Abstract
Scp160p is a multiple KH-domain RNA-binding protein in yeast known to associate with polyribosomes as an mRNP component, although its biological role remains unclear. As a genetic approach to examine Scp160p function, we applied an ethyl methanesulfonate (EMS) screen for loci synthetically lethal with scp160 loss, and identified a single candidate gene, EAP1, whose protein product functions in translation as an eIF4E-binding protein, with additional uncharacterized spindle pole body functions. To reconfirm scp160/eap1 synthetic lethality, we constructed a strain null for both genes, supported by an SCP160 maintenance plasmid. We used this strain to establish a quantitative assay for both Scp160p and Eap1p functions in vivo, and applied this assay to demonstrate that Y109A EAP1, a previously described allele of EAP1 that cannot bind eIF4E, is markedly impaired with regard to its SCP160-related activity. In addition, we explored the possibility of physical interaction between Eap1p and Scp160p, and discovered that Eap1p associates with Scp160p-containing complexes in an RNA-dependent manner. Finally, we probed the impact of EAP1 loss on Scp160p, and vice versa, and found that loss of each gene resulted in a significant change in either the complex associations or subcellular distribution of the other protein. These results clearly support the hypothesis that Scp160p plays a role in translation, demonstrate that the interaction of SCP160 and EAP1 is biologically significant, and provide important tools for future studies of the in vivo functions of both genes.
INTRODUCTION
The gene SCP160 encodes a 160 kDa protein in yeast originally hypothesized to function in the maintenance of ploidy (1), due in part to its null phenotype, which includes abnormal cell size and shape, increased DNA content and missegregation of genetic markers through meiosis (1). A number of more recent biochemical studies have demonstrated the encoded gene product, Scp160p, to be an RNA-binding protein (2) that associates in vivo with a specific subset of mRNAs (3) as a component of both soluble and membrane-associated mRNPs and polyribosomes (2,4–6). Target messages specifically associated with Scp160p include DHH1, an RNA helicase that functions in mRNA decapping (7), BIK1, a component of the kinetochore–microtubule binding interface (8), and NAM8, an RNA-binding protein required for the meiosis-specific splicing of MER2 and MER3 (9). Subcellular localization studies, using both indirect immunofluorescence and green fluorescent protein-tagged alleles, have demonstrated that the majority of Scp160p is cytoplasmic, with significant enrichment around the nuclear envelope and rough endoplasmic reticulum (1,5,6), consistent with the distribution of polyribosomes in yeast. Combined, these data implicate Scp160p for a role in cytoplasmic mRNA metabolism, potentially including translation, although the precise biological role of Scp160p remains unknown.
Biochemical studies of Scp160p-containing mRNP complexes, which migrate by native size exclusion chromatography at >1300 kDa, have revealed the presence of a large number of other proteins, including Pab1p and Bfr1p (4,5). The identities of most of the other proteins in these complexes remain unknown. Similarly, little is currently known about the structure of Scp160p, although amino acid sequence homology studies have demonstrated the presence of 14 copies of the hnRNP K homology (10), or KH domain (2,4,5,11). Indeed, Scp160p bears greatest similarity to a large family of multiple KH domain proteins collectively known as vigilins. First identified in chicken (12), vigilin orthologs have now been found in human (13), Xenopus laevis (14), Drosophila melanogaster (15,16), Caenorhabdtis elegans (2), Schizo saccharomyces pombe (GenPept 7493335), Neurospora crassa (GenPept 7899383) and Saccharomyces cerevisiae (1,2,11). Although all vigilin proteins reported to date appear to bind nucleic acid, in most cases both the type of nucleic acid bound and the functional significance of the interaction remain unclear. One notable exception is Xenopus vigilin, which was demonstrated not only to bind specifically to a defined sequence in the 3′-untranslated region of the vitellogenin message, but also to inhibit cleavage of that sequence by the mRNA endonuclease polysomal ribonuclease 1 (17). Whether Scp160p functions in a similar manner with regard to its target messages is a subject of current study.
As an independent approach to explore the function of Scp160p in vivo, we conducted a genetic screen to identify additional loci in the yeast genome that, when mutated, are lethal in combination with scp160 loss. This approach, termed a synthetic lethal screen, is a powerful tool to help define the function of any given non-essential gene in yeast (18,19). The rationale is 2-fold. First, by definition, the loci identified in a synthetic lethal screen interact in some functionally significant way with the gene of interest, albeit not always directly or physically. Often these are either genes of overlapping function, or genes of distinct function that nonetheless contribute to the same pathway or biological process as the gene of interest. Secondly, once a synthetic lethal relationship has been defined between two non-essential genes, strains null for either gene provide a genetic background in which the second gene is rendered essential, offering a powerful tool for further genetic or structure/function studies. We report here the results of an ethyl methanesulfonate (EMS) synthetic lethal screen for SCP160, from which we identified a single candidate gene: EAP1.
EAP1 is a non-essential gene in yeast whose product, Eap1p, was identified by Cosentino et al. (20) as a novel protein that binds directly to eIF4E, the cap-binding subunit of the cap-dependent translation initiation complex, eIF4F. Almost simultaneously, an independent function for Eap1p was discovered by Chial et al., who identified EAP1 in the course of a synthetic lethal screen with ndc1-1 (21), a cold-sensitive allele of the essential NDC1 gene required for spindle pole body duplication and function (22). The independence of these functions was most clearly demonstrated by studies involving the Y109A allele of EAP1, which fully rescued the synthetic lethal phenotype of ndc1-1/Δeap1 yeast (21), despite the fact that this substitution replaces a key residue in the eIF4E-binding domain of Eap1p, thereby precluding its ability to bind eIF4E both in vitro and in vivo (20).
The results reported here demonstrate four important points with regard to both SCP160 and EAP1. First, loss of both genes in yeast is a synthetically lethal event, implicating a functional relationship, either direct or indirect, between the two gene products. Considering what is already known about the role of Eap1p in both translation and spindle pole body function, this relationship both supports and extends previous hypotheses concerning the function of Scp160p in vivo. Secondly, the Y109A allele of Eap1p, which no longer binds eIF4E, is largely impaired in its ability to complement scp160/eap1 synthetic lethality. This result clearly supports the hypothesis that Scp160p plays a role in cytoplasmic mRNA metabolism, including translation. Thirdly, Eap1p associates with at least a subset of Scp160p-containing complexes in an RNA-dependent manner, although these data in no way implicate a direct physical interaction between the two proteins. Finally, eap1 loss results in a subtle but reproducible redistribution of Scp160p from predominantly large to both large and smaller complexes in yeast, while scp160 loss results in an apparently complete loss of Eap1p from the membrane pellets but not from the soluble portion of fractionated lysates. Together, these data reconfirm the functional significance of the interaction between SCP160 and EAP1, and offer clues to the associations and mechanisms of action for both proteins.
