-step (20 to 65% [wt/wt]) sucrose gradient over a 65% (wt/wt) 2-ml sucrose pad with each step prepared in 50 mM HEPES, pH 7.5, 150 mM NaCl, and 2 mM EDTA. The gradients were centrifuged at 38,000 rpm in an SW41 swinging-bucket rotor (Beckman) at 4°C for 12 h. Fractions (∼0.85 ml) were collected from the top of the gradient. Proteins from each fraction were precipitated with 10% trichloroacetic acid, and the protein pellets were washed with ice-cold acetone, resuspended in SDS sample buffer, and subjected to immunoblotting analyses.
Yeast mating and other assays.
Plate mating tests were carried out as described previously (19). Quantitative mating assays were carried out by a filter assay as described previously (21). β-Galactosidase activities were measured as described previously (19), with Miller units defined as follows: (OD420 × 1,000)/(OD600 × t × V), where t is the time in minutes and V is the volume in milliliters. Halo assays to test cell growth inhibition in response to α mating factor were performed as described previously (21).
Photomicroscopy.
Cells were grown to mid-log phase and fixed with formaldehyde at a final concentration of 3.7% with 150 mM NaCl. The cells were viewed with a microscope equipped with Nomarski optics, and microscopic photographs were acquired with a 100× objective using a Micro Max camera (Princeton Instruments Inc.) with Northern Eclipse imaging software (Empix Imaging Inc.) and processed using Adobe Photoshop for Macintosh.
RESULTS
Ste50p is a phosphoprotein in vivo and is phosphorylated on multiple serine/threonine residues.
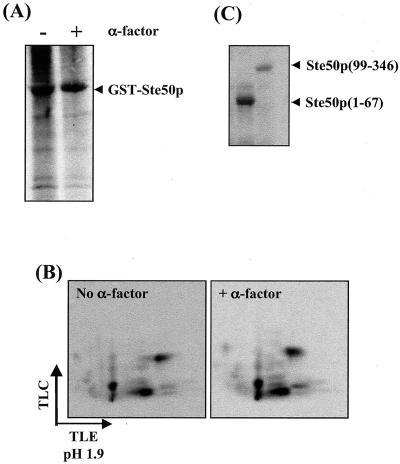
The functions of many signaling components are regulated through phosphorylation. Ste50p has been shown to be one of the regulators of the MAPKKK Ste11p and is involved in all the signal transduction pathways that need the function of Ste11p (17, 28, 35, 36, 49). To examine if Ste50p is phosphorylated in vivo, yeast cells expressing Ste50p as a GST fusion were metabolically labeled with [32P]orthophosphate. GST-Ste50p was then purified and analyzed by SDS-PAGE and autoradiography. As shown in Fig. 1, Ste50p was phosphorylated in vivo. The extent of phosphorylation of Ste50p was not significantly affected by treatment with pheromone at 5 μM. Tryptic phosphopeptide mapping revealed several labeled spots, indicating that Ste50p is phosphorylated on multiple sites in vivo, as the GST protein alone is not labeled (8, 33). Phosphoamino acid analysis indicated that the phosphorylation occurred exclusively on serine/threonine residues (data not shown). Fragmentation of Ste50p showed that both the N terminus (aa 1 to 67) and the C terminus (aa 99 to 346) were labeled in vivo (Fig. 1C). The N terminus of Ste50p contains all nine serine/threonine residues within the core SAM domain (32, 44), and they are located in a single 53-residue tryptic peptide (see Fig. 5C).
FIG. 1.
Ste50p is phosphorylated in vivo. (A) GST-Ste50p was expressed in a Δste50 strain, metabolically labeled with [32P]orthophosphate in the presence (+) or absence (−) of 5 μM α mating factor for 1 h, purified with glutathione-Sepharose, and resolved by SDS-PAGE. (B) Labeled GST-Ste50p from the gel in panel A was digested with trypsin and subjected to 2-D phosphopeptide mapping analysis. TLC, thin-layer chromatography; TLE, thin-layer electrophoresis. (C) Both the N- and C-terminal fragments are phosphorylated in vivo. Constructs expressing the N-terminal (aa 1 to 67) and C-terminal (aa 99 to 346) portions of Ste50p were expressed as GST fusions and analyzed as described for panel A.
FIG. 5.
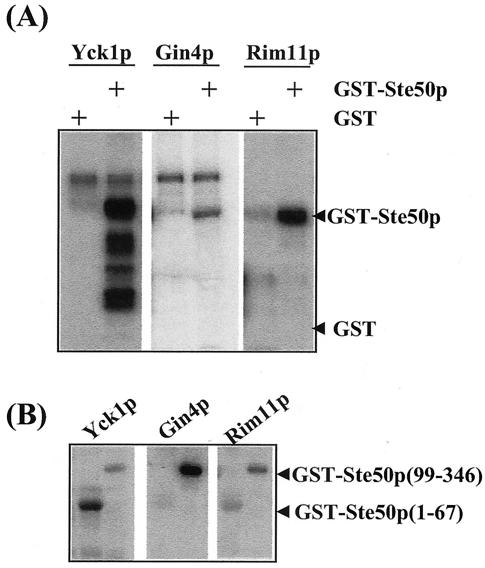
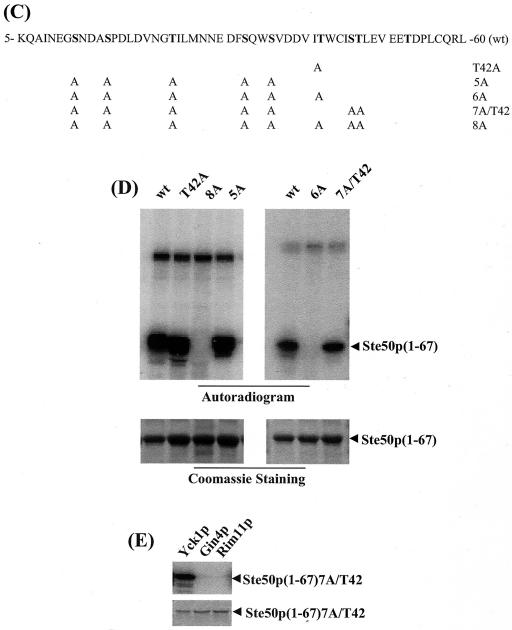
Protein kinases Yck1p, Gin4p, and Rim11p phosphorylate GST-Ste50p in vitro, and threonine 42 of Ste50p is phosphorylated in vitro by Yck1p. (A) Protein kinases Yck1p, Gin4p, and Rim11p isolated from yeast as GST fusions were tested for the ability to phosphorylate purified GST-Ste50p in vitro. The positions of phosphorylated GST-Ste50p and GST are indicated. +, present. (B) The N-terminal fragment of Ste50p is phosphorylated by Yck1p. The N-terminal and C-terminal fragments of Ste50p were expressed and purified from bacteria and used as substrates in vitro for the protein kinases indicated. The positions of the N-terminal and C-terminal fragments of Ste50p as GST fusions are indicated. (C) Amino acid sequence of the N-terminal fragment of Ste50p. The alanine substitutions are indicated, and the names of the corresponding mutants are indicated on the right. wt, wild type. (D) The N-terminal fragment (aa 1 to 67) of Ste50p with various alanine substitutions was purified from bacteria and used as a substrate in the in vitro kinase assay with Yck1p. The 32P-labeled GST-Ste50p(1-67) is shown in the autoradiogram above, and the amounts of protein used in the kinase assay are shown as Coomassie staining below. (E) Threonine 42 of Ste50p is not phosphorylated either by Gin4p or Rim11p. GST-Ste50p(1-67)7A/T42 was used as a substrate in the in vitro kinase assays with Yck1p, Gin4p, and Rim11p under the same conditions as for panel D. The autoradiogram (top) and the input protein indicated by Coomassie staining (bottom) are shown.
