Abstract
The immunoglobulin-like family of CD66 antigens, present on human neutrophils and epithelial cells, are used as receptors for adhesins expressed by the pathogenic Neisseriae. N. gonorrhoeae strain MS11 can express 11 isoforms of these adhesins, called opacity-related (Opa) proteins. Each MS11 Opa protein recognizes a distinct spectrum of CD66 receptors. CD66–Opa binding is mediated by the NH2-terminal domain of the receptor and occurs through protein–protein interactions. In this report, we have investigated the molecular basis for the binding between the CD66 and Opa protein families by mapping amino acids in CD66 receptors that determine Opa protein binding. We performed homologue scanning mutagenesis between CD66e, which binds multiple Opa variants, and CD66b, which binds none, and tested both loss-of-function by CD66e and gain-of-function by CD66b in solution assays and in assays involving full-length receptors expressed by epithelial cells. We found that three residues in the CD66e N-domain are required for maximal Opa protein receptor activity. Opa proteins that recognize the same spectrum of native CD66 molecules showed differential binding of receptors with submaximal activity, indicating that the binding characteristics of these Opa proteins are actually slightly different. These data provide a first step toward resolving the structural requirements for Opa–CD66 interaction.
Keywords: Neisseria gonorrhoeae, carcinoembryonic antigen, bacterial adhesion, opacity protein, mutagenesis
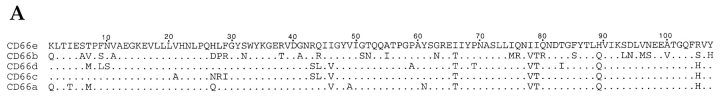
The causative agent of gonorrhea, Neisseria gonorrhoeae (Ngo),1 is an obligate human bacterial pathogen. The bacteria are thought to colonize the urogenital mucosa using different adhesins in a sequential manner. Initial attachment is mediated by gonococcal pili. Subsequently, tighter adherence to the mucosal epithelial cells is provided by bacterial adhesins called opacity-related (Opa) proteins 1 2. Both pathogenic Neisseria species, Ngo and Neisseria meningitidis (Nme), contain multiple opa genes. The expression of each opa gene can be turned on or off independently (phase variation) through a process of slipped-strand mispairing 3. Members of the Opa protein family are highly homologous, except for variable sequence domains present in three of the four surface exposed loops 4. Opa proteins were shown to account for the cell tropisms displayed by Ngo for human neutrophils and epithelial cells 5. Two types of Opa protein receptors have been identified. One is the heparan-sulfate proteoglycan receptor (HSPG), present on epithelial cells, which mediates binding and internalization of one particular Opa variant of Ngo strain MS11 (OpaA 6 7). The other receptor type include members of the carcinoembryonic antigen (CEA) or CD66 family, present on neutrophils and epithelial cells; they are recognized by multiple Opa variants of Ngo and Nme 8 9 10 11. The CD66 glycoprotein receptors belong to the Ig superfamily (IgSF): the molecules consist of one NH2-terminal Ig variable–like domain (N-domain) plus a varying number of Ig constant–like domains linked to the plasma membrane through a transmembrane domain or a glycosyl phosphatidylinositol moiety 12 13. The proteins were discovered decades ago when certain family members (CD66e [CEA] and CD66c) were found to be highly overexpressed in a large number of tumors; CEA has since become the prototype tumor marker. The function of CD66 glycoproteins in normal tissue or in tumorigenesis is still unclear, although the demonstration that these molecules function in vitro as cell adhesion molecules (CAMs) may indicate that they contribute to tissue architecture or to other cell–cell interactions 14. Interestingly, certain CD66 glycoproteins are subverted as receptors for bacterial (Escherichia coli or Salmonella expressing type I pili 15) or viral (murine coronaviruses 16) pathogens; recently, the pathogenic Neisseriae have been added to this list. To date, five members of the CD66 family (CD66a–e) have been studied for their interaction with gonococcal Opa proteins. We and others have found that this interaction is highly differential, i.e., some CD66 family members are recognized by four Opa variants while other CD66 receptors are recognized by none (CD66b), six (CD66a), or nine (CD66e) different Opa variants of Ngo strain MS11 10 11 17. We recently showed that the differential interaction observed with native receptors expressed on epithelial cells is mimicked by the binding pattern of recombinant CD66 N-domains produced in E. coli 18, indicating that the Opa–CD66 interaction is mediated by protein sequences in CD66 N-domains. It is remarkable that, despite extensive sequence similarity among the CD66a–e N-domains (71–90% amino acid sequence identity; see Fig. 1 A), the N-domains are recognized by different groups of Opa proteins. In our effort to understand the differential Opa–CD66 interactions, we investigated the molecular basis for the binding between the Opa and CD66 protein families. Homologue scanning mutagenesis, a strategy chosen to preserve structural integrity of mutant molecules 19, was used to map key residues in CD66 required for Opa protein binding. Residues within the N-domain of CD66e (which binds most Opa variants) were switched to homologous amino acids of the N-domain of CD66b (which does not bind any Opa variant). The key residues identified through loss-of-function mutations were confirmed by gain-of-function experiments, whereby the CD66b protein was converted into a functional receptor for Opa variants.
Figure 1.
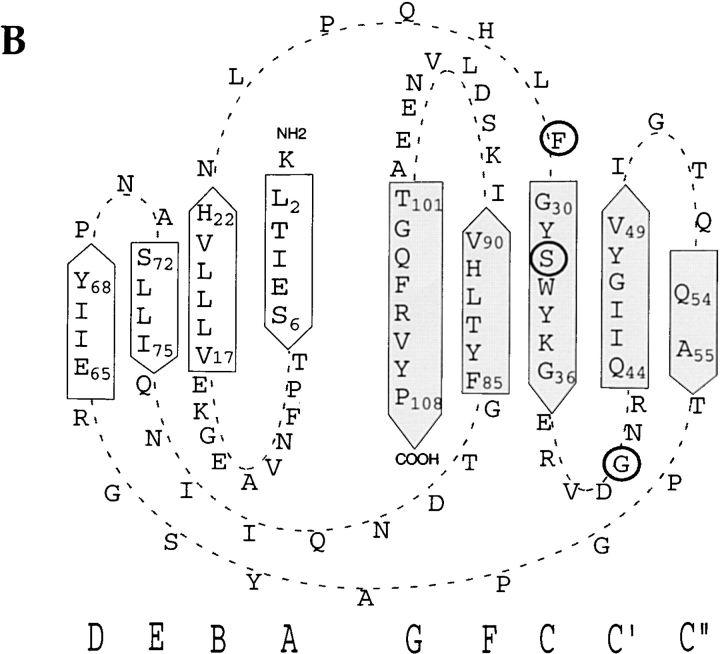
Protein sequence comparisons of CD66 N-domains. (A) Amino acid alignment of CD66 N-domains, deduced from DNA sequencing of N-domain clones as used in this and our previous study (reference 18). Dots indicate residues identical to CD66e. (B) Diagrammatic figure of the fold of the CD66e domain as predicted by Bates et al. (reference 32) and the PredictProtein computer program (reference 33). Strand designation is indicated by capitals below the diagram. Residues determining Opa protein binding as identified in this study are indicated by circles.
