Abstract
The capacity of activated T cells to alter their cytokine expression profiles after migration into an effector site has not previously been defined. We addressed this issue by paired daughter analysis of a type 1–polarized CD8+ effector T cell population freshly isolated from lung parenchyma of influenza virus–infected mice. Single T cells were activated to divide in vitro; individual daughter cells were then micromanipulated into secondary cultures with and without added IL-4 to assess their potential to express type 2 cytokine genes. The resultant subclones were analyzed for type 1 and 2 cytokine mRNAs at day 6–7. When the most activated (CD44highCD11ahigh) CD8+ subpopulation from infected lung was compared with naive or resting (CD44lowCD11alow) CD8+ cells from infected lung and from normal lymph nodes (LNs), both clonogenicity and plasticity of the cytokine response were highest in the LN population and lowest in the activated lung population, correlating inversely with effector function. Multipotential cells were nevertheless detected among clonogenic CD44highCD11ahigh lung cells at 30–50% of the frequency in normal LNs. The data indicate that activated CD8+ T cells can retain the ability to proliferate and express new cytokine genes in response to local stimuli after recruitment to an effector site.
Keywords: T lymphocyte subsets, interferon γ, interleukin 4, differentiation, influenza virus
Naive CD4+ and CD8+ T cells have the potential to express many different cytokine genes in various combinations, depending on signals received during and after primary activation 1 2 3 4. This has been demonstrated most directly in experiments where sublines or subclones of a single parental cell displayed different cytokine profiles when expanded in different culture conditions 5 6 7. Although these studies do not exclude genetic and developmental effects on the propensity to express some cytokine genes 8 9 10, they suggest that most, if not all, naive CD4+ and CD8+ T cells can respond flexibly to stimulation conditions in the periphery. Many factors can influence the cytokine profile of a responding T cell population, but the most striking effects are exerted by the cytokines IL-12 and IFN-γ, which promote the expression of IFN-γ and other type 1 cytokines, and IL-4, which promotes the expression of IL-4 and other type 2 cytokines 11 12 13 14.
Several groups have shown that CD4+ T cell populations activated in vitro progressively lose the ability to change their cytokine profile when transferred from type 1 to type 2 polarizing conditions or vice versa 15 16 17 18 19. These data and other work showing that many long-term CD4+ and CD8+ T cell clones also display stable cytokine profiles 3 6 16 20 suggest that activated T cells ultimately become irreversibly committed to the expression of a particular cytokine profile in vitro. A molecular basis for this loss of multipotentiality has been suggested by observations that type 2–polarized cells lose IL-12Rβ2 expression and type 1–polarized cells, while retaining surface IL-4R, lose IL-4 responsiveness 21 22 23 24. Stable demethylation of cytokine gene promoters, changes in chromatin structure, and induction of type-specific transcription factor expression probably also contribute to the maintenance of cytokine gene expression patterns under different stimulation conditions 25 26 27 28 29 30 31 32.
Less is known about the stability of T cell cytokine profiles in vivo, but the available data indicate that established T cell cytokine responses are more difficult to deviate than primary responses 33 34 35 36 37. Memory CD4+ and CD8+ T cell populations expressed their original cytokine profiles when restimulated after many weeks in vivo in the absence of antigen 18 38 39, suggesting that these gene expression patterns are “imprinted” during priming. Mocci and Coffman found that CD4+ T cells of memory (CD62Llow) phenotype isolated from Leishmania major–infected mice retained their type 1 profile when exposed to IL-4 in vitro, although undifferentiated L. major–reactive cells persisted in the naive CD62Lhigh fraction 36. Reports that allergic desensitization and other protocols shift the IL-4/IFN-γ ratio produced by antigen-specific T cells show that redirection of cytokine responses can occur in vivo 35 40 41 42. However, it is not known whether this occurs by replacing or reprogramming existing effector cells.
This uncertainty highlights a major limitation of population studies, both in vivo and in vitro. Since the fate of every cell cannot be monitored, it is usually unclear whether any change in cytokine profile is due to reprogramming of the cells or to selective outgrowth by a minor subpopulation. Similarly, the apparent stability of a population phenotype might reflect true commitment or other mechanisms, such as selective apoptosis 43, or a dominant cross-regulatory effect of cytokines from some of the cells on the response of other multipotential cells in the population 44. These limitations can be overcome by single-cell protocols in which the survival and cytokine profile of each cell or its progeny are assessed. Paired daughter analysis in particular can be used to identify and quantify multipotential cells by measuring the ability of daughters of a single cell to respond differently to different stimuli 7 45.
Here we report the use of paired daughter analysis to determine whether multipotential cells persist in a CD8+ effector T cell population ex vivo. The analyzed population comprised activated CD8+ cells freshly isolated from the lung parenchyma of influenza virus–infected mice at the peak of the local response. This population had previously been shown to synthesize IFN-γ with little or no detectable IL-4 both ex vivo and after in vitro restimulation with virus-infected cells 46 47 48 49. As shown here, a significant proportion of these cells retained the potential to respond to IL-4 by expressing IL-4 and/or IL-10. These findings suggest that activated T cells can leave the priming LN before undergoing irreversible commitment and retain the capacity to modulate their phenotype in response to local conditions at the effector site.
Materials and Methods
Mice and Influenza Infection.
Female 6–10-wk-old BALB/c mice purchased from the Animal Resources Centre (Perth, Western Australia) were used in all experiments. Mice were anesthetized with ether or Penthrane (Abbott Australasia Pty. Ltd.) and infected intranasally with 50 μl of PBS containing 5 × 105 PFU of the reassortant influenza A virus Mem71 (H3/N1). The viral stock, prepared in embryonated chicken eggs, was provided by Dr. Lorena Brown (University of Melbourne, Victoria, Australia).
T Cell Preparation.
Normal LN cell suspensions were prepared from pooled paraaortic, axillary, and inguinal LNs of uninfected mice by forcing through stainless steel mesh, and centrifugation over Ficoll-Paque (Amersham Pharmacia Biotech). To prepare lung cell suspensions, mice were killed 7 d after influenza infection, perfused in situ with balanced salt solution to remove blood cells, and the lungs were minced and digested with collagenase and DNase as described previously 49. Digested lung was passed through stainless steel mesh, and parenchymal cells were isolated by centrifugation over a gradient of Percoll (Amersham Pharmacia Biotech). Cells were stained for flow cytometry with PE-conjugated anti-CD8 mAb (53.6; Becton Dickinson) and biotinylated anti-CD44 mAb (IM7.8; PharMingen), followed by streptavidin-Tricolor (Caltag) and FITC-conjugated anti-CD11a mAb (Sigma Chemical Co.), then resuspended in balanced salt solution with 1 μg/ml propidium iodide. CD8+CD44low CD11alow and CD8+CD44highCD11ahigh cells were isolated using a FACS Vantage™ (Becton Dickinson) with Lysys II software, setting a small forward scatter/side scatter gate to collect cells from normal LNs and a large gate to include both small lymphocytes and larger activated cells from lung. Dead and damaged cells were excluded on the basis of propidium iodide uptake and low forward scatter. By reanalysis, purity of the initial bulk-sorted cells for any of the tested staining parameters was generally >96%. Cells were then resorted using the same parameters and deposited using an automated cell deposition unit directly into microwells for RNA extraction or cloning.
On average in this study, CD8+ cells with the naive phenotype CD44lowCD11alow from LNs of untreated mice (“normal LNlow cells”) comprised 40.5% (n = 2) of the CD8+ LN cell pool. CD8+ cells comprised 18.1% ± 4.5 of the 15.2 ± 7.6 × 106 lung leukocytes recovered per mouse at day 7 after infection. Of the CD8+ fraction, 32.8% ± 4.4 were defined as CD44lowCD11alow (“influenza lunglow”) and 37.0% ± 5.3 were defined as CD44high CD11ahigh (“influenza lunghigh”), as illustrated previously 49.
T Cell Cloning and Subcloning.