Threonine 42 is important for Ste50p function in the pheromone response pathway.
Given the functional importance (17, 35, 49) of the SAM domain (amino acid residues 32 to 100) and the fact that this region of Ste50p is phosphorylated in vivo, site-directed mutagenesis was performed to change all of the S/T residues in the SAM domain to A. To assess the role of the potential phosphorylation site(s), the single S/T-to-A mutants, expressed as GST fusions under the control of the CUP1 promoter on a 2μm plasmid, were first tested for the ability to suppress the mating defect of a Δste20 Δste50 strain.
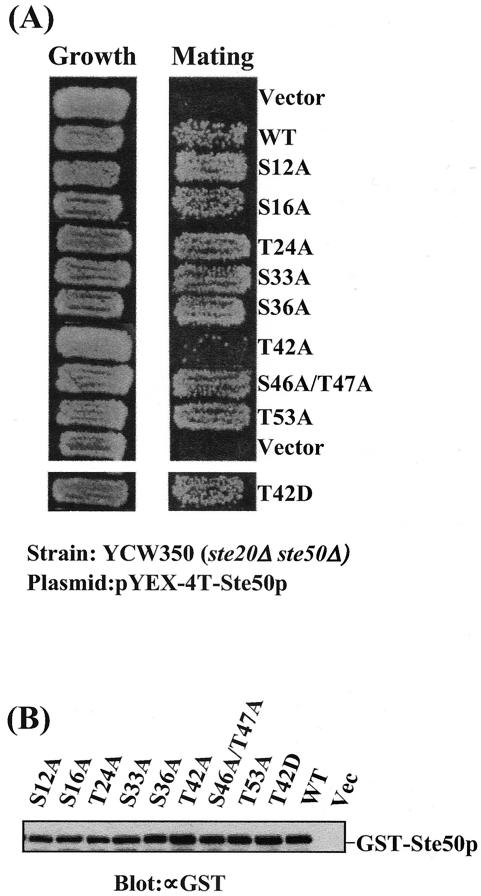
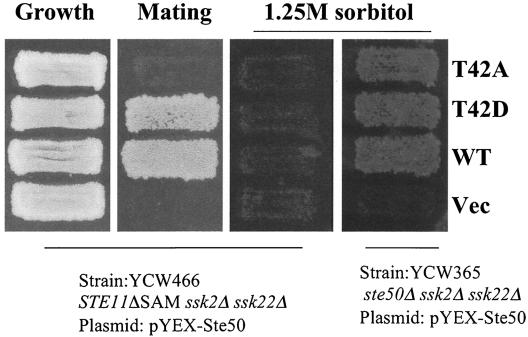
Both Ste20p and Ste50p are regulators of Ste11p MAPKKK. Deletion of STE50 results in reduced mating, but the cells are far from sterile, whereas Δste20 Δste50 strains are completely sterile. However, overexpression of Ste50p has been shown to partially suppress the mating defect of Δste20 mutants (52), and thus, the Δste20 Δste50 strain provides a sensitive assay for Ste50p function. We analyzed the S/T-to-A mutants of Ste50p for the ability to permit mating in the Δste20 Δste50 (YCW350) genetic background, since the mating response in this strain depends solely on the function of Ste50p. As shown in Fig. 2A, the yeast strain YCW350 (Δste20 Δste50) transformed with only the vector plasmid showed no detectable mating, whereas overexpression of wild-type Ste50p, as expected, increases the mating to a readily detectable level. All but one of the S/T-to-A mutants showed no significant difference in the ability to confer mating competence compared to the level of mating generated by wild-type Ste50p. However, the T42A mutant was significantly impaired in its ability to permit mating in the Δste20 Δste50 strain. In contrast, the T42D mutant allowed mating at a level comparable to that permitted by wild-type Ste50p. The phenotype observed was not due to different protein concentrations, because the steady-state levels of both mutant and wild-type Ste50p were very similar, as judged by Western blot analysis (Fig. 2B). Quantitative mating analysis revealed that the Ste50-T42Ap mutant caused a >30-fold decrease, compared to wild-type Ste50p, in suppression of the mating defect of the Δste20 Δste50 strain (Table 2), while the T42D mutant functioned essentially like wild-type Ste50p. These observations suggest that T42 may need to be phosphorylated for Ste50p to perform its function in the mating pathway.
FIG. 2.
Replacement of threonine 42 of Ste50p with alanine abolishes its ability to suppress the Δste20 defect in mating. (A) Yeast strain YCW350 (MATa Δste20 Δste50) was transformed with the various STE50 alleles indicated as GST fusion constructs under the control of the CUP1 promoter and tested for both growth (left) and ability to mate with tester strain DC17 (MATα his1) by plate mating assay. The mating was performed for 8 h at 30°C, and diploids were then selected on minimal medium (right). (B) Western blot analysis of the expression of the various Ste50p constructs. Yeast cell extracts from strains used in panel A were prepared, resolved on SDS-PAGE, and probed with an anti-GST antibody (∝GST).
TABLE 2.
Influence of Thr42 of Ste50p on the mating response of yeast with Δste20 or STE11 ΔSAM
| Relevant genotype | Mating efficiencya
|
|
|---|---|---|
| Δste20 Δste50 | STE11ΔSAM Δssk2 Δssk22 | |
| Vector | <0.1 × 10−5 | 3.0 × 10−5 ± 0.4 × 10−5 |
| STE50 | 34.0 × 10−5 ± 2.0 × 10−5 | 450.0 × 10−5 ± 40.0 × 10−5 |
| STE50-T42A | 1.0 × 10−5 ± 0.5 × 10−5 | 6.6 × 10−5 ± 0.5 × 10−5 |
| STE50-T42D | 40.0 × 10−5 ± 2.0 × 10−5 | 360.0 × 10−5 ± 30.0 × 10−5 |
Mating efficiency is the number of diploids formed with the MATα tester strain DC17 cells divided by the number of input MATa cells in the mating mixture at the begining of the experiment multiplied by 100. The data are expressed as the averages of three repetitions, with standard deviations indicated. The strains used were YCW350 (MATa Δste20 Δste50) and YCW466 (MATa STE11ΔSAM Δssk2 Δssk22) transformed with the STE50 alleles indicated. All STE50 alleles were expressed as GST fusions from the CUP1 promoter from the 2μm plasmid.