Materials and Methods
Bacterial Strains.
N. gonorrhoeae MS11 variants were propagated on gonococcal clear typing agar 20. Wild-type variants MS11mk expressing chromosomally encoded Opa proteins were provided by J. Swanson and are designated according to Swanson et al. 21 by capitals, e.g., OpaA–OpaK. Recombinant MS11 Opa variants 5 were a gift of T.F. Meyer (Max-Planck Institut für Biologie, Tübingen, Germany). Opa protein expression was verified by SDS-PAGE and immunoblotting of bacterial lysates followed by detection with anti-Opa antibody 4B12 21. Only nonpiliated bacteria were used. For experiments, bacteria were grown for 3 h in 10 ml Hepes medium (10 mM Hepes, 145 mM NaCl, 5 mM KCl, 0.5 mM MgCl2, 1 mM CaCl2, 5 mM glucose, and 1.5% proteose peptone no. 3 [Difco]), pH 7.4, in a gyratory shaker at 37°C. Bacterial suspensions were pelleted and resuspended in 1 ml Hepes buffer (Hepes medium without proteose peptone).
Construction of Mutant CD66 N-domains.
The construction of 6xHis-tagged N-domains of CD66e and CD66b in the pRSET-A vector was described previously 18. Mutations were introduced in CD66 N-domains by a modification of the procedure of Picard et al. 22. In brief, a mutagenic primer was designed containing the desired base changes flanked by at least 12 perfectly matched bases both upstream and downstream of the mutation (a list of primers is available on request). A megaprimer was generated by PCR using the mutagenic primer and a common vector-based 3′ primer (pRSET-rev) with the CD66 N-domain construct as template. The pRSET-rev primer was removed by passing the reaction mixture through a 100-kD Centricon device (5 min at 3,000 g; Amicon). A second PCR was performed with the same template plus 17 μl of the 50 μl of 100-kD Centricon retentate as 3′ primer and a common vector-based 5′ primer (pRSET-for). The resultant PCR product was cut with EcoRI and HindIII and ligated into pRSET-A. Constructs were electroporated into E. coli strain BL21 (DE3; Novagen). Mutations were verified by DNA sequencing through the entire N-domain insert. Primers were purchased from Genosys and restriction enzymes from New England Biolabs.
Binding of CD66 N-domains by Gonococci.
Cleared lysates of E. coli cells expressing the appropriate CD66 N-domain were prepared as described 18. Gonococci (108) in 200 μl Hepes buffer were incubated with 5–10 μl of cleared lysate for 20 min at 37°C. Bacteria were collected by centrifugation (5 min at 2,000 g), washed twice with 1 ml Hepes buffer, then solubilized in 30 μl SDS-PAGE sample buffer of which 2.5 μl was electrophoresed in 13.5% SDS-PAGE and transferred onto nitrocellulose. Bound CD66 N-domains were detected by anti-His antibody (1:15,000; Amersham Pharmacia Biotech) followed by peroxidase-conjugated protein A (1:20,000; Sigma Chemical Co.). Blots were developed using the enhanced chemiluminescence (ECL) protocol (Amersham Pharmacia Biotech). Documentation and quantification of bands were performed with an AlphaImager® 2000 Imaging system (Alpha Innotech).
Construction of CD66b cDNA Mutants.
CD66b cDNA in pUC118 was a gift from Motomu Kuroki (Fukuoka University, Fukuoka, Japan). The CD66b insert was subcloned into the eukaryotic expression vector pTracer-CMV2 (Invitrogen). All mutations were made first in the CD66b N-domain construct in pRSET-A as outlined above. To introduce mutated N-domains into the full-length CD66b cDNA, the BlpI site present at the start of the N-domain in CD66b cDNA was introduced in the CD66b N-domain construct in the pRSET-A vector by PCR. For introduction of the chimeric CD66e/b N-domain into CD66b, the BlpI site present in CD66b (GCTCAGC) was mutated to the BlpI site found in CD66e (GCTAAGC) by megaprimer PCR, in order to have the N-domain start with the CD66e-derived lysine. The resultant PCR product was cleaved with BlpI and NsiI (NsiI cuts at residue 71 in CD66 N-domains) and substituted with the fragment present in the full-length CD66b cDNA in pTracer-CMV2. To substitute the entire CD66b N-domain with the CD66e N-domain, a silent mutation containing a ClaI site was introduced at residue 113 just downstream of the N-domain in CD66b. The CD66e N-domain in pRSET-A was amplified with primers containing a BlpI and a ClaI site, respectively, cut and ligated into BlpI/ClaI-cut CD66b. The constructs were electroporated into E. coli DH5α. All mutations in the pTracer-CMV2-CD66b constructs were verified by DNA sequencing.
Transfection Procedure.
Chinese hamster ovary (CHO) cells (Pro5) were obtained from the American Type Culture Collection and were grown in RPMI 1640/5% FCS in 25-cm2 flasks to 50% confluency. Plasmid preparations of pTracer-CMV2-CD66b mutants were made by the Wizard miniprep procedure (Promega Corp.). Pro5 cells were incubated in 2 ml DMEM/10% Nu-serum (Collaborative Biomedical Products) containing 4 μg plasmid DNA and 0.2 mg/ml DEAE-dextran (M r 5 × 105; Amersham Pharmacia Biotech) for 4 h at 37°C. Cells were then shocked with 2.5 ml 10% DMSO in PBS for 1 min at room temperature, washed once with HBSS, and subsequently cultured overnight in RPMI 1640/5% FCS 23. The next day, the transfectants were trypsinized and seeded onto 12-mm-diameter circular glass coverslips in 24-well plates (105 cells per coverslip). Cells were cultured for 2–3 d before infection assays were carried out.
Infection Assay.
Gonococci (1.5 × 107) were added to 24-well plates containing the transfected cell cultures on coverslips, in 1 ml DMEM (without serum) for 45 min at 37°C and 5% CO2. Nonadherent bacteria were removed by three washes with HBSS. Infected cultures were fixed with 2% formaldehyde in PBS for at least 30 min.
Staining Procedures.