All cultures were performed in 15 μl volumes of supplemented DME containing 5 × 10−5 M 2-ME, 12.5% FCS, and 600 IU/ml recombinant human IL-2 (Cetus Corp.) in mAb-coated Terasaki microwells (Greiner Labortechnik) 50. For normal LN cells, microwells were coated with purified mAb to CD3∈ (145-2C11; 10 μg/ml), CD8 (53.6; 3 μg/ml), and CD11a (I21/7.7; 5 μg/ml). Antibody coating concentrations were altered to 3 μg/ml anti-CD3, 3 μg/ml anti-CD8, and 5 μg/ml anti-CD11a mAb for optimal cloning of influenza lunglow cells, and to 1 μg/ml anti-CD3, 5 μg/ml anti-CD8, and 5 μg/ml anti-CD11a mAb for influenza lunghigh cells.
For experiments where clones were generated under different conditions in parallel, all cultures were initiated with mAb and IL-2, then after 2 d, 5 μl medium was removed and replaced with 5 μl medium containing various combinations of IL-2 (final concentration 600 IU/ml), IL-4 (100 U/ml), and anti–IFN-γ mAb (supernatant of the hybridoma R4-6A2 at a concentration that reduced the activity of purified rIFN-γ by at least 30-fold in assays with WEHI-279 cells). For paired daughter analysis, cultures were initiated with mAb and IL-2, then checked microscopically for viable cells at day 2. Where a parent cell had divided one or two times, individual daughter or granddaughter cells were transferred by micromanipulation into new Terasaki wells coated with the same mAb as above: at least one cell was cultured with IL-2 and one with IL-2 plus 100 U/ml IL-4. After a total of 6 or 7 d, cultures were checked microscopically for clones or subclones, cell numbers were counted, and their RNA was extracted. Clone sizes of ≥200 cells were recorded as 200.
Reverse Transcription PCR.
Cells were lysed for reverse transcription (RT)1 using NP-40 by the method of Smith et al. 51, modified by combining the buffered saline solution and the lysis mix, and by including oligo-dT15 (18 μg/ml final concentration; Boehringer Mannheim) as a primer instead of random hexamers. Cells were sorted directly into 11 μl of combined buffered lysing solution, or clones and subclones were lysed in microwells by replacement of culture medium with 11 μl of buffered lysing solution. Cell lysates were heated to 65°C then quick-chilled, transferred to microfuge tubes containing 14 μl RT mix comprising 90 mM KCl, 18 mM Tris-HCl, pH 8.0, 12 mM MgCl2, 1.4 mM dithiothreitol, 700 μM of each dNTP, 10 U RNAsin, and 2 U AMV reverse transcriptase (Promega Corp.), and incubated at 42°C for 90 min. First strand cDNA products were diluted 1:2.4 in H2O, and 10 μl was added to 15 μl PCR mix consisting of 2.5 μl of 10× PCR buffer (500 mM KCl, 100 mM Tris-HCl, pH 8.0, 20 mM MgCl2), 0.25 μl of mixed dNTPs (20 mM; Amersham Pharmacia Biotech), 1 U Ampli-Taq polymerase (Perkin-Elmer), 1 μl of each external oligonucleotide primer (10 μM), and H2O. PCR samples were amplified for 40 cycles in the first round, then products were diluted 1:33 in H2O, and 2 μl was added to 23 μl PCR mix, as above, for the second round of 30 cycles of PCR with internal intron-spanning primer pairs. PCR cycles were one cycle of 5 min at 94°C, 60 s at 60°C, and 90 s at 70°C, with subsequent cycles of 45 s at 94°C, 60 s at 60°C, and 90 s at 70°C. Primer pairs were as described previously 7. First round amplifications were performed with the following combinations of primers: IL-4, IFN-γ, and CD3∈; IL-5 and IL-6; IL-2 and IL-10. In the second round, the respective first round products were amplified as follows: IL-4 and IFN-γ together, CD3∈ separately; IL-5 and IL-6 together; IL-2 and IL-10 separately. PCR products were separated by gel electrophoresis, visualized with ethidium bromide, and confirmed by Southern hybridization 50. All PCR runs included a titration of cloned CD3∈ and cytokine cDNA to monitor cDNA sensitivity (at least 10−16 g, usually 10−17 g, with no consistent differences between cytokines) and 10 negative control samples. No PCR products were detected in the negative control samples.
Clones were scored as positive for expression of a cytokine if the corresponding PCR product was identified unambiguously by electrophoretic migration at the appropriate size and Southern hybridization with a specific probe. PCR amplifications from clonal cDNA samples were repeated in several experiments. Overall, reproducibility of positive and negative results was 96% for IFN-γ (n = 306), 94% for IL-4 (n = 306), 100% for IL-2 (n = 23), and 98% for IL-10 (n = 129). Both losses and gains were observed on repetition, suggesting that variations were because the relevant cDNA was limiting in those samples.
Results
Isolation and Cloning of Activated CD8+ T Cells from Lung Parenchyma of Influenza Virus–infected Mice.
Previous experiments established that intranasal infection of BALB/c mice with the Mem71 reassortant influenza virus caused accumulation in the lung parenchyma of a population of activated CD8+ T cells that expressed mRNA for IFN-γ and other cytokines ex vivo, secreted high levels of IFN-γ in response to virus-infected cells in vitro, and contained virus-specific CTLs 46 48 49. At the peak of the cellular response at day 7, both cytokine production and lytic activity were markedly higher in CD8+ T cells from the lung than from the draining mediastinal LNs. Separation on the basis of high CD44 and CD11a expression achieved greater enrichment of this CD8+ effector activity from both lung and LNs than any other combination of activation markers tested 49. Therefore, these markers were used in the following experiments to distinguish the least activated (CD44lowCD11alow) and the most activated (CD44high CD11ahigh) lung parenchymal CD8+ T cells from 7-d influenza virus–infected mice (abbreviated as influenza lunglow and influenza lunghigh cells, respectively). Low expression of CD44 and CD11a was also used to define a control CD8+ population of naive phenotype from normal LNs (normal LNlow cells).
The basal patterns of expression of several cytokine mRNAs in multiple samples of 500 normal LNlow, influenza lunglow, or influenza lunghigh cells were analyzed by RT-PCR immediately after FACS® purification (Fig. 1), using methods previously shown to detect cytokine transcripts in single activated T cells 52 53. No cytokine expression was detected in any of the normal LN samples, whereas almost all samples of influenza lunglow and influenza lunghigh cells expressed IFN-γ mRNA. The influenza lunghigh population also frequently expressed IL-10 and occasionally one or more of the cytokines IL-2, IL-4, and IL-6. These data show the expected hierarchy of effector activity in the three populations and support earlier evidence that IFN-γ is a major product of CD8+ T cells in the response to influenza virus infection 46 47 49.
Figure 1.
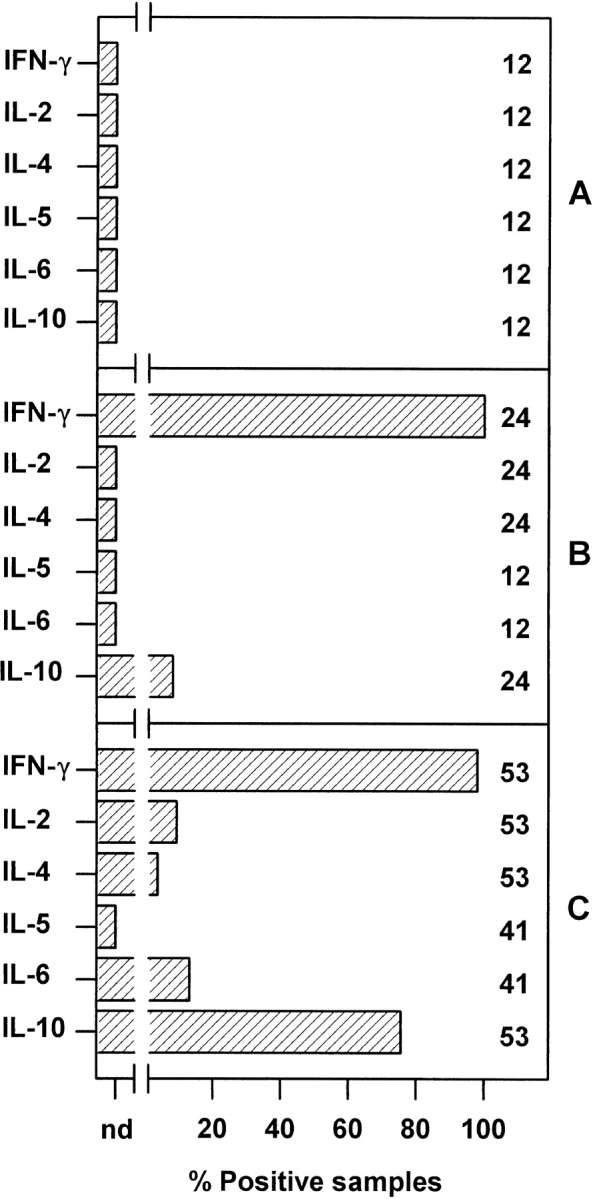
Cytokine mRNA expression by freshly isolated CD8+ cells from normal LNs and influenza virus–infected lung. RNA was prepared from multiple samples of 500 cells of (A) normal LNlow cells, (B) influenza lunglow cells, and (C) influenza lunghigh cells immediately after purification by FACS® and analyzed for CD3∈ and cytokine mRNA by RT-PCR. Data are shown for all samples that yielded a CD3∈ PCR product (89 of 90 samples) and pooled from at least two experiments (six samples per experiment). The number of samples tested is shown beside each bar; nd, not detected.
