Abstract
In macroautophagy, cytoplasmic components are delivered to lysosomes for degradation via autophagosomes that are formed by closure of cup-shaped isolation membranes. However, how the isolation membranes are formed is poorly understood. We recently found in yeast that a novel ubiquitin-like system, the Apg12-Apg5 conjugation system, is essential for autophagy. Here we show that mouse Apg12-Apg5 conjugate localizes to the isolation membranes in mouse embryonic stem cells. Using green fluorescent protein–tagged Apg5, we revealed that the cup-shaped isolation membrane is developed from a small crescent-shaped compartment. Apg5 localizes on the isolation membrane throughout its elongation process. To examine the role of Apg5, we generated Apg5-deficient embryonic stem cells, which showed defects in autophagosome formation. The covalent modification of Apg5 with Apg12 is not required for its membrane targeting, but is essential for involvement of Apg5 in elongation of the isolation membranes. We also show that Apg12-Apg5 is required for targeting of a mammalian Aut7/Apg8 homologue, LC3, to the isolation membranes. These results suggest that the Apg12-Apg5 conjugate plays essential roles in isolation membrane development.
Keywords: autophagy, ubiquitin-like protein, autophagosome, isolation membrane, gene targeting
Introduction
Most intracellular short-lived proteins are selectively degraded by the ubiquitin-proteosome pathway (Hershko and Ciechanover 1998), while most long-lived proteins are degraded in the lysosome. Macroautophagy, usually referred to simply as autophagy, is an intracellular bulk degradation system in which cytoplasmic components, including organelles, are directed to the lysosome/vacuole by a membrane-mediated process (Seglen and Bohley 1992; Dunn 1994). Cytoplasmic constituents are first enclosed by double- or multiple-membrane structures called autophagosomes. Eventually, autolysosomes are generated by the fusion of the outer membranes of the autophagosomes and lysosomes. Lysosomal hydrolases degrade the cytoplasm-derived contents of the autophagosome, together with its inner membrane. Autophagy is regulated by nutrients and hormones, and has been suggested to be essential for cellular homeostasis (Mortimore and Pösö 1987). In addition, autophagy is thought to be crucial for various physiological processes, such as cellular remodeling (Bolender and Weibel 1973; Masaki et al. 1987), differentiation (Tsukada and Ohsumi 1993), production of pulmonary surfactant (Hariri et al. 2000), and nonapoptotic cell death during embryogenesis (Clarke 1990). Defective autophagy may contribute to the pathogenesis of mammary tumors (Liang et al. 1999) and a specific type of myopathy (Nishino et al. 2000; Tanaka et al. 2000).
Autophagosomes are thought to be derived from cap-shaped cisternae known as isolation membranes. Electron microscopic analysis suggested that the isolation membranes appear to elongate with curvature and finally close to form autophagosomes with nearly constant diameter, 0.5–1.5 μm (Pfeifer 1987; Seglen and Bohley 1992). Development of the cisternae is a quite unique process compared with other known intracellular membrane dynamics. However, little is known about how the cup-shaped isolation membranes are formed.
In the yeast Saccharomyces cerevisiae, autophagy-defective (apg and aut) mutants were isolated (Tsukada and Ohsumi 1993; Thumm et al. 1994), and most of them were suggested to have defects in autophagosome formation (Klionsky and Ohsumi 1999). We recently found that a novel protein conjugation system is essential for yeast autophagy (Mizushima et al. 1998a). Two proteins, Apg12 and Apg5, are covalently attached in a manner similar to the ubiquitin conjugation system. The COOH-terminal glycine of Apg12 forms an isopeptide bond with the ε-amino group of a lysine residue in Apg5. This conjugation reaction requires ATP and two enzymes: Apg7 and Apg10 (Kim et al. 1999; Shintani et al. 1999; Tanida et al. 1999; Yuan et al. 1999). These discoveries expanded the field of ubiquitin-like protein conjugation systems (Hochstrasser 2000). The Apg12-Apg5 conjugate further interacts noncovalently with Apg16 (Mizushima et al. 1999). Analyses of an apg5 null mutant and a temperature-sensitive mutant suggested that Apg5 is required for formation of autophagosomes (George et al. 2000). However, its subcellular localization and molecular function are unknown.
We also demonstrated that the Apg12 conjugation system is conserved in human (Mizushima et al. 1998b). In the present study, to examine the role of Apg5, we created an Apg5 null mutant cell by the gene targeting method using mouse embryonic stem (ES) cells. The resulting clone clearly demonstrated that Apg5 is essential also for autophagy in mammals. By generating various stable transformants, we investigated the subcellular localization and function of Apg5, and the role of its modification by Apg12. This study also enabled us to identify the isolation membrane at early stages and visualize its development into autophagosome.
Materials and Methods
Plasmids
Mouse Apg5 cDNA was obtained by reverse transcription–PCR based on the sequences of expressed sequence tag clones (mv76e10.r1, me31a04.r1, mj23e09.r1). The cDNA sequence was deposited in the DDBJ/EMBL/GenBank databases (AB048349), which encodes for a 275 amino acids protein that is 97% identical to human Apg5 (Hammond et al. 1998; Mizushima et al. 1998b). Mouse APG5 genomic clones were isolated from a 129/Sv genomic library using the mouse Apg5 cDNA as a probe. A targeting vector was constructed by replacing a 5.6-kb BamHI-SpeI fragment including the putative second (containing the first ATG) and third exons with the neo-resistant cassette from pMCI-Neo. The herpes simplex thymidine kinase gene was inserted downstream of the short arm for negative selection against random integration of the vector (see Fig. 1). The mouse Apg5 cDNA was also subcloned into the SmaI site of a mammalian expression vector pCI-neo (Promega). The green fluorescent protein (GFP)–tagged Apg5 expression vector has been described previously (Mizushima et al. 1998b). Replacement of Lys130 to Arg (K130R) was performed using a Quick Change Site-directed Mutagenesis Kit (Stratagene).
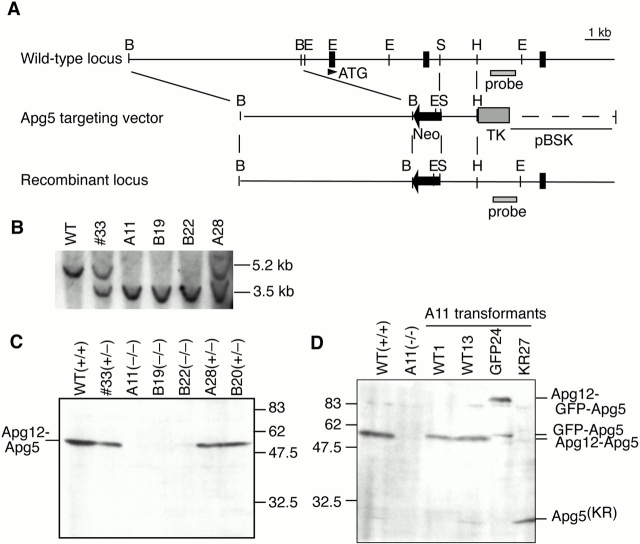
Figure 1.
Production of Apg5-deficient ES cells. (A) The restriction map of the wild-type APG5 allele, targeting construct, and mutated allele. Closed boxes indicate exons. Restriction enzymes: B, BamHI; E, EcoRI; S, SpeI; H, HindIII. (B) Southern blot analysis of wild-type ES cells (WT), an Apg5 single knockout clone (#33), three double knockout clones (A11, B19, and B22) and one single knockout clone obtained in the double knockout screening (A28). The probe indicated in A was used. (C) Immunoblot analysis of the ES clones. Total cell lysates were subjected to immunoblotting with anti–Apg5 antibody. Genotype of each clone is indicated. (D) Immunoblot analysis of stable transformants derived from the A11 clone with anti–Apg5 antibody.