Fixed infected cells were incubated with 0.5% Triton X-100 in PBS for 20 min and then blocked with 5% FCS in PBS for 1 h. Antibodies were diluted in PBS/0.05% Tween/0.5% FCS. To detect receptor expression, cells were stained with rabbit anti-CD66 antiserum (1:200; Dako) followed by Alexa 594–conjugated goat anti–rabbit (GAR) IgG (1:400; Molecular Probes, Inc.). To subsequently stain gonococci, coverslips were incubated with a mouse mAb against gonococcal LPS, generated in our laboratory by J. Swanson, followed by FITC-conjugated goat anti–mouse (GAM) IgG (1:400; Sigma Chemical Co.). When only receptor expression was evaluated, the permeabilization step with Triton X-100 was omitted. mAb Kat4C was provided by H. Turley (John Radcliffe Hospital, Oxford, UK). To distinguish extra- and intracellular bacteria, infected cell cultures on coverslips were subjected to a differential staining procedure as described previously 10 24. In brief, fixed cells were incubated successively with an anti-LPS mAb and a protein A–gold conjugate. The gold was enhanced by silver-staining to visualize extracellular bacteria, after which cells and intracellular bacteria were stained with 0.005% crystalviolet in H2O for 10 min.
Results
Identification of CD66e N-domain Residues Critical for Binding of Opa-expressing Gonococci.
The differential recognition of CD66 receptor N-domains by gonococcal Opa variants is shown in Table . CD66e is recognized by the majority of Opa variants, whereas CD66b does not bind any Opa variant. OpaB, OpaC, OpaG, and OpaI variants demonstrate the broadest recognition of CD66 receptors. Assays with previously constructed chimeric receptor N-domains consisting of the NH2-terminal half (residues 1–59) of CD66e fused to the COOH-terminal half of the CD66b N-domain and vice versa, located the critical domain for binding of OpaB, OpaC, OpaG, and OpaI variants to the first 59 residues of CD66e (18; compare binding of the chimeric N-domains CD66e/b and CD66b/e in Table ). Using homologue scanning mutagenesis, we exchanged regions and single residues between the NH2-terminal 59 residues of CD66e and CD66b (for sequence differences, see Fig. 1 A) and measured the ability of the mutant proteins to bind to Opa−, OpaB-, OpaC-, and OpaI-expressing gonococci in order to identify CD66 residues required for Opa protein binding. Since OpaG is nearly identical to OpaB 25, we did not include this Opa variant in our assays.
Table 1.
Recognition of CD66 Receptor N-domains by MS11 Opa Variants
| Receptor type | Opa variant | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| − | A | B | C | D | E | F | G | H | I | J | K | ||
| CD66e | − | − | + | + | + | + | + | + | + | + | + | − | |
| CD66a | − | − | + | + | − | − | + | + | + | + | − | − | |
| CD66c | − | − | + | + | − | − | − | + | − | + | − | − | |
| CD66d | − | − | + | + | − | − | − | + | − | + | − | − | |
| CD66b | − | − | − | − | − | − | − | − | − | − | − | − | |
| CD66b/e | − | − | − | − | − | − | − | − | − | − | − | − | |
| CD66e/b | − | − | + | + | − | − | − | + | − | + | − | − | |
MS11 Opa variants were incubated with E. coli lysates containing the indicated His-tagged CD66 N-domain and processed for immunoblotting. Blots were probed with anti-His antibody and peroxidase-conjugated protein A followed by ECL. When a significant signal was present on the blot, recognition was defined as +. CD66a–e are native N-domain sequences; CD66b/e and CD66e/b represent chimeric N-domains between CD66b and CD66e, as described in reference 18. Some of these data were shown previously in reference 18.
As is shown in Table and Fig. 2 A, mutations in the first 11 residues of the CD66e N-domain did not influence binding of any tested Opa variant. Mutation of the region comprising residues 27–29 (mut3) resulted in a complete loss of recognition by all variants. Evaluation of the individual residues in this region revealed that the loss of binding was caused by the single F29R mutation (mut6). The single mutation S32N (mut7) also abrogated all Opa binding. The double mutation G41A+Q44R (mut9) clearly diminished Opa binding, but when these mutations were tested individually, a difference between Opa variants was noted. For OpaI interaction, both the G41A and the Q44R mutations were deleterious. This was also the case for interaction with OpaC, although to a lesser extent. The Q44R mutation had a moderate effect on OpaB binding, whereas the G41A change had no effect at all. Mutations in the region between residues 51 and 55 caused only a moderate decrease in Opa binding. Thus, the two residues F29 and S32 in CD66e are critical for binding of all Opa variants, whereas residues G41 and Q44 are important to different extents for the various Opa proteins.
Table 2.
Binding of CD66e N-domain Mutants to MS11 Opa Variants
| mut | Mutations introduced in the CD66e N-domain | Binding by Opa variants | ||
|---|---|---|---|---|
| OpaB | OpaC | OpaI | ||
| None | ++ | ++ | ++ | |
| 1 | K1Q | ++ | ++ | ++ |
| 2 | S6A+T7V+F9S+V11A | ++ | ++ | ++ |
| 3 | H27D+L28P+F29R | − | − | − |
| 4 | H27D | ++ | ++ | ++ |
| 5 | L28P | ++ | ++ | ++ |
| 6 | F29R | − | − | − |
| 7 | S32N | − | − | − |
| 8 | R38T | ++ | ++ | ++ |
| 9 | G41A+Q44R | +/− | − | − |
| 10 | G41A | ++ | + | − |
| 11 | Q44R | + | +/− | +/− |
| 12 | G51S+T52N+A55I | + | + | + |
MS11 Opa variants were incubated with E. coli lysates containing the indicated mutant (mut) CD66e N-domain and processed for immunoblotting. Binding was calculated by determining the signal generated by the amount of His-tagged N-domain for each variant, compared with the signal generated in a lane where an amount of His-tagged N-domain was loaded representing 100% binding. Opa− variants did not bind any N-domain construct (data not shown). Results represent the mean of two to four independent experiments. ++, >50% binding; +, 20–50% binding; +/−, 5–20% binding; −, <5% binding.
Figure 2.