MATERIALS AND METHODS
Plasmids and yeast strains
All plasmid manipulations were performed according to standard techniques utilizing Escherichia coli strains XL1-Blue (Strategene, Inc.) and XL10-Gold (Stratagene, Inc.), as described previously (23). Plasmids used in this study are listed in Table 1.
Table 1. Plasmids used in this study.
| Plasmid code | Marker | Test allele | Type |
|---|---|---|---|
| JF3116 | URA3, ADE3 | SCP160 | 2µ |
| JF1102 | TRP1 | – | 2µ |
| JF4176 | LEU2 | – | CEN |
| JF2803 | TRP1 | SCP160 | 2µ |
| JF3782 | LEU2 | EAP1 | 2µ |
| JF3896 | LEU2 | HA-EAP1 | CEN |
| JF3902 | LEU2 | HA-Y109A.EAP1 | CEN |
The SCP160 maintenance plasmid JF3116 was constructed from YEPlac195 (24) by insertion of a 4.7 kb BamHI–PstI fragment encoding wild-type SCP160, as well as a PCR-amplified SacI fragment encoding the wild-type ADE3 gene. JF1102 refers to YEPlac195 with no additional inserts. JF4176 refers to pRS315 with no additional inserts (25). JF2803 was derived from YEPlac112 (24) by gap repair insertion of an N-terminally hemagglutinin (HA)-tagged allele of SCP160. JF3782 is a library plasmid derived from the YEp13-based yeast genomic library ATCC 37323, and carries a full-length copy of the wild-type EAP1 locus, in the absence of any full-length neighboring genes. JF3896 carries an HA-tagged allele of otherwise wild-type EAP1 in the pRS315 plasmid backbone, and JF3902 carries an HA-tagged allele of Y109A EAP1 in the same plasmid backbone. Both plasmids have been described previously (20), and were the generous gift of Drs Mark Winey and Alex Stemm-Wolfe (University of Colorado, Boulder, CO).
The yeast strains used in this study are listed in Table 2. Each of these strains was derived originally either from W303 (MATa ade2-1 his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 RAD5+, a generous gift from Dr R. Rothstein, Columbia University, NY) or from JJ52 (MATα gal7Δ102 ura3-52 trp1-289 ade1 lys1 leu2-3,112, which was a generous gift from Drs Mark Parthun and Judith Jaehning, University of Colorado Health Sciences Center). All studies comparing wild-type versus scp160-null cells were performed using diploid strains of W303-derived cells homozygous for a genomic scp160 deletion, that either did (JFy4100) or did not carry a plasmid-borne wild-type copy of SCP160 (JF3116, URA3), respectively. Due to concerns over potential and progressive aneuploidy in the scp160-null strains, these strains were always freshly generated from JFy4100 just prior to use by plasmid curing on medium containing 5-fluoro-orotic acid (5-FOA) (26). Similarly, all haploid strains genomically null for scp160 were generated by two-step gene replacement (26), with the introduction of an SCP160 maintenance plasmid into the strain prior to the genomic deletion event.
Table 2. Yeast strains used in this study.
| Strain code | Relevant genomic alleles | Plasmid(s) | Comments (parent strain) |
|---|---|---|---|
| JFy2806 | Δscp160 ade2– ade3– | SCP160 URA3 ADE3 (JF3116) | For synthetic lethal screen (W303) |
| JFy3400 | Δscp160 ade2– ade3–eap1– | SCP160 URA3 ADE3 (JF3116) | EMS derivative of JFy2806 (W303) |
| JFy3751 | Δscp160 ade2– ade3–eap1– | SCP160 URA3 ADE3 (JF3116) | Double backcrossed derivative of JFy3400 (W303) |
| JFy4100 | Δscp160/Δscp160 (diploid) | SCP160 URA3 ADE3 (JF3116) | For studies of SCP160+ versus scp160-null cells (W303) |
| JFy4247 | Δscp160 Δeap1 | SCP160 URA3 ADE3 (JF3116) | For quantitative test of plasmid loss (W303) |
| JFy4338 | FLAG.SCP160 | HA.EAP1 (JF3896, CEN) | To test Scp160p–Eap1p protein interaction (JJ52) |
| JFy4838 | native SCP160 | HA.EAP1 (JF3896, CEN) | Control for protein interaction studies (JJ52) |
| JFy4546 | Δscp160 Δeap1 | HA.SCP160 (JF2252, CEN) | Also has EAP1 URA3 plasmid JF3984 (W303) |
| JFy4522 | Δscp160 | HA.EAP1 (JF4323, CEN) | Also has SCP160 ADE3 URA3 plasmid JF3116 (W303) |
An EMS-based synthetic lethal screen using JFy2806 yeast genomically null for scp160
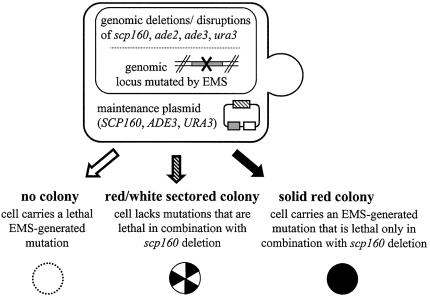
The haploid strain JFy2806 (MATa scp160Δ ade2-1 ade3Δ his3-11,15 leu2-3,112 ura3-1 trp1-1 can1-100 RAD5+) was generated from its W303 parent (the generous gift of Dr R. Rothstein, Columbia University, NY) by sequential deletion of both scp160 and ade3. Deletion of scp160 was accomplished by standard two-step gene replacement using the Alani–Kleckner cassette (27), with the introduction of an SCP160 maintenance plasmid following the genomic integration but preceding the genomic deletion event. The ade3 coding sequence was deleted by one-step gene replacement using the kanR cassette with selection for G418 resistance.
Prior to mutagenesis of JFy2806, an EMS kill curve was performed to determine the appropriate amount of drug (625 µl) needed to achieve an ∼80% kill rate. For the actual mutagenesis procedure, cells were grown for 48 h in SD-ura medium, to ensure the presence of the URA3 SCP160 maintenance plasmid (JF3116), at which point 2.5 ml of this culture was spun down, washed twice with 50 mM K3PO4 pH 7.0, and resuspended in a final volume of 10 ml of 50 mM K3PO4. Next, 625 µl of EMS was added, the cells were mixed by vortex agitation, and allowed to incubate for 30 min at 30°C, at which point 300 µl of 10% (w/v) sodium thiophosphate was added to deactivate the EMS and stop the mutagenesis. The cells were then spun down, washed twice with sterile water, and counted using a hemocytometer. Four hundred cells were spread onto each of twenty 10 cm YPD plates for a total of 8000 cells. Plates were incubated for ∼3 days, or until 2–3 mm diameter colonies were clearly visible and sectoring status could be easily scored by visual inspection. To intensify the red color, in some experiments the plates were stored at 4°C for 1–2 days prior to scoring. From the background of sectoring colonies, solid red colonies were picked and streaked onto fresh YPD. After 12 rounds of screening, as described above, only one mutant, designated 12007, was isolated as reproducibly failing to sector on YPD, yet exhibiting restored sectoring in the presence of an alternative (non-URA3) SCP160 maintenance plasmid (JF2803). Sectoring was not restored to this strain in the presence of the plasmid backbone alone (JF1102). This strain was designated JFy3400, and was backcrossed twice to the parental strain to remove unrelated EMS mutations prior to further analysis. The resulting strain was designated JFy3751.