To assess the potential of activated T cells to alter their cytokine profiles in response to different signals, a culture system was required in which isolated single cells would divide and could then be subcloned under different conditions before analysis of their cytokine profiles. Ideally cultures would lack accessory cells to enable identification of individual daughter cells for subcloning and to ensure that cytokine mRNAs measured by RT-PCR at the end of culture were derived from the progeny of those daughter cells. We have previously shown that stimulation with coimmobilized anti-CD3 (adsorbed to culture wells at 10 μg/ml), anti-CD8, and anti-CD11a mAb and IL-2 activated most naive CD8+ LN cells to form clones 7 50 54. When influenza lunghigh cells were cultured under these conditions, cloning frequencies and sizes were low. A series of optimization experiments showed that reduction of the plate-coating concentration of anti-CD3 mAb to 1 μg/ml increased cloning efficiencies of these cells to a mean of ∼25% (data not shown). No further improvement was obtained by varying the other mAb concentrations. Maximal cloning efficiencies of ∼75% were obtained for influenza lunglow cells using 3 μg/ml anti-CD3 mAb, comparable to those achieved for normal LN CD8+ T cells using 10 μg/ml (∼80%). Average clone sizes peaked at 6–7 d of culture, after which many clones died.
The ability of exogenous IL-4 to influence cytokine profile development in influenza lunghigh cells was tested by culturing individual cells with immobilized mAb and IL-2. After 2 d, cultures were supplemented with medium, IL-4, or IL-4 and anti–IFN-γ mAb, to mimic the protocol used in the paired daughter analyses shown below. All clones were counted and harvested on day 6 for RT-PCR analysis of their cytokine profiles (Fig. 2). Average cloning efficiencies, clone sizes, and frequencies of IFN-γ, IL-2, and IL-10 mRNA expression were comparable among the three sets of cultures (P > 0.05). However, addition of IL-4 at day 2 markedly increased the frequency of IL-4 expression (P < 0.01). No further increase was observed when anti–IFN-γ mAb was also added. This and other preliminary cloning experiments suggested that a significant proportion of influenza lunghigh and influenza lunglow cells could respond to IL-4 by expressing this cytokine, providing provisional evidence for the presence of multipotential cells in both populations. RT-PCR analyses of IL-5 and IL-6 expression by the clones in Fig. 2 and many other clones derived from influenza lunghigh or influenza lunglow cells in the presence or absence of exogenous IL-4 rarely detected either of these cytokine mRNAs, and are therefore not presented.
Figure 2.
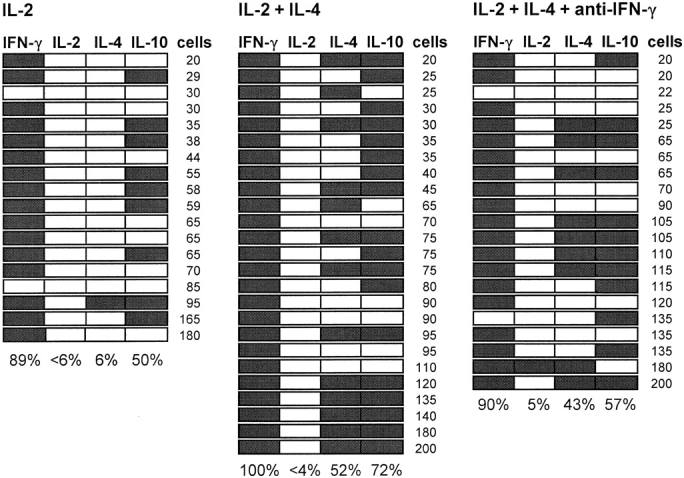
The effect of IL-4 on cytokine mRNA expression by clones of lung CD8+ CD44highCD11ahigh cells. Individual FACS®-purified CD8+ CD44highCD11ahigh cells from influenza virus–infected lung were deposited into wells coated with mAb to CD3 (1 μg/ml), CD8 (5 μg/ml), and CD11a (5 μg/ml) and containing culture medium with IL-2. After 2 d, 5 μl medium was removed from each culture and replaced with 5 μl medium containing IL-2, IL-2 + IL-4, or IL-2 + IL-4 + anti–IFN-γ mAb as indicated above the panels. Cloning efficiencies at day 6 were 16% (mean clone size 30 ± 39, n = 48), 15% (49 ± 50, n = 44), and 14% (52 ± 55, n = 41), respectively. The figure shows the results of RT-PCR analysis of all clones of ≥20 cells that yielded a CD3∈ PCR product (all but one clone). Detection of a cytokine PCR product is indicated by shading of the boxes. The number of cells in each clone is indicated at the right, and the frequency of expression of each cytokine is indicated at the bottom of the panels.
Paired Daughter Analysis of Lung CD8+ T Cells.
As shown above in Fig. 2, IL-4 mRNA–expressing clones could be generated from influenza lunghigh cells in the presence of exogenous IL-4. However, this experimental protocol did not distinguish whether these clones developed de novo from multipotential cells or whether they were expanded from committed cells that had been primed to produce this cytokine in vivo. To address this question directly, paired daughter analyses were performed in which CD8+ cells from normal LNs and infected lung were cultured with antireceptor mAb and IL-2. After 2 d, when significant numbers had undergone one or two divisions, individual daughter or granddaughter cells were transferred into new cultures with or without exogenous IL-4. Subclones were analyzed for CD3∈ and cytokine mRNAs 4–5 d later.
Table summarizes the mean cloning efficiencies, subclone sizes, and frequencies of CD3∈ and cytokine mRNA detection in subclones for all paired daughter analyses performed in this way with cells from normal LNs and influenza-infected lung. In general, influenza lunghigh cells subcloned at lower frequency and formed smaller subclones by day 6 than normal LNlow or influenza lunglow cells, indicating that even those influenza lunghigh cells that divided in the first 2 d were less able to undergo extended proliferation than the other populations. Most subclones of all cell types expressed IFN-γ mRNA, whereas IL-2 was preferentially expressed by normal LNlow and influenza lunglow subclones. As observed in our previous analysis of naive C57BL/6 CD8+ LN T cells 7, some subclones of all cell types expressed IL-4 and/or IL-10 mRNA in the absence of exogenous IL-4, but frequencies were significantly higher among subclones grown with IL-4. IL-4 expression frequencies were more strongly affected by exogenous IL-4 than expression frequencies of any of the other cytokines tested.
Table 1.