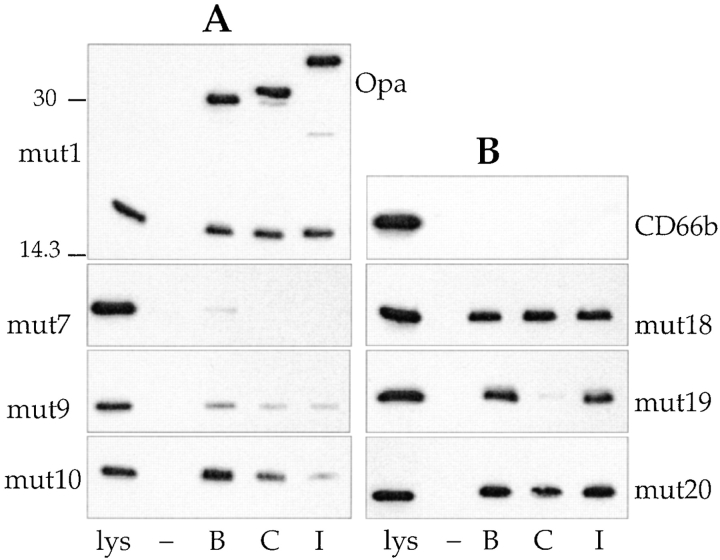
Binding of His-tagged mutant CD66 N-domains to MS11 Opa variants. Shown are representative immunoblots of lysates derived from bacteria that had been incubated with His-tagged CD66 N-domain mutants. The upper blot in A was probed with anti-His and anti-Opa antibody; other blots with anti-His antibody only. Position of Opa proteins is indicated on the right (Opa). Molecular mass standards (in kD) are indicated on the left of the top panel in A. Lanes labeled lys indicate E. coli lysate, containing the appropriate N-domain in an amount that would be seen on the blot if 100% of the N-domain present in the lysate was bound by the gonococci. Lanes labeled –, B, C, and I indicate amount of N-domain bound by Opa−, OpaB-, OpaC-, or OpaI-expressing gonococci, respectively.
Gain-of-function by the non-Opa Binding CD66 Family Member CD66b.
If the four residues indicated above were indeed responsible for the receptor function of CD66e, it should be possible to impart Opa binding properties to CD66b by introducing those four residues into CD66b. To test this concept, we changed the residues in question in the CD66b N-domain to the corresponding ones of CD66e in different combinations. As can be seen in Table , the presence of F29+S32 (mut13) did not result in detectable binding of any Opa variant. Residue F29 in mut13 is preceded by two CD66b-specific residues, D27 and P28, which may influence the correct conformation of F29. To address whether the F29 residue in a more “CD66e-like” environment would mediate Opa binding, we added residues H27 and L28 to mut13. This molecule (mut14) also failed to bind significantly to any Opa variant. The A41G+ R44Q mutant (mut15) did not bind any Opa variant either. We then added the G41 and Q44 residues individually to mut14, resulting in mut16 and mut17, and found that each addition slightly enhanced binding of all Opa variants compared with mut14. Only when both G41 and Q44 were present (mut18) was full binding activity by all Opa variants evident (Fig. 2 B). We then readdressed the question whether the origin of residues 27 and 28 played any role in Opa binding by construction of mut19–21. The presence of F29+S32+G41+Q44 was sufficient for binding of OpaB and OpaI variants; however, for OpaC binding an additional CD66e-derived residue (L28) was necessary, indicating a difference in binding characteristics among Opa variants (Fig. 2 B, and Table ). These data confirm the key role of four residues in the CD66e N-domain for receptor function of CD66e and again show a difference in binding characteristics within the OpaB, OpaC, and OpaI group of variants.
Table 3.
Binding of CD66b N-domain Mutants by MS11 Opa Variants
| mut | Mutations introduced in the N-domain of CD66b | Binding by Opa variants | ||
|---|---|---|---|---|
| OpaB | OpaC | OpaI | ||
| None | − | − | − | |
| 13 | R29F+N32S | − | − | − |
| 14 | D27H+P28L+R29F+N32S | − | − | − |
| 15 | A41G+R44Q | − | − | − |
| 16 | D27H+P28L+R29F+N32S+A41G | +/− | +/− | +/− |
| 17 | D27H+P28L+R29F+N32S+R44Q | +/− | +/− | +/− |
| 18 | D27H+P28L+R29F+N32S+A41G+R44Q | ++ | ++ | ++ |
| 19 | R29F+N32S+A41G+R44Q | ++ | − | + |
| 20 | P28L+R29F+N32S+A41G+R44Q | ++ | ++ | ++ |
| 21 | D27H+R29F+N32S+A41G+R44Q | ++ | − | ++ |
MS11 Opa variants were incubated with E. coli lysates containing the indicated mutant (mut) CD66b N-domain and processed for immunoblotting. Binding was calculated by determining the signal generated by the amount of His-tagged N-domain present for each variant, compared with the signal generated in a lane where an amount of His-tagged N-domain was loaded representing 100% binding. Opa− variants did not bind any N-domain construct (data not shown). Results represent the mean of two to four independent experiments. ++, >50% binding; +, 20–50% binding; +/−, 5–20% binding; −, <5% binding.
Expression and Recognition of Full-length CD66b Mutants on the Surface of Epithelial Cells.
To confirm that these results obtained with soluble recombinant His-tagged N-domains reflect interactions taking place at the cell surface, we introduced the identified key residues into full-length CD66b cDNA, transfected CHO cells with these constructs, and performed infection assays to determine Opa receptor activity. All transfectants stained positive with mAb Kat4C, which recognizes an epitope in internal CD66 domains (26; data not shown), as well as with a polyclonal anti-CD66 serum (Fig. 3), indicating that the receptors were expressed at the surface of the cells.
Figure 3.
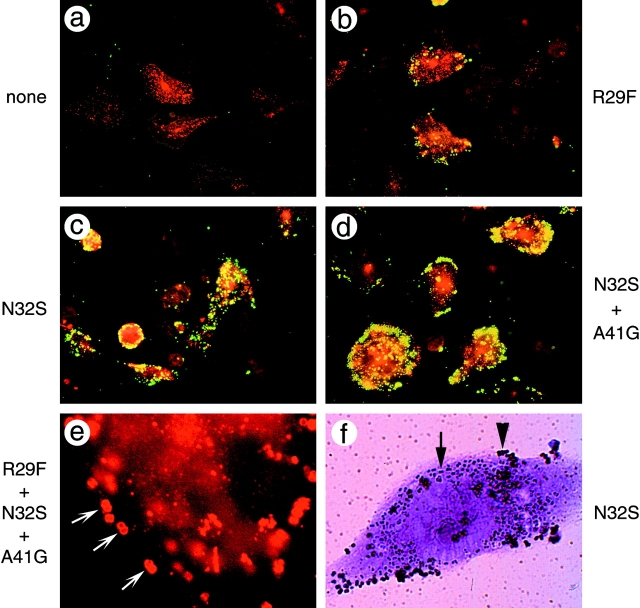
Photomicrographs of CHO cells transfected with different mutant CD66b receptors that were infected with OpaB-expressing gonococci for 45 min (a–e) or 90 min (f). The mutations introduced into CD66b are indicated next to the panels. (a–e) Permeabilized cells were stained for receptor expression with anti-CD66 antiserum plus GAR–Alexa 594 (red; a–e) and subsequently with an anti-LPS antibody plus GAM-FITC (green) to visualize bacteria (a–d). Images were obtained by one exposure through a double FITC/Texas Red bandpass optical filter using a 40× objective (a–d) or through a Texas Red bandpass filter using a 100× objective (e). Note the increase in number of associated bacteria (green) with the introduction of different CD66e residues into CD66b (a–d). At maximal interaction, receptor redistribution towards the sites of bacterial adhesion becomes apparent as footprint-like appearances (arrows in e). (f) Light micrograph of infected cells showing entry of OpaB-expressing gonococci into CHO cells expressing CD66b+N32S. Extracellular bacteria cells were visualized with an anti-LPS antibody followed by immunogold silver staining and can be distinguished by the dark rim of silver/gold precipitate covering the diplococci (arrowhead). Intracellular bacteria were stained with crystalviolet and are discerned by lack of a black outline and by their location in vacuolated intracellular compartments appearing as clear zones around the diplococci (arrow).