DNA sequencing of the EAP1 genomic locus
To identify the EMS mutation(s) present in the EAP1 locus of JFy3751 versus its unmutagenized parent (JFy2806), a 2570 bp fragment including the entire EAP1 coding sequence was PCR amplified from the genomic DNA of each strain using the primers EAP1F1 (5′-gcgcgaattcgacttatcattcactttggtttag-3′) and EAP1R1 (5′-gcgcgtcgacccagaacaagtggataaactaatcc-3′). These fragments were sequenced using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit with primers: EAP1F1, EAP1F2 (5′-gcgcgaattcaaaaatggaactcaacgacccttca-3′), EAP1F3 (5′-gaactttatcatttgaaaccatct-3′), EAP1F4 (5′-gattttgaagattggaaggcaaa-3′), EAP1F5 (5′-ttcgttgccttcattagataata-3′), EAP1F6 (5′-tgctcatcagggtccacaatt-3′), EAP1F7 (5′-ccttatccaaatatgatgctacaa-3′), EAP1F10 (5′-tcggtagcgtagaatcttcaa-3′), EAP1R1 and EAP1R10 (5′-ttcttgtccatctccagca-3′). All reactions were purified using Princeton Centrisep spin columns (Princeton Separations, Adelphia, NJ), and analyzed at the Iowa State University DNA Sequencing and Synthesis Facility (Ames, IA). The resulting EAP1 parent and mutant nucleotide sequences were aligned with the corresponding EAP1 sequence obtained from the Saccharomyces Genome Database (http://www.yeastgenome.org/) using the Local Alignment tool (available at http://pbil.univ-lyon1.fr/).
A quantitative test for SCP160 and EAP1 functions in vivo
The scp160/eap1 double null strain JFy4247 was generated by mating one haploid strain null for scp160 (JFy3607) with a haploid strain null for eap1 (JFy4084, the generous gift of Drs Alex Stemm-Wolf and Mark Winey, University of Colorado). The resulting diploid was sporulated, and the tetrads dissected and characterized as scp160/eap1 wild-type versus null via marker analysis and PCR amplification, as appropriate for each locus. A wild-type, plasmid-borne copy of SCP160 was maintained in the strain at all times.
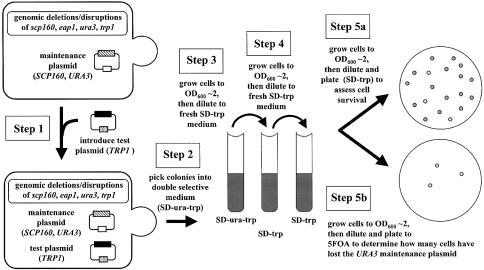
The in vivo function of any given test allele of SCP160 or EAP1 was assessed by introducing that allele into JFy4247 on a non-URA3 plasmid, either TRP1 or LEU2, in parallel with corresponding positive (SCP160+) and negative (plasmid backbone alone) controls. Transformants were then cultured through a fixed number of generations of growth in the absence of uracil selection so as to provide an opportunity for loss of the JF3116 (URA3, SCP160) maintenance plasmid. Prior to each step involving cell counting, the cultures were treated so as to minimize cell clumping. In brief, cells were placed in a 1.5 ml sterile snap-cap tube and centrifuged at 13 000 r.p.m. for 10 s. The medium was decanted and the cell pellet was resuspended in 1 ml of sterile water. Each cell suspension was then mixed by vigorous vortex agitation for 5 s, and 200 µl of the cell suspension was added to 1.8 ml of sterile separation solution [10 mM Tris pH 8, 500 mM EDTA, 1.2% polyoxyethylene sorbitan monolaurate (Tween-20)]. Samples were again mixed by vigorous vortex agitation for 10 s and readings were taken at OD600.
A total of 300 000 cells carrying both the test and maintenance plasmids (as demonstrated by growth on SD-trp-ura or SD-leu-ura medium) were inoculated into fresh SD-trp or SD-leu liquid medium. Samples were grown to an OD600 of 2–3, at which point 300 000 cells were transferred into fresh SD-trp or SD-leu liquid medium and allowed to grow again to an OD600 of 2–3. Finally, 300 cells from each culture were plated onto the appropriate single selective medium, to enable a measure of cell viability. To determine the percentage of yeast that had lost their SCP160 URA3 maintenance plasmid, appropriate numbers of cells were plated onto medium containing 5-FOA, which allowed only those cells that had lost their URA3 maintenance plasmid to grow. For yeast transformed with the positive control (wild-type SCP160) test plasmid, 300 versus 600 cells were inoculated per plate, and close to half resulted in 5-FOA-resistant colonies. For yeast transformed with the negative control test plasmids, 10 000 versus 50 000 cells were inoculated per plate, with few if any 5-FOA-resistant colonies appearing. For yeast transformed with the 2µ EAP1 or centromeric (CEN) HA-EAP1 and HA-EAP1.Y109A test plasmids, 1200 cells were inoculated per plate. These various numbers of cells plated from the different strains were each determined from pilot experiments to result in less than 300 5-FOA-resistant colonies growing per plate.
In all cases, plates were incubated at 34°C for 72 h prior to scoring, at which point the fraction of colonies observed versus cells plated was tabulated for each strain plated onto 5-FOA, normalized for culture viability, as determined from the corresponding single selective non-5-FOA plates. Differences between sample group means were determined using the two-sided t-test and considered statistically significant at a P-value of <0.05 using the MINITAB statistical program (State College, PA).
Affinity isolation of Scp160p-containing complexes
Scp160p-containing complexes were isolated from the yeast strain JFy4338 expressing both FLAG-Scp160p and HA-Eap1p by anti-FLAG affinity chromatography, as described previously (3). In brief, cells were grown to early log phase (OD600 ∼1) in rich medium, harvested by centrifugation, and lysed in T75 buffer (25 mM Tris pH 7.5, 75 mM NaCl) containing 30 mM EDTA to disrupt the polyribosomes. After centrifugation, the supernatant was collected and passed over an S-300 gel filtration column. The void material (>1300 kDa, derived from fractions 30–40 ml) was pooled, supplemented with FLAG peptide (Asp-Tyr-Lys-Asp-Asp-Asp-Asp-Lys, purchased from the Emory Microchemical Facility) at 2 µg/ml, and loaded onto a 1 ml M2 α-FLAG column (Sigma Aldrich). In experiments designed to test whether any interaction of Eap1p with Scp160p complexes was dependent on RNA, this void volume sample was also supplemented with RNase I (Promega) at 50 U/ml and incubated on ice for 30 min prior to loading. Finally, after washing the column with 100 ml of T75 buffer, the FLAG-Scp160p-containing complexes were eluted using 184 mg/ml FLAG peptide in 5 ml of T75 buffer. As a negative control, a parallel ‘mock’ isolation procedure was performed using strain JFy4848, which expresses HA-Eap1p in the presence of native, untagged Scp160p.