Efficiencies of T Cell Cloning and mRNA Detection among Subclones Obtained in Paired Daughter Analyses
| Parameter | Normal LNlow | Influenza lunglow | Influenza lunghigh | |||
|---|---|---|---|---|---|---|
| IL-2 | IL-2 + IL-4 | IL-2 | IL-2 + IL-4 | IL-2 | IL-2 + IL-4 | |
| Primary cultures | ||||||
| No. of parent cells cultured | 1,260 | 2,040 | 2,622 | |||
| % cultures with ≥2 cells at day 2 | 34 (17–48) | 7.8 (4.2–12) | 17 (16–22) | |||
| Secondary cultures | ||||||
| No. of cells transferred on day 2 | 259 | 264 | 168 | 117 | 322 | 293 |
| % cells that subcloned by day 6 | 81 (80–81) | 74 (70–80) | 79 (52–91) | 72 (48–82) | 45 (34–56) | 46 (39–57) |
| Mean subclone size on day 6 | 131 ± 60 | 89 ± 49 | 82 ± 61 | 68 ± 50 | 40 ± 49 | 43 ± 49 |
| No. of subclones assayed by RT-PCR | 112 | 114 | 88 | 92 | 89 | 94 |
| No. of CD3∈1 subclones | 109 | 108 | 85 | 91 | 87 | 92 |
| % IFN-γ1 subclones | 85 | 86 | 76 | 92 | 77 | 93 |
| % IL-2+ subclones | 33 | 49 | 8.2 | 11 | 0 | 1.1 |
| % IL-4+ subclones | 25 | 73 | 13 | 34 | 6.9 | 20 |
| % IL-10+ subclones | 18 | 51 | 9.4 | 23 | 21 | 36 |
| No. of informative subclone families | 93 | 71 | 74 | |||
Data are expressed as the weighted arithmetic mean of two experiments with normal LNlow cells, four experiments with influenza lunglow cells, and five experiments with influenza lunghigh cells with the range of values shown in parentheses. Subclone size is shown as the arithmetic mean ± SD. Cytokine-positive clones are those in which both CD3∈ and the indicated cytokine mRNAs were detected after RT-PCR. Informative subclone families are those in which CD3∈ mRNA was detected in at least one subclone grown in IL-2 and at least one grown in IL-2 + IL-4.
Fig. 3 shows the cytokine profiles of 12 families of subclones generated from each subset. Three or four subclones per family are illustrated in those cases where the original cell had divided twice by day 2 and more than two of its progeny yielded a CD3∈+ subclone. These examples illustrate several features of the whole data set. First, IFN-γ, IL-4, and IL-10 could be detected in almost any combination. Second, many subclones displayed identical cytokine profiles within a family regardless of culture conditions, while others displayed differences in the presence and absence of added IL-4; paired subclones grown in the same conditions also sometimes displayed different profiles. Third, IFN-γ expression was more frequently gained than lost when subclones were grown with IL-4. Finally, in many families where a subclone grown without IL-4 expressed IL-4 or IL-10, its sibling grown with IL-4 also produced this cytokine.
Figure 3.
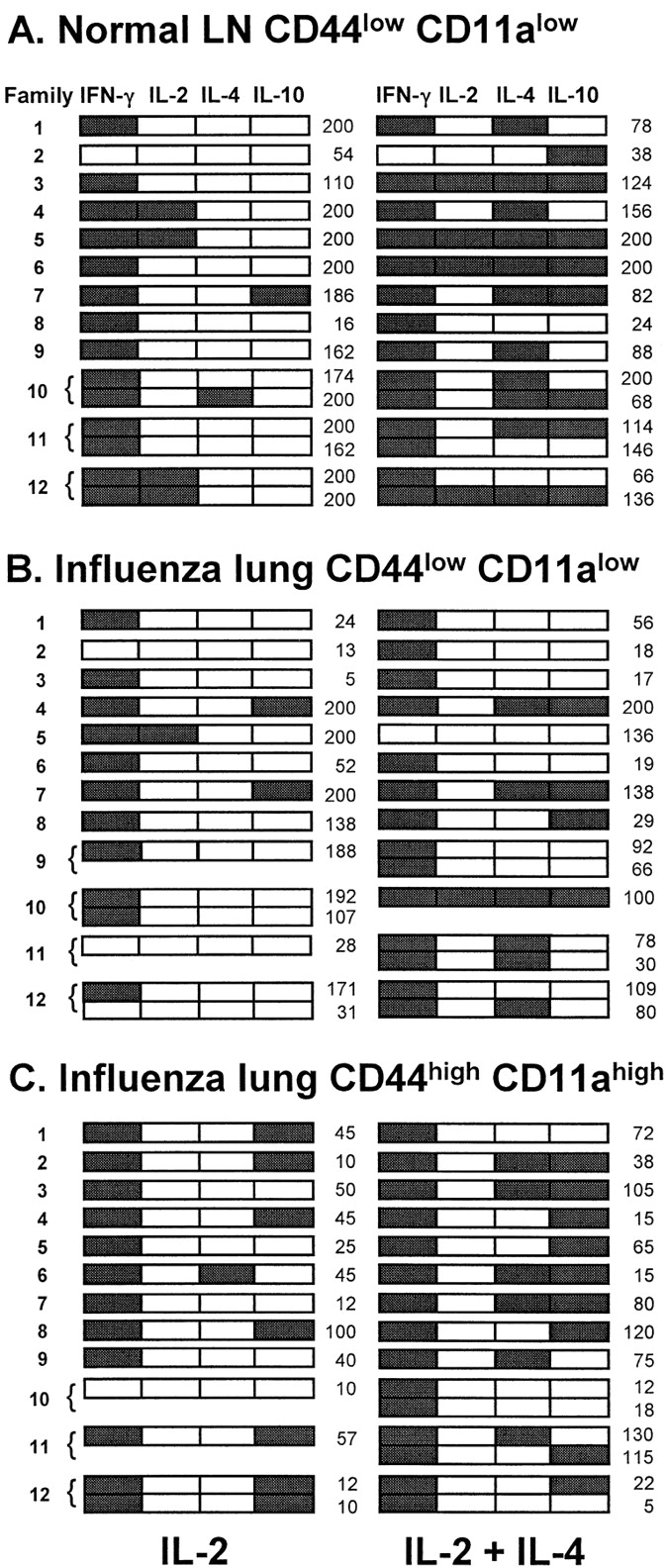
Cytokine mRNA expression patterns within families of subclones derived from normal LNlow cells, influenza lunglow cells, and influenza lunghigh cells after one or two divisions. In each group, cytokine profiles of 12 sequentially analyzed families from a single experiment are shown with subclones grown in IL-2 aligned with their siblings grown in IL-2 + IL-4. Brackets are used to group siblings grown in the same conditions. Detection of a cytokine PCR product is indicated by shading. The number of cells in each subclone is shown at the right of the panels.
The outcome of all paired daughter analyses for IL-4 and IL-10 expression is summarized in Table . Many families in all groups displayed a “−/−” phenotype for IL-4 expression, i.e., this cytokine was not expressed by subclones grown either in the absence or in the presence of IL-4. Several other families included subclones that expressed IL-4 in one (+/− and −/+ phenotypes) or both (+/+) types of culture. If detection of IL-4 mRNA were entirely dependent on addition of IL-4 to the cultures, the frequency of families with a −/+ phenotype would give a direct minimal estimate of the frequency of IL-4–inducible multipotential parent cells. However, in this study, there was a measurable level of IL-4 expression that occurred independently of exogenous IL-4, indicated by the frequency of +/− and +/+ families. Therefore, we estimated the frequency of multipotential parent cells by subtracting this IL-4–independent frequency from the sum of the −/+ and +/+ frequencies (Table ). Whether these calculations are based on expression of IL-4, IL-10, or both cytokines, they indicate a loss of multipotentiality as CD8+ T cells differentiate from the naive state in normal LNs to the activated state in the infected lung parenchyma.
Table 2.