Infection of the various transfectants with MS11 Opa variants was evaluated by double immunofluorescence staining. Bacteria were found only on receptor-positive cells, which comprised 50–75% of the total cell population. The results of these experiments are shown in Table , with Fig. 3 illustrating the designations used in Table . Surprisingly, a single mutation in CD66b (N32S) resulted in significant binding of OpaB (Fig. 3 c) and OpaI variants, but not of OpaC variants. The single mutations A41G and R29F resulted in low level binding of OpaB (Fig. 3 b) and OpaI variants, but not of OpaC variants. The single mutation R44Q conferred no detectable Opa binding properties upon CD66b. In addition, the double mutant R44Q+ N32S was indistinguishable from the N32S mutant, indicating that the R44Q mutation did not contribute to Opa binding. The double mutations R29F+N32S and N32S+ A41G resulted in binding of large numbers of OpaB, OpaC, and OpaI variants (Table , and Fig. 3 d). Addition of mutation P28L to N32S, to R29F+N32S, or to N32S+ A41G did not affect interaction with any Opa variant (data not shown). The triple mutant R29F+N32S+A41G showed strong binding of Opa variants: infections with OpaB and OpaI variants resulted in distinct redistribution of the receptors towards the sites of bacterial adhesion, resulting in footprint-like appearances in the microscope (Fig. 3 e). This phenomenon was not seen with OpaC variants. The triple mutant was indistinguishable from a CD66b receptor containing the chimeric CD66e/b or the native CD66e N-domain, indicating that maximal binding was achieved with the triple mutant (Table ). Opa− variants did not interact with any transfectant (data not shown). To determine whether the transfectants that bound Opa variants were also able to ingest them, we applied a differential staining procedure after infection of the cells. We found that in all transfectants that bound Opa variants, intracellular gonococci could readily be found (Fig. 3 f). This demonstrates that CD66b molecules, containing the appropriate Opa binding domain, can act as complete functional receptors for gonococcal Opa variants. In summary, our findings show that, when introduced in full-length receptors in CHO cells, the mutations R29F, N32S, and G41A each conferred significant receptor activity upon CD66b, with N32S being the most effective single mutation. Replacement of all three residues provided maximal adherence of Opa variants. These gain-of-function experiments largely confirm the results obtained with soluble receptor N-domains, although the requirements for binding appear to be less stringent. Furthermore, again using full-length receptor mutants a difference in binding characteristics was noted for variants expressing OpaC compared with OpaB and OpaI variants.
Table 4.
Recognition by Opa Variants of Mutant Full-length CD66b Receptors Expressed by CHO Cells
| Transfectant | Recognition by Opa variants | ||
|---|---|---|---|
| OpaB | OpaC | OpaI | |
| CD66b | − | − | − |
| R29F | +/− | − | +/− |
| N32S | + | − | + |
| A41G | +/− | − | +/− |
| R44Q | − | − | − |
| N32S+R29F | ++ | + | ++ |
| N32S+A41G | ++ | + | ++ |
| N32S+R44Q | + | − | + |
| RF29F+N32S+A41G | ++/fp | ++ | ++/fp |
| CD66e/b | ++/fp | ++ | ++/fp |
| CD66e | ++/fp | ++ | ++/fp |
Interaction with Opa variants was determined by evaluating the percentage of receptor-positive cells that had significant amounts of associated gonococci (>5 bacteria per cell). Presence of receptor and bacteria was determined by immunofluorescence microscopy. The first column indicates the mutations made in CD66b; CD66e/b and CD66e indicate that the entire CD66b N-domain was replaced with either the chimeric N-domain mentioned in Table or the entire native CD66e N-domain. The results were tabulated as follows: ++/fp, all receptor-positive cells were infected plus numerous footprints were present; ++, 60–90% of receptor-positive cells were infected; +, 30–60% of receptor-positive cells were infected; +/−, 10–30% of receptor-positive cells were infected; −, no infected cells present. Infection experiments with recombinant MS11 Opa variants (reference 5) resulted in identical recognition patterns as those shown here for wild-type MS11 Opa variants. Results represent the mean of two to four independent experiments.
Discussion
In this study, we have mapped residues on CD66 receptors that determine Opa protein binding. We have identified three key amino acid residues in CD66e required for maximal binding using homologue scanning mutagenesis and subsequent analysis of both loss-of-function and gain-of-function of CD66 mutants in binding and infection assays. Furthermore, Opa proteins B, C, and I, which bind to an identical spectrum of native CD66 receptors, were found to differ in their recognition of mutant CD66 N-domains and mutant receptors on cells, indicating that their binding characteristics may actually not be identical.
Neisserial Opa proteins are thought to span the bacterial outer membrane in an eight-stranded β-barrel conformation resulting in four extracellular loops. Three of the exposed loops consist of variable sequence domains while a fourth loop near the COOH terminus of the protein is highly conserved. The differential binding of Opa proteins to CD66 receptors is likely a reflection of the ability of these variable domains to interact with the receptors. As we pointed out previously 10, it is remarkable that Opa proteins, such as OpaB, OpaC, and OpaI, that contain heterologous variable domains recognize the same subgroup of CD66 family members. However, our present data showing differences in binding of mutant molecules by these Opa proteins may indicate that they actually bind with slightly different characteristics, which would be expected from proteins with such divergent binding domain sequences. The opa gene family is thought to have arisen from recent gene duplication events and genetic reassortment of variable sequence domains between members of the gene family 25 27 28 29. Interestingly, the CD66 family also appears to have arisen recently by duplication of at least one ancestral gene. Sequence comparisons between the rodent and primate CD66 gene families show higher interspecies than intraspecies variation, suggesting that the duplication occurred after mammalian radiation took place 30 31. In addition, the mutation rate in the CD66 N-domain exons is twice as high as that of the adjacent intron, suggesting that the CD66 family is still undergoing rapid evolution 30. It is tempting to speculate that the extensive evolution of the opa gene family in the strictly human pathogen Ngo has taken place in response to the rapidly evolving family of primate-specific CD66 molecules. The observed differential Opa–CD66 binding patterns may be a reflection of this process.