Phosphorylation of threonine 42 is required to suppress the mating defect of Ste11ΔSAMp.
The SAM domain of Ste11p is required for its proper function, as removal of this motif results in complete loss of Ste11p activity in the HOG pathway branch and greatly reduces the mating response (17, 35, 49). Intriguingly, the mating defect of Ste11ΔSAMp strains was sensitive to the expression level of Ste11p: the chromosomal-level expression of an integrated allele generated an extremely low mating level that was close to sterile, while overexpression led to an almost complete suppression of the mating defect (17, 49). We constructed a strain with an integrated allele of STE11 containing a deletion of aa 26 to 130 within the SAM domain (STE11ΔSAM). This strain had a very low level of mating response and behaved essentially as if it was sterile in plate mating assays. We found that overexpression of Ste50p from the CUP1 promoter as a GST fusion protein could partially suppress the mating defect of the STE11ΔSAM strain (Fig. 3, second panel from left), even though the Ste11ΔSAMp protein has no detectable interaction with Ste50p (17, 49). This suppression of the mating response requires the activity of Ste11ΔSAMp, since overexpression of Ste50p did not improve the mating of Δste11 cells (data not shown). However, overexpression of Ste50p was unable to make the osmosensitive ssk2 ssk22 STE11ΔSAM strain capable of growth on high-osmolarity medium (Fig. 3, third panel from left). Although overexpression of Ste50p resulted in partial suppression of the STE11ΔSAM mating defect, it did not allow a detectable pheromone-induced cell cycle arrest as measured by halo assay or permit either significant induction of a FUS1::LacZ reporter or typical pheromone-induced morphogenesis (data not shown).
FIG. 3.
Replacement of threonine 42 of Ste50p with alanine abolishes its ability to suppress the STE11ΔSAM defect in mating. Yeast strain YCW466 (MATa STE11ΔSAM Δssk2 Δssk22) was transformed with the STE50 alleles indicated and tested for growth (left), for the ability to mate with tester strain DC17 (MATα his1) by selecting for diploids on minimal medium (second panel from left), and for the ability to grow on high-osmolarity medium containing 1.25 M sorbitol (third panel from left). Both the T42A and T42D alleles of Ste50p complement wild-type (WT) Ste50p function to allow strain YCW365 (Δste50 Δssk2 Δssk22) to grow on high-osmolarity medium containing 1.25 M sorbitol (right). Vec, control vector.
The mechanism of suppression of the Ste11ΔSAMp signaling defect by overexpression of Ste50p is unclear. However, due to its sensitivity, this system was also used to assay the Ste50p mutants for the ability to suppress the signaling defect of Ste11ΔSAMp. All the S/T-to-A mutations except T42A were able to suppress the mating defect of Ste11ΔSAMp to a level identical to that generated by the wild-type Ste50p. However, the T42A mutation almost completely blocked the ability of Ste50p overproduction to suppress the mating defect of Ste11ΔSAMp (Fig. 3, second panel from left; Table 2). In contrast to the T42A substitution, the T42D mutation was able to suppress the Ste11ΔSAMp mating defect just like the wild-type Ste50p, suggesting that phosphorylation of Ste50p on T42 is required for the suppression function. Like the wild-type Ste50p, none of the Ste50p S/T-to-A mutants or the T42D mutant was able to suppress the Ste11ΔSAMp signaling defect in the HOG pathway, although they all complemented Ste50p function in the ssk2 ssk22 Δste50 strain and permitted growth on high-osmolarity media (Fig. 3, right panel). This is consistent with previous observations that the interaction between Ste11p and Ste50p through their respective SAM domains is absolutely required for HOG pathway function (17, 35, 49).
Threonine 42 preferentially regulates the function of Ste50p in the pheromone response pathway.
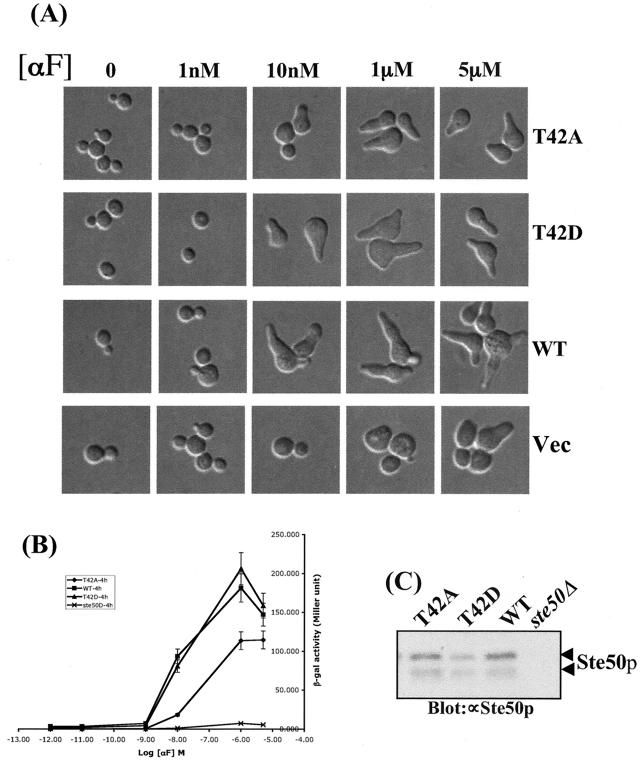
We next assessed the role of the potential regulatory phosphorylation site threonine 42 in functional-complementation assays. For these experiments, the T42A, T42D, and wild-type versions of Ste50p were expressed under the control of the STE50 promoter on a centromere (CEN) plasmid and tested for the ability to complement Δste50 in both the pheromone response and the HOG pathways. We found that the T42A mutant showed a defect in pheromone response as measured by FUS1::LacZ induction. Figure 4B shows that the Ste50-T42Ap mutant was less responsive than the wild-type protein, as indicated by the dose-response curve for FUS1::LacZ induction. In addition, the mating efficiency decreased ∼10-fold compared to the wild type. (Mating efficiencies were as follows: STE50, 13.8 ± 1.05; STE50-T42A, 1.70 ± 0.20; STE50-T42D, 18.0 ± 0.34; vector, <0.1 ± 0.05. The strain used was YCW315 [MAT a Δste50::TRP1] transformed with the STE50 alleles mentioned above. All STE50 alleles were carried on the CEN plasmid and expressed under the control of its own promoter. The mating efficiency is the number of diploids formed with the MATα tester strain DC17 cells divided by the number of plasmid-containing MATa cells in the mating mixture at the beginning of the experiment multiplied by 100. The data given are expressed as the averages of three repetitions ± standard deviations.) However, replacement of Thr42 with the phosphomimetic aspartic acid residue created no discernible phenotype, suggesting that phosphorylation of Thr42 is involved in the proper functioning of Ste50p. The phenotypes observed were not due to differences in protein concentrations, because the steady-state levels of both mutant and wild-type Ste50p proteins were very similar, as judged by Western blot analysis (Fig. 4C).
FIG. 4.