Decreased Responsiveness to IL-4 and Increased Commitment among Activated Cells Revealed by the Expression of IL-4 and IL-10 within Subclone Families
| Cells | No. | Observed IL-4 expression (expected) | Observed IL-10 expression (expected) | ||||
|---|---|---|---|---|---|---|---|
| IL-2 | IL-2 + IL-4 | IL-2 | IL-2 + IL-4 | ||||
| − | + | − | + | ||||
| Normal LNlow | 93 | − | 16 (15.4) | 52 (52.6) | − | 35 (31.4) | 38 (41.6) |
| + | 5 (5.6) | 20 (19.4) | + | 5 (8.6) | 15 (11.4) | ||
| P = 0.72 | P = 0.066 | ||||||
| Influenza lunglow | 71 | − | 38 (36.9) | 23 (24.1) | − | 49 (46.0) | 15 (18.0) |
| + | 5 (6.1) | 5 (3.9) | + | 2 (5.0) | 5 (2.0) | ||
| P = 0.46 | P = 0.007 | ||||||
| Influenza lunghigh | 74 | − | 56 (53.1) | 13 (15.9) | − | 38 (32.5) | 18 (23.5) |
| + | 1 (3.9) | 4 (1.2) | + | 5 (10.4) | 13 (7.5) | ||
| P = 0.002 | P = 0.003 | ||||||
The table shows the number of subclone families that displayed each possible combination of IL-4 or IL-10 expression by subclones grown in IL-2 alone or in IL-2 + IL-4. Numbers in parentheses are the expected number of families with each phenotype if expression is statistically independent in each subclone (determined from the product of the fraction of subclones grown with IL-2 that did/did not express the cytokine and the fraction of subclones grown with IL-2 + IL-4 that did/did not express the cytokine).
Table 3.
Estimated Frequencies of Multipotential Cells among CD8+ T Cells Isolated from Normal LNs and Influenza Virus–infected Lung
| Cytokines considered | Multipotential cells (%) | ||
|---|---|---|---|
| Normal LNlow | Influenza lunglow | Influenza lunghigh | |
| IL-4 | 51 | 25 | 17 |
| IL-10 | 35 | 18 | 17 |
| IL-4 and/or IL-10 | 45 | 30 | 24 |
Other analyses indicated a concomitant rise in the proportion of committed cells with differentiation from the naive to the activated state. For example, in all groups, 60% or more of subclones grown in IL-2 alone expressed IFN-γ without IL-4. Table summarizes the IFN-γ/IL-4 phenotypes of their siblings grown in IL-2 plus IL-4 and reveals that a markedly higher proportion displayed the same IFN-γ1 IL-4− phenotype in families derived from the most activated lung population than in families derived from normal LNs.
Table 4.
Increased Stability of the IFN-γ1 IL-4− Phenotype in Subclone Families Derived from Activated Cells
| Cells | IL-2 | IL-2 + IL-4 | |||
|---|---|---|---|---|---|
| IFN-γ1 IL-4− | IFN-γ1 IL-4− | IFN-γ2 IL-4− | IFN-γ2 IL-4+ | IFN-γ1 IL-4+ | |
| Normal LNlow | 56 | 12 | 0 | 3 | 41 |
| Influenza lunglow | 47 | 27 | 2 | 0 | 18 |
| Influenza lunghigh | 55 | 43 | 1 | 0 | 11 |
The table shows the breakdown of cytokine phenotypes among subclones grown with IL-2 + IL-4 in those families where subclones grown in IL-2 alone displayed a IFN-γ1 IL-4− phenotype. These families represent 60, 66, and 74%, respectively, of all normal LNlow, influenza lunglow, and influenza lunghigh families analyzed. Data shown as number of subclone families.
To address this issue in another way, the frequency data in Table were subjected to contingency table testing to determine whether expression of IL-4 or IL-10 by subclones grown with added IL-4 was statistically independent of expression of the same cytokine by their siblings grown without IL-4. As shown in the table, the frequencies of each IL-4 phenotype among normal LNlow and influenza lunglow subclone families were close to those expected by chance, indicating that IL-4 expression was independently regulated among subclones within these families. By the same criterion, IL-10 expression was also independently regulated within families of normal LNlow subclones. This was not the case for the other sets of data. The frequencies of IL-4 −/− and +/+ families of influenza lunghigh cells and the frequencies of IL-10 −/− and +/+ families of both influenza lunglow and influenza lunghigh cells were significantly higher than expected by chance. This suggests that some parents of these families were committed to the expression or nonexpression of IL-4 and/or IL-10; their daughter cells were therefore unaffected by exposure to IL-4.
Discussion
Here we show that an activated, type 1–polarized, effector population of CD8+ T cells isolated from the lungs of influenza virus–infected mice is not terminally differentiated. Although it includes cells that appear functionally committed, it also contains a significant proportion of cells able to proliferate and express IL-4 and/or IL-10 when exposed to IL-4 in vitro. This indicates that some CD8+ T cells in an effector site can retain the potential to alter their cytokine profile in response to local stimuli.
A key feature of the experimental design of this study was the analysis of paired daughter cells, in which the ability of sister cells to give rise to subclones with different cytokine profiles in different culture conditions demonstrates the multipotentiality of their parent. This protocol has several advantages over conventional bulk culture methods used by several groups to assess the stability of cytokine profiles 15 16 19 36. First, it allows the fate of every cultured cell to be monitored, so that selective growth can be distinguished from reprogramming in response to an exogenous stimulus. Second, it enables the frequency of multipotential (programmable) cells to be estimated in heterogeneous populations. In the present study, these estimates suggested a progressive decline in the frequency of multipotential cells from normal LNlow to influenza lunglow to influenza lunghigh cells (Table ), in parallel with the observed rise in their expression of cytokine genes in vivo (Fig. 1). Third, culture of single parent cells and subcloning of their progeny isolate these cells from the effects of cross-regulatory cytokines from other cells, which might override the differentiative effects of IL-4 in a bulk culture 44 and mask low frequencies of multipotential cells.
We have previously used paired daughter analysis to evaluate the multipotentiality of CD8+CD44low T cells from the LNs of normal C57BL/6 mice and found that almost 90% of cells were at least bipotential for the expression of six different cytokines; based on the expression of IL-4 alone, 50% of parent cells in that study were multipotential 7. A similar figure (51%) was obtained here for CD8+CD44lowCD11alow cells from LNs of normal BALB/c mice. Although it seems likely that most, if not all, naive CD8+ T cells are multipotential for expression of IL-4 and other cytokines, these minimal estimates establish a baseline for comparison with other populations. Subcloning efficiencies were high for normal LN cells in the presence and absence of IL-4, suggesting that the parent cells analyzed were representative of the larger pool and arguing against the selective expansion of committed cells by one or other culture condition.
This study is, to our knowledge, the first frequency analysis of multipotentiality in T cells activated in vivo and has yielded both expected and unexpected results. Not surprisingly, the two CD8+ populations isolated from 7-d influenza virus–infected lung parenchyma differed markedly by several criteria from their counterparts of naive phenotype isolated from normal LNs. Although the influenza lunglow population was selected for the same surface markers as the normal LNlow cells, their frequency of IFN-γ expression ex vivo was at least three orders of magnitude higher (as assessed in multiple 500-cell samples; Fig. 1). Their subcloning efficiency was almost as high as the LN population, but average subclone sizes and frequencies of IL-2, IL-4, and IL-10 expression were lower both in the absence and presence of exogenous IL-4. The influenza lunghigh population displayed greater differences from normal cells, including high expression of IL-10 as well as IFN-γ ex vivo, and yet lower subcloning efficiency, subclone size, and subclone frequencies of IL-2, IL-4, and IL-10 expression. The progressive loss of proliferative potential from normal LNlow to influenza lunglow to influenza lunghigh cells, in parallel with increased effector activity, mirrors the experience of many investigators that effector populations are more difficult to clone than naive populations, and probably reflects both senescence and increased sensitivity to activation-induced cell death 43 55 56. While the intermediate status of influenza lunglow cells implies that they are less differentiated than influenza lunghigh cells and therefore might be their precursors, the actual relationship between these two populations is not known.
