Abstract
The immunoreceptor tyrosine-based activation motifs (ITAMs) in the CD3 chains associated with the T cell receptor (TCR) are crucial for TCR signaling. To probe the role of the CD3γ–ITAM in T cell development, we created knock-in mice in which the CD3γ chain of the TCR complex is replaced by a mutant signaling-deficient CD3γ chain, lacking the CD3γ–ITAM. This mutation results in considerable impairment in positive selection in the polyclonal TCR repertoire. When CD3γ–ΔITAM mice are crossed to mice expressing transgenic F5 TCRs, their thymocytes are completely unable to perform positive selection in vivo in response to intrathymic ligands. Also, the in vitro positive selection response of double-positive (DP) thymocytes with F5–CD3γ–ΔITAM mutant receptors to their agonist ligand and many of its variants is severely impaired or abrogated. Yet, the binding and dissociation constants of agonist ligands for the F5 receptor are not affected by the CD3γ–ΔITAM mutation. Furthermore, DP thymocytes with mutant receptors can respond to agonist ligand with normal antigen sensitivity and to normal levels, as shown by their ability to induce CD69 up-regulation, TCR down-regulation, negative selection, and ZAP70 and c-Jun NH2-terminal kinase activation. In sharp contrast, induction of extracellular signal-regulated kinase (ERK) activation and linker for activation of T cells (LAT) phosphorylation are severely impaired in these cells. Together, these findings underscore that intrinsic properties of the TCR–CD3 complex regulate selection at the DP checkpoint. More importantly, this analysis provides the first direct genetic evidence for a role of the CD3γ–ITAM in TCR-driven thymocyte selection.
Keywords: thymocyte selection, CD3γ–ITAM, ITAM multiplicity, T cell activation, TCR signaling
Introduction
The CD3 chains associated with the TCR contain shared and semi-conserved immune receptor tyrosine-based activation motifs (ITAMs)* that link the TCR to the downstream signaling machinery by binding to key adaptor molecules and enzymes (1, 2; for reviews, see references 3 and 4). All of the signaling capacity of the TCR is contained within the CD3 chains (CD3γ, -δ, -ε, and -ζ). CD3γ, -δ, and -ε polypeptides each contain one and CD3ζ contains three ITAMs (YxxL/Ix6–8YxxL/I) within their cytoplasmic domains, and are assembled in each TCR complex as 3 dimers: γε, δε, and ζζ. Together, these dimers contribute 10 ITAMs to the TCR complex, and antigen receptor triggering results in phosphorylation of the tyrosine residues within the ITAMs by activated Src family kinases. The phosphorylation of ITAM tyrosine residues represents one of the earliest events in the TCR signaling cascade, permitting the recruitment of a number of SH2-domain–containing signaling proteins, of which the most crucial one is the Syk protein tyrosine kinase (PTK) ZAP70. SH2-domain–containing signaling proteins recruited to the TCR complex are subsequently phosphorylated and activated, allowing binding of additional signaling molecules to occur (for reviews, see references 3 and 5). It is clear therefore that the ITAMs are crucial for initiating TCR-mediated signaling, but the exact function of ITAM multiplicity remains to be elucidated.
The presence of multiple ITAMs within a single TCR complex may have several functions (for a review, see reference 6). CD3 components may make quantitative and partially overlapping contributions to TCR signaling, as the many ITAMs may provide the capacity to amplify signals required for reaching the activation threshold (2) through low affinity/avidity TCR–MHC interactions. In support of this view, a direct correlation between the number of CD3ζ–ITAMs and the efficiency of both positive and negative selection was observed in one particular TCR-transgenic model (7). Alternatively, supported by a number of studies (1, 8, 9) the individual ITAMs may function in signal discrimination. This may be the result of micro-heterogeneity in the amino acids flanking the phosphotyrosine residues within the ITAMs, which are able to specify interactions with distinct SH2-domain–containing signaling proteins (10, 11). Indeed, in vitro studies indicate that ITAMs derived from different CD3 chains bind preferentially to distinct signaling molecules, or bind with varying affinities (12–17; for a review, see reference 4). Certainly, the different hypotheses concerning the role of ITAM multiplicity are not mutually exclusive, and signal discrimination may be the result of quantitative differences in signal strength (6, 8, 18).
Most in vivo studies on the possible role of ITAM multiplicity in TCR signal specification have been performed on CD3ζ–ITAMs (2, 7, 8, 19, 20), and one on CD3ε–ITAM (21). In the present study, we addressed whether the CD3γ–ITAM performs a role in TCR signal specification. From our earlier studies in CD3γ-deficient mice, it is clear that the CD3γ chain is required for β-selection of DN thymocytes, most likely as a consequence of its crucial role in pre-TCR assembly (22). CD3γ-deficient mice therefore barely produce CD4+CD8+ (double positive [DP]) thymocytes and mature T cells, precluding an evaluation of the possible role of CD3γ at later stages of development. Moreover, as assembly of the mature TCR complex is severely hampered by absence of CD3γ (23), the exact contribution of CD3γ to selection of the TCR repertoire and mature T cell function is difficult to assess. Yet, when both the block in β-selection and TCR levels on DP thymocytes are rescued in CD3γ-deficient mice, through introduction of transgenic F5 TCRs, positive selection can still not occur (see Results). This suggests that CD3γ contributes specific signals required for positive selection, and this possibility was tested directly in knock-in mice expressing CD3γ chains lacking only the CD3γ–ITAM (23), leaving intact the contribution of CD3γ to assembly of the TCR–CD3 complex. These mice therefore have normal TCR expression levels, and we here show that this mutation profoundly affects positive selection, but not negative selection at the DP checkpoint.
During thymocyte selection, signals triggered by the TCR specify cell fate of immature thymocytes at the DP stage. Under certain conditions, TCR signals trigger apoptosis, resulting in negative selection of those DP thymocytes with self-reactive TCRs. Paradoxically, TCR signals can also trigger survival and further differentiation in DP thymocytes (positive selection) (for a review, see reference 24), and one of the central questions in the developmental biology of T cells is how the distinction between positive and negative selection signals is made. Both quantitative and qualitative models have been proposed, and models that merge the two concepts, such that quantitative differences in the strength of the stimulus can result in qualitative differences in the signals transduced (18; also reviewed in reference 6). In a strictly quantitative model, strong activation signals result in apoptosis (negative selection), and weaker signals in positive selection (25). The mitogen-activated protein kinase (MAPK) cascades have been suggested to be the “instruments” for dictating the qualitatively distinct biological outcomes that may result from quantitative differences in signal strength. Specifically, the ERK cascade has been implicated in positive selection (26–35), and the c-Jun NH2-terminal kinase (JNK) and p38 cascade in negative selection (18, 30, 36, 37). While it is to be expected that the decision between positive and negative selection is the consequence of engagement of different signaling pathways, it remains to be defined how individual TCRs link to distinct cascades. We here present evidence for a contribution of the signaling moiety of one of the TCR-associated CD3 chains, the CD3γ–ITAM, to signal specification at the DP checkpoint.
Materials and Methods
Mice.
Mice were maintained under specific pathogen-free conditions in the animal colony of the Netherlands Cancer Institute and analyzed at 6–8 wk of age unless indicated otherwise. CD3γ-deficient, CD3γ–ΔITAM mutant, and F5-recombination activating gene (RAG)−/− mice have been described in detail elsewhere (22, 23, 38). In all experiments in which the F5-transgenic TCR is introduced onto wild-type (WT) or mutant mice, the RAG-deficient background is used, to prevent expression of endogenous TCRs.
Flow Cytometry.
Single cell suspensions were prepared in PBA (1× PBS, 1% BSA, 0.02% NaN3), aliquoted into wells of a 96-well plate (105–106 cells/well), and pelleted at 200 g for 2 min. Cells were resuspended in 20 μl of anti-FcγRII/III (clone 2.4G2; final concentration 1 μg/ml) to reduce nonspecific staining, and incubated for 10 min at 4°C. Staining for cell-surface antigen expression was performed at saturating mAb concentrations for 20–30 min at 4°C. Cells were washed twice in 100 μl of PBA and incubated with second-step reagent if necessary. Finally, all samples were washed twice and resuspended in 100 μl of PBA. Cells were analyzed on a Becton Dickinson FACSCalibur™. Forward- and side-scatter gating and/or propidium iodide (PI) gating was used to exclude dead cells from the analysis.
Biotinylated, FITC-, PE-, or allophycocyanin (APC)-conjugated antibodies specific for murine CD4 (clone RM4–5), CD5 (clone 53–7.3), CD8α (clone 53–6.7), CD8β (clone 53–5.8), CD24 (HSA; clone M1/69), CD69 (clone H1.2F3), and TCRβ (clone H57–597) were obtained from BD PharMingen. R-PE anti–mouse CD4 (clone CT-CD4) was purchased from Caltag. Where appropriate, streptavidin (SA)-Tricolor or SA-PE (Caltag) were used as second-step reagents.
Preparation of PE- or APC-conjugated H-2Db-tetramers containing the nucleoprotein peptide (NP366–374(NT); ASNENMDAM) of the influenza A virus strain A/NT/60/68 has been described previously (39).
Fetal Thymic Organ Cultures.
Fetal thymic lobes were prepared from mice at day 14 of gestation. They were cultured on filter discs on gelfoam in Iscove's modified Dulbecco's medium supplemented with 10 mM Hepes buffer, nonessential amino acids, 4 mM l-glutamine, penicillin, streptomycin (all from GIBCO BRL), 5 × 10−5 M 2-β-mercaptoethanol, and 20% fetal calf serum. At the indicated time-points, single cell suspensions were prepared and thymocytes were examined by FACS® analysis.
T Cell Proliferation.
Lymph node T cells (2 × 105 cells/well) were prepared and cultured as described (23) with graded concentrations of the nominal peptide ASNENMDAM or a negative control peptide ASNANMDAM (this peptide variant lost H-2Db binding capacity) in a total volume of 200 μl complete medium (Iscove's modified Dulbecco's medium [GIBCO BRL] supplemented with 10% FCS [PAA Laboratories GmbH], 2 × 10−5 M 2-mercaptoethanol [Merck], 100 U/ml penicillin, and 100 μg/ml streptomycin) in a round-bottomed microtiter well at 37°C. After 3 d, cultures were pulsed with 0.5 μCi/well [3H]thymidine for 18 h and thymidine incorporation was measured by liquid scintillation counting. All assays were performed in triplicate and SDs were <10% of the mean.
MHC–TCR Dissociation Rates.
Determination of TCR–MHC off-rates were performed essentially as described previously (40). Briefly, thymocytes were stained with PE-conjugated H-2Db-ASNENMDAM-tetramers for 20 min at 4°C, washed once in PBA (1× PBS, 1% BSA, 0.02% NaN3), and subsequently, an excess of homologous unlabeled H-2Db monomers (10 μM) was added. Decay of H-2Db tetramer staining was analyzed by flow cytometry on electronically gated DP thymocytes at indicated times and plotted as the percentage of maximum staining as follows: (FIexp − FI0)/(FImax − FI0) × 100%. Simultaneous staining of H-2Db tetramers and 10 μM unlabeled homologous H-2Db monomers prevents the binding of tetrameric MHCs (not shown).