ES Cell Culture
R1 ES cells (a generous gift from Dr. Andras Nagy, Samuel Lunenfeld Research Institute, Toronto, Canada) were cultured on mitomycin C–treated embryonic fibroblasts, STO feeder cells (Lexicon Genetics Inc.), or gelatinized dish in a complete ES medium: high glucose Dulbecco's modified Eagle's medium supplemented with 20% FCS, 2 mM l-glutamine, 1× nonessential amino acids (GIBCO BRL), 1 μM 2-mercaptoethanol, antibiotics, and 1,000 U/ml leukemia inhibitory factor (Life Technologies, Inc.). For amino acid starvation, cells were cultured in Hanks' solution containing 10 mM Hepes, pH 7.5 (without amino acid and FCS).
Production of APG5−/− ES Cells and Stable Transformants
The linearized targeting vector (30 μg) was transfected into 107 R1 ES cells by electroporation using a Gene Pulser (Bio-Rad Laboratories) set at 270 V and 500 μF. The 0.25 mg/ml G418- and gancyclovir-selected clones (96 clones) were examined, and homologous recombination was detected in 13 clones. These clones were also tested for single integration by Southern blot analysis with a neo probe. To obtain APG5−/− cells, an APG5 +/− clone (#33) was grown in the ES medium containing 8 mg/ml G418. 3 of 123 clones selected were APG5−/− (A11, B19, B22). To obtain stable transformants, 8 × 106 ES cells were electroporated with 20 μg of circular or linearized expression vector, and with 2 μg of pPGKpurobpA when required. Cells were selected in the presence of 0.5–1 mg/ml G418 or 5–10 μg/ml puromycin.
Southern Blot Analysis
Genomic DNA was digested with EcoRI, and Southern blot was performed as described previously (Hatano et al. 1997). A 1-kb probe shown in Fig. 1 was labeled with digoxigenin by PCR using primers Pro1 (5′-CAATGCTTAATTTCAGCAAC-3′) and Pro2 (5′-AGGCTACTTTGGCAGTATATC-3′).
Antibodies
Anti–Apg5 antibody (SO4) against glutathione-S-transferase–fused human Apg5 was prepared by immunization of rabbits and affinity purified. Anti–mouse Apg12 antibodies (NM2) against a synthetic peptide corresponding to the NH2-terminal 14 amino acids of mouse Apg12 and an additional Cys (MSEDSEVVLQLPSAC) was prepared by immunization of rabbits as previously described (Yoshimori et al. 2000) and affinity purified. Anti–LC3 antibody against recombinant LC3 (Kabeya et al. 2000), anti–rat Lgp85 antibody (Okazaki et al. 1992), and anti–rat aldolase antibody (Kominami et al. 1983) were previously described. For immunofluorescence microscopy, fluorolink Cy5-labeled goat anti–rabbit IgG antibody (Amersham Pharmacia Biotech) was used. Rabbit polyclonal anti–GFP antibody (CLONTECH Laboratories, Inc.) was used for immunoelectron microscopy.
Preparation of Whole-Cell Lysates and Western Blotting
Whole-cell lysates were prepared with a lysis buffer (2% NP-40, 0.2% SDS in PBS supplemented with protease inhibitors). Western blotting was performed as previously described (Mizushima et al. 1998b).
Electron Microscopy
Conventional electron microscopy was performed as described previously (Yoshimori et al. 2000), except that ES cells grown on gelatinized plastic coverslips were prefixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, for 2 h.
For immunoelectron microscopy, the pre-embedding silver enhancement immunogold method was performed as previously described (Yoshimori et al. 2000), with a slight modification. ES cells cultured on gelatin-coated plastic coverslips were fixed in 4% paraformaldehyde in 0.1 M cacodylate buffer, pH 7.4, for 2 h. The cells were washed in the buffer three times, dipped in phosphate buffer (PB) containing 15% glycerol and 35% sucrose, frozen and thawed using liquid nitrogen, and then incubated in PB containing 0.005% saponin, 10% BSA, 10% normal goat serum, and 0.1% cold water fish skin gelatin for blocking for 30 min. Cells were then treated with rabbit IgG against GFP (diluted 500×) in the blocking solution, overnight. Then, the cells were washed in PB containing 0.005% saponin for 10 min six times, and incubated with goat anti–rabbit IgG that was conjugated to colloidal gold (1.4-nm diameter) in the blocking solution for 2 h. Cells were washed with PB for 10 min six times, and fixed with 1% glutaraldehyde in PB for 10 min. After washing, the gold labeling was intensified by using a silver enhancement kit for 6 min at 20°C in the dark. After washing in distilled water, cells were post-fixed in 0.5% OsO4 for 90 min at 4°C, washed in distilled water, incubated with 50% ethanol for 10 min, and stained with 2% uranyl acetate in 70% ethanol for 2 h. The cells were further dehydrated with a graded series of ethanol and embedded in epoxy resin. Ultra-thin sections were doubly stained with uranyl acetate and lead citrate.
Morphometric analysis was performed with NIH image software. 30 cell sections were analyzed. Autophagosomes were defined as double- or multiple-membrane structures surrounding undigested cytoplasmic constituents. Autolysosomes were defined as single membrane structures containing cytoplasmic components at various stages of degradation.
Bulk Protein Degradation Assay
Degradation of long-lived proteins was measured based on a standard method (Ogier-Denis et al. 1996). ES cells maintained on gelatinized plates without feeder cells were plated at 2 × 105 cells/ well in gelatinized 24-well plates and cultured in complete ES medium for 24 h. Cells were then labeled for 24 h with the same medium containing 1.5 μCi/ml l-[14C] valine (Moravek Biochemicals Inc.). After three rinses with PBS, cells were incubated in either complete ES medium or Hanks' solution containing 0.1% BSA and 10 mM cold valine. When required, 1 mM chloroquine, 0.1 μM bafilomycin A1, or 10 mM 3-methyladenine (3-MA) was added. After the 1-h incubation, the medium was replaced with identical fresh medium and incubated for additional 2 h. The medium was precipitated in 10% TCA, and TCA-soluble radioactivity was measured. Total-cell radioactivity was measured after lysis with 0.1 M NaOH. [14C] Valine release was calculated as a percentage of the radioactivity in the TCA-soluble supernatant to the total cell radioactivity.
Fluorescence Microscopy
ES cells grown on gelatinized coverslips were fixed and, if necessary, stained with affinity-purified anti–mouse Apg12 antibody (200× dilution) or anti–LC3 antibody (200× dilution), and then examined under a fluorescence laser scanning confocal microscope, LSM510 (Carl Zeiss, Inc.), as previously described (Yoshimori et al. 2000). Time-lapse video microscopy was performed at 37°C with a DeltaVision microscope system (Applied Precision Inc.) equipped with a ΔTC3 culture dish system (Bioptechs) for temperature control.
Online Supplemental Material
A supplemental figure is supplied in which subcellular fractionation analysis demonstrates that most Apg12-Apg5 conjugates are recovered in the cytosolic fraction (Fig. S1). The QuickTime movies from which the images of Fig. 7 A were taken are also available. APG5−/− cells expressing GFP-Apg5 (videos 1–3) or GFP-Apg5K130R (video 4) were cultured in Hanks' solution for 1 h, and GFP images were taken every 3.8 s. They are animated at 15 images/s. Online supplemental materials can be found at http://www.jcb.org/cgi/content/full/152/4/657/DC1.
