Abstract
The complete sequence of the double-stranded DNA (dsDNA) genome of the Salmonella enterica serovar Typhimurium ST64B bacteriophage was determined. The 40,149-bp genomic sequence of ST64B has an overall G+C content of 51.3% and is distinct from that of P22. The genome architecture is similar to that of the lambdoid phages, particularly that of coliphage λ. Most of the putative tail genes showed sequence similarity to tail genes of Mu, a nonlambdoid phage. In addition, it is likely that these tail genes are not expressed due to insertions of fragments of genes related to virulence within some of the open reading frames. This, together with the inability of ST64B to produce plaques on a wide range of isolates, suggests that ST64B is a defective phage. In contrast to the tail genes, most of the head genes showed similarity to those of the lambdoid phages HK97 and HK022, but these head genes also have significant sequence similarities to those of several other dsDNA phages infecting diverse bacterial hosts, including Escherichia, Pseudomonas, Agrobacterium, Caulobacter, Mesorhizobium, and Streptomyces. This suggests that ST64B is a genetic mosaic that has acquired significant portions of its genome from sources outside the genus Salmonella.
Natural relatives of bacteriophage λ have been isolated from various sources, with most of them growing on Escherichia coli and a few on Salmonella enterica serovar Typhimurium (P22, L, LP7, ES18, and ST64T) (3, 12). The best studied of the temperate phages is bacteriophage λ that infects E. coli strains, and it is the archetype of the lambdoid phages. However, the best-studied Salmonella-infecting phage is P22 that is related to phage λ. The major aspects of the life cycles of λ and P22 are used as reference points for comparative studies with newly isolated bacterial viruses. Phage ST64B, like phage ST64T, was isolated from S. enterica serovar Typhimurium DT 64 strain 2558 (13). These phages were induced by mitomycin C, and after CsCl gradient centrifugation, two well-separated bands were evident; the phages were designated ST64T and ST64B to reflect their relative banding positions in the CsCl gradient. Purification of genomic DNA from the purified particles from each band established that these phages were distinct. ST64B could not be propagated on any strains tested, but ST64T could mediate phage type conversion, serotype conversion, and generalized transduction (11, 12, 13). The ST64B genomic sequence was 40,149 bp in size, with an overall G+C content of 51.3%, which is less than the 52% G+C content of ST64B's S. enterica host. The amino acid sequences of the inferred proteins in ST64B were used in searches for similarity to other sequences present in databases by using the BlastP algorithm (1).
Genomic similarity to other lambdoid phages.
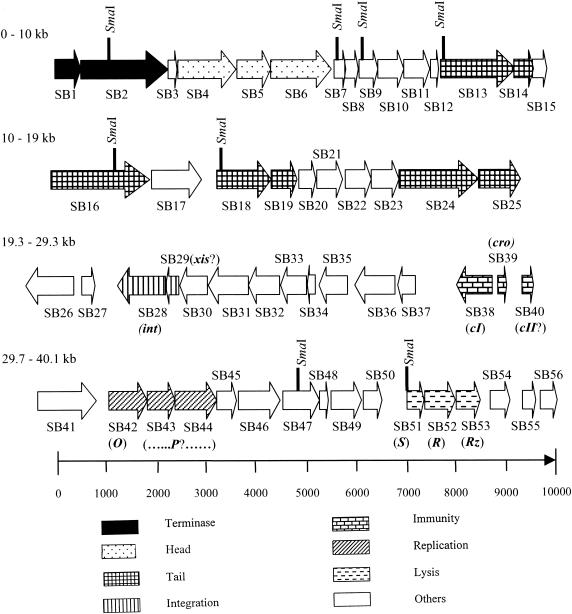
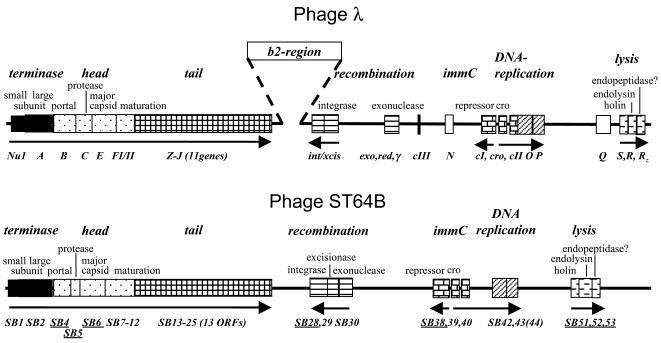
The genomic architecture of ST64B (Fig. 1) is similar to that of phage λ. This is illustrated by comparison of the genetic architectures of the ST64B and λ phages (Fig. 2). Phages ST64T and ST64B, despite being residents in the same S. enterica serovar Typhimurium DT 64 strain 2558, share almost no significant sequence identity (12); however, they share the same modular genetic architecture. In view of this finding, the λ nomenclature was adopted for ST64B open reading frames (ORFs) where possible, with the ORF designations prefixed SB (for ST64B) and the ORFs numbered consecutively from left to right, independent of orientation (Fig. 1). In general, ST64B putative genes showed only weak similarities to genes in the National Center for Biotechnology Information database. However, directions and orders of many ORFs showed that ST64B is a member of the lambdoid phage family.