Phosphorylation of threonine 42 of Ste50p is required for the proper pheromone response. Yeast strain YCW393 (MATa Δste50::TRP1 FUS1::LacZ::LEU2 sst1::hisG) was transformed with STE50 alleles expressed from its own promoter on a CEN plasmid or with a control vector. Exponentially growing cells were treated with the indicated concentrations of α mating factor (αF) for 4 h. (A and B) Cell morphology was monitored by microscopy (A), and mating-specific transcriptional activation was measured by β-galactosidase assays (B) as described in Materials and Methods. Each data point represents the average of triplicates ± standard deviation. (C) The steady-state level of Ste50p protein before induction was analyzed by Western blotting with an anti-Ste50p (∝Ste50p) polyclonal antibody. The arrowheads indicate Ste50p; the lower band is likely to be a degradation product.
Threonine 42 is phosphorylated by CKI in vitro.
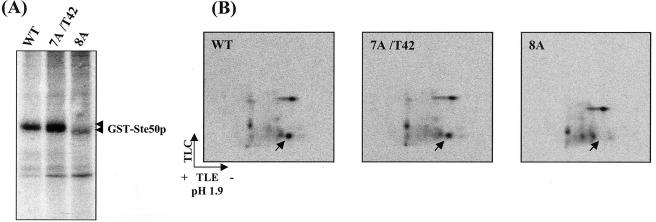
To identify the kinase that is responsible for the phosphorylation of Thr42 of Ste50p, we first identified the protein kinase(s) that could phosphorylate Ste50p in vitro. Ste50p was expressed and purified as a GST fusion protein from E. coli and used as a substrate in a kinase assay screen with a yeast kinase library (see Materials and Methods). The protein kinases from the library were purified from yeast as pools of eight kinases. The positive pools were then deconvoluted to identify candidate kinases. This in vitro kinase screening identified Yck1p, Gin4p, and Rim11p as kinases capable of phosphorylating Ste50p in vitro (Fig. 5A). Further analysis showed that Yck1p phosphorylated both the N-terminal portion (aa 1 to 67) and the C-terminal portion (aa 99 to 346) of Ste50p, whereas Gin4p phosphorylated only the C-terminal portion. Rim11p phosphorylated both the N- and C-terminal fragments of Ste50p but gave a relatively weak signal on the N terminus of the protein (Fig. 5B). These results identified Yck1p and/or Rim11p as a candidate kinase for the phosphorylation of Thr42 of Ste50p. Sequence analysis indicated that Thr42 matches well with the canonical consensus sequence (D/E)n-X-X-S/T (where D/E is aspartic acid or glutamic acid, S/T is serine or threonine, X is any amino acid residue, and n = 1 to 3) for CKI phosphorylation (reference 45 and references therein). To determine if Thr42 of Ste50p is phosphorylated by Yck1p in vitro, the aa 1 to 67 fragment of Ste50p, which contains all the S/T residues in the SAM domain (Fig. 5C), was subcloned from the various mutants. These fragments were expressed, purified from E. coli, and used as substrates in the in vitro kinase assays with Yck1p. The results of the kinase assays are summarized in Fig. 5D. The fragment aa 1 to 67 of wild-type Ste50p was phosphorylated extensively by Yck1p, indicating that it is a good in vitro substrate for the kinase. In contrast, the phosphorylation was totally abolished when the first eight S/T residues in this fragment were mutated to alanine residues (8A), indicating that the Yck1p phosphorylation site(s) is among the first eight S/T residues. The 8A construct also served as a negative control for the kinase assay to show that Yck1p does not phosphorylate the GST part of the fusion protein. In contrast, the Ste50p fragment with all of the first eight S/T residues except Thr42 mutated to alanine residues (7A/T42) was a good substrate for Yck1p, indicating that the Thr42 residue of Ste50p represents a good Yck1p phosphorylation site in vitro. To check if Thr42 was the sole Yck1p phosphorylation site, the Ste50p fragment (aa 1 to 67) with only the T42A mutation was used in the kinase assay. The Ste50(1-67)T42Ap fragment was efficiently phosphorylated by Yck1p, indicating that Thr42 is not the sole Yck1p phosphorylation site in this region of Ste50p. Other Yck1p in vitro phosphorylation sites are limited to the first five S/T residues, since the Ste50(1-67)p fragment with the first six S/T residues (including Thr42) mutated to alanine was not phosphorylated by Yck1p. Neither Gin4p nor Rim11p phosphorylated the T42 site of Ste50 in vitro using Ste50(1-67)7A/T42p as a substrate (Fig. 5E), indicating that T42 is phosphorylated specifically by Yck1p in vitro, although there is still the possibility that Rim11p phosphorylates other S/T sites within the first 67 aa of Ste50p.
Although the screen identified only Yck1p as a Ste50p in vitro kinase, three other casein kinase isomers (Yck2p, Yck3p, and Hrr25p) exist in S. cerevisiae. We asked whether these other casein kinase isomers could also phosphorylate Ste50p in vitro. Yck2p and Hrr25 (Yck3p was found not to be expressed in the library) were individually purified from the yeast expression library and used in the kinase assay and were found to effectively phosphorylate both the full-length GST-Ste50p and the N-terminal region (aa 1 to 67) of Ste50p in vitro. These kinases also phosphorylated GST-Ste50(1-67)7A/T42p, but not GST-Ste50(1-67)8Ap, in vitro (data not shown).
Ste50p is phosphorylated on threonine 42 in vivo.
To confirm that Thr42 was phosphorylated in vivo, the 8A and 7A/T42 mutants, as well as the wild-type Ste50p, were expressed as GST fusion proteins in a yeast strain with STE50 deleted and then subjected to [32P]orthophosphate metabolic labeling. The labeled Ste50p was purified, resolved by SDS-PAGE (Fig. 6A), and analyzed by tryptic phosphopeptide mapping. As shown in Fig. 6B, a single phosphopeptide that was observed on the maps of both the wild-type and the 7A/T42 mutant was absent from the map of the 8A mutant. Since the only difference between the 8A and 7A/T42 mutants was that Thr42 in the 7A/T42 mutant was replaced by a nonphosphorylatable alanine in the 8A mutant, Thr42 of Ste50p was the only residue responsible for the appearance of the phosphopeptide that was absent in the map obtained from the 8A mutant. This result strongly suggests that Thr42 of Ste50p was phosphorylated in vivo, most likely by a CKI isomer.
FIG. 6.
Ste50p is phosphorylated on threonine 42 in vivo. Ste50p wild type (WT) and the 7A/T42 and 8A mutants were expressed as GST fusion proteins and metabolically labeled with [32P]orthophosphate and purified with glutathione-Sepharose beads. (A) The labeled Ste50p was resolved on SDS-PAGE; the arrowheads indicate GST-Ste50p. (B) The bands corresponding to Ste50p in panel A were excised, digested with trypsin, and subjected to 2-D phosphopeptide mapping. The arrows indicate the positions of a phosphopeptide which is present in the maps of both wild-type Ste50p and the 7A/T42 mutant but absent from the map of the 8A mutant.
T42D does not bypass the requirement for CKI activity for efficient mating.