The loss of proliferative potential from the naive state to the activated effector state introduces an important consideration in interpreting the concomitant decline in multipotential cell frequency observed here. Since the assessment of multipotentiality depended on cloning and subcloning parent cells, no information is available about the fraction of influenza lung cells, especially those of CD44high CD11ahigh phenotype, that failed to proliferate in these cultures. Other experiments will be necessary to determine whether individual activated T cells can change their cytokine profile without cell division. However, since prolonged survival and clonal expansion are likely to improve the chances of de novo induction of cytokine gene expression by exogenous IL-4, as recently shown for naive CD4+ T cells 26 57, we expect multipotential cells to be found preferentially among the clonogenic cells. If this is the case, the differences in multipotentiality between influenza lunghigh cells and the other two CD8+ populations analyzed here would be compounded by their differences in clonogenicity and would therefore be greater than indicated by the estimates in Table .
The detection of IL-4 and IL-10 mRNAs at higher frequencies in subclones grown with IL-4 than in those grown without IL-4 was not due to any detectable enhancement of subclone size. Although mean subclone sizes varied between the three populations, they were not significantly affected within each population by exogenous IL-4 (Table ). Moreover, the minimum, median, and mean subclone sizes of IL-4+ and IL-10+ subclones were generally lower among those grown with IL-4 than those grown without IL-4 (data not shown). This suggests that IL-4 increased the probability of expression of these cytokines at the single-cell level, accelerating the onset of their expression as a clone expands 58.
In addition to demonstrating that some cells retain their multipotentiality after activation in vivo, this study provided evidence for a concomitant increase in the proportion of committed cells. Sharing of the IFN-γ+ IL-4− phenotype by siblings grown in the absence and presence of added IL-4 was progressively more common among families derived from normal LNlow, influenza lunglow, and influenza lunghigh cells. Statistical analysis also suggested that some influenza lunghigh cells were committed to IL-4 or IL-10 expression or nonexpression. It is not possible from this analysis to conclude whether a given family whose subclones displayed the same IL-4 or IL-10 phenotype in the absence and presence of exogenous IL-4 (−/− or +/+ phenotypes, as shown in Table ) was derived from a committed parent. However, χ2 testing indicated that influenza lunghigh cells gave rise to such families at a significantly higher frequency than expected by chance. By contrast, a similar analysis of the data for normal LNlow cells suggested that cytokine expression was regulated independently in related subclones, as expected if most of these cells are multipotential. The result for influenza lunglow cells was mixed, indicating independent regulation of IL-4 expression but higher than expected occurrence of −/− and +/+ families for IL-10 expression.
Collectively, these data indicate substantial heterogeneity within the most activated fraction of lung CD8+ cells, both in proliferative potential and in their flexibility to change cytokine profile in response to exogenous IL-4. This is despite the fact that the CD44highCD11ahigh fraction contains essentially all the IFN-γ–producing effector cell activity detected in the lung CD8+ fraction at this stage of influenza infection 48 49 and might be expected to comprise terminally differentiated type 1 cells.
Influenza lunghigh cells are likely to be heterogeneous in other ways not assessed here. Most importantly, since these experiments have not yet been done with populations selected for peptide–MHC tetramer binding, the antigen specificity of the cells we have analyzed is not known and may include a range of TCR affinities for several influenza epitopes as well as irrelevant specificities. However, in experiments by other groups with the A/HKx31 influenza virus strain, cells specific for the immunodominant nucleoprotein epitope comprised 3–12% of CD8+ cells in the bronchoalveolar lavage at day 7 59 and ∼20% of CD8+ T cells and 94% of CD44highCD8+ T cells isolated from lung tissue at day 8 60. Virus-specific cells are therefore likely to be a substantial fraction of the CD44highCD11ahighCD8+ T cells isolated from lung tissue at the peak of the response in our study. When methods become available to monitor both antigen specificity and relative avidity in polyclonal systems, it will be possible to determine whether multipotentiality persists among the most reactive cells or is limited to those receiving the weakest antigenic signals. In addition, isolation of antigen-specific T cells will allow multipotentiality to be analyzed in the small populations of cells that persist in the lung after the peak of the primary response and in secondary lymphoid organs as memory cells.
The proliferative history of the cells isolated from infected lung is also unknown. Others have shown marked heterogeneity in cell division number among responding CD4+ T cells in draining LNs in the first few days after immunization 61. We expect that both the influenza virus–specific and other activated T cells found in the lung 7 d after infection would also vary in their history of division before and after recruitment to the lung. It is not yet known whether there is any relationship between division history and multipotentiality. Previous studies of peptide-stimulated transgenic CD4+ T cells have generally shown a loss of flexibility at the population level by the second or third cycle of in vitro stimulation, but proliferative responses and division numbers were not reported and these findings cannot be readily extrapolated to the CD8+ T cell response to a replicating antigen in vivo where clonal burst sizes are likely to be higher. However, our previous observation that multipotential cells persisted in some type 1 CD8+ T cell clones through as many as 13–15 cell divisions over 7 d in vitro raises the possibility that even the most responsive, influenza virus–specific CD8+ T cells activated over the same time frame in vivo might also retain some flexibility in their cytokine profiles.
The molecular mechanisms underlying cytokine profile commitment in T cells are beginning to be unraveled. Loss of responsiveness to the counter-regulatory cytokines IL-12 and IL-4 is one mechanism that perpetuates existing cytokine profiles 21 22 23 24, but it is not yet known whether the reported changes in IL-12 receptor expression and IL-4 receptor function are causally or temporally associated with commitment. Similarly, while certain signal transducers and transcriptional activators selectively activate or antagonize type 1 or type 2 cytokine expression 29 30 31 32, their relationship to the loss of multipotentiality is not clear. Changes in CpG methylation and chromatin conformation of cytokine and other regulatory gene promoters are likely to be important events in enabling transcription 25 27 28. Recent work showing that demethylation of the IFN-γ promoter after primary activation of CD8+ T cells can be faithfully inherited through at least 16 cell divisions 27 suggests demethylation as a stable marker of gene activation. Again, however, its relationship to commitment is unknown. Key issues are whether this or any other candidate commitment event is irreversible and whether it can be permanently prevented from activating a silent cytokine gene.
In conclusion, we have found that the activated effector CD8+ T cell population in the lungs of influenza virus–infected mice contains both committed and multipotential cells. The presence of multipotential cells indicates that cytokine profiles are not irreversibly programmed during priming in the draining LN. Until now, it has been unclear whether differences in T cell cytokine profiles sometimes noted between tissue sites during an immune response were entirely due to preferential migration of differentiated effector cells 48 62 63 64. Our finding that some activated T cells can proliferate and express new cytokine genes in response to exogenous signals after recruitment to an effector site allows the possibility of substantial reshaping of the T cell response by the local environment. It remains to be seen whether this flexibility can be exploited to alleviate pathological cytokine responses.
Acknowledgments
We thank Dr. Lorena Brown for the generous provision of virus stock and continuing advice on the influenza infection model, Dr. Michael Rudd for advice and assistance with the model, Grace Chojnowski and Paula Hall for assistance with flow cytometry, and Dr. David Fitzpatrick and Prof. Michael Good for valuable comments on the manuscript.
This work was supported by the National Health and Medical Research Council of Australia, the Cooperative Research Centre for Vaccine Technology, and the Queensland Institute of Medical Research Trust.