FIG. 1.
Schematic representation of phage ST64B genome showing putative ORFs, with EcoRI and SmaI restriction sites shown. The ORFs are labeled SB1 to SB56. The map is circular, with the end of the map (40,149 bp) joined to the start of the map. Phage λ nomenclature was used to designate a few ORFs (in parentheses and italics) where possible.
FIG. 2.
Comparison of genetic maps of phages λ and ST64B. The maps are drawn to scale. The ORFs included in the map of ST64B are those whose inferred gene products function like the corresponding gene products of λ. The underlined ORFs show sufficient similarity with the corresponding genes of lambdoid phages, and they are considered homologues, whereas the other ORFs whose functions are deduced from similarities of their products to gene products of distant phages or bacteria are not underlined and are considered to be orthologues. The figure indicates high similarity between λ and ST64B ORF positions, the sizes of most of the corresponding genes, and their directions of transcription.
Significant identity of ST64B genes involved in replication and morphogenesis to similar genes from diverse bacterial and phage groups.
During its evolution, ST64B may have acquired a number of genes from outside the family Enterobacteriaceae. The putative terminase small subunit SB1 gene product has significant sequence identity to putative terminase small subunits of Lactococcus lactis phage BK5-T (34%) and prophage pi3 (29%). This is also evident for the SB2 gene product (putative large terminase), which shows significant identity to similar proteins from Pseudomonas aeruginosa phage D3 (46%), Staphylococcus aureus phage phiSLT (30%), and E. coli phage CP-933C (30%) and to a phage-like protein of Haemophilus influenzae (31%; accession no. AAF27357) and phage-like sequences from Clostridium acetobutylicum (29%; accession no. NP_348518). Moreover, the SB2 gene product has lower identities (24 to 28%) to similar proteins from other phages, including Staphylococcus aureus phiPV83, Lactococcus casei phage A2, and Lactococcus lactis bIL285, as well as lambdoid phages HK022 and HK97.
In addition to putative gene products involved in ST64B replication, genes involved in morphogenesis have significant homologies outside the family Enterobacteriaceae. The SB5 (putative prohead protease) product has 37% identity to a Brevibacterium flavum bacteriophage BFK20 protein and 43% identity to prohead proteases of Rhodobacter capsulatus (accession no. AAF13181) and Caulobacter crescentus (accession no. AAK24750) phages. In addition, it has 35% sequence identity to the prohead proteases of HK022 and HK97.