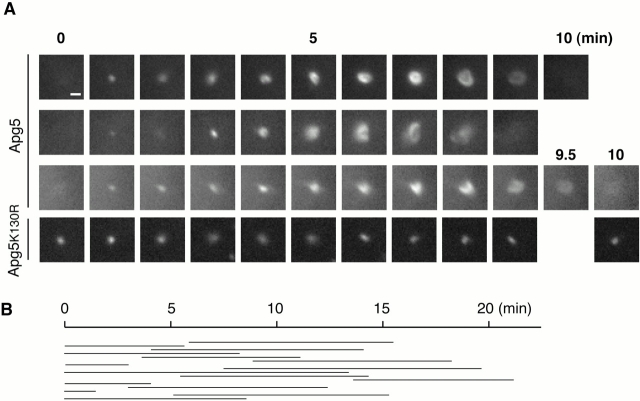
Figure 7.
Formation of autophagosomes is traced with GFP-Apg5. (A) Sequential frames (1-min intervals) of three newly generated GFP-Apg5 spots, or a GFP-Apg5K130R spot. GFP24 or GKR-1 cells were cultured in Hanks' solution for 1 h and directly observed by time-lapse video microscopy. Bars, 1 μm. See supplemental videos of GFP-Apg5 (videos 1–3) and GFP-Apg5K130R (video 4). (B) Duration of each spot of GFP-Apg5 staining was measured for 15 cases that showed circularization in the same field.
Results
Production of Apg5-deficient Cells
To produce a null mutation in the mouse APG5 gene, we targeted the mouse APG5 gene of R1 ES cells, using a targeting vector shown in Fig. 1 A. By subjecting an APG5 +/− ES clone (#33) to growth selection in an elevated concentration of G418, three Apg5-deficient (APG5−/−) clones (A11, B19, and B22) were obtained (Fig. 1 B). An anti–Apg5 antibody recognized a 56-kD protein in the wild-type ES cells (Fig. 1 C). In addition, a very faint band at 30 kD, corresponding to Apg5 monomer, was observed. The 56-kD band was also reactive with anti–mouse Apg12 antibodies (data not shown). These data suggest that Apg5 exists almost completely in the Apg12-conjugated form in the wild-type ES cells. The amount of the Apg12-Apg5 conjugate decreased in the heterozygous (APG5 +/−) mutant, and was not detected at all in the three independent APG5−/− clones (Fig. 1 C). Taken together, these data confirm that the targeted ES cells contain a null mutation in the APG5 gene.
We then generated several stable transformants by transfecting A11 cells with Apg5 cDNAs (Fig. 1 D). In two transformants with wild-type Apg5 cDNA (WT1 and WT13), exogenous Apg5, of which expression was a little weaker than in wild-type cells, was conjugated with Apg12 almost completely. The Apg5K130R mutant was unable to be conjugated (KR27), confirming our previous observation that Lys130 is an acceptor residue for Apg12 conjugation (Mizushima et al. 1998b). GFP-fused Apg5 was expressed (GFP24) at a similar level as in wild-type cells, most of which is conjugated with Apg12.
APG5−/− Cells Exhibit a Block in the Autophagic Pathway
APG5−/− cells were found to grow at the same rate as wild-type cells and form colonies with normal morphology (data not shown). Since three APG5−/− clones (A11, B19, and B22) showed similar phenotypes, we used A11 for further studies unless otherwise indicated.
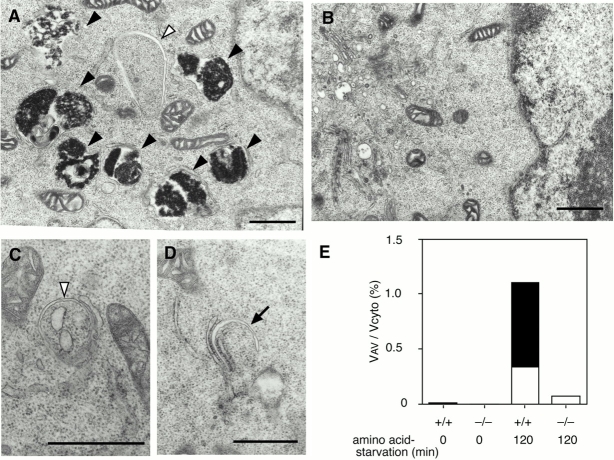
Autophagy can be well induced by amino acid starvation in wild-type ES cells (Fig. 2 A) as other cultured cells. 2 h after amino acid withdrawal, autophagic vacuoles occupied ∼1.1% of the total cytoplasmic volume (Fig. 2 E). Autolysosomes were observed more frequently than autophagosomes (autophagosome 0.33%, autolysosome 0.77%). In contrast, no autolysosomes were observed at all in APG5−/− cells, even under amino acid starvation conditions (Fig. 2B and Fig. E). Some autophagosome-like structures were induced by starvation (Fig. 2C and Fig. D), but these structures were significantly fewer in number than autophagosomes in wild-type cells (Fig. 2 E). Other organelles, such as mitochondria, the endoplasmic reticulum, the Golgi apparatus, lysosomes, and multivesicular bodies, appeared normal. These results suggest that, in APG5−/− cells, autophagic degradation does not occur and autophagosome formation is impaired.
Figure 2.
APG5−/− cells exhibit a block in the autophagic pathway. Wild-type ES cells (A) and APG5−/− cells (B–D) were cultured in Hanks' solution for 2 h, and then fixed and subjected to conventional electron microscopic analysis. The isolation membrane (arrow), autophagosomes or autophagosome-like structures (open arrowheads), and autolysosomes (closed arrowheads) are indicated. (C and D) Rarely detected autophagosome-like structures in APG5−/− cells. As often observed in wild-type cells, isolation membranes develop between cisternae of the ER (C and D). Bars, 1 μm. (E) Morphometric analysis of wild-type (+/+) and APG5−/− (−/−) ES cells before or after amino acid withdrawal. Ratio of total area of autophagosomes (open column) and autolysosomes (closed column) to the total cytoplasmic area is shown.
Bulk Protein Degradation Is Reduced in Apg5-deficient Cells
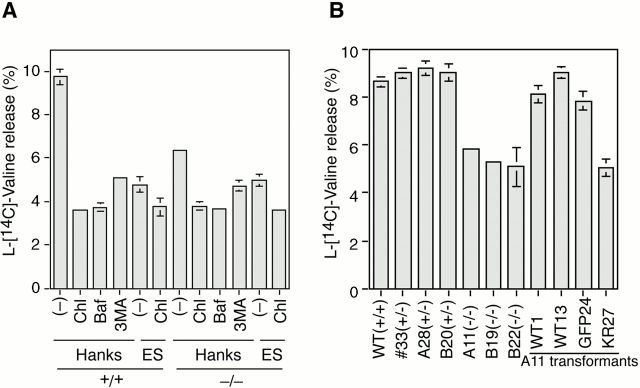
Autophagy is thought to be a major pathway for degradation of cytoplasmic proteins. We assessed bulk degradation of long-lived proteins by measuring the release of TCA-soluble [14C] valine from cells. Lysosomal protein degradation was estimated by the difference between the [14C] valine release from cells treated with and without lysosomotropic reagents such as chloroquine and bafilomycin A1, a vacuolar H+-ATPase inhibitor. Lysosomal protein degradation induced by amino acid starvation in APG5−/− cells was significantly reduced to less than half of that in APG5 +/+ and APG5 +/− cells (Fig. 3A and Fig. B). Apg5 cDNA transformants (WT1, WT13, and GFP24) restored protein degradation, confirming the specificity of Apg5 deficiency (Fig. 3 B). Cells grown in nutrient-rich medium showed a very low level of basal lysosomal degradation, with no significant difference between APG5 +/+ and APG5−/− cells. These data indicate that autophagy is indeed a major route for lysosomal protein degradation. In addition, in the KR27 clone, protein degradation was not restored, suggesting that Apg12 conjugation of Apg5 is essential for autophagic degradation.