Footnotes
1used in this paper: RT, reverse transcription
A. Doyle's present address is Peptech Limited, Locked Bag 2053, North Ryde, New South Wales 2113, Australia.
References
- Hsieh C.-S., Heimberger A.B., Gold J.S., O'Garra A., Murphy K.M. Differential regulation of T helper phenotype development by interleukins 4 and 10 in an αβ T-cell-receptor transgenic system. Proc. Natl. Acad. Sci. USA. 1992;89:6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seder R.A., Paul W.E., Davis M.M., Fazekas de St. Groth B. The presence of interleukin 4 during in vitro priming determines the lymphokine-producing potential of CD4+ T cells from T cell receptor transgenic mice. J. Exp. Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manetti R., Gerosa F., Giudizi M.G., Biagiotti R., Parronchi P., Piccinni M.-P., Sampognaro S., Maggi E., Romagnani S., Trinchieri G. Interleukin 12 induces stable priming for interferon γ (IFN-γ) production during differentiation of human T helper (Th) cells and transient IFN-γ production in established Th2 cell clones. J. Exp. Med. 1994;179:1273–1283. doi: 10.1084/jem.179.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbas A.K., Murphy K.M., Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–793. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- Röcken M., Saurat J.-H., Hauser C. A common precursor for CD4+ T cells producing IL-2 or IL-4. J. Immunol. 1992;148:1031–1036. [PubMed] [Google Scholar]
- Sad S., Mosmann T.R. Single IL-2-secreting precursor CD4 T cell can develop into either Th1 or Th2 cytokine secretion phenotype. J. Immunol. 1994;153:3514–3522. [PubMed] [Google Scholar]
- Kelso A., Groves P. A single peripheral CD8+ T cell can give rise to progeny expressing type 1 and/or type 2 cytokine genes and can retain its multipotentiality through many cell divisions. Proc. Natl. Acad. Sci. USA. 1997;94:8070–8075. doi: 10.1073/pnas.94.15.8070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C.-S., Macatonia S.E., O'Garra A., Murphy K.M. T cell genetic background determines default T helper phenotype development in vitro. J. Exp. Med. 1995;181:713–721. doi: 10.1084/jem.181.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bix M., Wang Z.-E., Thiel B., Schork N.J., Locksley R.M. Genetic regulation of commitment to interleukin 4 production by a CD4+ T cell–intrinsic mechanism. J. Exp. Med. 1998;188:2289–2299. doi: 10.1084/jem.188.12.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guler M.L., Gorham J.D., Dietrich W.F., Murphy T.L., Steen R.G., Parvin C.A., Fenoglio D., Grupe A., Pelt G., Murphy K.M. Tpm1, a locus controlling IL-12 responsiveness, acts by a cell-autonomous mechanism. J. Immunol. 1999;162:1339–1347. [PubMed] [Google Scholar]
- Seder R.A., Paul W.E. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu. Rev. Immunol. 1994;12:635–673. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- Croft M., Carter L., Swain S.L., Dutton R.W. Generation of polarized antigen-specific CD8 effector populationsreciprocal action of interleukin (IL)-4 and IL-12 in promoting type 2 versus type 1 cytokine profiles. J. Exp. Med. 1994;180:1715–1728. doi: 10.1084/jem.180.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Le Gros G., Bachmann M., Lamers M.C., Bluethmann H., Köhler G. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 1993;362:245–248. doi: 10.1038/362245a0. [DOI] [PubMed] [Google Scholar]
- Magram J., Connaughton S.E., Warrier R.R., Carvajal D.M., Wu C.Y., Ferrante J., Stewart C., Sarmiento U., Faherty D.A., Gately M.K. IL-12-deficient mice are defective in IFNγ production and type 1 cytokine responses. Immunity. 1996;4:471–481. doi: 10.1016/s1074-7613(00)80413-6. [DOI] [PubMed] [Google Scholar]
- Perez V.L., Lederer J.A., Lichtman A.H., Abbas A.K. Stability of Th1 and Th2 populations. Int. Immunol. 1995;7:869–875. doi: 10.1093/intimm/7.5.869. [DOI] [PubMed] [Google Scholar]
- Murphy E., Shibuya K., Hosken N., Openshaw P., Maino V., Davis K., Murphy K., O'Garra A. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J. Exp. Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sornasse T., Larenas P.V., Davis K.A., de Vries J.E., Yssel H. Differentiation and stability of T helper 1 and 2 cells derived from naive human neonatal CD4+ T cells, analyzed at the single-cell level. J. Exp. Med. 1996;184:473–483. doi: 10.1084/jem.184.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu-Li J., Huang H., Ryan J., Paul W.E. In differentiated CD4+ T cells, interleukin 4 production is cytokine-autonomous, whereas interferon γ production is cytokine-dependent. Proc. Natl. Acad. Sci. USA. 1997;94:3189–3194. doi: 10.1073/pnas.94.7.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenmacher M., Löhning M., Scheffold A., Richter A., Miltenyi S., Schmitz J., Radbruch A. Commitment of individual Th1-like lymphocytes to expression of IFN-γ versus IL-4 and IL-10selective induction of IL-10 by sequential stimulation of naive Th cells with IL-12 and IL-4. J. Immunol. 1998;161:2825–2832. [PubMed] [Google Scholar]
- Sad S., Marcotte R., Mosmann T.R. Cytokine-induced differentiation of precursor mouse CD8+ T cells into cytotoxic CD8+ T cells secreting Th1 or Th2 cytokines. Immunity. 1995;2:271–279. doi: 10.1016/1074-7613(95)90051-9. [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Jacobson N.G., Dighe A.S., Gubler U., Murphy K.M. Developmental commitment to the Th2 lineage by extinction of IL-12 signaling. Immunity. 1995;2:665–675. doi: 10.1016/1074-7613(95)90011-x. [DOI] [PubMed] [Google Scholar]
- Szabo S.J., Dighe A.S., Gubler U., Murphy K.M. Regulation of the interleukin (IL)-12R β2 subunit expression in developing T helper 1 (Th1) and Th2 cells. J. Exp. Med. 1997;185:817–824. doi: 10.1084/jem.185.5.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogge L., Barberis-Maino L., Biffi M., Passini N., Presky D.H., Gubler U., Sinigaglia F. Selective expression of an interleukin-12 receptor component by human T helper 1 cells. J. Exp. Med. 1997;185:825–831. doi: 10.1084/jem.185.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Paul W.E. Impaired interleukin 4 signaling in T helper type 1 cells. J. Exp. Med. 1998;187:1305–1313. doi: 10.1084/jem.187.8.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick D.R., Shirley K.M., McDonald L.E., Bielefeldt-Ohmann H., Kay G.F., Kelso A. Distinct methylation of the interferon γ (IFN-γ) and interleukin 3 (IL-3) genes in newly activated primary CD8+ T lymphocytesregional IFN-γ promoter demethylation and mRNA expression are heritable in CD44highCD8+ T cells. J. Exp. Med. 1998;188:103–117. doi: 10.1084/jem.188.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird J.J., Brown D.R., Mullen A.C., Moskowitz N.H., Mahowald M.A., Sider J.R., Gajewski T.F., Wang C.-R., Reiner S.L. Helper T cell differentiation is controlled by the cell cycle. Immunity. 1998;9:229–237. doi: 10.1016/s1074-7613(00)80605-6. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D.R., Shirley K.M., Kelso A. Cutting edgestable epigenetic inheritance of regional IFN-γ promoter demethylation in CD44highCD8+ T lymphocytes. J. Immunol. 1999;162:5053–5057. [PubMed] [Google Scholar]
- Agarwal S., Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- Zheng W., Flavell R.A. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- Ho I.-C., Lo D., Glimcher L.H. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4–dependent and –independent mechanisms. J. Exp. Med. 1998;188:1859–1866. doi: 10.1084/jem.188.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W., Ranganath S.H., Weindel K., Bhattacharya D., Murphy T.L., Sha W.C., Murphy K.M. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- Li B., Tournier C., Davis R.J., Flavell R.A. Regulation of IL-4 expression by the transcription factor JunB during T helper cell differentiation. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:420–432. doi: 10.1093/emboj/18.2.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelman F.D., Madden K.B., Cheever A.W., Katona I.M., Morris S.C., Gately M.K., Hubbard B.R., Gause W.C., Urban J.F. Effects of interleukin 12 on immune responses and host protection in mice infected with intestinal nematode parasites. J. Exp. Med. 1994;179:1563–1572. doi: 10.1084/jem.179.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiner S.L., Locksley R.M. The regulation of immunity to Leishmania major . Annu. Rev. Immunol. 1995;13:151–177. doi: 10.1146/annurev.iy.13.040195.001055. [DOI] [PubMed] [Google Scholar]