Native size exclusion chromatography
Soluble lysates of yeast were size fractionated using an S-300 gel filtration column (Pharmacia) attached to an FPLC, essentially as described previously (4,5). One liter of yeast culture was grown to early log phase (OD600 = 1.0–1.5), harvested by centrifugation, and washed twice in T75 buffer (25 mM Tris pH 7.5, 75 mM NaCl). The cells were lysed by vortex agitation with an equal volume of acid-washed 0.5 mm glass beads in 4 ml of T75 buffer containing 30 mM EDTA. Next, the lysate was centrifuged at 3000 r.p.m. for 10 min at 4°C and the supernatant spun a second time at 14 000 r.p.m. for 10 min at 4°C. Finally, the resultant supernanant was passed through a 0.2 µm syringe filter and applied to the S-300 column in T75 buffer, with 2 ml fractions collected representing elution volumes from 30 to 70 ml.
Fractionation of soluble and membrane-associated populations of Scp160p
The soluble and membrane-associated pools of Scp160p were fractionated by differential centrifugation, as described previously (3,5). Cells were lysed by vortex agitation with an equal volume of acid-washed 0.5 mm glass beads in 1× polyribosome lysis buffer (25 mM Tris–HCl pH 7.2, 50 mM KCl, 30 mM MgCl2, 5 mM β-mercaptoethanol, 100 µg/ml cycloheximide). Each lysate was transferred to a clean microfuge tube and centrifuged at 3000 r.p.m. (in an Eppendorf 5415C microfuge) for 5 min at 4°C to remove glass beads and cell debris. The supernatant from each sample was then transferred to a fresh tube, with or without RNase I (Promega) treatment (50 U/ml at 4°C for 30 min), and centrifuged at 12 000 r.p.m. for 10 min at 4°C. The supernatant resulting from this high speed spin contained the soluble pool of Scp160p. The pellet contained the membrane-associated pool. Finally, the membrane pellet was solubilized by resuspension in lysis buffer containing 1.5% Triton X-100, with a 10 min incubation (on ice), followed by a 10 min, 4°C spin at 14 000 r.p.m. to remove residual insolubles.
Sucrose gradient fractionation of yeast lysates
Sucrose gradient fractionation of soluble cell lysates was performed as described previously (3–5). In brief, a 100 ml culture of yeast was grown in rich (YPD) medium to early log phase (OD600 ∼1.5), at which point cycloheximide (Sigma) was added directly to the culture to a final concentration of 100 µg/ml. The culture was incubated on ice for 25 min, and cells were harvested by centrifugation (4000 r.p.m. for 10 min). Following two washes in 10 ml of water containing 100 µg/ml cycloheximide, cells were lysed as described above in 1× polyribosome lysis buffer. Each lysate was transferred to a clean microfuge tube and centrifuged in an Eppendorf microfuge (Model 5415C) at 6500 r.p.m. for 10 min at 4°C, followed by a second centrifugation of the supernatant at 14 000 r.p.m., also for 10 min at 4°C. Finally, 100 µl of supernatant were either pre-treated with RNase I (Promega) at a concentration of 50 U/ml for 30 min at 4°C prior to loading, or loaded directly onto a 15–45% sucrose gradient (11 ml) made using a Gradient Master automatic system. Each gradient was centrifuged in an SW41ti rotor (Beckman) at 39 000 r.p.m. for 2.5 h at 4°C, after which fractions were harvested using an Isco gradient fractionator. Gradient RNA profiles were monitored by following absorbance at 254 nm.
Sucrose gradient fractionation of membrane-associated Scp160p complexes was achieved using a high speed centrifugation (12 000 r.p.m.) to separate membrane pellets, as described above. Prior to loading onto the sucrose gradient, these pellets were resuspended in lysis buffer containing 1.5% Triton-X and incubated for 10 min (on ice), then centrifuged a second time (10 min, 4°C) at 14 000 r.p.m. to remove insolubles.
RESULTS
EAP1 loss is synthetically lethal with loss of SCP160 in yeast
As a genetic approach to identify other genes in yeast whose products interact physically and/or functionally with Scp160p, we conducted an EMS synthetic lethal screen (Fig. 1) (18,19) using a strain genomically null for scp160, but supported by a maintenance plasmid encoding wild-type SCP160 as well as the markers URA3 and ADE3 (JF3116). As described in Materials and Methods, after 12 rounds of screening, we identified a single positive clone (JFy3400) in which SCP160 had been rendered essential, as demonstrated by lack of colony sectoring on rich medium, unless a second (non-URA3) SCP160-encoding plasmid was provided. Backcrossing JFy3400 to the unmutagenized parent strain produced diploids in which SCP160 was no longer essential, demonstrating that the EMS-derived mutation was recessive. Dissection of the sporulated diploids produced tetrad sets in which the essential nature of SCP160 segregated 2:2, thereby confirming that the EMS-derived mutation affected a single genomic locus. A second backcross (resulting in strain JFy3751) was performed to remove any remaining unrelated EMS mutations.
Figure 1.
A synthetic lethal screen using scp160-null cells. As illustrated, any cell that carries an EMS-generated mutation that is synthetically lethal with scp160 loss will result in a totally red, non-sectoring colony.
The identity of the EMS-mutated locus in JFy3751 was defined by a standard complementation cloning procedure using a high copy number wild-type yeast genomic library (ATCC 37323). Of approximately 5000 library transformants screened, representing approximately one haploid yeast genome equivalent, six colonies were identified that demonstrated plasmid-dependent restoration of red/white colony sectoring. The library plasmids from these cells were rescued by standard procedures (26), reconfirmed by transformation into JFy3751, and the genomic inserts sequenced from both ends using primers that bound within the plasmid backbone. The sequences obtained were compared against the S.cerevisiae genome database (http://www.yeastgenome.org/) to determine the genomic fragments present. In all cases, the genomic fragments identified within a given plasmid were indeed contiguous in the genome, and plasmid insert sizes predicted from the database search agreed with the results of restriction enzyme digestion analyses of the plasmids.
Of the six plasmids obtained, one (JF3778) carried a wild-type copy of SCP160, and four (JF3777, 3781, 3782 and 3785) carried varying genomic fragments that each included a full-length copy of the gene EAP1. The sixth plasmid carried a large genomic insert including multiple genes (but not EAP1 or SCP160), which will be the subject of future study.