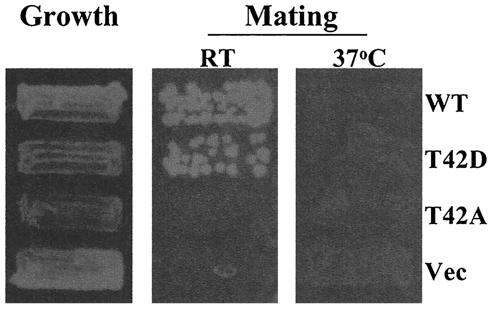
The above-mentioned data show that Thr42 is phosphorylated by CKI and that the Ste50p T42A mutant is unable to suppress the mating defect of strains with STE11ΔSAM, suggesting that phosphorylation of Ste50p on Thr42 is required for the suppression effect. To determine if the phosphomimetic mutant (T42D) of Ste50p is able to bypass the requirement for yeast CKI activity in this process, we constructed a yeast strain with the genotype STE11ΔSAM ssk1 Δyck1 Δyck2ts. This strain (YCW1151) was temperature sensitive for growth at 37°C and had a nearly sterile phenotype at room temperature, as judged by plate mating assays; SSK1 was also deleted to make the strain hyperosmotically sensitive, which can be useful for other assays. This strain was transformed with the overexpression constructs STE50, STE50-T42A, and STE50-T42D, as well as a control vector, and the transformants were tested for their mating responses. As expected, both wild-type Ste50p and T42D suppressed the mating defect of the strain at 25°C, whereas the T42A mutant, like the control vector, failed to do so. At the nonpermissive temperature (37°C), the T42D mutant should be able to mate if phosphorylation of Ste50p Thr42 by YCK activity was the only requirement for the suppression of the mating defect of STE11ΔSAM. However, we found that Ste50-T42Dp was unable to give rise to a detectable level of mating, as judged by the plate mating assay (Fig. 7). This result is consistent with the observation that CKI activities are involved in multiple physiological processes, including mating (5, 16), some of which are collectively essential in S. cerevisiae (38, 47).
FIG. 7.
Replacement of T42 of Ste50p with aspartic acid does not bypass the requirement for Yck1p and/or Yck2p to suppress the mating defect of STE11ΔSAM. Yeast YCW1151 (MATa ura3 leu2 his3 Δssk1::KanR yck1 yck2 [supi]ts STE11ΔSAM) was transformed with various STE50 alleles (described in Fig. 3) and control plasmid (Vec) as indicated. The cells were tested for the ability to mate both at 25 (room temperature [RT]) and 37°C with the tester strain DC16 (MATa his1). For mating at 37°C, both the tester and the strain to be tested were preincubated for 2 h before the two strains were mixed to start the mating process at the same temperature for five more hours and then selected for diploids on a minimal medium plate. For mating at 25°C, the mating was allowed to proceed for 5 h at 25°C, and diploids were then selected at 37°C. WT, wild type.
The T42A mutation alters the subcellular fractionation profile of Ste50p.
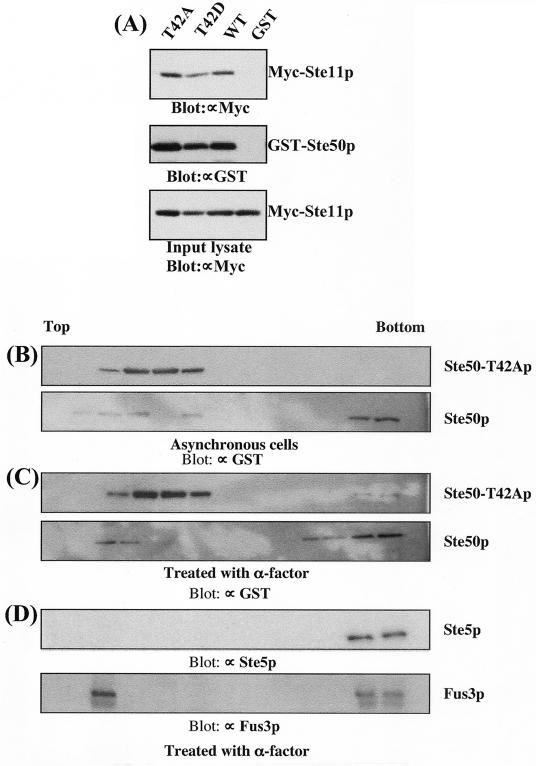
Since T42 is located within the N-terminal SAM domain core, we used a GST resin pull-down assay to examine whether the observed defect in the mating response was due to altered interaction of Ste50-T42Ap with Ste11p. To this end, the wild-type, T42A, and T42D versions of Ste50p expressed as GST fusion proteins, together with a control vector, were cotransformed with Myc-tagged Ste11p (49). The GST fusion proteins were purified on glutathione-Sepharose beads, and the presence of Ste11p was analyzed by Western blotting using an anti-Myc antibody (9E10). As shown in Fig. 8A, there was no significant difference in the amount of Myc-tagged Ste11p brought down by the wild type or the Thr42 mutants (either T42A or T42D) of Ste50p. This suggests that phosphorylation of Thr42 does not play a major role in modulating the interaction of Ste50p with Ste11p.
FIG. 8.
The T42A mutation retained the ability to interact with Ste11p but altered the subcellular fractionation profile of Ste50p. (A) T42A of Ste50p does not alter the association between Ste50p and Ste11p. Yeast strain YCW315 (MATa Δste50::TRP1) was cotransformed with pCW199 (Myc-tagged Ste11p) (49) and various STE50 alleles as GST fusions as indicated. Total cell extracts were prepared from ∼109 exponentially growing cells and incubated with glutathione-Sepharose beads (GSB). The beads were washed, and bound proteins were analyzed by immunoblotting them with an anti-Myc (∝Myc) (9E10) monoclonal antibody for the presence of Ste11p (top) and an anti-GST (∝GST) antibody for GST-Ste50p (middle). The presence of Myc-Ste11p in all the samples at similar levels before GSB pull-down is shown in the bottom blot, which represents ∼1/10 of the cell extract used in the pull-down experiment. (B to D) Total cell extracts from YCW315 expressing ether GST-Ste50p or GST-Ste50-T42Ap were fractionated on sucrose gradients. Proteins from each fraction were subjected to immunoblot analyses with the indicated antibodies.
However, signaling protein complexes usually involve multiple interactions among their components, so we asked whether the mutation had any influence on the formation of Ste50p-containing protein complexes in vivo. We fractionated cell extracts using sucrose velocity gradients and analyzed the protein distributions by immunoblotting. Comparative analysis of the fractionation profiles showed that the distribution of GST-Ste50-T42Ap was significantly different from that of the GST-Ste50 protein. Fractionation of the cell extracts prepared from asynchronously growing cells revealed that both GST-Ste50p and GST-Ste50-T42Ap were found in the light pool near the top of the gradient but that only the wild-type protein was found with heavy protein complexes at the bottom of the gradient (Fig. 8B). These differences became even more apparent when we analyzed the distribution of the GST-Ste50p-containing protein complexes prepared from pheromone-treated cells. Pheromone treatment resulted in more of the wild-type Ste50p pool shifting to the bottom of the density gradient. In contrast, pheromone treatment did not efficiently recruit the mutant Ste50-T42Ap to these heavy protein complexes; the majority of the T42A substituted protein remained at the top of the gradient (Fig. 8C). We also examined the distributions of both Ste5p and Fus3p from pheromone-treated cell extracts in the density gradient. As shown in Fig. 8D, this heavy fraction includes much of the Fus3p and essentially all of the Ste5p. These changes in the cosedimentation profiles suggest that the mutant Ste50p may be ineffectively associated with pheromone signaling complexes.