TCR Down-regulation.
Total thymocytes (105 cells/well) were stimulated with indicated amounts of soluble H-2Db-ASNENMDAM-tetramers in a total volume of 200 μl complete medium in a flat-bottomed microtiter well at 37°C. After 1 or 5 h, cells were stained with antibody combinations allowing gating on the CD4+CD8+ T cells and simultaneously determining the levels of TCRβ expression. 100% value corresponds to the mean fluorescence of the TCRβ staining on unstimulated DP thymocytes.
In Vitro Thymocyte Stimulation Assays.
DP thymocytes were purified by panning total thymocytes on anti-CD4 (20 μg/ml) coated petri dishes twice, and purity was >98%, as determined by flow cytometry. TCR levels were equal on the CD69-negative sorted DP cells (see Figs. 4 B and 5 A). Stimulation of DP thymocytes in dispersed culture were essentially performed as described (41), with the exception that the LEC B cell lymphoma line (42, 43) was used as a source of antigen presenting cells. Briefly, graded concentrations of the immuno-dominant peptide of the influenza A virus ASNENMDAM and a panel of 8 variant ligands were added individually to purified DP thymocytes (1 × 105 cells/well) in the presence of LEC cells (0.5 × 105 cells/well) in a total volume of 200 μl complete medium in a flat-bottomed microtiter well at 37°C. After 18–24 h of culture, analysis of CD69 up-regulation, CD4 and CD8 expression, and thymocyte cell death by propidium iodide staining was performed. It should be noted that the absolute numbers of CD8 cells (confirmed to express both CD8α and CD8β) increased in a peptide-dependent fashion, and therefore does not merely represent selective survival.
Figure 4.
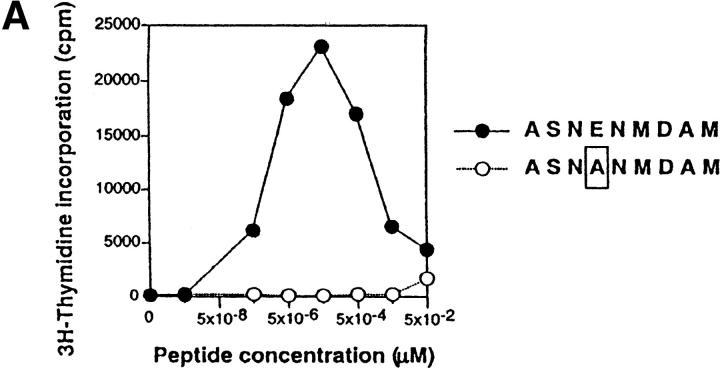
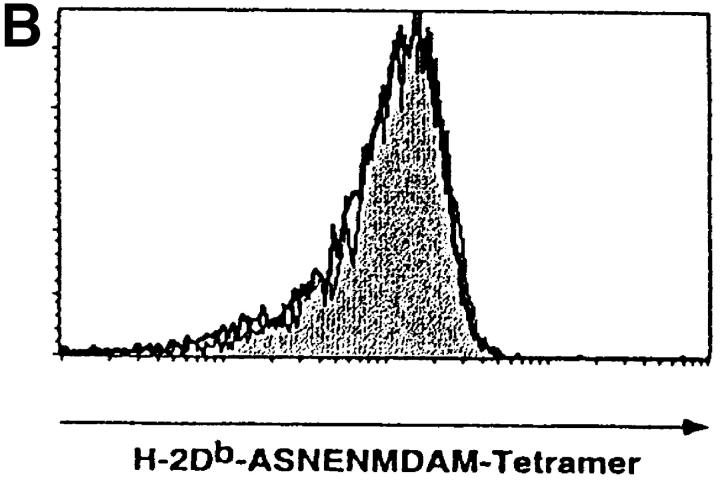
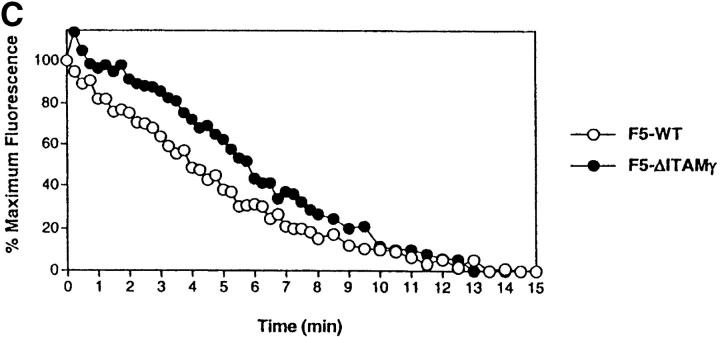
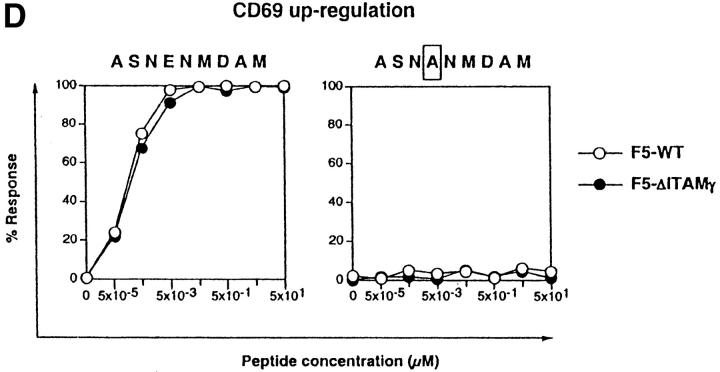
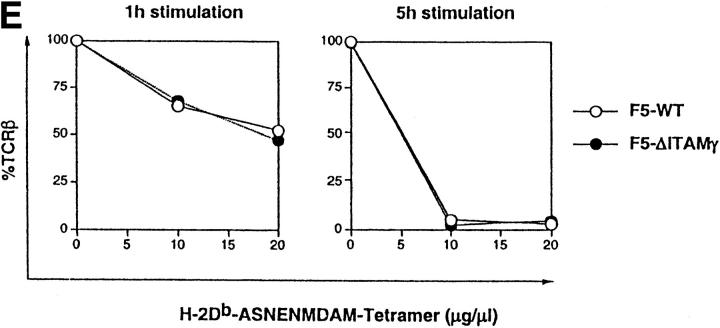
Loss of the CD3γ–ITAM in mice with F5 TCRs does not affect ligand binding and off-rate, nor does it affect CD69 up-regulation and TCR down-regulation. (A) F5–WT lymph node cells were stimulated with graded concentrations of the nominal peptide ASNENMDAM (filled circles) or the negative control peptide ASNANMDAM mutated in the critical MHC-anchor residue at position 4 (open circles). After 3 d, proliferation was assessed by pulsing cultures with [3H]thymidine for 18 h. Data correspond to the mean of triplicate cultures. (B) Sorted DP thymocytes from 6–8-wk-old F5–WT (white histogram, thick line) and F5–ΔITAMγ (gray histogram, thin line) mice were analyzed for binding to H2-Db-ASNENMDAM-tetramers. Note that this analysis is for electronically gated CD69-negative DP cells, whereas the TCRβ staining shown in Fig. 2 is for all DP cells. (C) Comparison of the off-rate of H2-Db-ASNENMDAM-tetramers to F5–WT (open circles) and F5–ΔITAMγ mutant TCRs (filled circles). Fluorescence intensity corresponds to: FIexp − FI0. (D) Purified DP thymocytes of F5–WT (open circles) and F5–ΔITAMγ mice (filled circles) were incubated overnight in the presence of LEC cells as APCs with the indicated concentrations of the immuno-dominant peptide ASNENMDAM (left panel) or a negative control peptide ASNANMDAM (right panel). Cells were analyzed for up-regulation of the early activation marker CD69, and are presented as percentage response (see Materials and Methods). For the actual histograms of CD69 expression, see Fig. 5 A. (E) Sorted DP thymocytes of F5–WT (open circles) and F5–ΔITAMγ mice (filled circles) were analyzed by flow cytometry for the expression of TCRβ in response to exposure of indicated amounts of H2-Db-ASNENMDAM tetramers for 1 h (left panel) or 5 h (right panel). 100% value corresponds to the mean fluorescence of the TCRβ staining on unstimulated DP thymocytes.
Because some reactivity against the LEC cell line was observed without adding peptide, and because some F5–WT DP thymocytes developed overnight into CD8 single-positive (SP) thymocytes without exposure to any exogenous added peptide, CD69 expression, generation of CD8 SP thymocytes, and cell death of DP thymocytes are expressed as “% response”. Calculations were performed as described (41), by correcting for this background by subtraction of the response obtained with LEC cells without peptide added. “100% response” represents the CD69 up-regulation by all cells, the maximal induction of CD8 SP thymocytes by the nominal peptide ASNENMDAM in the experiment, or the maximal cell death induced by the agonist peptide ASNENMDAM in the experiment. As CD69 expression on DP thymocytes was analyzed by flow cytometry only on viable cells, and because a significant proportion of the DP thymocytes died during the overnight suspension culture without (and to a greater extent with) peptide exposure, we corrected for this by assuming that all DP thymocytes induced to die by peptide exposure also expressed CD69 (expression of CD69 precedes both positive and negative selection). Calculation: percentage CD69 induced on DP thymocytes = (percentage viable DP cells × percentage CD69 measured by flow cytometry) + (percentage of dead DP cells).
Immunoblotting and Kinase Activity Assays.