To identify the relevant EMS mutation(s) within EAP1 in the strain JFy3751, a 2570 bp fragment of genomic DNA including EAP1 was PCR amplified, as described in Materials and Methods, and sequenced by standard procedures. As a control, the corresponding genomic fragment was amplified and sequenced from the unmutagenized parent strain (JFy2806). A comparison of the sequences obtained from these strains identified a single point mutation within the JFy3751 EAP1 locus: a C to T transition at the first position of codon 397, thereby changing a CAG (glutamine) residue to TAG (stop), effectively cutting the 632 codon open reading frame almost in half. Whether the truncated polypeptide fragment is expressed and remains stable is not known.
Finally, to confirm that actual loss (and not just point mutation) of EAP1 was synthetically lethal with SCP160 loss, we generated a strain carrying genomic deletions of both loci (see Materials and Methods), supported by a maintenance plasmid encoding wild-type Scp160p (JF3116). As expected, this strain (JFy4247) demonstrated a complete inability to survive in the absence of the maintenance plasmid, unless some alternative plasmid encoding either Scp160p or Eap1p was provided (Table 3). These data not only confirmed the synthetic lethality of scp160 and eap1, but they also further offered the basis for a quantitative test of in vivo function for modified alleles of both genes.
Table 3. Relative plasmid loss as a measure of in vivo function of SCP160 and EAP1 in yeast.
| Test plasmid | Allele | Relative plasmid loss (n) |
|---|---|---|
| pJF2803 (2µ) | SCP160 | 100.00 ± 31.46 (27) |
| pJF1102 (2µ) | – | 0.05 ± 0.06 (52) |
| pJF4176 (CEN) | – | 0.16 ± 0.30 (24) |
| pJF3782 (2µ) | EAP1 | 82.16 ±j121.13 (30) |
| pJF3896 (CEN) | HA-EAP1 | 68.31 ± 15.59 (34) |
| pJF3902 (CEN) | HA-EAP1.Y109A | 18.05 ± 6.36 (38) |
High plasmid loss values indicate that a given test plasmid is sufficient to relieve the essential nature of the SCP160 maintenance plasmid in the eap1/scp160 double null strain, while low plasmid loss values indicate that a given test plasmid is not sufficient to relieve the essential nature of the SCP160 maintenance plasmid in this strain. All values were normalized to the degree of plasmid loss seen when the test plasmid encoded a wild-type allele of SCP160.
A quantitative test of in vivo function for alleles of SCP160 and EAP1
The general strategy enabling quantitative analysis of in vivo function for modified alleles of SCP160 and EAP1 is illustrated in Figure 2, and described in Materials and Methods. As explained above, the basis for this strategy is the inability of yeast genomically null for both scp160 and eap1 to survive the loss of an SCP160 maintenance plasmid unless an alternative source of functional SCP160 or EAP1 is provided. If a partially functional allele of either SCP160 or EAP1 is provided, an intermediate degree of maintenance plasmid loss is observed. Since the maintenance plasmid also carries a URA3 marker, the degree of presence or absence of this plasmid in a large population of cells can be easily scored by plating a defined number of cells onto pairs of plates that do and do not contain 5-FOA, respectively. Since only those cells that have lost the URA3 marker (and therefore the maintenance plasmid) will survive in the presence of the drug, the ratio of colonies that appear on the two media represents a quantitative measure of the degree of maintenance plasmid loss in the population. By including both positive (wild-type SCP160 or EAP1 on the test plasmid) and negative (empty plasmid backbone) controls in each experiment, the relative degree of in vivo function for any modified allele is easily ascertained.
Figure 2.
A quantitative test of in vivo function for modified alleles of SCP160 and/or EAP1. As illustrated, the relative functional capacity of any given allele of SCP160 or EAP1 is assessed in terms of its ability to enable survival of a double null strain in the absence of its SCP160 maintenance plasmid.
The first five rows of Table 3 present data demonstrating the results of applying this analysis to various test plasmids encoding either wild-type SCP160 (JF2803) or EAP1 (JF3782 or JF3896), versus the corresponding empty plasmid backbones (JF1102 or JF4176). A difference of almost three orders of magnitude separates the positive versus negative controls. In contrast, there is no statistically significant difference evident between test plasmids encoding wild-type Scp160p versus Eap1p, regardless of the presence or absence of an epitope tag (HA), or the high (2µ) versus low (CEN) copy number status of the plasmid.
eIF4E binding is an important aspect of EAP1 function with regard to SCP160
As explained above, the EAP1 gene product encodes at least two distinct functions, one relevant to translation initiation that involves binding to eIF4E (20), and the other relevant to spindle pole body function that is independent of eIF4E binding (21). To determine whether the SCP160-relevant function of EAP1 requires eIF4E binding, we applied our plasmid loss system to a test plasmid (JF3902) encoding the Y109A allele of HA-EAP1. As mentioned above, the Y109A substitution interrupts the Eap1p eIF4E-binding domain, thereby eliminating eIF4E binding both in vitro and in vivo (20). As demonstrated in Table 3 (rows 5 and 6), although this single point mutation did not completely eliminate the ability of the HA-EAP1.Y109A test plasmid to complement scp160/ eap1 synthetic lethality, apparent function of the mutant allele was lowered relative to its wild-type counterpart by almost a factor of 4.
Eap1p associates with Scp160p-containing complexes
The observation that two genes are synthetically lethal in yeast implicates a functional relationship between the two genes and/or their encoded products, but does not necessarily reflect a stable physical association between the encoded gene products. To test whether the Eap1p protein associates physically with Scp160p-containing complexes, we generated a strain of yeast co-expressing FLAG-tagged Scp160p with HA-tagged Eap1p (JFy4338). Anti-FLAG affinity isolation of the Scp160p-containing complexes from these cells, as described in Materials and Methods, resulted in samples that also contained HA-Eap1p, as demonstrated by western blot analysis (Fig. 3, top two panels). When the samples were pre-treated with RNase, however, the association disappeared (Fig. 3, middle two panels), indicating the RNA-dependent nature of the interaction. As a control, mock anti-FLAG affinity isolation procedures were also performed using samples prepared from cells (JFy4838) expressing HA-tagged Eap1p with native, untagged Scp160p. As expected, although HA-Eap1p protein was present in the void, flow through and first wash fractions, no HA-Eap1p signal was detected in the elution fractions from these control lysates (Fig. 3, bottom two panels), demonstrating the specificity of the isolation procedure, and of the HA-Eap1p–FLAG-Scp160p association illustrated in the top set of panels.
Figure 3.
Eap1p physically associates with Scp160p-containing complexes in an RNA-dependent manner. Top two panels: Scp160p-containing complexes were affinity isolated as described in Materials and Methods from lysates of yeast co-expressing FLAG-Scp160p and HA-Eap1p, and subjected to western blot analysis with both anti-FLAG (top panel) and anti-HA (bottom panel) monoclonal antibodies. As illustrated, although the affinity isolation procedure was directed against the FLAG-Scp160p protein alone, HA-Eap1p protein co-eluted from the affinity column. Middle two panels: Scp160p-containing complexes were affinity isolated and characterized as described above, except that samples were pre-treated with RNase prior to the affinity isolation procedure. As illustrated, the association between Scp160p and Eap1p was RNase sensitive. Bottom two panels: as a control for specificity of the isolation and elution procedure, a parallel isolation procedure was performed on lysates of yeast expressing HA-Eap1p with native, untagged Scp160p. As illustrated, although HA-Eap1p protein was present in the lysates (void, flow through and first wash fractions), no HA-Eap1 signal eluted from the anti-FLAG affinity column. Although the results of only single representative experiments are presented, both test and control experiments were each repeated at least three times with indistinguishable results.