DISCUSSION
MAP3K nodes are important for the regulation of MAPK modules. One of the best-studied MAP3K proteins is Ste11p, which is shared by several signaling pathways in the yeast S. cerevisiae (24, 28, 34). Ste50p is an important regulator of Ste11p function (17, 35, 36, 49). Despite this regulatory role played by Ste50p in the MAPK cascades that share Ste11p, little is known about the regulation of Ste50p itself. In an attempt to gain some insight into the role that phosphorylation may play in modulating the function of Ste50p, we established by 2-D tryptic phosphopeptide mapping analysis that Ste50p is phosphorylated on multiple serine/threonine residues in vivo. One of these sites of phosphorylation is a target for CKI activity, and this site plays a role in the regulation of Ste50p function.
Using a yeast protein kinase library (55), we identified several protein kinases, including Yck1p, that can phosphorylate GST-Ste50p in vitro. It is evident that our screen may not have identified all possible protein kinases that could phosphorylate GST-Ste50p in vitro for two reasons: first, not all the protein kinases are present or expressed in the library; second, expression of multiple kinase clones as a pool may lead to the loss or underrepresentation of those clones that grow slowly. For example, Yck3p was not found in the library, and while Yck2p and Hrr25p were present in the library and were active as individual proteins, they were not picked up in the first round of the in vitro kinase screen using the pools of eight kinases. Nevertheless, this genomewide screen provides an approach to identify candidate kinases for a specific substrate in the absence of genetic or biological information.
We focused our study on the N-terminal portion of Ste50p, since it contains the functionally important and relatively well-studied SAM domain (17, 32, 35, 36, 49). The in vitro kinase assay established that Thr42 of Ste50p was phosphorylated by CKI. Although initially only Yck1p was identified as a relevant kinase, further analysis using other purified CKI isomers (Yck2p and Hrr25p) indicated that all yeast CKI isomers tested were able to phosphorylate GST-Ste50p in vitro. It is not surprising that CKI isomers lack specificity in the in vitro assay. It is believed that regulation of the specificity of this family of kinases is controlled in part by their spatial localization and proximity to substrates (13). Thus, more detailed in vivo studies in which both spatial organization and temporal regulation of CKI activity is maintained will be necessary to address the question of which YckI isomer(s) is responsible for the phosphorylation of T42 of Ste50p in vivo. However, it is perhaps significant that Yck1p and/or Yck2p is localized to the plasma membrane, where the mating response signals are initiated.
The CKI family of protein kinases is a group of highly related, ubiquitously expressed serine/threonine kinases found in all eukaryotic organisms from protozoa to mammals. With a wide subcellular distribution that includes the plasma-organelle membranes, the cytoplasm, and the nucleus, CKI family members have been implicated in diverse roles that control critical physiological processes, such as Wnt signaling, circadian rhythms, DNA repair, and nuclear import (31, 45, 56). There are at least seven CKI isomers that have been characterized in mammals (13, 45). Recently, it has been elegantly shown that CKIα, in concert with the glycogen synthase kinase-3 protein kinase, is responsible for the hyperphosphorylation of β-catenin, which is required to target β-catenin for degradation in the Wnt signaling pathway. CKI is required for the “priming” phosphorylation event (1, 53). CKI has also been shown to phosphorylate the NF-AT4 transcription factor to mask its nuclear import signal and thus regulate its subcellular localization (56).
The yeast S. cerevisiae has four CKI gene homologues: HRR25 and YCK3 are an essential pair that are involved in processes such as DNA repair and protein vesicle transport (47), whereas the YCK1 and YCK2 genes, which provide all the plasma membrane CKI activity, comprise another pair encoding an essential function (38). Yck1p and Yck2p have been shown to associate with the plasma membrane, to localize to sites of polarized growth, and to act in morphogenesis and septin organization (37, 38). It has been shown that yeast Yck1p and Yck2p are responsible for the phosphorylation of plasma membrane-localized proteins, including the H+-ATPase (10) and the α-factor receptor, Ste2p (16). The phosphorylation of Ste2p provides the signal for ubiquitination and internalization of the receptor (16). Yeast cells containing Ste2p variants that have C-terminal phosphorylation sites mutated show mating response defects, such as increased sensitivity to α-factor and moderately decreased mating efficiency (5). Similarly, the signal for ubiquitination and subsequent internalization of Ste3p, the a-factor receptor, is also mediated by Yck1p/Yck2p phosphorylation within the PEST-like sequence of the receptor. In this case, however, the phosphorylation is constitutive and independent of ligand binding (11, 30). Overexpression of Yck1p has also been reported to interfere with pheromone-induced cell cycle arrest in wild-type yeast cells (4).
It appears that residue Thr42 of Ste50p needs to be phosphorylated for a proper mating response, since the T42A mutation causes a defect in mating. The defect is not likely due merely to a structural change, since T42D-containing proteins behave indistinguishably from wild-type proteins. Furthermore, in yeast strain YCW365 (Δssk2 Δssk22 Δste50), both the T42A and T42D alleles complement Ste50p function in the HOG pathway in a manner indistinguishable from that of the wild type, suggesting that the interaction between Ste11p and Ste50p is not affected. This notion is supported by the fact that both the T42A and T42D versions of Ste50p can pull down Ste11p as well as the wild-type protein does. The residue Thr42 of Ste50p is located in the first helix of the SAM domain in the alignment with the structure of the Eph receptor SAM domain (43). Although the residue at this position is not highly conserved at the primary sequence level, many SAM domains from a variety of organisms have an acidic glutamic or aspartic acid residue at the Thr42-equivalent position (44). Interestingly, T42A, but not T42D, is severely defective in its ability to suppress the mating defect of Ste11ΔSAMp, which has no detectable interaction with Ste50p (17, 35, 49), suggesting that phosphorylation of Ste50p at T42 may be required to maintain or amplify a weak mating signal generated from the crippled Ste11p kinase. There is no suppression observed with a ste11 null mutation through overexpression of Ste50p, suggesting that the suppression is not taking place at a step downstream of Ste11p.