Figure 3.
Starvation-induced lysosomal protein degradation is reduced in APG5−/−cells. (A) APG5 +/+ and APG5−/− ES cells were labeled with l-[14C] valine for 24 h, and degradation of long-lived proteins during a 2-h incubation in Hanks' solution or the complete ES medium was measured as described in Materials and Methods. Chloroquine (Chl), bafilomycin A1 (Baf), or 3-MA was added as indicated. Data are the mean ± SD of triplicates from representative experiments. (B) ES clones and transformants were labeled with l-[14C] valine for 24 h and degradation of long-lived proteins during a 2-h incubation in Hanks' solution was measured.
3-MA, a commonly used inhibitor of autophagic sequestration (Seglen and Gordon 1982), reduced bulk degradation during amino acid deprivation even more markedly than did the disruption of the APG5 gene (Fig. 3 A). Furthermore, the effect of 3-MA was still observed in autophagy-negative APG5−/− cells, suggesting that 3-MA may have some additional effects on protein degradation other than autophagy.
Apg5 Colocalizes with Punctate LC3 Structures
Since the electron microscopic analysis of APG5−/− cells suggested that Apg5 acts at an early stage of autophagosome formation, we examined the subcellular distribution of Apg5 in wild-type ES cells. The Apg12-Apg5 conjugates were recovered primarily in the cytosolic fraction, with a very small portion found in the membrane fraction (see Fig. S1 in the Online Supplement). The total amount and subcellular distribution of the Apg12-Apg5 conjugates did not change significantly after amino acid starvation.
We further examined the localization of Apg5 by confocal microscopy. Bulk protein degradation (Fig. 3 B) and autolysosome formation (see Fig. 6) were restored in the GFP24 clone, indicating that GFP-fused Apg5 (GFP-Apg5) was functional and that the physiological localization of Apg5 can be indicated by observing the GFP signal. Under nutrient-rich conditions, most GFP-Apg5 was found to distribute evenly throughout the cytoplasm, with few punctate spots (Fig. 4, 0 min). 30 min after the medium was replaced with amino acid–free medium, the number of the punctate structures increased, which continued at 60 and 120 min. These GFP-Apg5 punctate spots were overlapped with anti–Apg12 antibody staining, indicating that GFP-Apg5 on these structures were conjugated with Apg12 (Fig. 5 A). 3-MA and wortmannin, an inhibitor of phosphatidylinositol 3-kinases, which were known to inhibit autophagy, suppressed GFP-Apg5 spot induction during amino acid deprivation (Fig. 4). APG5 +/+ cells stably expressing GFP alone displayed only cytoplasmic staining (data not shown).
Figure 6.
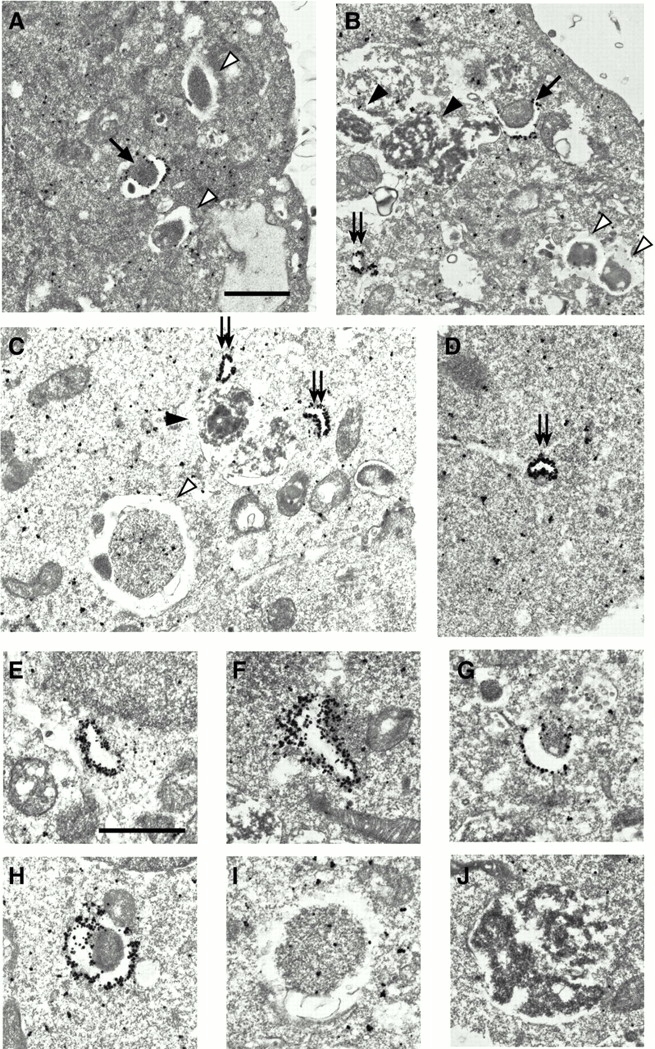
GFP-Apg5 is present on isolation membranes. GFP24 (A–C, E–J) or GKR-1 (D) cells were cultured in Hanks' solution for 2 h and fixed. The localization of GFP-Apg5 was examined by silver-enhanced immunogold electron microscopy using an anti–GFP antibody. In B, an isolation membrane is enclosing a mitochondrion. The isolation membranes (arrows), autophagosomes (open arrowheads) and autolysosomes (closed arrowheads) are indicated. Double arrows indicate small membrane compartments to which GFP-Apg5 extensively localizes. The typical images of these structures are shown at higher magnification (E–J). Bars, 1 μm.
Figure 4.
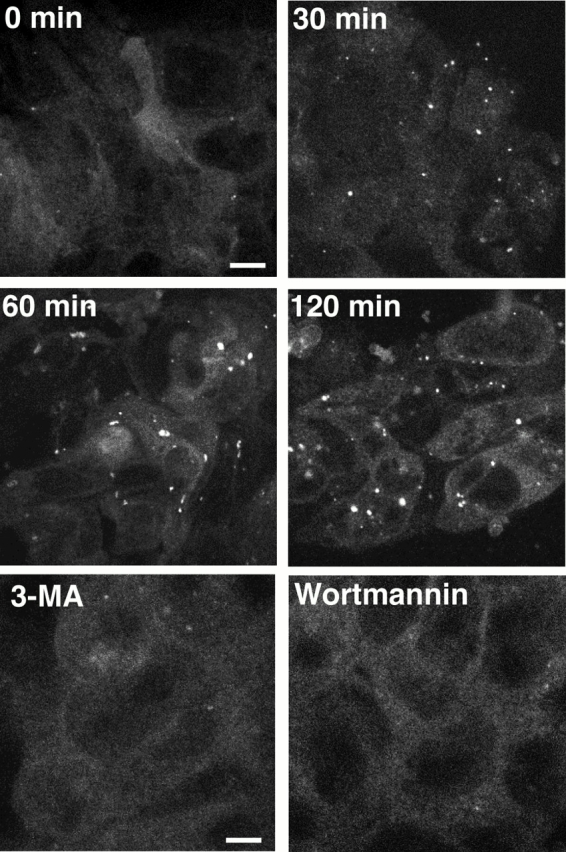
Punctate signals of GFP-Apg5 increase under starvation conditions. (A) GFP24 cells were cultured in Hanks' solution for the times indicated or in Hanks' solution with 10 mM 3-MA or 100 nM wortmannin for 120 min and examined by confocal microscopy. Bars, 10 μm.