- Nabors G.S., Afonso L.C.C., Farrell J.P., Scott P. Switch from a type 2 to a type 1 T helper cell response and cure of established Leishmania major infection in mice is induced by combined therapy with interleukin 12 and Pentostam. Proc. Natl. Acad. Sci. USA. 1995;92:3142–3146. doi: 10.1073/pnas.92.8.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocci S., Coffman R.L. The mechanism of in vitro T helper cell type 1 to T helper cell type 2 switching in highly polarized Leishmania major-specific T cell populations. J. Immunol. 1997;158:1559–1564. [PubMed] [Google Scholar]
- Trembleau S., Penna G., Gregori S., Gately M.K., Adorini L. Deviation of pancreas-infiltrating cells to Th2 by interleukin-12 antagonist administration inhibits autoimmune diabetes. Eur. J. Immunol. 1997;27:2330–2339. doi: 10.1002/eji.1830270930. [DOI] [PubMed] [Google Scholar]
- Swain S.L. Generation and in vivo persistence of polarized Th1 and Th2 memory cells. Immunity. 1994;1:543–552. doi: 10.1016/1074-7613(94)90044-2. [DOI] [PubMed] [Google Scholar]
- Cerwenka A., Carter L.L., Reome J.B., Swain S.L., Dutton R.W. In vivo persistence of CD8 polarized T cell subsets producing type 1 or type 2 cytokines. J. Immunol. 1998;161:97–105. [PubMed] [Google Scholar]
- Secrist H., Chelen C.J., Wen Y., Marshall J.D., Umetsu D.T. Allergen immunotherapy decreases interleukin 4 production in CD4+ T cells from allergic individuals. J. Exp. Med. 1993;178:2123–2130. doi: 10.1084/jem.178.6.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellinghausen I., Metz G., Enk A.H., Christmann S., Knop J., Saloga J. Insect venom immunotherapy induces interleukin-10 production and a Th2-to-Th1 shift, and changes surface marker expression in venom-allergic subjects. Eur. J. Immunol. 1997;27:1131–1139. doi: 10.1002/eji.1830270513. [DOI] [PubMed] [Google Scholar]
- Racke M.K., Bonomo A., Scott D.E., Cannella B., Levine A., Raine C.S., Shevach E.M., Röcken M. Cytokine-induced immune deviation as a therapy for inflammatory autoimmune disease. J. Exp. Med. 1994;180:1961–1966. doi: 10.1084/jem.180.5.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadhachary A.S., Salgame P. CD95 mediated T cell apoptosis and its relevance to immune deviation. Oncogene. 1998;17:3271–3276. doi: 10.1038/sj.onc.1202572. [DOI] [PubMed] [Google Scholar]
- Nakamura T., Lee R.K., Nam S.Y., Podack E.R., Bottomly K., Flavell R.A. Roles of IL-4 and IFN-γ in stabilizing the T helper cell type 1 and 2 phenotype. J. Immunol. 1997;158:2648–2653. [PubMed] [Google Scholar]
- Metcalf D. Clonal analysis of proliferation and differentiation of paired daughter cellsaction of granulocyte-macrophage colony-stimulating factor on granulocyte-macrophage precursors. Proc. Natl. Acad. Sci. USA. 1980;77:5327–5330. doi: 10.1073/pnas.77.9.5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N., Brown L., Jackson D., Kelso A. Novel features of the respiratory tract T-cell response to influenza virus infectionlung T cells increase expression of γ interferon mRNA in vivo and maintain high levels of mRNA expression for interleukin-5 (IL-5) and IL-10. J. Virol. 1994;68:7575–7581. doi: 10.1128/jvi.68.11.7575-7581.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N., Kelso A. In vivo blockade of γ interferon affects the influenza virus-induced humoral and the local cellular immune response in lung tissue. J. Virol. 1996;70:4411–4418. doi: 10.1128/jvi.70.7.4411-4418.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarth N., Kelso A. Functionally distinct T cells in three compartments of the respiratory tract after influenza virus infection. Eur. J. Immunol. 1996;26:2189–2197. doi: 10.1002/eji.1830260934. [DOI] [PubMed] [Google Scholar]
- Baumgarth N., Egerton M., Kelso A. Activated T cells from draining lymph nodes and an effector site differ in their responses to TCR stimulation. J. Immunol. 1997;159:1182–1191. [PubMed] [Google Scholar]
- Maraskovsky E., Troutt A.B., Kelso A. Co-engagement of CD3 with LFA-1 or ICAM-1 adhesion molecules enhances the frequency of activation of single murine CD4+ and CD8+ T cells and induces synthesis of IL-3 and IFN-γ but not IL-4 or IL-6. Int. Immunol. 1992;4:475–485. doi: 10.1093/intimm/4.4.475. [DOI] [PubMed] [Google Scholar]
- Smith K.G.C., Nossal G.J.V., Tarlinton D.M. FAS is highly expressed in the germinal center but is not required for regulation of the B-cell response to antigen. Proc. Natl. Acad. Sci. USA. 1995;92:11628–11632. doi: 10.1073/pnas.92.25.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso A., Groves P., Troutt A.B., Francis K. Evidence for the stochastic acquisition of cytokine profile by CD4+ T cells activated in a T helper type 2-like response in vivo . Eur. J. Immunol. 1995;25:1168–1175. doi: 10.1002/eji.1830250506. [DOI] [PubMed] [Google Scholar]
- Kelso A., Groves P., Ramm L., Doyle A.G. Single-cell analysis by RT-PCR reveals differential expression of multiple type 1 and 2 cytokine genes among cells within polarized CD4+ T cell populations. Int. Immunol. 1999;11:617–621. doi: 10.1093/intimm/11.4.617. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D.R., Kelso A. Dissociated expression of granulocyte-macrophage CSF and IL-3 in short-term T cell clones from normal mice. J. Immunol. 1995;155:5140–5150. [PubMed] [Google Scholar]
- Weng N.P., Hathcock K.S., Hodes R.J. Regulation of telomere length and telomerase in T and B cellsa mechanism for maintaining replicative potential. Immunity. 1998;9:151–157. doi: 10.1016/s1074-7613(00)80597-x. [DOI] [PubMed] [Google Scholar]
- Zheng L., Fisher G., Miller R.E., Peschon J., Lynch D.H., Lenardo M.J. Induction of apoptosis in mature T cells by tumour necrosis factor. Nature. 1995;377:348–351. doi: 10.1038/377348a0. [DOI] [PubMed] [Google Scholar]
- Gett A.V., Hodgkin P.D. Cell division regulates the T cell cytokine repertoire, revealing a mechanism underlying immune class regulation. Proc. Natl. Acad. Sci. USA. 1998;95:9488–9493. doi: 10.1073/pnas.95.16.9488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelso A. Educating T cellsearly events in the differentiation and commitment of cytokine-producing CD4+ and CD8+ T cells. Springer Sem. Immunopathol. 1999;In press doi: 10.1007/BF00812255. [DOI] [PubMed] [Google Scholar]
- Flynn K.J., Belz G.T., Altman J.D., Ahmed R., Woodland D.L., Doherty P.C. Virus-specific CD8+ T cells in primary and secondary influenza pneumonia. Immunity. 1998;8:683–691. doi: 10.1016/s1074-7613(00)80573-7. [DOI] [PubMed] [Google Scholar]
- Haanen J.B.A.G., Toebes M., Cordaro T.A., Wolkers M.C., Kruisbeek A.M., Schumacher T.N.M. Systemic T cell expansion during localized viral infection. Eur. J. Immunol. 1999;29:1168–1174. doi: 10.1002/(SICI)1521-4141(199904)29:04<1168::AID-IMMU1168>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Gudmundsdottir H., Wells A.D., Turka L.A. Dynamics and requirements of T cell clonal expansion in vivo at the single-cell leveleffector function is linked to proliferative capacity. J. Immunol. 1999;162:5212–5223. [PubMed] [Google Scholar]
- Austrup F., Vestweber D., Borges E., Löhning M., Bräuer R., Herz U., Renz H., Hallmann R., Scheffold A., Radbruch A., Hamann A. P- and E-selectin mediate recruitment of T-helper-1 but not T-helper-2 cells into inflamed tissues. Nature. 1997;385:81–83. doi: 10.1038/385081a0. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Mackay C.R., Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T helper 2 cells. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- Cerwenka A., Morgan T.M., Harmsen A.G., Dutton R.W. Migration kinetics and final destination of type 1 and type 2 CD8 effector cells predict protection against pulmonary virus infection. J. Exp. Med. 1999;189:423–434. doi: 10.1084/jem.189.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]