Impact of eap1 loss on Scp160p, and of scp160 loss on Eap1p
To test the functional significance of Scp160p–Eap1p physical association, we explored the effect of eap1 loss on three biochemical/cell biological properties of Scp160p, and vice versa: (i) formation of >1300 kDa complexes; (2) association with polyribosomes; and (iii) distribution between the soluble and membrane-associated subcellular pools. As illustrated in the top two panels of Figure 4, the signals of FLAG-tagged Scp160p and HA-tagged Eap1p co-expressed in otherwise wild-type yeast demonstrated essentially indistinguishable patterns upon fractionation through an S-300 gel filtration column; both eluted predominantly with the void volume (≥1300 kDa ). This elution pattern for Scp160p is RNA dependent, as has been demonstrated previously (4,5), suggesting that the Scp160p-containing complexes are mRNPs. Loss of eap1 resulted in a marked extension of the HA-Scp160p ‘tail’ eluting from the column (Fig. 4, third panel down), while loss of scp160 resulted in little if any change in the elution pattern of HA-Eap1p (Fig. 4, bottom panel). These results suggest that in the absence of eap1, at least some Scp160p complexes are partially impaired, either in terms of the extent of their assembly or in terms of their stability. No parallel impact of scp160 loss was evident on Eap1p complexes.
Figure 4.
Size estimation of Scp160p- and Eap1p-containing complexes using S-300 gel filtration column chromatography. Lysates of cells expressing the indicated alleles of either epitope-tagged or null SCP160 and/or EAP1 were subjected to S-300 gel filtration column chromatography, as described in Materials and Methods, followed by western blot analysis of the fractions using the indicated anti-tag antibodies (FLAG or HA). Each panel represents the corresponding sections of two gels juxtaposed, one loaded with samples representing column elution fractions from 30 to 50 ml, and the other representing fractions from 50 to 70 ml. The position of the void volume for this column, representing complexes of ≥1300 kDa, is indicated. Although the results of only single representative experiments are presented, both test and control experiments were each repeated at least three times with indistinguishable results.
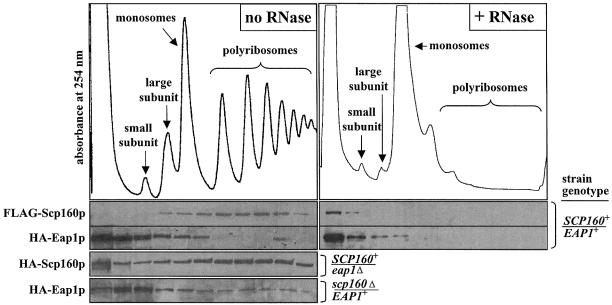
With regard to polyribosome association, as measured by sucrose gradient subcellular fractionation, we observed that, as previously reported, FLAG-Scp160p co-fractionated predominantly with polyribosomes (2,4–6), while HA-Eap1p co-fractionated predominantly with smaller complexes and perhaps with ribosomal subunits and/or monosomes (Fig. 5, upper left set of panels). Collapse of the polyribosomes by pre-treatment with RNase (see Materials and Methods) moved the FLAG-Scp160p signal to the top of the gradient (Fig. 5, right set of panels), as previously reported (4,5). RNase pre-treatment had little apparent effect on distribution of the HA-Eap1p signal, which was already concentrated predominantly near the top of the gradient (Fig. 5, right set of panels). Loss of eap1 again appeared partially to impair or destabilize at least a subset of Scp160p complexes, although the majority of Scp160p clearly remained associated with very large complexes consistent with polyribosomes (Fig. 5, lower left panels). As above, little if any parallel impact of scp160 loss was evident on the size distribution of the majority of Eap1p complexes, although some subtle changes at both the high and low ends of the gradient were seen (Fig. 5, left bottom panel).
Figure 5.
Analysis of Scp160p- and Eap1p-containing complexes using sucrose gradient fractionation. Left panels: lysates of cells expressing the indicated alleles of either epitope-tagged or null SCP160 and/or EAP1 were subjected to sucrose gradient fractionation, as described in Materials and Methods, followed by western blot analysis of the fractions using the indicated anti-tag antibodies (FLAG or HA). The top panel illustrates a representative tracing of the UV (254 nm) absorption profile of the gradient fractions from wild-type cells, with the positions of the small and large ribosomal subunits, monosomes and polysomes indicated. Although the results of only single representative experiments are presented, both test and control experiments were each repeated at least three times with indistinguishable results. Right panels: lysates of yeast expressing tagged alleles of both SCP160 and EAP1 were fractionated and analyzed as described above, except that the samples were pre-treated with RNase prior to loading onto the sucrose gradient.
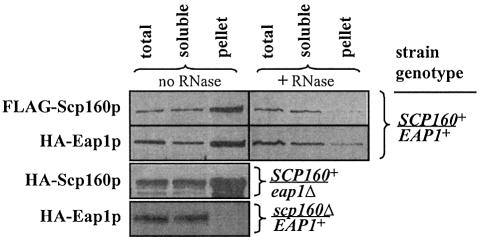
Finally, with regard to the distribution of Scp160p and Eap1p between the soluble and membrane-associated subcellular pools, in yeast expressing wild-type alleles of both genes, both tagged proteins fractionated predominantly although not exclusively with the membrane-associated pool (Fig. 6, upper left panels) and, for both proteins, the membrane association was RNase sensitive (Fig. 6, right panels). Loss of eap1 caused no significant change in the apparent distribution of HA-Scp160p (Fig. 6, left panel 3), while loss of scp160 resulted in a complete loss of HA-Eap1p from the membrane-associated pool (Fig. 6, left panel 4).
Figure 6.
Distribution of Scp160p and Eap1p between the soluble and membrane-associated subcellular pools. Left panels: lysates of cells expressing the indicated alleles of either epitope-tagged or null SCP160 and/or EAP1 were fractionated into total versus soluble and membrane-associated pools by differential centrifugation, as described in Materials and Methods, followed by western blot analysis of the fractions using the indicated anti-tag antibodies (FLAG or HA). Although the results of only single representative experiments are presented, both test and control experiments were each repeated at least three times with indistinguishable results. Right panels: lysates of yeast expressing tagged alleles of both SCP160 and EAP1 were fractionated and analyzed as described above, except that the samples were pre-treated with RNase prior to centrifugation.