TCR stimulation of purified DP thymocytes was performed by incubating cells with 10 μg/ml biotinylated anti-TCRβ (clone H57–597) mAbs for 20 min at 4°C followed by cross-linking with 20 μg/ml Avidin D (Vector Laboratories) at 37°C, or by incubating with 5 μg/ml ASNENMDAM peptide together with 10 μg/ml anti-CD28 (clone 37.51) at 37°C for the times indicated. Subsequently, whole-cell lysates were prepared in lysis buffer (1% NP-40, 20 mM Tris-HCl pH 8.0, 140 mM NaCl, 10% glycerol, 2 mM EDTA, 20 mM NaF, 1 mM Na3VO4, and a cocktail of protease inhibitors [Boehringer]) and proteins were resolved by SDS-PAGE and transferred onto nitrocellulose membranes. Kinase activation was analyzed by immunoblotting using anti-phosphoERK (New England Biolabs, Inc.) or anti-phosphoTyrosine (clone 4G10; Upstate Biotechnology) antibodies, followed by horseradish peroxidase–conjugated secondary antibodies (Dako), and developing the blots by chemiluminescence (Pierce Chemical Co.). Total levels of ERK and linker for activation of T cells (LAT) expression were measured by reprobing the blots with anti-ERK (Santa Cruz Biotechnology, Inc.) or anti-LAT (Upstate Biotechnology, Inc.) antibodies. JNK activity was determined by incubating cell extracts (25 μg) with 10 μg GST-c-Jun bound to glutathione-agarose beads in dilution buffer (25 mM Hepes, pH 7.6, 2.5 mM MgCl2, 0.05 mM EDTA, 0.025% Triton X-100, 0.5 mM DTT, 0.1 mM Na3VO4, 20 mM β-glycerophosphate, 4 mM p-nitrophenyl phosphate, and a cocktail of protease inhibitors) overnight at 4°C. Subsequently, precipitates were washed five times in washing buffer (20 mM Hepes, pH 7.6, 50 mM NaCl, 2.5 mM MgCl2, 0.1 mM EDTA, 0.05% Triton X-100, 0.1 mM Na3VO4, 20 mM β-glycerophosphate, 4 mM p-nitrophenyl phosphate) and resuspended in 30 μl kinase buffer (20 mM Hepes, pH 7.6, 20 mM MgCl2, 2 mM DTT, 0.1 mM Na3VO4, 20 mM β-glycerophosphate, 4 mM p-nitrophenyl phosphate) containing 20 μM ATP and 3 μCi γ-[32P]ATP for 20 min at 37°C. After four washes in cold washing buffer, GST-c-Jun was eluted in Laemmli buffer and resolved on 12% SDS-PAGE. The phosphorylated GST-c-Jun fusion protein was visualized and quantified using a PhosphorImager. Total levels of JNK were measured by immunoblotting with anti-JNK1/2 antibodies (Santa Cruz Biotechnology, Inc.).
Detection of ZAP70 Tyrosine Phosphorylation.
TCR stimulation of purified DP thymocytes was performed in stimulation media (RPMI containing 1 mM vanadate and 20 mM HEPES pH 7.0). Cells were incubated at 108 cells/ml with 150 μg/ml anti-TCRβ (clone H57–597) antibodies for 15 min on ice. Subsequently, cells were washed twice in stimulation media followed by cross-linking with 100 μg/ml goat anti-hamster (ICN Biomedicals/Cappel) at 37°C for the indicated times. After stimulation, cells were lysed at 107 cells/ml in Triton X-100 lysis buffer for 20 min on ice and the detergent extracts were clarified for 10 min at 14,000 rpm as described (44). For immunoprecipitations, lysates were incubated for 2 h with anti-ZAP70 antibodies preadsorbed to Protein A sepharose beads. The beads were washed three times in lysis buffer following which the immune complexes were eluted by boiling for 5 min in SDS sample buffer. Immune complexes were resolved by SDS-PAGE, transferred to Immobilon-P (Millipore) membranes, and immunoblotted with anti-phosphoTyrosine antibodies (clone 4G10; Transduction Laboratories) or rabbit anti-ZAP70 antibodies. Bound antibodies were detected by incubation with either anti–mouse-horseradish peroxidase (HRP) or protein A-HRP, followed by enhanced chemiluminecence (Renaissance; NEN Life Science Products).
Results
Loss of the CD3γ -ITAM Impairs Positive Selection.
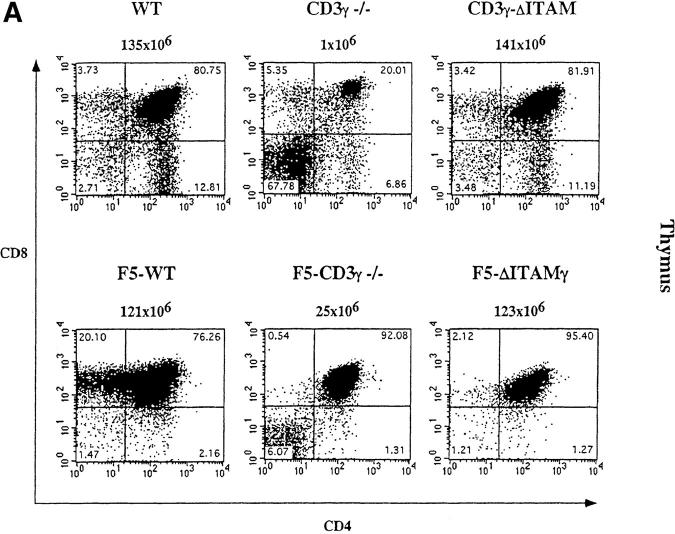
Mice lacking the CD3γ chain of the TCR-CD3 complex have a severe defect in β-selection, and therefore produce few DP thymocytes (22; and Fig. 1 A, top). This is presumably a consequence of defective pre-TCR assembly (unpublished data), and precludes assessment of whether the CD3γ chain has any role in selection at the DP stage of thymocyte development. As the block in β-selection in various models of defective pre-TCR function can be bypassed by introduction of transgenic TCRs (45), we explored whether this might also apply to CD3γ-deficient pre-TCRs. To this end, F5 TCRαβ transgenes (38) were introduced into CD3γ null mutant mice, and mice were also crossed to the RAG-1–deficient background, to prevent expression of endogenous TCR chains (Fig. 1). F5 TCRs are specific for the dominant NP epitope of influenza A virus, which is recognized in association with H-2Db, and therefore skew positive selection toward the CD8 compartment (38). When crossed to RAG-1–deficient mice, positive selection of CD8 cells is virtually absolute for WT F5 TCRs (F5–WT; Fig. 1 A, bottom), with hardly any cells progressing to the CD4 compartment.
Figure 1.
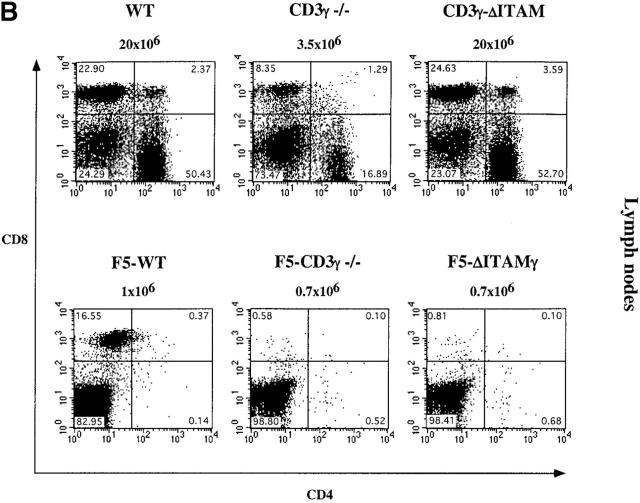
The ITAM of the CD3γ chain plays a major role in thymocyte selection. Total thymocytes (A) and lymph node cells (B) from 6–8-wk-old WT, CD3γ−/−, CD3γ–ΔITAM, F5–WT, F5–CD3γ−/−, and F5–ΔITAMγ mice were monitored for the expression of CD4 versus CD8. The percentage of cells within each quadrant is indicated. The absolute number of thymocytes and lymph node cells detected in the different genotypes is depicted above the corresponding dot display.
Introduction of F5-transgenic TCRs into CD3γ-deficient mice crossed to the RAG−/− background (F5–CD3γ−/− mice) clearly has the ability to bypass the β-selection defect: thymic cellularity is partially restored, the percentage of DP cells reaches normal levels, and the DN compartment is reduced in size (Fig. 1 A, bottom panel). These findings confirm earlier studies on restoration of β-selection by transgenic TCRs (45), and presumably reflect that αβ and γδ TCRs can partially replace pre-TCRs in β-selection. Also TCR levels in the DP compartment of F5–CD3γ−/− mice are considerably increased when compared with the CD3γ−/− mice (Fig. 2 A), as is CD5 expression (Fig. 2 B), which is up-regulated during the DN to DP transition. Yet, important differences remain, as F5–CD3γ−/− mice completely lack CD8 SP cells in the thymus (Fig. 1 A, bottom), as well as in the periphery (Fig. 1 B, bottom), and lag behind in down-regulating CD24 (Fig. 2 B). These results therefore suggest a crucial role for the CD3γ chain during positive selection.
Figure 2.