Figure 5.
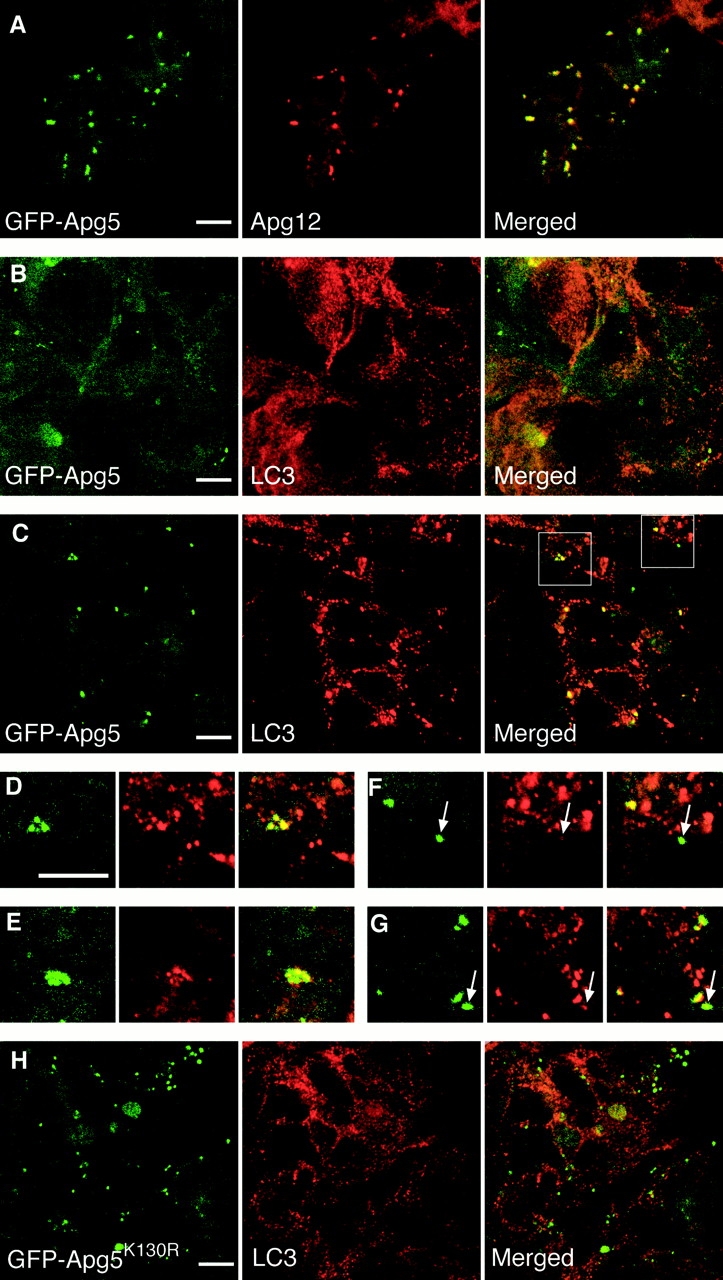
Apg12-Apg5 colocalizes with part of LC3. GFP24 (A–G) or GKR1 (H) cells were cultured in complete ES medium (B) or in Hanks' solution for 2 h (A and C–H). The cells were fixed, permeabilized, and subjected to immunofluorescence confocal microscopy using anti–mouse Apg12 antibody (A) or anti–LC3 antibody (B–H) and Cy5-conjugated goat anti–rabbit IgG antibody. GFP-Apg5 labeling (left), Apg12 or LC3 staining (middle), and merged images (right) are shown. (D and F) Higher magnifications of images shown in C. E and G are from another image. (F and G) Arrows indicate structures showing high levels of GFP-Apg5 but low levels of LC3. Bars, 10 μm.
We next counterstained endogenous LC3 to use as a specific marker for autophagosome. LC3 is a mammalian homologue of Aut7/Apg8 (Lang et al. 1998; Kirisako et al. 1999) and is the only mammalian protein that has been identified to localize to the autophagosome membrane as well as to the cytosol (Kabeya et al. 2000). In cells grown in complete medium, LC3 was stained as a cytoplasmic pattern with very fine punctate structures throughout the cytoplasm (Fig. 5 B), as reported previously (Kabeya et al. 2000). Amino acid starvation induced large dots of LC3 staining, which could represent autophagic vacuoles (Fig. 5 C). In the starved cells, GFP-Apg5 well colocalized with LC3 structures (Fig. 5, C–G), although some GFP-Apg5 spots were very weak for LC3 staining (Fig. 5F and Fig. G, arrows). On the other hand, there were many spots that were positive for LC3 staining but negative for GFP-Apg5. The colocalization of Apg5 with part of LC3 suggests that Apg12-Apg5 is targeted to certain restricted structures in the autophagic pathway.
Apg5 Localizes to Isolation Membranes
To examine the localization of Apg5 in more detail, we carried out immunoelectron microscopy using an anti–GFP antibody. Under nonstarvation conditions, silver-enhanced gold particles showed mostly cytoplasmic distribution (data not shown). In cells subjected to a 2-h starvation period, gold particles were found extensively associated with isolation membranes (Fig. 6A, Fig. B, and Fig. G). Their distribution was asymmetric; most of the gold particles were found on the outer side of isolation membranes, with a few on the inner membrane. In contrast to the isolation membrane, almost no GFP-Apg5 signal was detected on the autophagosome and autolysosome membranes (Fig. 6, A–C, I, and J). Apg5 associated with an isolation membrane that was close to completing circularization (Fig. 6 H), suggesting that Apg12-Apg5 detaches from the membrane immediately before or after autophagosome formation is completed.
In addition to typical isolation membranes, there were some smaller structures that were labeled throughout with gold particles (Fig. 6B and Fig. C, double arrows, and E and F). These single membrane-bound compartments adopted a crescent shape and were induced by amino acid starvation. Occasionally, small vesicles were observed inside these compartments. The curved morphology suggests that these structures represent the early stage of the development of isolation membrane.
To determine whether the isolation membranes are derived from the newly generated, Apg5-associated small compartment, we carried out time-lapse video microscopy. Small dots of GFP-Apg5 appeared and elongated slowly, and then bent and became semi-spherical structures (Fig. 7 A and videos 1–3). Finally, the GFP signal disappeared when the structure was close to forming a complete spherical body. Such a pattern of development is consistent with the electron microscopic images (Fig. 6, E–J). The time-lapse microscopy showed that the GFP-Apg5 spots were transient, appearing at random times during amino acid starvation and disappearing immediately before or after circularization, never forming any stable structures (Fig. 7 B). The average lifetime of the GFP-Apg5 structures was 9.7 ± 1.8 min.
Apg12 Conjugation Is Not Required for Membrane Targeting of Apg5, but Is Essential for Membrane Elongation
To investigate the role of Apg12 conjugation of Apg5, we generated an APG5−/− clone expressing GFP-Apg5K130R (GKR-1). Western blot analysis confirmed that GFP-Apg5K130R was not conjugated with Apg12 (data not shown). In amino acid–starved GKR-1 cells, numerous punctate GFP signals were observed (Fig. 5 H). The GFP-Apg5K130R dots were slightly increased in number, about twice as many as those in starved wild-type (GFP24) cells. Apg12 was not recruited to the GFP-Apg5K130R spots (data not shown). Immunoelectron microscopy demonstrated that the crescent-shaped compartments were generated, to which GFP-Apg5K130R localized (Fig. 6 D). These results clearly indicate that Apg12 conjugation is not required for membrane association of Apg5. However, autolysosomes were not detected in starved GKR-1 cells. Time-lapse video microscopy demonstrated that the GFP-Apg5K130R-associated spots did not mature into cup-shaped structures and basically remained as punctate spots (Fig. 7 A and video 4). These results suggest that Apg12 conjugation is required for involvement of Apg5 in elongation of the membrane to form cup-shaped isolation membrane and autophagosome. This is in agreement with the data that bulk protein degradation was not restored in KR27 cells (Fig. 3 B).