DISCUSSION
We report four important points with regard to the genes SCP160 and EAP1 in yeast. First, loss of both genes in yeast is a synthetically lethal event. This point is not surprising, considering that both gene products have previously been implicated for roles in some aspect of cytoplasmic mRNA metabolism, although the observation of synthetic lethality serves to strengthen the argument that these roles must overlap in some functionally significant way. Considering that Eap1p is known to serve a general role in translation, competing with eIF4G for binding to eIF4E, and thereby inhibiting cap-dependent translation initiation (20), while Scp160p has been demonstrated to associate with only a small subset of mRNAs in the cytoplasm and on polyribosomes (3), the mechanism of functional interaction between these two proteins that would lead to their synthetic lethality is not immediately obvious. One possibility stems from the fact that eIF4G is known to bind to the poly(A)-binding protein Pab1p (28,29), a component of Scp160p-associated complexes (4). One could speculate that the lack of both Eap1p and Scp160p in this system may result in some loss of translational regulation, perhaps leading to the abnormal expression of one or more genes, causing lethality to the cell. The observation that Eap1p physically associates with at least a subset of Scp160p-containing complexes further suggests that Eap1p may influence translational initiation of at least some of the mRNAs in these complexes, although other interpretations also remain viable, as will be discussed below. Uncertainties remain concerning the synthesis, folding and stability of the truncated protein. Nonetheless, it is tempting to use the position of the EMS-generated premature stop codon (N397X) reported here as a clue to discerning the nature of the functional interaction between Scp160p and Eap1p. The point of truncation occurs C-terminal to both the eIF4E-binding domain (residues 109–115) and the potential bipartite nuclear localization sequence (residues 183–197), but N-terminal of a noted Walker A motif (residues 440–449) and a series of proline-rich regions (residues 452–457, 482–485 and 536–541) (20).
One model that may explain the synthetic lethal relationship between EAP1 and SCP160 involves the potentially abnormal expression of target genes normally regulated by Scp160p. If Scp160p acts to mediate the expression of other genes, then the reliance of a Δscp160 cell on EAP1 could also result from the effect of a downstream gene. While there are countless possibilities, one gene stands out as particularly noteworthy: NDC1.
The ndc1-1 mutant allele is synthetically lethal with EAP1 (21). Overexpression of wild-type Ndc1p has been demonstrated to cause spindle pole body duplication defects indistinguishable from those observed in ndc1-1 mutant cells (30). It has been hypothesized that gene products affecting Ndc1p expression levels may, if mutated, cause phenotypes similar to those observed with the ndc1-1 allele (21). Thus, if Scp160p is required to maintain a proper dosage of Ndc1p, then an SCP160-null cell could have a phenotype similar to that caused by the ndc1-1 mutation, and hence require EAP1 for survival. Indeed, using RT–PCR and NDC1-specific primers, we have demonstrated that NDC1 mRNA is significantly enriched in Scp160p-associated complexes (data not shown). The potential role of NDC1 in mediating EAP1/SCP160 synthetic lethality will be a focus of future work.
The second key point demonstrated here is that the eIF4E-binding capacity of Eap1p is important, although not absolutely essential, to its SCP160-related function. This conclusion stems from the observation that the Y109A allele of Eap1p, which no longer binds eIF4E in vitro or in vivo (20), is ∼4-fold less effective at complementing scp160/eap1 synthetic lethality than is its wild-type counterpart. This result clearly supports the hypothesis that Scp160p plays a role in cytoplasmic mRNA metabolism, most probably including translation. However, the fact that Y109A Eap1p retains almost 25% of its ability to complement scp160/eap1 synthetic lethality implies either that the Y109A substitution does not completely impede the function of Eap1 in translation, or that some other aspect of Eap1p function, independent of translation, is involved. Considering that missegregation of genetic markers through meiosis is one aspect of the SCP160-null phenotype, it seems possible that the spindle pole body function of Eap1p (21) may be relevant. Clearly, other functions, unknown at this time, may also be involved.
Thirdly, as mentioned above, Eap1p physically associates with at least a subset of Scp160p-containing complexes, and that association is RNA dependent. This conclusion stems from the observation that HA-tagged Eap1 co-purifies with anti-tag affinity isolated FLAG-Scp160p, and that this co-purification is disrupted by pre-treatment of the sample with RNase. It is important to note that co-purification from a cell lysate, as was demonstrated here, does not necessarily imply a direct binding between the two proteins; their association may be mediated by other factors. Indeed, the RNA-dependent nature of the association implies either that both Scp160p and Eap1p associate with overlapping sets of RNAs, or that RNA binding by one (or both) of the proteins otherwise impacts the affinity of that protein for its partner(s).
Considering the semi-quantitative nature of the assay utilized here, it is impossible to estimate the stoichiometries of association between Scp160p and Eap1p. However, considering the differential profiles observed for Scp160p and Eap1p by sucrose gradient ultracentrifugation, it appears that only a fractional component of either protein pool would be in a position to associate with the other. Whether those molecules of Eap1p that do associate with Scp160p are in any way structurally or functionally distinct from the remainder of the Eap1p pool remains to be determined.
The final point presented here deals with the size distribution of Scp160p and Eap1p-containing complexes, and the impact of loss of each protein on complexes involving the other. As described above, loss of eap1 resulted in a notable shift in the size profile of Scp160p-containing complexes, as seen by both size exclusion column chromatography and sucrose gradient ultracentrifugation. Indeed, although some Scp160p complexes remained as large as those seen in the presence of EAP1, a significant ‘tail’ of smaller complexes was seen, stretching all the way to ∼440 kDa, which is the migration position of native monomeric Scp160p (4). Loss of eap1 had no major impact on the apparent distribution of Scp160p between the soluble and membrane-associated pools. The change in apparent size distribution of Scp160p complexes in the absence of EAP1 could reflect either impaired assembly or partial destabilization.
Loss of scp160 resulted in only minor if any changes in the apparent size distribution of Eap1p complexes in yeast, consistent with the scenario raised above, namely that only a fraction of Eap1p associates with Scp160p-containing complexes. However, the striking loss of Eap1p from membrane pellets in the absence of Scp160p suggests that while the soluble fraction may contain both Scp160p-associated and Scp160p-independent pools of Eap1p, the membrane pellet contains exclusively Eap1p that is associated with Scp160p. Considering that RNase pre-treatment also resulted in a significant loss of HA-Eap1p signal from the membrane pellet of fractionated cells, it appears likely that RNase pre-treatment simply uncoupled Eap1p from the Scp160p complexes that held it there.
Finally, embedded within the science presented here is a powerful assay that utilizes the relative degree of SCP160 maintenance plasmid loss to provide a quantitative measure of in vivo function for test alleles of either SCP160 or EAP1. While this assay was applied in only a limited manner here, enabling comparison of the functional capacities of high copy versus low copy (tagged) versus a single mutated allele of EAP1, this approach could be applied easily to enable a more thorough structure/function analysis of both proteins.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Anita Corbett and to the members of her laboratory, especially Adam Berger, who provided invaluable advice and support throughout the course of this project. This work was supported in part by funds from the Emory University Research Committee (to J.L.F.K.) and in part by funds from the National Science Foundation (award 0112911 to J.L.F.K.). B.A.M. was supported in part by a summer award from Pfizer Global Research and Development, and in part by the Howard Hughes Medical Institute through its support of the Summer Undergraduate Research Experience (SURE) program at Emory.