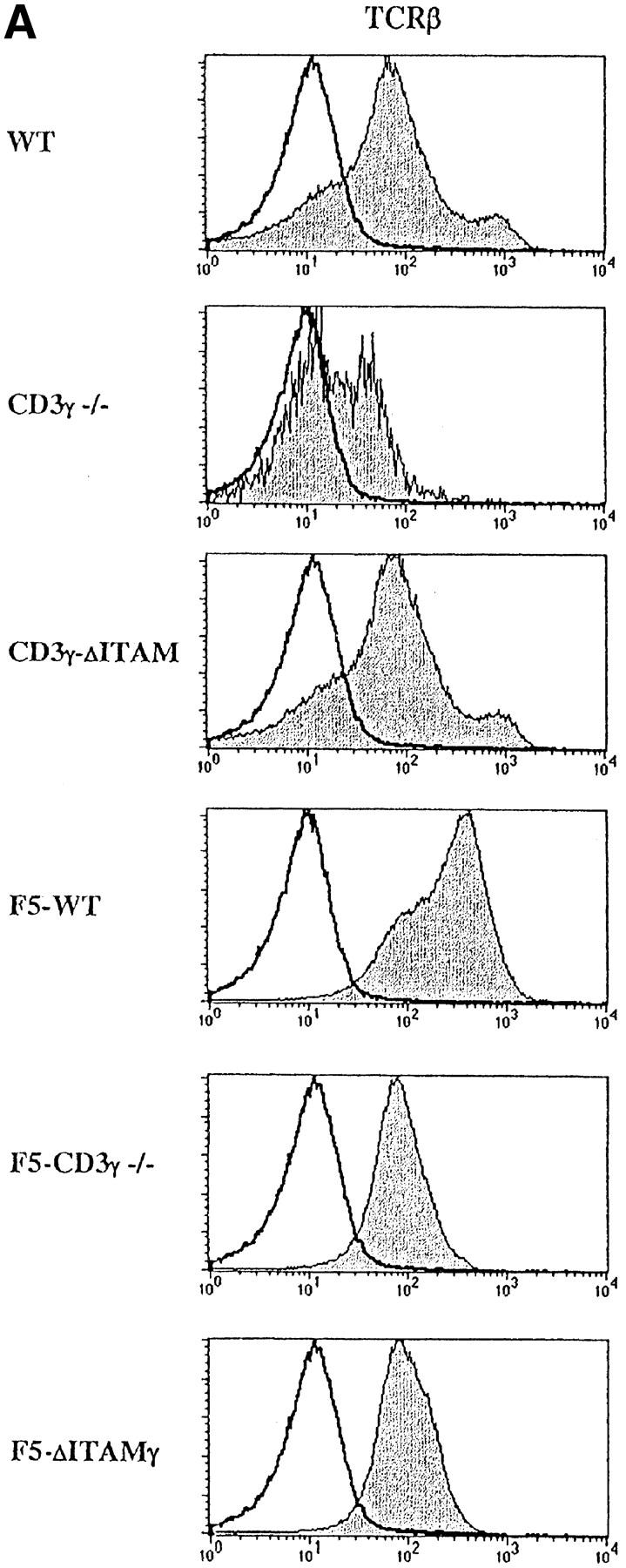
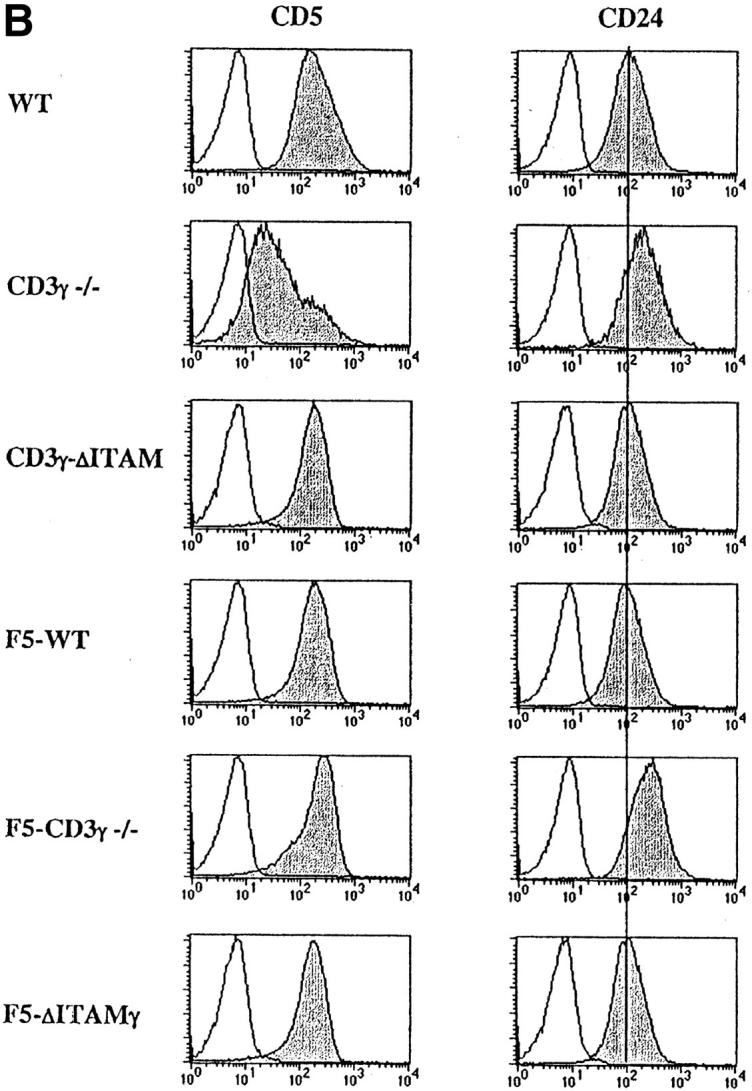
Analysis of TCR-, CD5-, and CD24- expression on DP thymocytes. Expression of cell surface TCRβ (A), CD5 and CD24 (B) (gray), or an irrelevant mAb (white) was analyzed within the electronically gated CD4+CD8+ DP compartment derived from 6–8-wk-old WT, CD3γ−/−, CD3γ–ΔITAM, F5–WT, F5–CD3γ−/−, and F5–ΔITAMγ mice.
As the CD3γ protein is clearly required for efficient cell surface expression of the mature TCR (22, 23; Fig. 2 A), besides endowing this receptor with signaling capacity, quantitative or qualitative causes for the positive selection defect in F5–CD3γ−/− mice must be distinguished. DP thymocytes of knock-in mice expressing an ITAM signaling-deficient CD3γ chain exhibit normal cell surface levels of the TCRβ protein (23; Fig. 2 A), allowing an opportunity to discriminate between these two options. Therefore, CD3γ–ΔITAM mice were crossed to F5–RAG−/− mice. Strikingly, loss of only the CD3γ–ITAM completely abolished positive selection in F5–RAG−/− mice (further referred to as F5–ΔITAMγ mutants), as no CD8 SP thymocyte subset could be detected (Fig. 1 A, bottom). This observation documents that the endogenous thymic peptide repertoire is unable to trigger positive selection signals through F5–ΔITAMγ mutant TCRs. Further corroboration of this profound defect is seen in the peripheral T cell compartment: lymph nodes from F5–ΔITAMγ mutant mice completely lack T cells (Fig. 1 B, bottom panel), whereas F5–WT mice have the expected skewed presence of mature CD8 T cells.
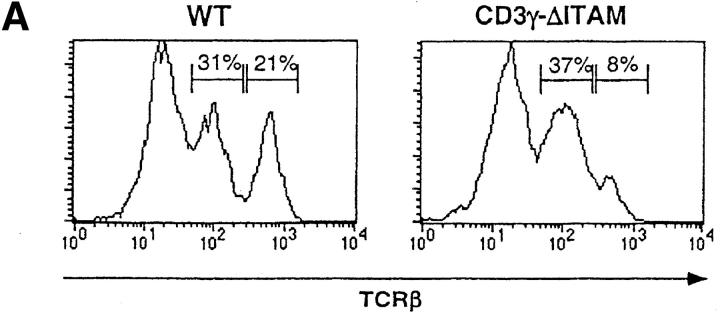
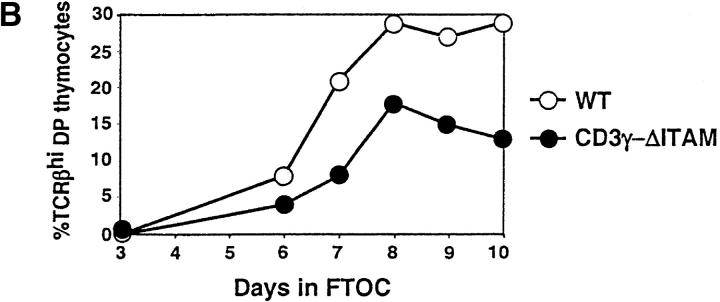
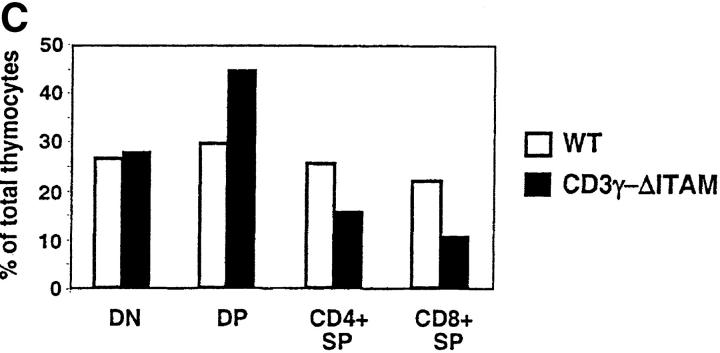
In contrast to F5–ΔITAMγ mice, CD3γ–ΔITAM mutants with a polyclonal TCR repertoire have normal thymic cellularity, and the distribution of the CD4 and CD8 coreceptors over the different thymic and lymph node populations is also comparable to WT mice (Fig. 1, A and B, top). At face value, it therefore appears that thymocyte selection of a polyclonal repertoire is unaffected by absence of a CD3γ–ITAM. We argued, however, that the snapshot phenotypical evaluation of selection taken in adult mice may not accurately reflect the dynamics of selection: in a pool of DP thymocytes engaged in a wide range of diverse signaling events, the consequences of a mutation in a single signaling molecule might easily be obscured. In a dynamic evaluation of selection, on the other hand, possible disturbances may be revealed. We therefore compared the ability of WT and CD3γ–ΔITAM fetal thymocytes from mice with a polyclonal TCR repertoire to perform positive selection in fetal thymic organ cultures (FTOCs), and clear differences emerged. First, the frequency of thymocytes expressing high levels of TCR was reduced at least twofold in the CD3γ–ΔITAM mutants (Fig. 3 A), at all time points tested (Fig. 3 B). Second, the percentage of SP cells generated is decreased, with a concomitant increase in DP thymocytes (Fig. 3 C). Thus, positive selection occurs less efficiently in multiple TCRs lacking the CD3γ–ITAM. At the same time, compensatory effects likely related to TCR affinity and off-rate clearly can occur, as SP do accumulate in a polyclonal TCR repertoire (Fig. 1). Nevertheless, for some TCRs, represented by the F5, an intact repertoire of ITAMs is required (Figs. 1 and 3).
Figure 3.
Positive selection occurs less efficiently in TCRs lacking the CD3γ–ITAM. Day 14 fetal thymic lobes of WT and CD3γ–ΔITAM mice were cultured in FTOCs, and analyzed over time for the generation of thymocytes expressing TCRβ (A and B), and CD4 and CD8 (C). A histogram for the TCRβ staining on day 7 of the FTOC is shown in panel A, as well as the gates for distinguishing TCRβ-hi and TCRβ-lo cells. A time course for the development of cells expressing high levels of TCRβ is shown in B, and the percentages of DN, DP, and SP cells on day 7 of the FTOC are shown in C. For SP cells, only those also expressing TCR are shown. Data are representative of three separate experiments.
Together, these findings support a role for the CD3γ–ITAM at the DP checkpoint of thymocyte selection for some TCRs. Further dissection of the possible contribution of CD3γ–ITAM requires experiments with a transgenic TCR, such that interactions of the mutant and WT receptors with defined ligands can be compared. The remainder of this study is therefore performed with F5–ΔITAMγ mutant DP thymocytes.
Loss of the CD3γ–ITAM Does Not Affect Ligand Interaction, TCR Down-regulation, or CD69 Up-regulation.
Why might F5–ΔITAMγ mutant receptors not respond to the endogenous peptide repertoire in the thymus by triggering the positive selection program (Fig. 1)? Two overall possibilities need to be distinguished: either the CD3γ–ITAM provides prerequisite signaling information for positive selection of F5 TCRs and some other TCRs (Fig. 3), or F5–ΔITAMγ mutant receptors fail to bind to peptide ligands, or bind with insufficient affinity. To formally exclude any (highly unlikely) effects of the CD3γ–ITAM mutation on peptide-ligand binding, we first compared the binding characteristics of F5–WT and F5–ΔITAMγ mutant receptors to their nominal ligand. We also addressed the possibility that the CD3γ–ITAM mutation selectively destroys some responses (i.e., positive selection), but not others in DP thymocytes. If so, this would then support a model in which the CD3γ–ITAM contributes to signal specification.
The F5 TCR recognizes the ASNENMDAM epitope of the influenza A nucleoprotein, as published previously (38) and illustrated in Fig. 4 A, in the proliferative response of F5–WT lymph node T cells to this peptide. To address whether the CD3γ–ITAM mutation has any effect on ASNENMDAM binding, we compared binding of MHC-ASNENMDAM-tetramers to F5–WT and F5–ΔITAMγ mutant receptors in the CD69-negative DP compartment. As expected, we found tetramer binding to be entirely overlapping (Fig. 4 B). Furthermore, the F5–WT and F5–ΔITAMγ mutant TCRs have comparable off-rates for this ligand (Fig. 4 C): if anything, binding of the nominal epitope to the F5–ΔITAMγ mutant receptor has a somewhat slower off-rate than that for the F5–WT receptor.