According to the predicted structure of the CD66e N-domain 32, the residues we have identified as important for Opa protein binding, F29, S32, and G41, are located in exposed loops and strands of the GFCC′C′′ face of the CD66e N-domain (Fig. 1 B). This face of the molecule is not covered by carbohydrate, in contrast to the ABED face, as predicted by a low resolution model for CD66e 34, and would therefore be accessible for protein–protein interactions. The key role of S32 and G41 in binding of Opa proteins is supported by the fact that these residues are conserved among Opa-binding CD66 molecules (Fig. 1 A). Residue F29 is conserved in three out of four of the Opa-binding CD66 molecules (CD66a, d, and e), whereas CD66c contains an I at residue 29. Possibly hydrophobic residues such as F or I at that position support Opa protein binding, while a charged residue, such as R present in CD66b, does not. The role of residues L28 and Q44 is probably minor, since they are not required for Opa recognition of the native molecule, although their presence enhances binding of the soluble N-domain. Several ligand and viral binding sites on IgSF members have been found on the GFC face of the ligand-binding domain 35 36, indicating that this domain face is positioned favorably to serve as a ligand-binding platform for IgSF members.
Homologue scanning mutagenesis does not address the role of conserved residues among the two homologous proteins in ligand binding. The finding that single, relatively conservative mutations in CD66b, such as N32S and A41G, were sufficient to induce receptor function in CD66b may suggest that CD66b contains conserved residues participating in Opa protein binding. To test whether the actual binding site is comprised in the first 59 residues of CD66e, we constructed a truncated, His-tagged CD66e molecule (residues 1–59) and tested it for binding to Opa variants. This molecule was well expressed by E. coli but failed to bind significantly to any Opa variant (data not shown), indicating that other sites within the N-domain may be required for binding or that the peptide did not adopt the correct conformation for binding activity. The finding that single mutations in CD66b result in functional receptor activity could suggest that in vivo isoforms of CD66b exist that will be recognized by Opa variants. Evidence to support this concept comes from analysis of CD66b cDNAs cloned from normal white blood cells and leukemic cells. These cDNAs differed in two base pairs in the coding region, resulting in two amino acid differences, one in the N-domain (R80K) and one in the COOH-terminal M-domain (V288L) 37 38. Cloning of another CD66 family member, CD66d, by two different groups resulted in proteins differing in two residues 39 40. These phenomena fit with the notion that CD66 molecules are subject to sequence variation associated with rapid evolution.
CD66 family members are capable of mediating homophilic and heterophilic intercellular adhesion, like many other IgSF proteins 41. Binding between CAMs is usually of very low affinity, but due to the highly multimeric nature of cell–cell adhesion, sufficient avidity can be achieved to allow detection of the interaction between cells. The weakness of CAM interactions is illustrated by the difficulties in detecting binding of purified, monomeric forms of CAMs. This difficulty arises because binding assays require separation and washing steps, during which time weakly interacting molecules dissociate 42. This phenomenon may explain our observation that binding of Opa variants to soluble CD66b N-domain mutants required more CD66e-derived residues than binding to cell surface CD66 receptors. If each mutation introduced into CD66b enhances binding affinity between Opa and CD66b, as suggested by our infection assay data, then the threshold level of affinity necessary for detection will be reached sooner for the infection assay than for binding in solution. Alternatively, the level of multimerization may be important for Opa–CD66 binding, as has been shown for the binding of IgSF member intercellular adhesion molecule 1 (ICAM-1) to its receptor LFA-1. Recombinant ICAM-1 exists as monomers in solution, and direct binding to LFA-1 has been impossible to detect. Only when ICAM-1 was modified to induce dimerization could LFA-1 binding be detected 43 44. CD66 family members can exist as dimers in the plasma membrane of eukaryotic cells 45, and recombinant CD66e N-domains have been shown to form oligomers in solution 46. Receptor dimers will more likely be found on the surface of cells, where the receptor concentration is higher than in solution. Thus, if Opa binding requires a receptor dimer, one would expect Opa variants to bind more readily to receptor-expressing cells than to soluble receptors. Another possible explanation for the discrepancy between the two binding detection methods is that in the solution assay the receptor binding domain exists as an unglycosylated, single domain, while in the infection assay the binding domain is presented in the context of a complete, glycosylated molecule. Although Opa binding does not require carbohydrate 18, the presence of sugar moieties may influence the strength of adhesion, as has been observed for the interaction between CD2 and CD58. Human CD2 has a single carbohydrate addition site in its Ig variable–like N-domain that is absolutely required for binding to its normal ligand, CD58. Evidence from solution structure of this carbohydrate chain in relation to the GFC binding face indicates that the glycan is not itself situated in the binding face but is required to balance an unfavorable negative charge in order to maintain an active binding configuration 47. In fact, CD66 N-domains contain a potential glycosylation site at residue 70 (Fig. 1), which corresponds exactly to the structural position of the glycosylation site affecting ligand binding ability of human CD2 41. Mutation of this site in CD66e influences CD66e homophilic interactions, which are based on protein–protein interactions, indicating that in CD66e also the degree of glycosylation can influence binding events mediated through CD66 protein sequences 41. Our data stress the importance of evaluating binding events in different assays, since differences between assays can reveal further details of the molecular interaction between ligands. Regardless of the molecular basis for the observed discrepancy, both assays show clearly that OpaC binding requires more CD66e-derived residues than OpaB or OpaI, which may indicate that OpaC binding of the receptor is of lower affinity than binding of OpaB or OpaI.
In conclusion, mapping of key residues in CD66 required for recognition by the various gonococcal Opa adhesins indicates that single amino acid residues in CD66 receptors determine Opa protein binding. These results may provide a first step towards resolving the structural requirements for the Opa–CD66 receptor interaction and thereby help the development of infection inhibitory strategies, and may provide insights into the function of CD66 molecules in normal tissue and in carcinogenesis.
Acknowledgments
We thank Drs. Jos van Putten, Byron Caughey, and Jim Fox for helpful comments on the manuscript; Jim Fox for help with the PredictProtein program; and Gary Hettrick for assistance in preparation of the figures.