Targeting of LC3 to Isolation Membrane Depends on Apg12-Apg5
We observed some autophagosome-like structures in amino acid–starved APG5−/− cells (Fig. 2). We thus determined whether LC3 normally localized to these membranes. In amino acid–starved wild-type ES cells, LC3 staining was observed on both inner and outer membranes of autophagosomes (Fig. 8A and Fig. B) and isolation membranes (C), as well as in the cytosol. However, no LC3 staining was detected on the autophagosome-like structures in APG5−/− cells (Fig. 8D and Fig. E). In immunofluorescence microscopy, large dots of LC3 staining were not observed in starved APG5−/− cells (data not shown), in contrast to wild-type cells (Fig. 5 C). We also examined the localization of LC3 in the GKR-1 clone. While GFP-Apg5K130R localized to punctate spots as mentioned above, LC3 did not colocalize with the GFP-Apg5K130R spots and retained its cytoplasmic staining during starvation (Fig. 5 H). Taken together, these results suggest that Apg5 and its modification with Apg12 play important roles in recruitment of LC3 to isolation membranes.
Figure 8.
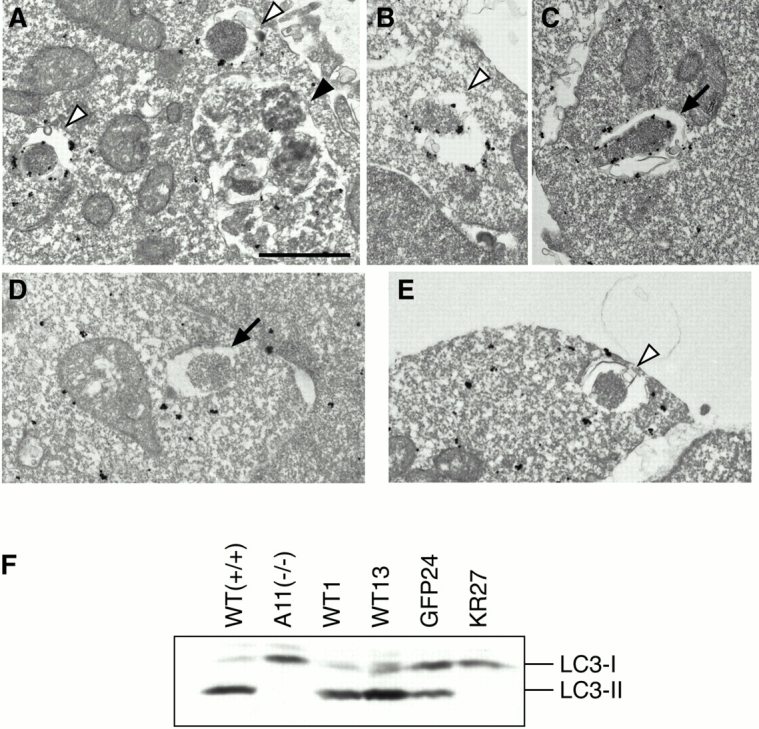
LC3 does not target to membrane in APG5−/− cells. Wild-type ES cells (A–C) and APG5−/− cells (D and E) were cultured in Hanks' solution for 2 h and fixed. Localization of LC3 was examined by silver-enhanced immunogold electron microscopy using an antibody against recombinant LC3. The isolation membrane (arrow), autophagosomes (open arrowheads) and autolysosome (closed arrowhead) are indicated. Bar, 1 μm. (F) Total lysates from APG5 +/+ and APG5 −/− cells and their various stable transformants cultured in Hanks' solution for 2 h were subjected to immunoblot analysis with anti–LC3 antibody. Positions of LC3-I and LC3-II are indicated.
We previously demonstrated that LC3 is post-translationally modified to generate LC3-I, and then LC3-II. LC3 on autophagic vacuoles is in the LC3-II form, while LC3-I is cytosolic (Kabeya et al. 2000). In amino acid–starved ES cells, significant amounts of LC3-II were generated. However, LC3-II was not detected in APG5 −/− cells (Fig. 8 F). LC3-II generation was restored in the stable transformants with wild-type Apg5 cDNA, but not with the Apg5K130R mutant. These biochemical data confirmed that without Apg12-Apg5, LC3 could not target to the membranes.
Discussion
We have genetically dissected autophagic process in yeast and now are a getting better understanding of the process. However, many questions remain to be addressed. How the isolation membranes are formed is one of the important yet poorly understood subjects. In this study, we demonstrated that the cup-shaped isolation membranes are developed from the crescent-shaped small membrane compartments using Apg5 as a marker molecule in mouse cells. Serial sections revealed that the crescent-shaped structures were not parts of mature isolation membranes (data not shown). These structures have never been recognized unless labeled with GFP-Apg5, but they are likely to be direct precursors of the autophagosome, as they were shown to mature into cup-shaped structures (Fig. 7). We were able to visualize the autophagosome formation in real time for the first time. The results directly evidenced the elongation model of isolation membrane, which has been hypothesized hitherto based on electron microscopic observations. The video microscopic analysis clearly demonstrated that the cisternae membranes elongate, bend, and form spherical autophagosomes, which is a quite unique membrane movement within cells. The molecular mechanism for such membrane dynamics is interesting, and studies on Apg proteins on the membrane would provide us further insights.
The existence of autophagosome precursors was proposed earlier by Seglen 1987 and Fengsrud et al. 1995. They observed thick, osmiophilic membrane multilayers in isolated hepatocytes by conventional electron microscopy, and postulated that they were autophagosome precursors. These structures, termed phagophores, were morphologically different in some respects from the Apg5-associated compartment. However, these two structures might be essentially the same, with such differences due to different fixation methods and different cell types. Our direct evidence indicated that isolation membranes are maturated from the small compartments, not simply derived from large preexisting membranes, although we do not know at this stage whether the small compartments are formed de novo or derived from some membrane sources. Many questions regarding autophagosome formation would be clarified by using Apg5 as a marker for autophagosome precursors. For example, our results indicate that phosphatidylinositol 3-kinase activity is required for generation of the Apg5-associated small compartment rather than its maturation into autophagosome (Fig. 4). Isolation and characterization of these compartments would provide evidences about the origin of autophagosomal membrane, which has been the subject of controversy (Hirsimäki and Reunanen 1980; Dunn 1990; Yamamoto et al. 1990; Ueno et al. 1991).
During development of the isolation membranes, the distribution pattern of Apg5 becomes very characteristic. Most Apg5 associates with the outer side of the isolation membranes (Fig. 6), in contrast to the symmetrical distribution of LC3 (Fig. 8). Previous freeze-replica ultrastructural analysis pointed out differences between the outer and inner membranes of autophagosomes (Réz and Meldolesi 1980; Hirsimäki et al. 1982; Baba et al. 1995). The inner membrane is almost free of intramembrane particles, while the outer membrane has a small, but significant, number of particles. The unique distribution of Apg5 is the first evidence that the molecular composition of the two membranes is different. It further suggests that such molecular asymmetry is already generated before completion of autophagosome formation. Apg5 must dissociate from the membrane immediately before or after completion of autophagosome formation, since almost no Apg5 was detected on the autophagosomes and autolysosomes (Fig. 9 A).
Figure 9.