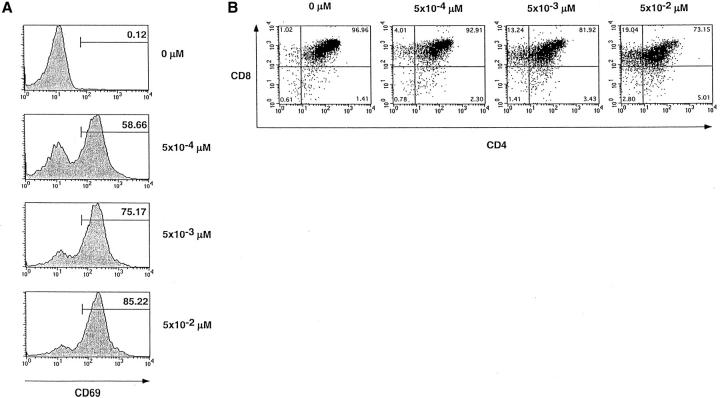
We then asked whether we could identify any parameters which would illustrate that F5–ΔITAMγ mutant receptors cannot only bind to their ligand, but also respond to bound ligand by triggering appearance of activation markers. Importantly, both the F5–WT and the F5–ΔITAMγ mutant receptor can respond to the ASNENMDAM epitope, as shown by the observation that DP thymocytes from both up-regulate CD69 when activated by ASNENMDAM (Fig. 4 D; for the actual CD69 expression data on F5–ΔITAMγ DP thymocytes, see Fig. 5 A), and do so with comparable sensitivity, in agreement with their similar binding kinetics. Induction of this early activation marker is therefore not affected by the CD3γ–ITAM mutation. We also tested the effects of ASNENMDAM binding on internalization of F5–WT and F5–ΔITAMγ mutant TCRs. TCR down-modulation represents another early marker for response to TCR triggering, although its physiological relevance is unclear. We previously showed, in mice with a polyclonal TCR repertoire, that the CD3γ chain is required for both anti-CD3ε- and PMA-triggered TCR down-regulation (23), but the CD3γ–ITAM is dispensable. When ligand-triggered TCR internalization is compared between F5–WT and F5–ΔITAMγ mutant DP thymocytes, it is clear that this ITAM is not required: down-modulation of TCR expression occurs with similar kinetics and to a comparable extent in the F5–WT and F5–ΔITAMγ mutant receptors (Fig. 4 E).
Figure 5.
CD69 upregulation and induction of CD8 SP thymocytes. Representation of the actual flow cytometry observations from which the percentage response data in Figs. 4 D and 6 are derived. Sorted F5–ΔITAMγ DP thymocytes were stimulated overnight in dispersed cultures with graded concentrations of the agonist peptide ASNENMDAM, and viable cells were stained for (A) CD69 or (B) CD4 versus CD8.
Together, these findings document that F5–ΔITAMγ mutant receptors can both bind to and respond to the agonist peptide for the F5 TCR, with equal binding kinetics and antigen sensitivity. It is therefore very striking that F5–ΔITAMγ mutant receptors cannot identify, among the broad range of intrathymic peptides it interacts with in vivo, any ligands at all that bind with appropriate kinetics to allow positive selection to occur. These findings suggest that lack of CD3γ results in quantitative or qualitative differences in signaling which, for F5 TCRs and at least for some other (unidentified) TCRs (see Fig. 3), result in a selective defect in positive selection.
Ex Vivo Triggering of F5–ΔITAMγ Mutant Receptors Can Induce Negative Selection, but Positive Selection Is Severely Impaired or Abrogated.
Upregulation of CD69 precedes both positive and negative selection in DP thymocytes (41; and references therein). The observation that CD69 up-regulation was normal in thymocytes expressing F5–ΔITAMγ mutant receptors (Fig. 4 D) raises the question whether agonist ligand can also induce the signals required for positive and negative selection. We therefore tested whether the ASNENMDAM epitope could induce positive selection in DP thymocytes with F5–ΔITAMγ mutant TCRs, as judged by its ability to induce appearance of CD8 single-positive (CD8 SP) cells in an overnight culture system, previously described (41). In this setting, DP thymocytes from different origins can be exposed to defined ligands presented by a homogeneous antigen-presenting cell population, such that their ability to undergo the early manifestations of selection events can be directly compared. An example of the actual flow cytometry data of F5–ΔITAMγ mutant DP thymocytes responding to the agonist ASNENMDAM ligand by induction of CD8 SP is shown in Fig. 5 B.
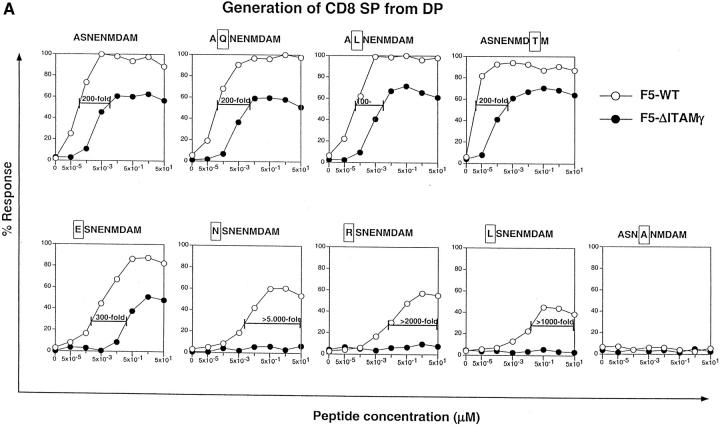
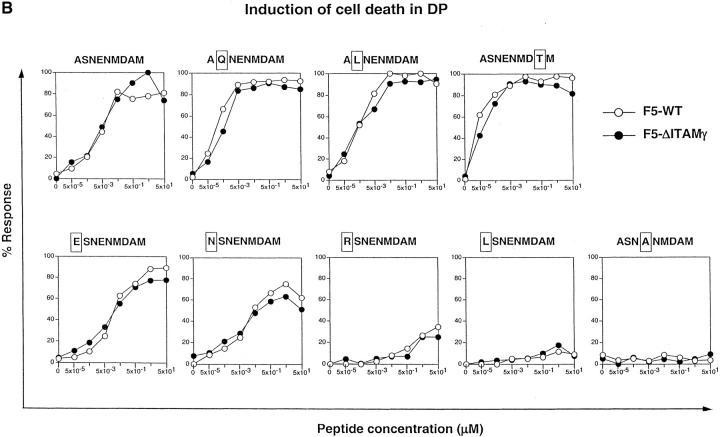
The results unequivocally show (Fig. 6 A) that this agonist ligand for the F5 TCR can induce phenotypical positive selection in DP thymocytes from F5–ΔITAMγ mutant mice in vitro, but important differences between the F5–WT and F5–ΔITAMγ mutant TCRs remain. First, at least 200-fold more ASNENMDAM peptide is required for inducing a phenotypical positive selection response in DP cells with F5–ΔITAMγ mutant TCRs, and the maximal number of CD8 SP cells generated by thymocytes with F5–ΔITAMγ mutant TCRs never reaches more than 50–60% of that generated by F5–WT cells (Fig. 6 A). Second, this pattern of responsiveness (i.e., reduced antigen sensitivity and lower levels of induced CD8 SP cells) is reproduced for three ASNENMDAM variants with nonconservative mutations at the nonanchor residue positions 2 or 8 (Fig. 6 A, top panels). Also a variant peptide with a replacement at position 1 exhibits this reduced ability to induce positive selection in F5–ΔITAMγ mutant DP thymocytes (Fig. 6 A, bottom left). Finally, for three additional variant peptides with mutations at position 1, the ability to induce CD8 SP cells through F5–ΔITAMγ mutant receptors is completely abrogated (Fig. 6 A, remaining bottom panels; responsiveness to the non-MHC binding variant ASNANMDAM is shown as a negative control). Thus, we have identified, among a small panel of seven variants of the ASNENMDAM epitope, four with a severely reduced ability to induce CD8 SP cells in the F5–ΔITAMγ mutant DP thymocytes (accompanied by a reduced maximal response), and three that are completely unable to do so. Although these in vitro data represent only the initiation of the positive selection response, they mirror the observations in the in situ thymus (see Fig. 1 A), and document that the F5–ΔITAMγ mutation severely affects positive selection for multiple ligands.
Figure 6.
F5–ΔITAMγ mutant receptors are fully capable of transmitting signals for negative selection in response to ASNENMDAM, but are impaired in triggering signals for positive selection. Purified DP thymocytes of F5–WT (open circles) and F5–ΔITAMγ mice (filled circles) were incubated overnight in the presence of LEC cells as APCs with the indicated concentrations of the nominal peptide ASNENMDAM and seven of its variants, or with the null-peptide ASNANMDAM (bottom right panel) as a negative control. Cells were analyzed (A) for the percentage of CD8 SP thymocytes as a readout system indicative for induction of positive selection or (B) cell death by propidium iodide staining as a readout system indicative for induction of negative selection. Data shown is presented as the percentage of response calculated as described in Materials and Methods.
Does the CD3γ–ITAM mutation also affect the other cell-fate decision that DP thymocytes make on the basis of signals from their TCR, i.e., negative selection? One would predict, on the basis of kinetic models for thymocyte selection, that ligands impaired in induction of positive selection, would be even less capable of inducing negative selection signals. Quite in contrast to this prediction however, we find that ASNENMDAM does induce negative selection in both F5–WT and F5–ΔITAMγ mutant DP thymocytes, with completely identical ligand sensitivity and to the same extent (Fig. 6 B). Furthermore, ASNENMDAM does not stand alone in its ability to induce negative selection in a normal manner, as also its variants with replacements at positions 1, 2, and 8 induce negative selection through F5–WT and F5–ΔITAMγ mutant TCRs (Fig. 6 B). These experiments therefore uncouple, through a single ITAM mutation, signals inducing the genetic program for negative selection from that inducing positive selection for this TCR.
Distinct Effects of the CD3γ–ITAM Mutation on ERK, JNK, and ZAP70 Activation, and on LAT Phosphorylation.