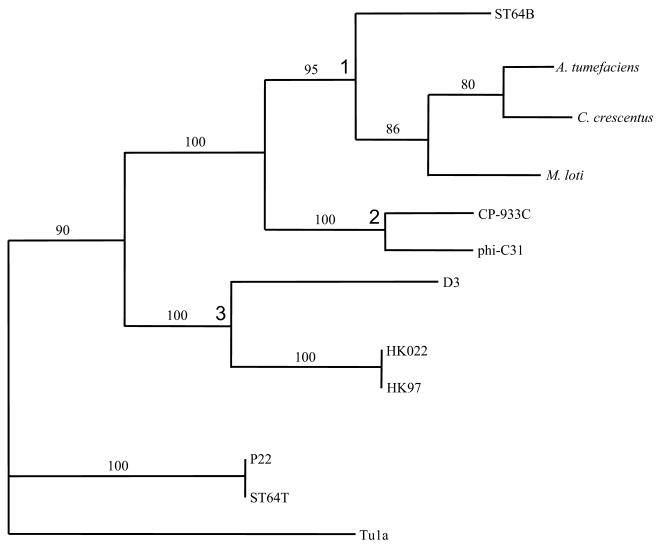
Initial BlastP analysis of the putative major capsid protein, SB6, revealed 43 and 37% sequence identities to major capsid proteins of Streptomyces phage φC31 and E. coli phage CP-933C, respectively. In view of this observation, an alignment of the putative major head proteins from a number of phages with high BlastP scores was performed using ClustalW in BioEdit version 4.8.10 (4) (data not shown). Maximum parsimony analysis of the 509-amino-acid consensus alignment was performed with 39 characters excluded; however, 422 characters were parsimony informative with the use of PAUP* version 4.05 (16). A consensus of two equally parsimonious trees suggested that the putative major head protein of ST64B was related to the major head proteins of phages found in Agrobacterium tumefaciens, Caulobacter crescentus, and Mesorhizobium loti at a confidence level of 95% as shown in branch 1 of Fig. 3. The branch 2 phages were related to the branch 1 phages at a confidence level of 100% pseudo replicates. However, ST64B was more distantly related to the branch 3 lambdoid phages D3, HK022, and HK97 at a confidence level of 90%. Indeed, BlastP search also revealed that the major head protein of ST64B has closer identity to the major head protein of φC31 than to those of HK97 and HK022. The most parsimonious tree of the same topology requires a total of 100 changes. A partial sequence of the major head protein of phage Tu1a, a T4 type phage, was employed as an out-group.
FIG. 3.
Phylogenetic dendrogram of putative and actual major head proteins of phages and prophages based upon the maximum parsimony analysis. Phage Tula (accession no. AF221994), a T4 type coliphage, was used to root the tree. Branch numbers are shown in boldface. Bootstrap values are indicated on branches. Where bacterial species designations are given, these data relate to putative phage-related proteins from these organisms. The accession numbers for the members of this tree are as follows: AAK86763 (Agrobacterium tumefaciens phage-like protein), AAK24747 (Caulobacter crescentus phage-like protein), AAG55944 (E. coli phage CP-933C), AAD38957 (Pseudomonas aeruginosa phage D3), AAF30354 (E. coli phage HK022), AAF31098 (E. coli phage HK97), NP_108602 (Mesorhizobium loti phage-like protein), NP_047927 (Streptomyces phage φC31), AAF75047 (Salmonella phage P22), AY052766 (Salmonella phage ST64T), and AF221994 (E. coli phage Tula). This tree indicates that the major head protein of ST64B is closely related to the prophage-related proteins in Agrobacterium tumefaciens, Caulobacter crescentus, and Mesorhizobium loti at a bootstrap confidence level of 95% pseudo replicates and is more distantly related to proteins of phages HK97, HK022, and D3. It is clearly shown that ST64B is not related to ST64T (isolated from the same S. enterica serovar Typhimurium DT 64 2558 strain) with regard to the major capsid protein.
In keeping with the overall lambdoid genetic architecture of the ST64B genome, the ST64B capsid genes in general resemble those of the lambdoid phages HK97, HK022, and D3.
Does ST64B have a functional tail?
Most of the ST64B putative tail genes showed similarity to those of phage Mu (a nonlambdoid phage), with two putative tail genes showing similarity to the putative tail genes of S. enterica serovar Typhimurium phages Fels-2, Fels-1, and Gifsy-2. Although ST64B has many putative tail genes, suggesting that it should have a longer tail than P22 and ST64T, icosahedral heads with short tails were observed when phage lysate containing both ST64B and ST64T was examined by transmission electron microscopy (11). Additionally, when the CsCl gradient-purified ST64B lysate was examined, the majority of phages showed icosahedral heads with no visible tails, and a few phages with short tails were present. Because ST64B and ST64T are carried by the same S. enterica serovar Typhimurium DT 64 2558 strain (11) and ST64B could not be propagated on any strain tested, it was not possible to test a pure lysate of ST64B. This may explain why a few phages with short tails were observed under transmission electron microscopy: they may be ST64T contaminants. ST64B bands at the bottom (more-dense location) on a CsCl gradient (13). This observation is consistent with the prediction that this phage does not have a tail. In addition, ST64B failed to produce plaques on many diverse Salmonella and E. coli strains (11, 13). It is possible that the complete phage, ST64T, that propagates autonomously in S. enterica serovar Typhimurium DT 64 strain 2558 (12, 13) trans-activates the defective genome of ST64B and compensates for some ST64B deficiencies other than the interrupted tail genes, allowing partially complete virions to be produced. In any case, it is unknown whether the ST64B putative tail genes are expressed.