REFERENCES
- 1.Wintersberger U., Kuhne,C. and Karwan,A. (1995) Scp160p, a new yeast protein associated with the nuclear membrane and the endoplasmic reticulum, is necessary for maintenance of exact ploidy. Yeast, 11, 929–944. [DOI] [PubMed] [Google Scholar]
- 2.Weber V., Wernitznig,A., Hager,G., Harata,M., Frank,P. and Wintersberger,U. (1997) Purification and nucleic-acid-binding properties of a Saccharomyces cerevisiae protein involved in the control of ploidy. Eur. J. Biochem., 249, 309–317. [DOI] [PubMed] [Google Scholar]
- 3.Li A.-M., Watson,A. and Fridovich-Keil,J.L. (2003) Scp160p associates with specific mRNAs in yeast. Nucleic Acids Res., 31, 1830–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lang B.D. and Fridovich-Keil,J.L. (2000) Scp160p, a multiple KH-domain protein, is a component of mRNP complexes in yeast. Nucleic Acids Res., 28, 1576–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lang B.D., Li,A.-M., Black-Brewster,H.D. and Fridovich-Keil,J.L. (2001) The brefeldin A resistance protein Bfr1p is a component of polyribosome-associated mRNP complexes in yeast. Nucleic Acids Res., 29, 2567–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frey S., Pool,M. and Seedorf,M. (2001) Scp160p, an RNA-binding, polysome-associated protein, localizes to the endoplasmic reticulum of Saccharomyces cerevisiae in a microtubule-dependent manner. J. Biol. Chem., 276, 15905–15912. [DOI] [PubMed] [Google Scholar]
- 7.Coller J.M., Tucker,M., Sheth,U., Valencia-Sanchez,M.A. and Parker,R. (2001) The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA, 7, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin H., de Carvalho,P., Kho,D., Tai,C.Y., Pierre,P., Fink,G.R. and Pellman,D. (2001) Polyploids require Bik1 for kinetochore–microtubule attachment. J. Cell Biol., 155, 1173–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spingola M. and Ares,M.J. (2000) A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol. Cell, 6, 329–338. [DOI] [PubMed] [Google Scholar]
- 10.Siomi H., Matunis,M.J., Michael,W.M. and Dreyfuss,G. (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res., 21, 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Currie J.R. and Brown,W.T. (1999) KH domain-containing proteins of yeast: absence of a fragile X gene homologue. Am. J. Med. Genet., 84, 272–276. [PubMed] [Google Scholar]
- 12.Schmidt C., Henkel,B., Poschl,E., Zorbas,H., Purschke,W.G., Gloe,T.R. and Muller,P.K. (1992) Complete cDNA sequence of chicken vigilin, a novel protein with amplified and evolutionary conserved domains. Eur. J. Biochem., 206, 625–634. [DOI] [PubMed] [Google Scholar]
- 13.Plenz G., Kugler,S., Schnittger,S., Rieder,H., Fonatsch,C. and Muller,P.K. (1994) The human vigilin gene: identification, chromosomal localization and expression pattern. Hum. Genet., 93, 575–582. [DOI] [PubMed] [Google Scholar]
- 14.Dodson R.E. and Shapiro,D.J. (1997) Vigilin, a ubiquitous protein with 14 K homology domains, is the estrogen-inducible vitellogenin mRNA 3′-untranslated region-binding protein. J. Biol. Chem., 272, 12249–12252. [DOI] [PubMed] [Google Scholar]
- 15.Cortes A., Huertas,D., Fanti,L., Pimpinelli,S., Marsellach,F.X., Pina,B. and Azorin,F. (1999) DDP1, a single-stranded nucleic acid-binding protein of Drosophila, associates with pericentric heterochromatin and is functionally homologous to the yeast Scp160p, which is involved in the control of cell ploidy. EMBO J., 18, 3820–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortes A. and Azorin,F. (2000) DDP1, a heterochromatin-associated multi-KH-domain protein of Drosophila melanogaster, interacts specifically with centromeric satellite DNA sequences. Mol. Cell. Biol., 20, 3860–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cunningham K.S., Dodson,R.E., Nagel,M.A., Shapiro,D.J. and Schoenberg,D.R. (2000) Vigilin binding selectively inhibits cleavage of the vitellogenin mRNA 3′ untranslated region by the mRNA endonuclease polysomal ribonuclease 1. Proc. Natl Acad. Sci. USA, 97, 12498–12502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bender A. and Pringle,J.R. (1991) Use of a screen for synthetic lethal and multicopy suppresser mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costigan C., Gehrung,S. and Snyder,M. (1992) A synthetic lethal screen identifies SLK1, a novel protein kinase homologue implicated in yeast cell morphogenesis and cell growth. Mol. Cell. Biol., 12, 1162–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cosentino G.P., Schmelzle,T., Haghighat,A., Helliwell,S.B., Hall,M.N. and Sonenberg,N. (2000) Eap1p, a novel eukaryotic translation initiation factor 4E-associated protein in Saccharomyces cerevisiae. Mol. Cell. Biol., 20, 4604–4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chial H.J., Stemm-Wolf,A.J., McBratney,S. and Winey,M. (2000) Yeast Eap1p, an eIF4E-associated protein, has a separate function involving genetic stability. Curr. Biol., 10, 1519–1522. [DOI] [PubMed] [Google Scholar]
- 22.Chial H.J., Rout,M.P., Giddings,T.H. and Winey,M. (1998) Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol., 143, 1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 24.Gietz R.D. and Sugino,A. (1988) New yeast–Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene, 74, 527–534. [DOI] [PubMed] [Google Scholar]
- 25.Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guthrie C. and Fink,G.R. (1991) Guide to yeast genetics and molecular biology. Methods Enzymol., 194, 281–329. [PubMed] [Google Scholar]
- 27.Alani E., Cao,L. and Kleckner,N. (1987) A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics, 116, 541–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kessler S.H. and Sach,A.B. (1998) RNA recognition motif 2 of yeast Pab1 is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol., 18, 51–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von der Haar T., Ball,P.D. and McCarthy,J.E. (2000) Stabilization of eukaryotic initiation factor 4E binding to the mRNA 5′-cap by domains of eIF4G. J. Biol. Chem., 275, 30551–30555. [DOI] [PubMed] [Google Scholar]
- 30.Chial H., Giddings,T.H., Siewert,E.A., Hoyt,M.A. and Winey,M. (1999) Altered dosage of the Saccharomyces cerevisiae spindle pole body duplication gene, NDC1, leads to aneuploidy and polyploidy. Proc. Natl Acad. Sci. USA, 96, 10200–10205. [DOI] [PMC free article] [PubMed] [Google Scholar]