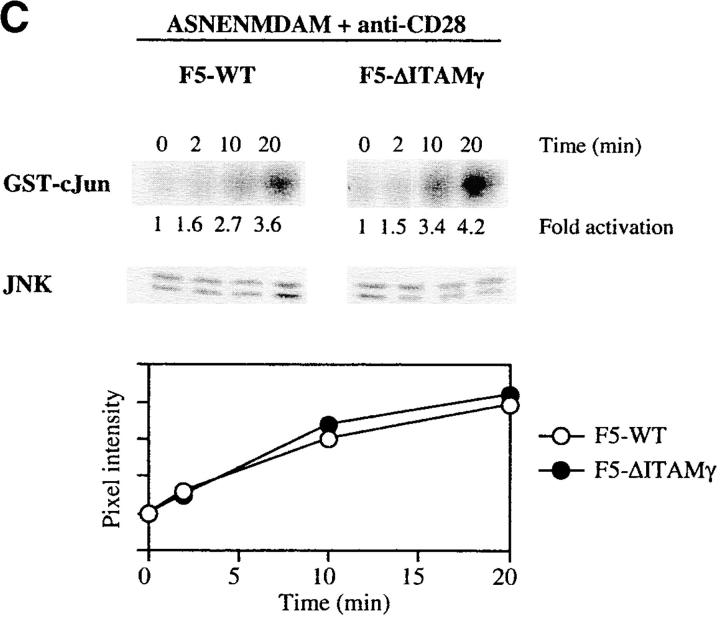
In search of a molecular explanation for these findings, we compared the ability of the WT and ΔITAMγ mutant F5 receptors to activate a number of signaling cascades in response to the agonist ligand and cross-linking TCR antibodies. Both genetic and biochemical studies have implicated activation of the Ras/Raf/MEK/ERK cascade in positive selection (26–35), so we evaluated whether ERK activation might be impaired as a consequence of this ΔITAMγ mutation. Under the short-term culture conditions applied for such signaling studies, a very dramatic effect of the mutation is observed: the amount of phosphorylated ERK that could be detected following ligation of the TCR by anti-TCRβ mAb (Fig. 7 A) is reduced to negligible levels by loss of the CD3γ–ITAM.
Figure 7.
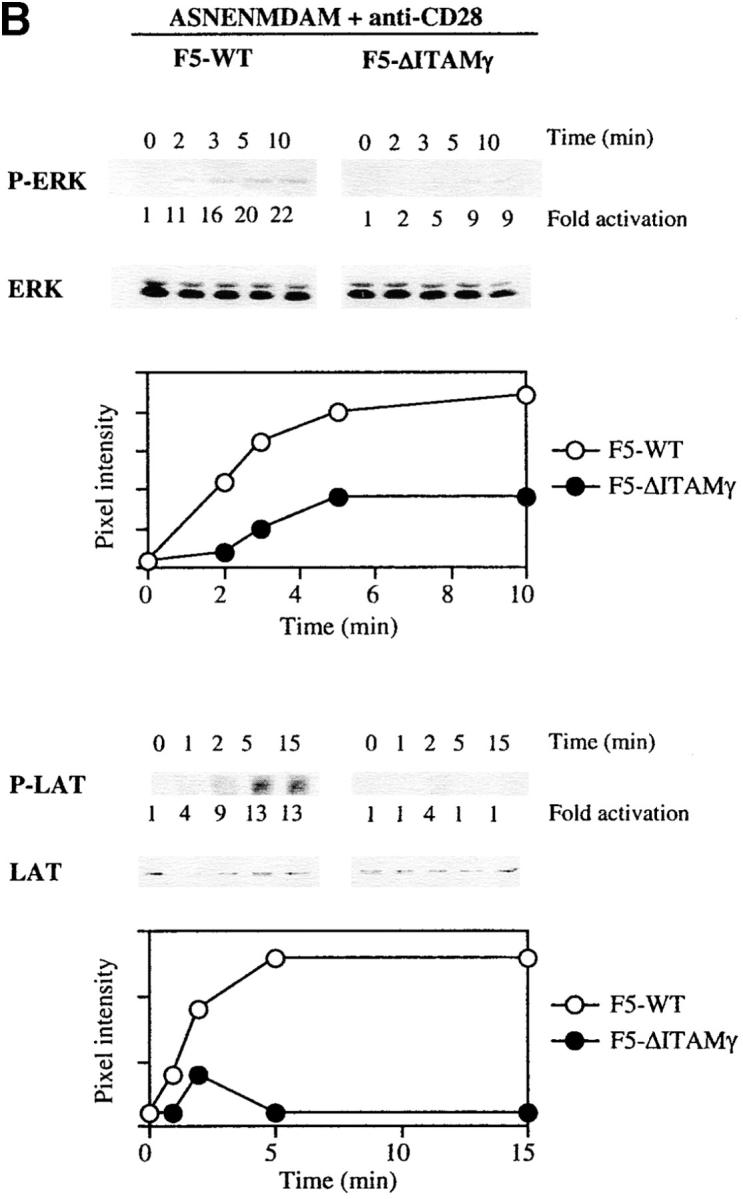
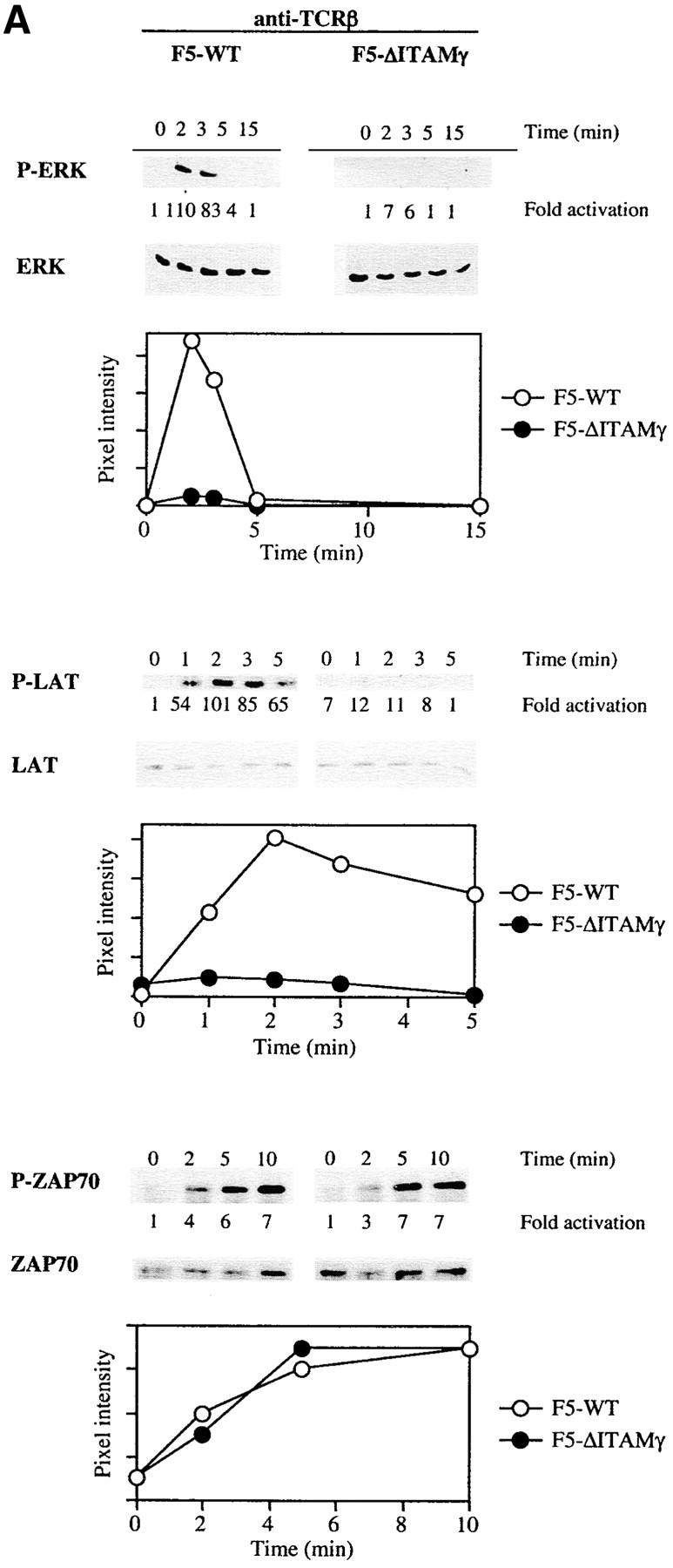
Activation of LAT and ERK but not JNK is strongly reduced in the absence of a signaling deficient CD3γ chain. DP thymocytes from 6–8-wk-old F5–WT and F5–ΔITAMγ mutant mice were stimulated with cross-linked anti-TCRβ (A) or the agonist peptide ASNENMDAM and anti-CD28 (B and C). At the indicated times, induction of ERK, LAT, and ZAP70 phosphorylation was assessed by immunoblotting of whole-cell lysates or ZAP70 immunoprecipitates (A and B), while JNK activity was measured using an in vitro kinase assay (C). JNK and ZAP70 experiments were performed only with anti-TCRβ. Protein loading was analyzed by immunoblotting with anti-ERK, anti-LAT, anti-ZAP70 (A and B), or anti-JNK (C) mAbs.
We also evaluated phosphorylation of the adaptor protein LAT, as it represents one of the most upstream mediators implicated in positive selection (34, 35) probably due to its requirement for complete activation of the Ras-ERK pathway (46, 47). Like activation of ERK, LAT phosphorylation is dramatically impaired in the absence of a signaling-competent CD3γ chain (Fig. 7 A). This defect in LAT phosphorylation corroborates the ERK results. To the extent that antibody-mediated cross-linking of TCRs approximates perhaps most closely the conditions triggered by the intrathymic peptide repertoire (certainly not physically, but at least in polyclonality), these findings are consistent with the complete absence of positive selection in the in situ thymus (see Fig. 1 A). Experiments in which TCR triggering was achieved with ASNENMDAM peptide (Fig. 7 B) are in complete agreement with these antibody-mediated TCR-triggering experiments, in that defects in ERK activation and LAT phosphorylation are again revealed.
In sharp contrast, activation of the JNK cascade is unimpaired by this mutation (Fig. 7 C). Activation of the JNK and p38 pathways has been linked to negative selection (18, 30, 36, 37), and the completely overlapping ability of F5–ΔITAMγ mutant and F5–WT TCRs to induce JNK activation is fully consistent with the normal negative selection phenotype in F5–ΔITAMγ thymocytes (Fig. 6 B). Also ZAP70 activation (Fig. 7 A) is entirely unaffected by the CD3γ–ITAM mutation, leaving to be defined which upstream events result in the abrogation of LAT phosphorylation. Taken together, these findings indicate that absence of CD3γ–ITAM in thymocytes with F5 TCRs selectively alters ERK activation and LAT phosphorylation, and this defect may explain the blockade of positive selection in mice lacking the CD3γ–ITAM. This analysis therefore provides the first genetic evidence that the CD3γ–ITAM contributes in a quantitative or qualitative manner to TCR signal specification.