Integration site of ST64B.
Analysis of the ST64B sequence revealed the phage-bacterium junction 162 bp downstream from the putative gene int (SB28). ST64B therefore integrates within the tRNA gene serU, which encodes a tRNA for serine, immediately upstream from the umuCD operon. Integration of phages within or close to tRNAs is not uncommon. In addition, many pathogenicity islands insert at sites close to tRNAs (2, 7, 15). Comparison of the nucleotide sequences of ST64B and the ST64B attP/attB clones amplified from S. enterica serovar Typhimurium DT 64 strain 2558 and S. enterica serovar Typhimurium LT2 (accession number AE008788) revealed a sequence shared by all three genomes (data not shown). The attB/attP clone showed perfect homology with ST64B left of the junction and almost perfect homology with both the S. enterica serovar Typhimurium DT 64 and LT2 sequences. The LT2 sequence, however, has a C deletion at position 269 at the right of the junction.
Fragments of genes associated with virulence genes.
Interestingly, ST64B carries fragments of genes involved in the virulence of Salmonella. Two of the genes, sopE and sspH2 (Salmonella-secreted protein H2) are associated with the SPI-1 and SPI-2 virulence-associated type III secretion systems (8, 9). Virulence-associated type III secretion systems permit gram-negative pathogens to translocate effector proteins directly into the host (6). A phage, SopEφ, that encodes a type III secretion system substrate, SopE, was identified (10). It was also observed that the sopE gene is flanked by sequences resembling tail and tail fiber genes of P2-like phages (5). Indeed, fragments of sopE genes identified in ST64B were found within the putative tail and tail fiber genes. The third virulence-associated gene (orgA) fragment found in ST64B was also found within a tail gene. However, the 3′ end of the sspH2 gene was not found within a tail gene. Although the sspH2 and the orgA genes found in ST64B did not have the core sequences for virulence and the sopE gene fragments were small, their presence in this phage supports the notion of horizontal gene transfer mediated by temperate phages. For example, effector proteins that evolved from other bacterial pathogens may become integrated into the genome of a temperate phage, and integration of this phage into the genome of a bacterium may provide these proteins to any susceptible pathogen bearing the type III secretion system (10). Moreover, both SPI-1 and SPI-2 are approximately 40 kb, a size which can be transduced by generalized transducing phages (9, 14), and ST64B integrates into the tRNA gene, a target for integration of many pathogenicity islands.
The immC region.
The SB38 gene product appears to be a putative CI repressor that shows 42 and 26% sequence identities to the CI repressor proteins of lambdoid phages HK022 and D3, respectively. Similar to what has been observed in the case of the replication and morphogenesis putative products, this gene product showed significant sequence identities (30, 28, 27, and 26%) to phage-like repressor proteins found in Xylella fastidiosa (accession no. XF1650), Haemophilus influenzae (accession no. A64031), Neisseria meningitidis (accession no. CAB71959), and Streptococcus pyogenes (accession no. NP_269132), respectively.
The putative SB39 product is a Cro-like protein encoded on the strand opposite that encoding the putative CI. It bears significant (34%) sequence identity to a probable DNA-binding protein of Streptomyces coelicolor (accession no. CAB39703) and 32% identity to probable members of a family of transcriptional regulators, MerR and LacI in Sinorhizobium meliloti (accession no. CAC49253). The finding that both SB38 and SB39 gene products have helix-turn-helix motifs and the relative map positions of these genes on the ST64B genome suggest strongly that SB38 and SB39 gene products are CI and Cro orthologues, respectively, in ST64B (Fig. 2). Likewise, the map position and size of SB40 suggest that it encodes a CII-like (λ CII orthologue) gene product (Fig. 2). However, no possible CII-binding site (TTGCN6TTGC) was found to be associated with this ORF. The absence of a CII-binding site similar to that found in λ indicates that the putative CII protein of ST64B is distinct from both the P22 C1 and λ CII proteins. This region encompassing the putative cI-, cro-, and cII-like genes is likely to represent the immunity C (immC) region of ST64B. Cloning of this region and expression in a number of S. enterica strains has resulted in phage type conversion of these strains; this further confirms the likelihood that these ORFs constitute a functional immC region (M. W. Heuzenroeder and C. P. Tucker, unpublished results).
Role of ST64B in the evolution of the genus Salmonella.