Footnotes
1used in this paper: CAM, cell adhesion molecule; CHO, Chinese hamster ovary; CEA, carcinoembryonic antigen; GAM, goat anti–mouse; GAR, goat anti–rabbit; ICAM, intercellular adhesion molecule; IgSF, immunoglobulin superfamily; mut, mutant; N-domain, NH2-terminal Ig variable–like domain; Ngo, Neisseria gonorrhoeae; Nme, Neisseria meningitidis; Opa, opacity-related
References
- Van Putten J.P.M., Duensing T.D. Infection of mucosal epithelial cells by Neisseria gonorrhoeae . Rev. Med. Microbiol. 1997;8:51–59. [Google Scholar]
- Meyer T.F. Pathogenic Neisseria-interplay between pro- and eukaryotic worlds. Folia Microbiol. (Praha) 1998;43:311–319. doi: 10.1007/BF02818617. [DOI] [PubMed] [Google Scholar]
- Murphy G.L., Connell T.D., Barritt D.S., Koomey M., Cannon J.G. Phase variation of gonococcal protein IIregulation of gene expression by slipped-strand mispairing of a repetitive DNA sequence. Cell. 1989;56:539–547. doi: 10.1016/0092-8674(89)90577-1. [DOI] [PubMed] [Google Scholar]
- Malorny B., Morelli G., Kusecek B., Kolberg J., Achtmann M. Sequence diversity, predicted two-dimensional protein structure, and epitope mapping of Neisserial Opa proteins. J. Bacteriol. 1998;180:1323–1330. doi: 10.1128/jb.180.5.1323-1330.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupsch E.-M., Knepper B., Kuroki T., Heuer I., Meyer T.F. Variable opacity (Opa) outer membrane proteins account for the cell tropisms displayed by Neisseria gonorrhoeae for human leukocytes and epithelial cells. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:641–650. doi: 10.1002/j.1460-2075.1993.tb05697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Putten J.P.M., Paul S.M. Binding of syndecan-like cell surface proteoglycan receptors is required for Neisseria gonorrhoeae entry into human mucosal cells. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:2144–2154. doi: 10.1002/j.1460-2075.1995.tb07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Belland R.J., Wilson J., Swanson J. Adherence of pilus− Opa+ gonococci to epithelial cells in vitro involves heparan sulphate. J. Exp. Med. 1995;182:511–517. doi: 10.1084/jem.182.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Gotschlich E.C. CGM1a antigen of neutrophils, a receptor of gonococcal opacity proteins. Proc. Natl. Acad. Sci. USA. 1996;93:14851–14856. doi: 10.1073/pnas.93.25.14851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virji M., Makepeace K., Ferguson D.J.P., Watt S.M. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 1996;22:941–950. doi: 10.1046/j.1365-2958.1996.01551.x. [DOI] [PubMed] [Google Scholar]
- Bos M.P., Grunert F., Belland R.J. Differential recognition of members of the carcinoembryonic antigen family by Opa variants of Neisseria gonorrhoeae . Infect. Immun. 1997;65:2353–2361. doi: 10.1128/iai.65.6.2353-2361.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray-Owen S.D., Lorenzen D.R., Haude A., Meyer T.F., Dehio C. Differential Opa specificities for CD66 receptors influence tissue interactions and cellular response to Neisseria gonorrhoeae . Mol. Microbiol. 1997;26:971–980. doi: 10.1046/j.1365-2958.1997.6342006.x. [DOI] [PubMed] [Google Scholar]
- Thompson J.A., Grunert F., Zimmermann W. Carcinoembryonic antigen familymolecular biology and clinical perspectives. J. Clin. Lab. Anal. 1991;5:344–366. doi: 10.1002/jcla.1860050510. [DOI] [PubMed] [Google Scholar]
- Obrink B. CEA adhesion moleculesmultifunctional proteins with signal-regulatory properties. Curr. Opin. Cell Biol. 1997;9:616–626. doi: 10.1016/S0955-0674(97)80114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchimol S., Fuks A., Jothy S., Beachemin N., Shirota K., Stanners C.P. Carcinoembryonic antigen, a human tumor marker, functions as an intercellular adhesion molecule. Cell. 1989;57:327–334. doi: 10.1016/0092-8674(89)90970-7. [DOI] [PubMed] [Google Scholar]
- Leusch H.G., Hefta S.A., Drzeniek Z., Hummel K., Markos-Puusztai Z., Wagener C. Binding of Escherichia coli and Salmonella strains to members of the carcinoembryonic antigen familydifferential binding inhibited by aromatic α-glycosides of mannose. Infect. Immun. 1990;59:2051–2057. doi: 10.1128/iai.59.6.2051-2057.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dveksler G.S., Dieffenbach C.W., Cardellichio C.B., McCuaig K., Pensiero M.N., Jiang G.S., Beauchemin N., Holmes K.V. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 1993;67:1–8. doi: 10.1128/jvi.67.1.1-8.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Grunert F., Medina-Marino A., Gotschlich E.C. Several carcinoembryonic antigens (CD66) serve as receptors for gonococcal opacity proteins. J. Exp. Med. 1997;185:1557–1564. doi: 10.1084/jem.185.9.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos M.P., Kuroki M., Krop-Watorek A., Hogan D., Belland R.J. CD66 receptor specificity exhibited by neisserial Opa variants is controlled by protein determinants in CD66 N-domains. Proc. Natl. Acad. Sci. USA. 1998;95:9584–9589. doi: 10.1073/pnas.95.16.9584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B.C., Jhurani P., Ng P., Wells J.A. Receptor and antibody epitopes in human growth hormone identified by homolog-scanning mutagenesis. Science. 1989;243:1330–1336. doi: 10.1126/science.2466339. [DOI] [PubMed] [Google Scholar]
- Swanson J. Studies on gonococcus infection. XII. Colony color and opacity variants of gonococci. Infect. Immun. 1978;19:320–331. doi: 10.1128/iai.19.1.320-331.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J., Barrera O., Sola J., Boslego J. Expression of outer membrane protein II by gonococci in experimental gonorrhea. J. Exp. Med. 1988;168:2121–2129. doi: 10.1084/jem.168.6.2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard V., Ersdal-Badju E., Lu A., Bock S.C. A rapid and efficient one-tube PCR based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 1994;22:2587–2591. doi: 10.1093/nar/22.13.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruffo A. Transient expression of proteins using COS cells. In: Ausubel F.M., editor. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; New York: 1997. pp. 16.13.1–16.13.7. [DOI] [PubMed] [Google Scholar]
- Van Putten J.P.M., Weel J.F.L., Grassmé H.U.C. Measurements of invasion by antibody labeling and electron microscopy. Methods Enzymol. 1994;236:420–437. doi: 10.1016/0076-6879(94)36031-6. [DOI] [PubMed] [Google Scholar]