Our subcellular-fractionation results suggest that Ste50-T42Ap has an altered ability to form heavy protein complexes compared to the wild-type protein (Fig. 8). Such alteration may result in less Ste50-T42Ap localizing to the pheromone signaling protein core complex and thus lead to inefficient signaling. Overall, it appears that phosphorylation of residue Thr42 is important for the function of Ste50p in the mating response pathway, but intriguingly, the role of Ste50p in the HOG pathway does not appear to be influenced by the phosphorylation state of T42. This suggests that although the role of Ste50p is to serve as a coactivator of Ste11p in both pathways, posttranslational modifications of the protein can alter its relative effectiveness in one pathway or the other. Although our metabolic-labeling result indicated that the general phosphorylation of Ste50p did not seem to be affected by pheromone treatment and appeared to be constitutive, we cannot rule out the possibility that some phosphorylation site(s) may be dynamically regulated. Because the Yck1 and Yck2 proteins exhibit localization to sites of polarized growth that overlap with the observed localization of proteins like Ste20p, it is possible that the modulation of Ste50p function through casein kinase phosphorylation of Thr42 acts to fine tune the activation of Ste11p in the mating pathway. Yck1p and Yck2p also phosphorylate pheromone receptors and signal their internalization, which is necessary for proper mating functions, such as orientation or reorientation of mating projections (16, 46). Although the low level of expression of endogenous Ste50p and Ste11p has precluded their direct cellular localization, they function as parts of complexes that are localized to mating projections.
Sharing components among different MAPK cascades is a common phenomenon in eukaryotes. Although our understanding of why this architecture has evolved and how these shared components are differentially regulated is far from clear, it is generally accepted that cells use the sharing of MAPK cascade components to coordinate physiological processes among different pathways. It is conceivable that fine tuning of the signal distribution of the shared components is required for the differential regulation of specific pathways. In this case, the Ste50p function in the mating pathway may be fine tuned by yeast CKI-directed phosphorylation.
Acknowledgments
We thank H. Zhu and M. Snyder for the yeast kinase library and L. Robinson for strains.
G.J. was the recipient of a Deutsche Forschungsgemeinschaft postdoctoral fellowship and an NRC/NSERC postdoctoral fellowship. M.Z. was the recipient of an NRC/NSERC postdoctoral fellowship. This work was supported by the NRC/GHI and by grants from the Cancer Research Society to S.M. and MOP 85769 to D.Y.T. and M.W.
Footnotes
This is publication number 46158 of the National Research Council of Canada.
REFERENCES
- 1.Amit, S., A. Hatzubai, Y. Birman, J. S. Andersen, E. Ben-Shushan, M. Mann, Y. Ben-Neriah, and I. Alkalay. 2002. Axin-mediated CKI phosphorylation of beta-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev. 16:1066-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banuett, F. 1998. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 62:249-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyle, W. J., P. V. D. Geer, and T. Hunter. 1991. Phosphopeptide mapping and phosphoamino acid analysis by 2-dimensional separation on thin layer cellulose plates. Methods Enzymol. 201:110-149. [DOI] [PubMed] [Google Scholar]
- 4.Burchett, S., A. Scott, B. Errede, and H. G. Dohlman. 2001. Identification of novel pheromone-response regulators through systematic overexpression of 120 protein kinases in yeast. J. Biol. Chem. 276:26472-26478. [DOI] [PubMed] [Google Scholar]
- 5.Chen, Q., and J. B. Konopka. 1996. Regulation of the G-protein-coupled alpha-factor pheromone receptor by phosphorylation. Mol. Cell. Biol. 16:247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cobb, M. H. 1999. MAP kinase pathways. Prog. Biophys. Mol. Biol. 71:479-500. [DOI] [PubMed] [Google Scholar]
- 7.Cook, J. G., L. Bardwell, and J. Thorner. 1997. Inhibitory and activating functions for MAPK Kss1 in the S. cerevisiae filamentous-growth signalling pathway. Nature 390:85-88. [DOI] [PubMed] [Google Scholar]
- 8.Drogen, F., S. M. O'Rourke, V. M. Stucke, M. Jaquenoud, A. M. Neiman, and M. Peter. 2000. Phosphorylation of the MEKK Ste11p by the PAK-like kinase Ste20p is required for MAP kinase signaling in vivo. Curr. Biol. 10:630-639. [DOI] [PubMed] [Google Scholar]
- 9.Elion, E. A. 2000. Pheromone response, mating and cell biology. Curr. Opin. Microbiol. 3:573-581. [DOI] [PubMed] [Google Scholar]
- 10.Estrada, E., P. Agostinis, J. R. Vandenheede, J. Goris, W. Merlevede, J. Francois, A. Goffeau, and M. Ghislain. 1996. Phosphorylation of yeast plasma membrane H+-ATPase by casein kinase I. J. Biol. Chem. 271:32064-32072. [DOI] [PubMed] [Google Scholar]
- 11.Feng, Y., and N. G. Davis. 2000. Akr1p and the type I casein kinases act prior to the ubiquitination step of yeast endocytosis: Akr1p is required for kinase localization to the plasma membrane. Mol. Cell. Biol. 20:5350-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopalbhai, K., and S. Meloche. 1998. Repression of mitogen-activated protein kinases ERK1/ERK2 activity by a protein tyrosine phosphatase in rat fibroblasts transformed by upstream oncoproteins. J. Cell Physiol. 174:35-47. [DOI] [PubMed] [Google Scholar]
- 13.Gross, S. D., and R. A. Anderson. 1998. Casein kinase I: spatial organization and positioning of a multifunctional protein kinase family. Cell Signal. 10:699-711. [DOI] [PubMed] [Google Scholar]
- 14.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herskowitz, I. 1995. MAP kinase pathways in yeast: for mating and more. Cell 80:187-197. [DOI] [PubMed] [Google Scholar]
- 16.Hicke, L., B. Zanolari, and H. Riezman. 1998. Cytoplasmic tail phosphorylation of the alpha-factor receptor is required for its ubiquitination and internalization. J. Cell Biol. 141:349-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jansen, G., F. Buhring, C. P. Hollenberg, and M. Ramezani Rad. 2001. Mutations in the SAM domain of STE50 differentially influence the MAPK-mediated pathways for mating, filamentous growth and osmotolerance in Saccharomyces cerevisiae. Mol. Genet. Genomics 265:102-117. [DOI] [PubMed] [Google Scholar]
- 18.Leberer, E., D. Dignard, D. Harcus, D. Y. Thomas, and M. Whiteway. 1992. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 11:4815-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leberer, E., D. Dignard, L. Hougan, D. Y. Thomas, and M. Whiteway. 1992. Dominant-negative mutants of a yeast G-protein β subunit identify two functional regions involved in pheromone signalling. EMBO J. 11:4805-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leberer, E., D. Y. Thomas, and M. Whiteway. 1997. Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev. 7:59-66. [DOI] [PubMed] [Google Scholar]