The genome of ST64B is clearly a mosaic, composed of putative genes with similarities to genes from phages of diverse bacterial groups, including gram-positive organisms. Mosaicism is clearly seen in the tail genes, with the majority of ST64B tail genes having identity to those of phage Mu and the remaining two putative tail genes having similarity to those of phages Fels-1, Fels-2, and Gifsy-2. Although the capsid genes showed most similarity to those of the lambdoid phages HK97, HK022, and D3, significant sequence similarity to capsid genes from outside the genus Salmonella and the family Enterobacteriaceae was also observed. In contrast, all three putative lysis genes of ST64B had strong sequence identity (93 to 100%) to those of the S. enterica serovar Typhimurium Fels-1 phage. This finding, together with the observation of fragments of putative virulence determinant genes usually found on the bacterial chromosome, suggests that ST64B could have or has played a role in the transfer of virulence determinants in the past. It is unlikely that ST64B is able to mediate transduction, since the tail genes are unlikely to be functional due to the insertion of the virulence gene fragments. These fragments, together with other ST64B genes with identity to genes from diverse sources, could act as targets for the integration of new virulence factors or marker rescue. The precise role ST64B is playing or has played in the evolution and epidemiology of Salmonella is yet to be determined.
Nucleotide sequence accession number.
The complete nucleotide sequence of bacteriophage ST64B has been deposited in GenBank and assigned accession number AY055382.
Acknowledgments
We thank Arthur Mangos and Lesley Rawlings for assistance with sequencing and phylogenetic analyses, respectively. The “Online Analysis Tools” website of Andrew Kropinski was invaluable for the analysis of sequence data. The assistance of the Australian Salmonella Reference Centre in phage typing and interpretation of results is gratefully acknowledged.
Finally, we thank the Australian Agency for International Development (AusAID) and the Rural Industries Research and Development Corporation (chicken meat) for their generous financial support.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Blum, G., M. Ott, A. Lischewski, A. Ritter, H. Imrich, H. Tschape, and J. Hacker. 1994. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect. Immun. 62:606-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, A. 1994. Comparative molecular biology of lambdoid phages. Annu. Rev. Microbiol. 48:193-222. [DOI] [PubMed] [Google Scholar]
- 4.Hall, T. A. 1999. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 5.Hardt, W.-D., H. Urlaub, and J. E. Galan. 1998. A substrate of the centisome 63 type III protein secretion system of Salmonella typhimurium is encoded by a cryptic bacteriophage. Proc. Natl. Acad. Sci. USA 95:2574-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hueck, C. J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McDaniel, T. K., K. G. Javis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miao, E. A., C. A. Scherer, R. M. Tsolis, R. A. Kingsley, L. G. Adams, A. J. Baumler, and S. I. Miller. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850-864. [DOI] [PubMed] [Google Scholar]
- 9.Mills, D. M., V. Bajaj, and C. A. Lee. 1995. A 40 kb chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol. Microbiol. 15:749-759. [DOI] [PubMed] [Google Scholar]
- 10.Mirold, S., W. Rabsch, M. Rohde, S. Stender, H. Tschape, H. Russmann, E. Igwe, and W. D. Hardt. 1999. Isolation of a temperate bacteriophage encoding the type III effector protein SopE from an epidemic Salmonella typhimurium strain. Proc. Natl. Acad. Sci. USA 96:9863-9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mmolawa, P. T. 2002. Ph.D. thesis. University of Adelaide, Adelaide, Australia.
- 12.Mmolawa, P. T., H. Schmieger, C. P. Tucker, and M. W. Heuzenroeder. 2003. Genomic structure of the Salmonella enterica serovar Typhimurium DT 64 bacteriophage ST64T: evidence for modular genetic architecture. J. Bacteriol. 185:3473-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mmolawa, P. T., R. Willmore, C. J. Thomas, and M. W. Heuzenroeder. 2002. Temperate phages in Salmonella enterica serovar Typhimurium: implications for epidemiology. Int. J. Med. Microbiol. 291:633-644. [DOI] [PubMed] [Google Scholar]
- 14.Shea, J. E., M. Hensel, C. Gleeson, and D. W. Holden. 1996. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun, J., M. Inouye, and S. Inouye. 1991. Association of a retro-element with a P4-like cryptic prophage (retronphage phi R73) integrated into the selenocystyl tRNA gene of Escherichia coli. J. Bacteriol. 173:4171-4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swofford, D. 1993. PAUP—phylogenetic analysis using parsimony, 3.1 ed. Natural History Survey, Champaign, Ill.