Discussion
Our results document that the CD3γ–ΔITAM mutation destroys the ability of F5 TCRs to be positively selected by any of the peptides represented among the thymic peptide repertoire in vivo, and severely reduced its ability to respond to multiple synthetic peptide agonists in vitro. This mutation also results in considerable impairment in positive selection in the polyclonal TCR repertoire. These observations are remarkable for at least three reasons. First, the CD3γ–ΔITAM mutation is unlikely to affect TCR-ligand binding, and was in fact shown not to for the nominal ligand ASNENMDAM for the F5 TCR (see Fig. 4, B and C). Second, effect of the CD3γ–ΔITAM mutation is limited to positive selection, as all other hallmarks of TCR responsiveness in DP thymocytes were not affected i.e., CD69 up-regulation, TCR down-regulation, and negative selection. Finally, in a receptor with 9 additional ITAMs (i.e., 2 in the CD3δε heterodimer, 1 in the CD3γε heterodimer, and 6 in the CD3ζζ homodimer), redundancy in ITAM function would have been a reasonable prediction, certainly on the basis of earlier studies about the amplification function of ITAM multiplicity. Instead, these results suggest that at least for some TCRs, for which the F5 TCR is the prototype, an intact repertoire of CD3-ITAMs is required for positive selection to occur. Other receptors, on the other hand, can still undergo positive selection without the CD3γ–ITAM. It will be of interest to define the particular qualities (affinity, off-rate) that renders a given TCR sensitive to loss of certain ITAMs. In addition, it remains to be established how the dramatic effect of the CD3γ–ΔITAM mutation on positive selection of F5 TCRs is linked to quantitative and/or qualitative changes in signaling events.
These results complement other studies which addressed the role of ITAM multiplicity in thymocyte selection (7;19–21): for CD3ζ- and CD3ε-ITAMs, the data supported either a quantitative contribution of ITAM diversity in T cell maturation, or no role at all. The results differ, dependent on the TCR studied. The selective effect of CD3γ–ΔITAM on thymocyte selection by F5 TCRs in this study can be explained either by considering a quantitative contribution of CD3γ–ITAMs to TCR signaling that results in a qualitatively different repertoire, or reflect a unique role for the CD3γ–ITAM in TCR-driven selection. This latter possibility seems unlikely, as adult mice with the CD3γ–ITAM mutation and a polyclonal TCR repertoire have an apparently normal positive selection phenotype (see Fig. 1). At the same time, we also predict that there will be “holes” in the TCR repertoire of non-TCR transgenic CD3γ–ΔITAM mutant mice, as there clearly are multiple TCRs that are not indifferent to loss of the CD3γ–ITAM: a dynamic evaluation of T cell development in FTOCs reveals at least a twofold reduction in the efficiency of positive selection (Fig. 3). These findings are not at variance with the seemingly normal positive selection phenotype in adult mice, as in FTOCs, development from only a limited precursor pool provides a more stringent test of the capacity of a polyclonal set of precursors to be positively selected. Together, the results indicate that different TCRs interpret the qualitative contributions to signal discrimination from ITAMs differently.
Another remarkable finding in the above studies is that F5–ΔITAMγ mutant receptors cannot identify, in the sea of thymic peptides they can interact with in vivo, even a single peptide that induces positive selection, yet positive selection can be induced (though in a severely “crippled” manner) by the agonist ligand for the F5 receptor, and by a number of its variants in vitro. Are these observations at variance with the observation that positively selecting ligands normally bear little resemblance to the cognate peptide for a given receptor (48)? It is difficult to answer this question, in view of the fact that the culture conditions used for the positive selection experiments probably do not reflect the peptide concentrations present in the thymic microenvironment. Nevertheless, it should be noted that peptide binding and off-rate are not affected by loss of CD3γ–ITAM, nor are early hallmarks of TCR-ligand interactions, such as CD69 up-regulation and TCR down-regulation (Fig. 4). More importantly, our findings underscore that the decision for triggering signaling pathways required for positive selection is not dictated by the binding kinetics of a given TCR-ligand interaction alone, but also by intrinsic properties of the TCR-CD3 complex, in this case, modulated in its cytoplasmic domains by removal of the CD3γ–ITAM.
Two precedents for mutations in the TCR-CD3 complex that regulate positive selection without affecting ligand binding were reported last year. In one study (35), the CD3δ chain was shown to have a role, independent of its ITAM, in coupling the TCR to phosphorylation of LAT and CD3ζ and activation of ERK, and in induction of positive selection. In mice expressing mutant TCRs lacking the TCRα chain connecting peptide domain (α-CPM), positive selection is also abrogated (34), and this defect was found to be associated with impaired recruitment to lipid rafts of activated PTKs Lck and ZAP-70, and of phosphorylated CD3ζ and LAT. Interestingly, the α-CPM mutation specifically disrupts association of CD3δ with the TCR complex, providing an explanation for the finding that both loss of CD3δ and loss of α-CPM result in impairment of activation of the same upstream signaling components, and that both have a defect in positive selection.
The specific effect of the CD3γ–ΔITAM mutation on positive selection by the F5 TCR is associated with a selective defect in activation of the ERK cascade, previously implicated in positive selection (26–35). JNK activation, on the other hand, is not affected by the CD3γ–ΔITAM mutation, nor is negative selection, in accordance with the previously found association between JNK and p38 activation and negative selection (18, 30, 36, 37). Although these findings are only correlative, they do suggest that the JNK and ERK signaling pathways can bifurcate upstream of a given ITAM. As ERK is a downstream target of Ras, it is interesting that the most upstream defect associated with loss of CD3γ–ITAM is an almost complete abrogation of LAT phosphorylation. Phosho-LAT recruits multiple signaling molecules to the plasma membrane, including the Grb2-Sos complex, such that Sos can then activate Ras (45, 46, 49). Defective LAT phosphorylation may thus result in an impairment of Ras activation, providing one mechanism through which ERK activation may have been reduced. Other possibilities need to be considered, however, as another pathway for Ras activation has been proposed (50): phospho-LAT also recruits PLCγ1 and the Gads-SLP-76 complex, and these appear required for optimal Ras activation as well (50). Interestingly, the RasGRP has been implicated in this pathway, and RasGRP-deficient mice have a severe block in positive selection (51). These different options illustrate that further dissection of how loss of the CD3γ–ITAM affects the very first steps in TCR-induced phosphorylation and recruitment of signaling proteins to the “signalosome,” is needed. Nevertheless, we do not observe any modulations in ZAP70 activation, unlike the findings in the α-CPM mutants (34), but in keeping with earlier observations in ZAP70-deficient mice, in which both positive and negative selection are affected. The positive selection defects in α-CPM mutant mice (34) and CD3δ-deficient mice (35) are also associated with very early modulations in TCR signaling, i.e., in recruitment of phospho-CD3ζ and phospho-LAT to lipid rafts.
Summarizing, the results define one ITAM as a tool for linking a given TCR to the distinct signaling pathways it needs to engage in order to trigger paradoxically both negative and positive selection at the DP checkpoint. It will be of interest to exploit the present model for searching for specific downstream events associated with selection. Positive selection must be associated with activation of specific genes, different from those promoting negative selection, either because of triggering of specific sets of transcription factors, or because the levels of transcription factors triggered by positively selecting signals are different from those triggered by negative selecting signals. Having now available a model system that selectively impairs positive selection while leaving negative selection of DP thymocytes intact, provides the opportunity to search for such events.
Acknowledgments
We would like to thank H. Spits, T. Schumacher, D. Wiest, and D. Kappes for helpful discussions and critically reviewing the manuscript, D. Kioussis for making available the F5–TCR transgenics, K. de Visser for her help in some experiments, M. Toebes, M. Hoffmann, M. Beumkes, E. Noteboom, and A. Pfauth for their excellent technical assistance, and M.A. van Halem for editing the manuscript.
M.C. Haks and E. Pépin were supported by grant RG0335/1998-M from the Human Frontier Science Program Organization (HFSPO).
Footnotes
Abbreviations used in this paper: DP, double positive; ERK, extracellular signal-regulated kinase; FTOC, fetal thymic organ culture; ITAM, immunoreceptor tyrosine-based activation motif; JNK, c-Jun NH2-terminal kinase; LAT, linker for activation of T cells; RAG, recombination activating gene; SP, single positive; WT, wild-type.