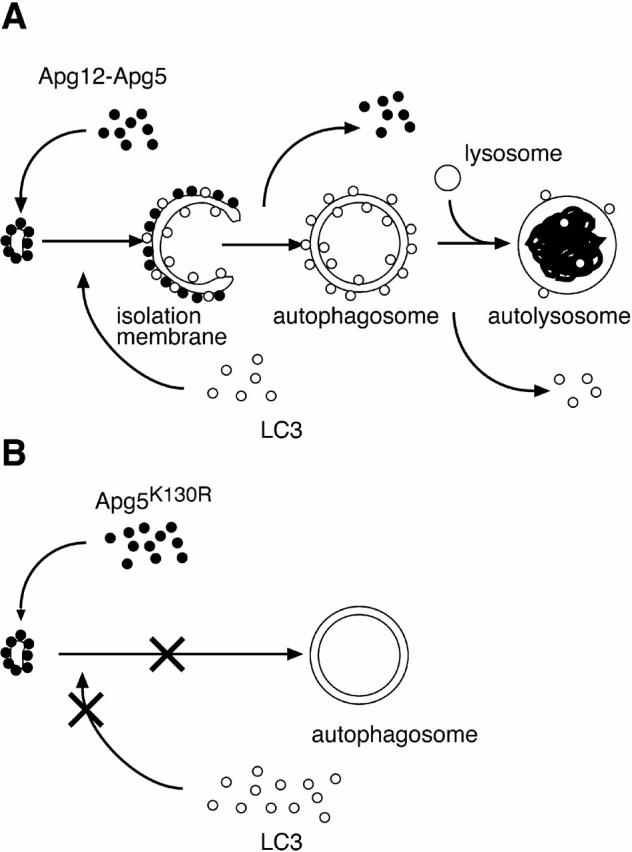
Model of subcellular localization of Apg12-Apg5 and LC3, and role of Apg12 conjugation in autophagosome formation. (A) Apg12-Apg5 conjugate localizes to the crescent-shaped autophagosome precursors. While these structures elongate and maturate into cup-shaped isolation membranes, LC3 is recruited to the membrane in the Apg5-dependent manner and Apg5 changes its localization to the outer side of the membrane. Apg5 plays an essential role in this membrane development. Immediately before or after the completion of autophagosome formation, Apg5 detaches from the membrane. Some LC3 also dissociate from the autophagosomal membrane thereafter. (B) Apg12 conjugation is not required for membrane targeting of Apg5, but it is essential for maturation of the isolation membrane into autophagosome and recruitment of LC3 to the membrane.
Based on the localization of Apg5, we postulate that Apg5 functions during development of isolation membranes. This hypothesis was supported by the phenotype of the Apg5 null mutant cells: the number of the isolation membranes and autophagosomes is reduced, and the rarely observed isolation membrane- and autophagosome-like structures are not labeled with LC3. We also examined the role of Apg12 conjugation by generating the stable clones expressing only conjugation-deficient Apg5K130R. Some ubiquitin-like modification systems affect intracellular localization of substrate proteins. For example, protein ubiquitination is accompanied by proteasome targeting or endocytosis (Hershko and Ciechanover 1998). SUMO-1 modification of Ran-GAP1 leads it to the nuclear pore complex (Hochstrasser 2000). However, our data suggest that Apg12 is not required for membrane association of Apg5. Rather, Apg12 conjugation of Apg5 is required for dynamic change of membrane morphology. We concluded that Apg5, by forming a conjugate with Apg12, directs elongation of the isolation membranes from the small crescent compartments (Fig. 9 B). How Apg5 is involved in membrane elongation is not known, but at least there are two possibilities. First, Apg12-Apg5 transports membranes or lipids from somewhere to the isolation membranes. Second, Apg12-Apg5 association with the membrane regulates acquisition of additional membranes. We showed in the present study that cytosolic LC3 targets to the isolation membranes in an Apg12-Apg5–dependent manner (Fig. 9 B). Therefore, membrane elongation would be related to recruitment of LC3 to the membrane. Whether there is a direct interaction between Apg12-Apg5 and LC3 is not clear; coimmunoprecipitation of these molecules has thus far been unsuccessful (data not shown).
This study provided us the first mammalian clone with genetically impaired autophagic activity. Although some autophagosome-like structures are occasionally seen in APG5−/− cells, LC3 staining revealed that they are not normal autophagosomes, suggesting that impaired autophagosome formation results in a complete block in the autophagic pathway. Based on the data in Fig. 3, autophagy would account for ∼60–70% of starvation-induced lysosomal protein degradation in ES cells. Lysosomal degradation is still clearly enhanced by starvation in APG5−/− cells. The process responsible for this is unknown. So far, at least two pathways other than macroautophagy have been proposed for delivery of cytoplasmic constituents to lysosomes. One is microautophagy, in which small portions of cytoplasm are sequestered into lysosomes by invagination of the lysosomal membrane (Marzella and Glaumann 1987). The other is chaperon-mediated autophagy, proposed by Dice 1990. Although it remains to be determined, these pathways may account for the remaining degradation activity of APG5−/− cells. In any case, this clone would be useful to study physiological meanings of autophagy in mammals, which would be more complex than in unicellular eukaryotes.
In conclusion, our present study suggests that Apg12-Apg5 localizes to autophagosome precursors, and plays an essential role in their development into autophagosomes, in cooperation with LC3. Establishment of the Apg5 null mutant cells not only demonstrated the indispensable role of Apg5, but also provided us new insights into the early stages of autophagosome formation that have been poorly understood both in yeast and mammals even since the discovery of autophagy in the 1960s.
Supplemental Material
Acknowledgments
We thank Dr. M. Kadowaki for helpful discussions and suggestions on the protein degradation assay and Dr. A. Bradley for kind donation of the pPGKpurobp1 plasmid. We also thank Drs. Y. Tanaka, M. Himeno, and E. Kominami for kind donation of antibodies, and S. Okamoto for antibody preparation.
This work was supported in part by grants-in-aid for Scientific Research from the Ministry of Education, Science, Culture and Sports of Japan.
Footnotes
The online version of this article contains supplemental material.
Abbreviations used in this paper: 3-MA, 3-methyladenine; ES cells, embryonic stem cells; GFP, green fluorescent protein.