- 21.Leberer, E., C. Wu, T. Leeuw, A. Fourest-Lieuvin, J. E. Segall, and D. Y. Thomas. 1997. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 16:83-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leeuw, T., A. Fourest-Lieuvin, C. Wu, J. Chenevert, K. Clark, M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Pheromone response in yeast: association of Bem1p with proteins of the MAP kinase cascade and actin. Science 270:1210-1213. [DOI] [PubMed] [Google Scholar]
- 23.Levin, D. E., and B. Errede. 1995. The proliferation of MAP kinase signaling pathways in yeast. Curr. Opin. Cell Biol. 7:197-202. [DOI] [PubMed] [Google Scholar]
- 24.Liu, H., C. Styles, and G. R. Fink. 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262:1741-1744. [DOI] [PubMed] [Google Scholar]
- 25.Madhani, H. D., and G. R. Fink. 1998. The riddle of MAP kinase signaling specificity. Trends Genet. 14:151-155. [DOI] [PubMed] [Google Scholar]
- 26.Madhani, H. D., C. A. Styles, and G. R. Fink. 1997. MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91:673-684. [DOI] [PubMed] [Google Scholar]
- 27.Marshall, C. J. 1995. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80:179-185. [DOI] [PubMed] [Google Scholar]
- 28.O'Rourke, S. M., and I. Herskowitz. 1998. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12:2874-2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan, X., T. Harashima, and J. Heitman. 2000. Signal transduction cascades regulating pseudohyphal differentiation of Saccharomyces cerevisiae. Curr. Opin. Microbiol. 3:567-572. [DOI] [PubMed] [Google Scholar]
- 30.Panek, H. R., J. D. Stepp, H. M. Engle, K. M. Marks, P. K. Tan, S. K. Lemmon, and L. C. Robinson. 1997. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel clathrin AP-like complex. EMBO J. 16:4194-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peters, J. M., R. M. McKay, J. P. McKay, and J. M. Graff. 1999. Casein kinase I transduces Wnt signals. Nature 401:345-350. [DOI] [PubMed] [Google Scholar]
- 32.Ponting, C. P. 1995. SAM: a novel motif in yeast sterile and Drosophila polyhomeotic proteins. Protein Sci. 4:1928-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Portela, P., S. Howell, S. Moreno, and S. Rossi. 2002. In vivo and in vitro phosphorylation of two isoforms of yeast pyruvate kinase by protein kinase A. J. Biol. Chem. 277:30477-30487. [DOI] [PubMed] [Google Scholar]
- 34.Posas, F., and H. Saito. 1997. Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276:1702-1705. [DOI] [PubMed] [Google Scholar]
- 35.Posas, F., E. A. Witten, and H. Saito. 1998. Requirement of STE50 for osmostress-induced activation of the STE11 mitogen-activated protein kinase kinase kinase in the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 18:5788-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramezani Rad, M., G. Jansen, F. Buhring, and C. P. Hollenberg. 1998. Ste50p is involved in regulating filamentous growth in the yeast Saccharomyces cerevisiae and associates with Ste11p. Mol. Gen. Genet. 259:29-38. [DOI] [PubMed] [Google Scholar]
- 37.Robinson, L. C., C. Bradley, J. D. Bryan, A. Jerome, Y. Kweon, and H. R. Panek. 1999. The Yck2 yeast casein kinase 1 isoform shows cell cycle-specific localization to sites of polarized growth and is required for proper septin organization. Mol. Biol. Cell. 10:1077-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson, L. C., E. J. Hubbard, P. R. Graves, A. A. DePaoli-Roach, P. J. Roach, C. Kung, D. W. Haas, C. H. Hagedorn, M. Goebl, M. R. Culbertson, et al. 1992. Yeast casein kinase I homologues: an essential gene pair. Proc. Natl. Acad. Sci. USA 89:28-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson, M. J., and M. H. Cobb. 1997. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 9:180-186. [DOI] [PubMed] [Google Scholar]
- 40.Rose, M. D., F. Winston, and P. Hieter. 1990. Methods in yeast genetics. A laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Saiki, R. J., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487-491. [DOI] [PubMed] [Google Scholar]
- 42.Schaeffer, H. J., and M. J. Weber. 1999. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol. Cell. Biol. 19:2435-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sprague, G. F., Jr. 1998. Control of MAP kinase signaling specificity or how not to go HOG wild. Genes Dev. 12:2817-2820. [DOI] [PubMed] [Google Scholar]
- 44.Stapleton, D., I. Balan, T. Pawson, and F. Sicheri. 1999. The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6:44-49. [DOI] [PubMed] [Google Scholar]
- 45.Tobin, A. B. 2002. Are we beta-ARKing up the wrong tree? Casein kinase 1 alpha provides an additional pathway for GPCR phosphorylation. Trends Pharmacol. Sci. 23:337-343. [DOI] [PubMed] [Google Scholar]
- 46.Vallier, L. G., J. E. Segall, and M. Snyder. 2002. The alpha-factor receptor C-terminus is important for mating projection formation and orientation in Saccharomyces cerevisiae. Cell Motil. Cytoskeleton 53:251-266. [DOI] [PubMed] [Google Scholar]
- 47.Wang, X., M. F. Hoekstra, A. J. DeMaggio, N. Dhillon, A. Vancura, J. Kuret, G. C. Johnston, and R. A. Singer. 1996. Prenylated isoforms of yeast casein kinase I, including the novel Yck3p, suppress the gcs1 blockage of cell proliferation from stationary phase. Mol. Cell. Biol. 16:5375-5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Widmann, C., S. Gibson, M. B. Jarpe, and G. L. Johnson. 1999. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79:143-180. [DOI] [PubMed] [Google Scholar]
- 49.Wu, C., E. Leberer, D. Y. Thomas, and M. Whiteway. 1999. Functional characterization of the interaction of Ste50p with Ste11p MAPKKK in Saccharomyces cerevisiae. Mol. Biol. Cell. 10:2425-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu, C., V. Lytvyn, D. Y. Thomas, and E. Leberer. 1997. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J. Biol. Chem. 272:30623-30626. [DOI] [PubMed] [Google Scholar]
- 51.Wu, C., M. Whiteway, D. Y. Thomas, and E. Leberer. 1995. Molecular characterization of Ste20p, a potential mitogen-activated protein or extracellular signal-regulated kinase kinase (MEK) kinase kinase from Saccharomyces cerevisiae. J. Biol. Chem. 270:15984-15992. [DOI] [PubMed] [Google Scholar]
- 52.Xu, G., G. Jansen, D. Y. Thomas, C. P. Hollenberg, and M. Ramezani Rad. 1996. Ste50p sustains mating pheromone-induced signal transduction in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 20:773-783. [DOI] [PubMed] [Google Scholar]
- 53.Yanagawa, S., Y. Matsuda, J. S. Lee, H. Matsubayashi, S. Sese, T. Kadowaki, and A. Ishimoto. 2002. Casein kinase I phosphorylates the Armadillo protein and induces its degradation in Drosophila. EMBO J. 21:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou, Z., A. Gartner, R. Cade, G. Ammerer, and B. Errede. 1993. Pheromone-induced signal transduction in Saccharomyces cerevisiae requires the sequential function of three protein kinases. Mol. Cell. Biol. 13:2069-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu, H., J. F. Klemic, S. Chang, P. Bertone, A. Casamayor, K. G. Klemic, D. Smith, M. Gerstein, M. A. Reed, and M. Snyder. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26:283-289. [DOI] [PubMed] [Google Scholar]
- 56.Zhu, J., F. Shibasaki, R. Price, J. C. Guillemot, T. Yano, V. Dotsch, G. Wagner, P. Ferrara, and F. McKeon. 1998. Intramolecular masking of nuclear import signal on NF-AT4 by casein kinase I and MEKK1. Cell 93:851-861. [DOI] [PubMed] [Google Scholar]