References
- 1.Letourneur, F., and R.D. Klausner. 1992. Activation of T cells by a tyrosine kinase activation domain in the cytoplasmic tail of CD3 ɛ. Science. 255:79–82. [DOI] [PubMed] [Google Scholar]
- 2.Irving, B.A., A.C. Chan, and A. Weiss. 1993. Functional characterization of a signal transducing motif present in the T cell antigen receptor ζ chain. J. Exp. Med. 177:1093–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cambier, J.C. 1995. Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM). J. Immunol. 155:3281–3285. [PubMed] [Google Scholar]
- 4.Wange, R.L., and L.E. Samelson. 1996. Complex complexes: signaling at the TCR. Immunity. 5:197–205. [DOI] [PubMed] [Google Scholar]
- 5.Tybulewicz, V.L.J. 1998. Analysis of antigen receptor signalling using mouse gene targeting. Curr. Opin. Cell Biol. 10:195–204. [DOI] [PubMed] [Google Scholar]
- 6.Love, P.E., and E.W. Shores. 2000. ITAM multiplicity and thymocyte selection: how low can you go? Immunity. 12:591–597. [DOI] [PubMed] [Google Scholar]
- 7.Shores, E.W., T. Tran, A. Grinberg, C.L. Sommers, H. Shen, and P.E. Love. 1997. Role of the multiple T cell receptor (TCR)-ζ chain signaling motifs in selection of the T cell repertoire. J. Exp. Med. 185:893–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Combadière, B., M. Freedman, L. Chen, E.W. Shores, P. Love, and M.J. Lenardo. 1996. Qualitative and quantitative contributions of the T cell receptor ζ chain to mature T cell apoptosis. J. Exp. Med. 183:2109–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen, W.A., C.M. Pleiman, P. Beaufils, A.-M.K. Wegener, B. Malissen, and J.C. Cambier. 1997. Qualitatively distinct signaling through T cell antigen receptor subunits. Eur. J. Immunol. 27:707–716. [DOI] [PubMed] [Google Scholar]
- 10.Songyang, Z., S.E. Shoelson, M. Chaudhuri, G. Gish, T. Pawson, W.G. Haser, F. King, T. Roberts, S. Ratnofsky, R.J. Lechleider, et al. 1993. SH2 domains recognize specific phosphopeptide sequences. Cell. 72:767–778. [DOI] [PubMed] [Google Scholar]
- 11.Songyang, Z., S.E. Shoelson, J. McGlade, P. Olivier, T. Pawson, X.R. Bustelo, M. Barbacid, H. Sabe, H. Hanafusa, T. Yi, et al. 1994. Specific motifs recognized by the SH2 domains of Csk, 3BP2, fps/fes, GRB-2, HCP, SHC, Syk, and Vav. Mol. Cell. Biol. 14:2777–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osman, N., H. Turner, S. Lucas, K. Reif, and D.A. Cantrell. 1996. The protein interactions of the immunoglobulin receptor family tyrosine-based activation motifs present in the T cell receptor ζ subunits and the CD3 γ, δ and ɛ chains. Eur. J. Immunol. 26:1063–1068. [DOI] [PubMed] [Google Scholar]
- 13.Exley, M., L. Varticovski, M. Peter, J. Sancho, and C. Terhorst. 1994. Association of phosphatidylinositol 3-kinase with a specific sequence of the T cell receptor ζ chain is dependent on T cell activation. J. Biol. Chem. 269:15140–15146. [PubMed] [Google Scholar]
- 14.Isakov, N., R.L. Wange, W.H. Burgess, J.D. Watts, R. Aebersold, and L.E. Samelson. 1995. ZAP-70 binding specificity to T cell receptor tyrosine-based activation motifs: the tandem SH2 domains of ZAP-70 bind distinct tyrosine-based activation motifs with varying affinity. J. Exp. Med. 181:375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, S.A., C.M. Pleiman, L. Pao, J. Schneringer, K. Hippen, and J.C. Cambier. 1995. Phosphorylated immunoreceptor signaling motifs (ITAMs) exhibit unique abilities to bind and activate Lyn and Syk tyrosine kinases. J. Immunol. 155:4596–4603. [PubMed] [Google Scholar]
- 16.Ravichandran, K.S., K.K. Lee, Z. Songyang, L.C. Cantley, P. Burn, and S.J. Burakoff. 1993. Interaction of Shc with the ζ chain of the T cell receptor upon T cell activation. Science. 262:902–905. [DOI] [PubMed] [Google Scholar]
- 17.Cambier, J.C., and S.A. Johnson. 1995. Differential binding activity of ARH1/TAM motifs. Immunol. Lett. 44:77–80. [DOI] [PubMed] [Google Scholar]
- 18.Gong, Q., A.M. Cheng, A.M. Akk, J. Alberola-Ila, G. Gong, T. Pawson, and A.C. Chan. 2001. Disruption of T cell signaling networks and development by Grb2 haploid insufficiency. Nat. Immunol. 2:29–36. [DOI] [PubMed] [Google Scholar]
- 19.Ardouin, L., C. Boyer, A. Gillet, J. Trucy, A.-M. Bernard, J. Nunes, J. Delon, A. Trautmann, H.-T. He, B. Malissen, and M. Malissen. 1999. Crippling of CD3-ζ ITAMs does not impair T cell receptor signaling. Immunity. 10:409–420. [DOI] [PubMed] [Google Scholar]
- 20.Van Oers, N.S.C., P.E. Love, E.W. Shores, and A. Weiss. 1998. Regulation of TCR signal transduction in murine thymocytes by multiple TCR ζ-chain signaling motifs. J. Immunol. 160:163–170. [PubMed] [Google Scholar]
- 21.Sommers, C.L., J.B. Dejarnette, K. Huang, J. Lee, D. El-Khoury, E.W. Shores, and P.E. Love. 2000. Function of CD3ɛ-mediated signals in T cell development. J. Exp. Med. 192:913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haks, M.C., P. Krimpenfort, J. Borst, and A.M. Kruisbeek. 1998. The CD3γ chain is essential for development of both the TCRαβ and TCRγδ lineages. EMBO J. 17:1871–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haks, M.C., T.A. Cordaro, J.H.N. Van den Brakel, J.B.A.G. Haanen, E.F.R. De Vries, J. Borst, P. Krimpenfort, and A.M. Kruisbeek. 2001. A redundant role of the CD3γ-immunoreceptor tyrosine-based activation motif in mature T cell function. J. Immunol. 166:2576–2588. [DOI] [PubMed] [Google Scholar]
- 24.Sebzda, E., S. Mariathasan, T. Ohteki, R. Jones, M.F. Bachmann, and P. Ohashi. 1999. Selection of the T cell repertoire. Annu. Rev. Immunol. 17:829–874. [DOI] [PubMed] [Google Scholar]
- 25.Sebzda, E., V.A. Wallace, J. Mayer, R.S.M. Yeung, T.W. Mak, and P.S. Ohashi. 1994. Positive and negative thymocyte selection induced by different concentrations of a single peptide. Science. 263:1615–1618. [DOI] [PubMed] [Google Scholar]
- 26.Swat, W., Y. Shinkai, H.-L. Cheng, L. Davidson, and F.W. Alt. 1996. Activated Ras signals differentiation and expansion of CD4+8+ thymocytes. Proc. Natl. Acad. Sci. USA. 93:4683–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alberola-Ila, J., K.A. Forbush, R. Seger, E.G. Krebs, and R.M. Perlmutter. 1995. Selective requirement for MAP kinase activation in thymocyte differentiation. Nature. 373:620–623. [DOI] [PubMed] [Google Scholar]
- 28.Pagès, G., S. Guérin, D. Grall, F. Bonino, A. Smith, F. Anjuere, P. Auberger, and J. Pouysségur. 1999. Defective thymocyte maturation in p44 MAP kinase (Erk 1) knockout mice. Science. 286:1374–1377. [DOI] [PubMed] [Google Scholar]
- 29.Sharp, L.L., D.A. Schwarz, C.M. Bott, C.J. Marshall, and S.M. Hedrick. 1997. The influence of the MAPK pathway on T cell lineage commitment. Immunity. 7:609–618. [DOI] [PubMed] [Google Scholar]
- 30.Sugawara, T., T. Moriguchi, E. Nishida, and Y. Takahama. 1998. Differential roles of ERK and p38 MAP kinase pathways in positive and negative selection of T lymphocytes. Immunity. 9:565–574. [DOI] [PubMed] [Google Scholar]
- 31.Alberola-Ila, J., K.A. Hogquist, K.A. Swan, M.J. Bevan, and R.M. Perlmutter. 1996. Positive and negative selection invoke distinct signaling pathways. J. Exp. Med. 184:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Shea, C.C., T. Crompton, I.R. Rosewell, A.C. Hayday, and M.J. Owen. 1996. Raf regulates positive selection. Eur. J. Immunol. 26:2350–2355. [DOI] [PubMed] [Google Scholar]
- 33.Swan, K.A., J. Alberola-Ila, J.A. Gross, M.W. Appleby, K.A. Forbush, J.F. Thomas, and R.M. Perlmutter. 1995. Involvement of p21ras distinguishes positive and negative selection in thymocytes. EMBO J. 14:276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werlen, G., B. Hausmann, and E. Palmer. 2000. A motif in the αβ T-cell receptor controls positive selection by modulating ERK activity. Nature. 406:422–426. [DOI] [PubMed] [Google Scholar]
- 35.Delgado, P., E. Fernández, V. Dave, D. Kappes, and B. Alarcón. 2000. CD3δ couples T-cell receptor signalling to ERK activation and thymocyte positive selection. Nature. 406:426–430. [DOI] [PubMed] [Google Scholar]
- 36.Rincón, M., A. Whitmarsh, D.D. Yang, L. Weiss, B. Dérijard, P. Jayaraj, R.J. Davis, and R.A. Flavell. 1998. The JNK pathway regulates the in vivo deletion of immature CD4+CD8+ thymocytes. J. Exp. Med. 188:1817–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sabapathy, K., Y. Hu, T. Kallunki, M. Schreiber, J.-P. David, W. Jochum, E.F. Wagner, and M. Karin. 1999. JNK2 is required for efficient T-cell activation and apoptosis but not for normal lymphocyte development. Curr. Biol. 9:116–125. [DOI] [PubMed] [Google Scholar]
- 38.Mamalaki, C., J. Elliott, T. Norton, N. Yannoutsos, A.R. Townsend, P. Chandler, E. Simpson, and D. Kioussis. 1993. Positive and negative selection in transgenic mice expressing a T-cell receptor specific for influenza nucleoprotein and endogenous superantigen. Dev. Immunol. 3:159–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haanen, J.B.A.G., M. Toebes, T.A. Cordaro, M.C. Wolkers, A.M. Kruisbeek, and T.N.M. Schumacher. 1999. Systemic T cell expansion during localized viral infection. Eur. J. Immunol. 29:1–7. [DOI] [PubMed] [Google Scholar]
- 40.Kessels, H.W.H.G., M.D. Van den Boom, H. Spits, E. Hooijberg, and T.N.M. Schumacher. 2000. Changing T cell specificity by retroviral T cell receptor display. Proc. Natl. Acad. Sci. USA. 97:14578–14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas, B., I. Stefanová, K. Yasutomo, N. Dautigny, and R.N. Germain. 1999. Divergent changes in the sensitivity of maturing T cells to structurally related ligands underlies formation of a useful T cell repertoire. Immunity. 10:367–376. [DOI] [PubMed] [Google Scholar]
- 42.Van Pel, A., F. Vessière, and T. Boon. 1983. Protection against two spontaneous mouse leukemias conferred by immunogenic variants obtained by mutagenesis. J. Exp. Med. 157:1992–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amsen, D., C. Revilla Calvo, B.A. Osborne, and A.M. Kruisbeek. 1999. Costimulatory signals are required for induction of transcription factor Nur77 during negative selection of CD4+CD8+ thymocytes. Proc. Natl. Acad. Sci. USA. 96:622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiest, D.L., L. Yuan, J. Jefferson, P. Benveniste, M. Tsokos, R.D. Klausner, L.H. Glimcher, L.E. Samelson, and A. Singer. 1993. Regulation of T cell receptor expression in immature CD4+CD8+ thymocytes by p56+lck tyrosine kinase: basis for differential signaling by CD4 and CD8 in immature thymocytes expressing both coreceptor molecules. J. Exp. Med. 178:1701–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Buer, J., I. Aifantis, J.P. DiSanto, H.J. Fehling, and H. Von Boehmer. 1997. Role of different T cell receptors in the development of pre-T cells. J. Exp. Med. 185:1541–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Finco, T.S., T. Kadlecek, W. Zhang, L.E. Samelson, and A. Weiss. 1998. LAT is required for TCR-mediated activation of PLCγ and the Ras pathway. Immunity. 9:617–626. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, W., J. Sloan-Lancaster, J. Kitchen, R.P. Trible, and L.E. Samelson. 1998. LAT: the ZAP-70 tyrosine kinase substrate that links T cell receptor to cellular activation. Cell. 92:83–92. [DOI] [PubMed] [Google Scholar]
- 48.Hogquist, K.A., S.C. Jameson, and M.J. Bevan. 1995. Strong agonist ligands for the T cell receptor do not mediate positive selection of functional CD8+ T cells. Immunity. 3:79–86. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, W., R.P. Trible, and L.E. Samelson. 1998. LAT palmitoylation: its essential role in membrane microdomain targeting and tyrosine phosphorylation during T cell activation. Immunity. 9:239–246. [DOI] [PubMed] [Google Scholar]
- 50.Yablonski, D., M.R. Kuhne, T. Kadlecek, and A. Weiss. 1998. Uncoupling of nonreceptor tyrosine kinases from PLC-γ1 in an SLP-76-deficient T cell. Science. 281:413–416. [DOI] [PubMed] [Google Scholar]
- 51.Dower, N.A., S.L. Stang, D.A. Bottorff, J.O. Ebinu, P. Dickie, H.L. Ostergaard, and J.C. Stone. 2000. RasGRP is essential for mouse thymocyte differentiation and RCR signaling. Nat. Immunol. 1:317–321. [DOI] [PubMed] [Google Scholar]