References
- Baba M., Ohusumi M., Ohsumi Y. Analysis of the membrane structure involved in autophagy in yeast by freeze-replica method. Cell Struct. Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- Bolender R.P., Weibel E.R. A morphometric study of the removal of phenobarbital-induced membranes from hepatocytes after cessation of treatment. J. Cell Biol. 1973;56:746–761. doi: 10.1083/jcb.56.3.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke P.G. Developmental cell deathmorphological diversity and multiple mechanisms. Anat. Embryol. 1990;181:195–213. doi: 10.1007/BF00174615. [DOI] [PubMed] [Google Scholar]
- Dice J.F. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biol. Sci. 1990;15:305–309. doi: 10.1016/0968-0004(90)90019-8. [DOI] [PubMed] [Google Scholar]
- Dunn W.A. Studies on the mechanisms of autophagyformation of the autophagic vacuole. J. Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn W.A. Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Fengsrud M., Roos N., Berg T., Liou W., Slot J.W., Seglen P.O. Ultrastructural and immunocytochemical characterization of autophagic vacuoles in isolated hepatocyteseffects of vinblastine and asparagine on vacuole distributions. Exp. Cell Res. 1995;221:504–519. doi: 10.1006/excr.1995.1402. [DOI] [PubMed] [Google Scholar]
- George M.D., Baba M., Scott S.V., Mizushima N., Garrison B.S., Ohsumi Y., Klionsky D.J. Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol. Biol. Cell. 2000;11:969–982. doi: 10.1091/mbc.11.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond E.M., Brunet C.L., Johnson G.D., Parkhill J., Milner A.E., Brady G., Gregory C.D., Grand R.J.A. Homology between a human apoptosis specific protein and the product of APG5, a gene involved in autophagy in yeast. FEBS Lett. 1998;425:391–395. doi: 10.1016/s0014-5793(98)00266-x. [DOI] [PubMed] [Google Scholar]
- Hariri M., Millane G., Guimond M.P., Guay G., Dennis J.W., Nabi I.R. Biogenesis of multilamellar bodies via autophagy. Mol. Biol. Cell. 2000;11:255–268. doi: 10.1091/mbc.11.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano M., Aoki T., Dezawa M., Yusa S., Iitsuka Y., Koseki H., Taniguchi M., Tokuhisa T. A novel pathogenesis of megacolon in Ncx/Hox11L.1 deficient mice. J. Clin. Invest. 1997;100:795–801. doi: 10.1172/JCI119593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hirsimäki P., Reunanen H. Studies on vinblastine-induced autophagocytosis in mouse liver. II. Origin of membranes and acquisition of acid phosphatase. Histochemistry. 1980;67:139–153. doi: 10.1007/BF00493232. [DOI] [PubMed] [Google Scholar]
- Hirsimäki Y., Hirsimäki P., Lounatmaa K. Vinblastine-induced autophagic vacuoles in mouse liver and Ehrlich ascites tumor cells as assessed by freeze-fracture electron microscopy. Eur. J. Cell Biol. 1982;27:298–301. [PubMed] [Google Scholar]
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2000;2:E153–E157. doi: 10.1038/35019643. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO (Eur. Mol. Biol. Organ.) J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Dalton V.M., Eggerton K.P., Scott S.V., Klionsky D.J. Apg7p/Cvt2p is required for the Cvt, macroautophagy, and peroxisome degradation pathway. Mol. Biol. Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell. Dev. Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Kominami E., Hashida S., Khairallah E.A., Katunuma N. Sequestration of cytoplasmic enzymes in an autophagic vacuole-lysosomal system induced by injection of leupeptin. J. Biol. Chem. 1983;258:6093–6100. [PubMed] [Google Scholar]
- Lang T., Schaeffeler E., Bernreuther D., Bredschneider M., Wolf D.H., Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X.H., Jackson S., Seaman M., Brown K., Kempkes B., Hibshoosh H., Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1 . Nature. 1999;402:672–676. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- Marzella L., Glaumann H. Autophagy, microautophagy and crinophagy as mechanisms for protein degradation. In: Glaumann H., Ballard F.J., editors. LysosomesTheir Role in Protein Breakdown. Academic Press; London, UK: 1987. pp. 319–367. [Google Scholar]
- Masaki R., Yamamoto A., Tashiro Y. Cytochrome P-450 and NADPH-cytochrome P-450 reductase are degraded in the autolysosomes in rat liver. J. Cell Biol. 1987;104:1207–1215. doi: 10.1083/jcb.104.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Ohsumi Y. Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:3888–3896. doi: 10.1093/emboj/18.14.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. A protein conjugation system essential for autophagy Nature 395 1998. 395 398a [DOI] [PubMed] [Google Scholar]
- Mizushima N., Sugita H., Yoshimori T., Ohsumi Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy J. Biol. Chem. 273 1998. 33889 33892b [DOI] [PubMed] [Google Scholar]
- Mortimore G.E., Pösö A.R. Intracellular protein catabolism and its control during nutrient deprivation and supply. Annu. Rev. Nutr. 1987;7:539–564. doi: 10.1146/annurev.nu.07.070187.002543. [DOI] [PubMed] [Google Scholar]
- Nishino I., Fu J., Tanji K., Yamada T., Shimojo S., Koori T., Mora M., Riggs J.E., Oh S.J., Koga Y. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- Ogier-Denis E., Houri J.J., Bauvy C., Codogno P. Guanine nucleotide exchange on heterotrimeric Gi3 protein controls autophagic sequestration in HT-29 cells. J. Biol. Chem. 1996;271:28593–28600. doi: 10.1074/jbc.271.45.28593. [DOI] [PubMed] [Google Scholar]
- Okazaki I., Himeno M., Ezaki J., Ishikawa T., Kato K. Purification and characterization of an 85 kDa sialoglycoprotein in rat liver lysosomal membranes. J. Biochem. (Tokyo) 1992;111:763–769. doi: 10.1093/oxfordjournals.jbchem.a123833. [DOI] [PubMed] [Google Scholar]
- Pfeifer U. Functional morphology of the lysosomal apparatus. In: Glaumann H., Ballard F.J., editors. LysosomesTheir Role in Protein Breakdown. Academic Press; London, UK: 1987. pp. 3–59. [Google Scholar]
- Réz G., Meldolesi J. Freeze-fracture of drug-induced autophagocytosis in the mouse exocrine pancreas. Lab. Invest. 1980;43:269–277. [PubMed] [Google Scholar]
- Seglen P.O. Regulation of autophagic protein degradation in isolated liver cells. In: Glaumann H., Ballard F.J., editors. LysosomesTheir Role in Protein Breakdown. Academic Press; London, UK: 1987. pp. 371–414. [Google Scholar]
- Seglen P.O., Bohley P. Autophagy and other vacuolar protein degradation mechanisms. Experientia. 1992;48:158–172. doi: 10.1007/BF01923509. [DOI] [PubMed] [Google Scholar]
- Seglen P.O., Gordon P.B. 3-Methyladeninespecific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc. Natl. Acad. Sci. USA. 1982;79:1889–1892. doi: 10.1073/pnas.79.6.1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Mizushima N., Ogawa Y., Matsuura A., Noda T., Ohsumi Y. Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:5234–5241. doi: 10.1093/emboj/18.19.5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Guhde G., Suter A., Eskelinen E.-L., Hartmann D., Lullmann-Rauch R., Janssen P.M.L., Blanz J., von Figura K., Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- Tanida I., Mizushima N., Kiyooka M., Ohsumi M., Ueno T., Ohsumi Y., Kominami E. Apg7p/Cvt2pa novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thumm M., Egner R., Koch B., Schlumpberger M., Straub M., Veenhuis M., Wolf D.H. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae . FEBS Lett. 1994;349:275–280. doi: 10.1016/0014-5793(94)00672-5. [DOI] [PubMed] [Google Scholar]
- Tsukada M., Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae . FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- Ueno T., Muno D., Kominami E. Membrane markers of endoplasmic reticulum preserved in autophagic vacuolar membranes isolated from leupeptin-administered rat liver. J. Biol. Chem. 1991;266:18995–18999. [PubMed] [Google Scholar]
- Yamamoto A., Masaki R., Tashiro Y. Characterization of the isolation membranes and the limiting membranes of autophagosomes in rat hepatocytes by lectin cytochemistry. J. Histochem. Cytochem. 1990;38:573–580. doi: 10.1177/38.4.2319125. [DOI] [PubMed] [Google Scholar]
- Yoshimori T., Yamagata F., Yamamoto A., Mizushima N., Kabeya Y., Nara A., Miwako I., Ohashi M., Ohsumi M., Ohsumi Y. The mouse SKD1, a homologue of yeast Vps4p, is required for normal endosomal trafficking and morphology in mammalian cells. Mol. Biol. Cell. 2000;11:747–763. doi: 10.1091/mbc.11.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Strømhaug P.E., Dunn W.A., Jr. Glucose-induced microautophagy of peroxysomes in Pichia pastoris requires a unique E1-like protein. Mol. Biol. Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.