- Bhat K.S., Gibbs C.P., Barrera O., Morrison S.G., Jähnig F., Stern A., Kupsch E.M., Meyer T.F., Swanson J. The opacity proteins of Neisseria gonorrhoeae strain MS11 are encoded by a family of 11 complete genes. Mol. Microbiol. 1991;5:1889–1901. doi: 10.1111/j.1365-2958.1991.tb00813.x. [DOI] [PubMed] [Google Scholar]
- Ruchaud-Sparagano M.H., Stocks S.C., Turley H., Dransfield I. Activation of neutrophil function via CD66differential effects upon β2 integrin mediated adhesion. Br. J. Haematol. 1997;98:612–620. doi: 10.1046/j.1365-2141.1997.2523070.x. [DOI] [PubMed] [Google Scholar]
- Hobbs M.M., Malorny B., Prasad P., Morelli G., Kusecek B., Heckels J.E., Cannon J.G., Achtmann M. Recombinational reassortment among opa genes from ET-37 complex Neisseria meningitidis isolates of diverse geographical origins. Microbiology. 1998;144:157–166. doi: 10.1099/00221287-144-1-157. [DOI] [PubMed] [Google Scholar]
- Connell T.D., Black W.J., Kawula T.H., Barritt D.S., Dempsey J.A., Kverneland K., Jr., Stephenson A., Schepart B.S., Murphy G.L., Cannon J.G. Recombination among protein II genes of Neisseria gonorrhoeae generates new coding sequences and increases structural variability in the protein II family. Mol. Microbiol. 1988;2:227–236. doi: 10.1111/j.1365-2958.1988.tb00024.x. [DOI] [PubMed] [Google Scholar]
- Van der Ley P. Three copies of a single protein II-encoding sequence in the genome of Neisseria gonorrhoeae JS3evidence for gene conversion and gene duplication. Mol. Microbiol. 1988;2:797–806. doi: 10.1111/j.1365-2958.1988.tb00091.x. [DOI] [PubMed] [Google Scholar]
- Rudert F., Zimmermann W., Thompson J.A. Intra- and interspecies analyses of the carcinoembryonic antigen (CEA) gene family reveal independent evolution in primates and rodents. J. Mol. Evol. 1989;29:126–134. doi: 10.1007/BF02100111. [DOI] [PubMed] [Google Scholar]
- Zimmermann W. The nature and expression of the rodent CEA familiesevolutionary considerations. In: Stanners C.P., editor. Cell Adhesion and Communication Mediated by the CEA Family. Harwood Academic Publishers; Amsterdam: 1998. pp. 31–55. [Google Scholar]
- Bates P.A., Luo J., Sternberg M.J. A predicted three-dimensional structure for the carcinoembryonic antigen (CEA) FEBS Lett. 1992;301:207–214. doi: 10.1016/0014-5793(92)81249-l. [DOI] [PubMed] [Google Scholar]
- Rost B., Casadio R., Fariselli P., Sander C. Transmembrane helices predicted at 95% accuracy. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M.K., Mayans M.O., Thornton J.D., Begent R.H.J., Keep P.A., Perkins S.J. Extended glycoprotein structure of the seven domains in human carcinoembryonic antigen by X-ray and neutron solution scattering and an automated curve fitting procedureimplications for cellular adhesion. J. Mol. Biol. 1996;259:718–736. doi: 10.1006/jmbi.1996.0353. [DOI] [PubMed] [Google Scholar]
- Wang J., Springer T.A. Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol. Rev. 1998;163:197–215. doi: 10.1111/j.1600-065x.1998.tb01198.x. [DOI] [PubMed] [Google Scholar]
- Davis S.J., Ikemizu S., Wild M.K., van der Merwe P.A. CD2 and the nature of protein interactions mediating cell-cell adhesion. Immunol. Rev. 1998;163:217–236. doi: 10.1111/j.1600-065x.1998.tb01199.x. [DOI] [PubMed] [Google Scholar]
- Arakawa F., Kuroki M., Misumi Y., Oikawa S., Nakazato H., Matsuoka Y. Characterization of a cDNA clone encoding a new species of the nonspecific cross-reacting antigen (NCA), a member of the CEA gene family. Biochem. Biophys. Res. Commun. 1990;106:1063–1071. doi: 10.1016/0006-291x(90)90975-s. [DOI] [PubMed] [Google Scholar]
- Berling B., Kolbinger F., Grunert F., Thompson J.A., Brombacher F., Buchegger F., von Kleist S., Zimmermann W. Cloning of a carcinoembryonic antigen gene family member expressed in leukocytes of chronic myeloid leukemia patients and bone marrow. Cancer Res. 1990;50:6534–6539. [PubMed] [Google Scholar]
- Nagel G., Grunert F., Kuijpers T.W., Watt S.M., Thompson J.A., Zimmerman W. Genomic organization, splice variants and expression of CGM1, a CD66-related member of the carcinoembryonic antigen gene family. Eur. J. Biochem. 1993;214:27–35. doi: 10.1111/j.1432-1033.1993.tb17892.x. [DOI] [PubMed] [Google Scholar]
- Kuroki M., Arakawa F., Matsuo Y., Oikawa S., Misumi Y., Nakazato H., Matsuoka Y. Molecular cloning of nonspecific cross-reacting antigens in human granulocytes. J. Biol. Chem. 1991;266:11810–11817. [PubMed] [Google Scholar]
- Stanners C.P., Fuks A. Properties of adhesion mediated by the human CEA family. In: Stanners C.P., editor. Cell Adhesion and Communication Mediated by the CEA Family. Harwood Academic Publishers; Amsterdam: 1998. pp. 57–71. [Google Scholar]
- Van der Merwe P.A., Barclay A.N. Transient intercellular adhesionthe importance of weak protein-protein interactions. Trends Biochem. Sci. 1994;19:354–358. doi: 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- Miller J., Knorr R., Ferrone M., Houdei R., Carron C.P., Dustin M.L. Intercellular adhesion molecule-1 dimerization and its consequences for adhesion mediated by lymphocyte function associated-1. J. Exp. Med. 1995;182:1231–1241. doi: 10.1084/jem.182.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly P.L., Woska J.R., Jr., Jeanfavre D.D., McNally E., Rothlein R., Bormann B. The native structure of intercellular adhesion molecule-1 (ICAM-1) is a dimer. J. Immunol. 1995;155:529–532. [PubMed] [Google Scholar]
- Hunter I., Sawa H., Edlund M., Obrink B. Evidence for regulated dimerization of cell-cell adhesion molecule (C-CAM) in epithelial cells. Biochem. J. 1996;320:847–853. doi: 10.1042/bj3200847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krop-Watorek A., Oikawa S., Oyama Y., Nakazato H. Oligomerization of N-terminal domain of carcinoembryonic antigen (CEA) expressed in Escherichia coli . Biochem. Biophys. Res. Commun. 1998;242:79–83. doi: 10.1006/bbrc.1997.7920. [DOI] [PubMed] [Google Scholar]
- Wyss D.F., Choi J.S., Li J., Knoppers M.H., Willis K.J., Arulanan A.R.N., Smolyar A., Reinherz E., Wagner G. Conformation and function of the N-linked glycan in the adhesion domain of human CD2. Science. 1995;269:1273–1278. doi: 10.1126/science.7544493. [DOI] [PubMed] [Google Scholar]