Summary
Long range interactions between distant regulatory elements, such as enhancers, and their target genes underlie the specificity of gene expression in many developmentally regulated gene families. NLI/Ldb1, a widely expressed nuclear factor, is a potential mediator of long range interactions. Here we show that NLI/Ldb1 and erythroid binding partners GATA-1/SCL/Lmo2 bind in vivo to the β-globin locus control region (LCR). The C-terminal LIM interaction domain of NLI is required for formation of the complex on chromatin. Loss of the LIM domain converts NLI into a dominant negative inhibitor of globin gene expression, and knock down of NLI using shRNA results in failure to activate β-globin expression. Kinetic studies reveal the NLI/GATA-1/SCL/LMO2 complex is detected at the β-globin promoter coincident with RNA pol II recruitment, β-globin transcription and chromatin loop formation during erythroid differentiation providing evidence that NLI may facilitate long range gene activation.
Introduction
In metazoans, regulatory elements such as enhancers and locus control regions (LCR) can be located at great distances from the genes they regulate. Recent evidence suggests that LCRs and their distant target genes come into close proximity during gene activation with the exclusion of intervening chromatin. For example, in the β-globin locus, the LCR is proposed to exist in a ‘chromatin hub’ with actively transcribing globin genes in erythroid cells (Tolhuis et al., 2002; Palstra et al., 2003). The erythroid activators EKLF, GATA-1 and FOG-1 are required for the close interactions (Drissen et al., 2004; Vakoc et al., 2005). The intra-chromosomal associations are not restricted to the globin loci but have been observed at the TH2 cytokine locus (Spilianakis and Flavell, 2004), the Igf2/H19 imprinted locus (Lopes et al., 2003), the androgen receptor locus (Wang et al., 2005) and at the Ig kappa gene (Liu and Garrard, 2005). Furthermore the close interactions can exist between chromosomes to regulate TH1 and TH2 cytokine and odorant receptor choice during differentiation (Spilianakis et al., 2005; Lomvardas et al., 2006). These diverse examples of close regulatory interactions between genes and distant enhancers raise the possibility that in addition to cell type specific activators, common participants might be important to form the associations.
The widely expressed nuclear protein NLI (Nuclear LIM Interactor, also known as Ldb1 and CLIM2) is the human homologue of the Drosophila melanogaster protein, Chip, which was identified in a genetic screen for factors involved in long range activation of the cut gene (Morcillo et al., 1996; Morcillo et al., 1997). Chip is a LIM domain binding protein with no known enzymatic or DNA binding activity that has been hypothesized to act as a protein binding interface facilitating multiple interactions that may serve to bring distant sequences into proximity (Jurata et al., 1998; Dorsett, 1999). In Drosophila, the N-terminus of Chip directly interacts with the GATA factor Pannier and might act as a bridge molecule to the bHLH heterodimeric Achaete/Scute complex allowing activation of proneural genes (Ramain et al., 2000). Using the conserved C-terminal LIM interaction domain (LID), NLI/Ldb1 can interact with LIM-homeodomain or LIM-only (Lmo) proteins, such as Lmo2 [for review see (Matthews and Visvader, 2003)]. Chip contains a second conserved C-terminal domain through which it interacts with other factors including homeoproteins and the Drosophila insulator protein Su(HW) (Torigoi et al., 2000). In addition, Chip and NLI/Ldb1 have conserved N-terminal self-interacting domains (Agulnick et al., 1996; Jurata and Gill, 1997; Torigoi et al., 2000). The null phenotype of the murine Chip homologue Ldb1 exhibits pleiotrophic effects during mouse development, including failure of erythroid differentiation, consistent with a broad role in gene expression in mammals (Mukhopadhyay et al., 2003).
Earlier work showed that NLI/Ldb1 participates in a DNA-binding complex formed in murine erythroid cell extracts along with the hematopoietic transcription factors GATA-1, Lmo2 and the SCL/Tal1 (hereafter referred to as SCL)-E2A heterodimer (Wadman et al., 1997). The complex binds a compound E box/GATA motif in vitro presumably through SCL/E2A and GATA-1 interactions respectively (Wadman et al., 1997; Cohen-Kaminsky et al., 1998). NLI/Ldb1 functions locally as a positive regulator of late erythroid gene expression through the promoters of the EKLF, c-Kit, 4.2 and glycophorin A genes (Anderson et al., 1998; Lecuyer et al., 2002; Xu et al., 2003; Lahlil et al., 2004). NLI/Ldb1 can also play a repressive role in gene expression along with Rb, ETO2 or BRG1 proteins (Vitelli et al., 2000; Goardon et al., 2006; Xu et al., 2006). The members of the E box/GATA complex can be detected in vivo at an autoregulatory site upstream of the GATA-1 gene (Valverde-Garduno et al., 2004). Only the GATA-1 portion of an E box/GATA motif and not the E box was critical for complex interaction at this site. The complex also assembles as a function of developmental stage at sites upstream of the α-globin genes (Anguita et al., 2004) and NLI protein was detected both at the β-globin LCR and gene after differentiation of erythroid MEL cells (Brand et al., 2004). However, no functional role for the E box/GATA complex in long range globin gene regulation has been uncovered.
Here we show that the NLI/GATA-1/SCL/Lmo2 complex binds in vivo to the human β-globin locus control region (LCR) which is located far upstream of the globin genes and is required for their high level expression in erythroid cells (Stamatoyannopoulos, 2005). The C-terminal LIM interaction domain of NLI is required for formation of the complex on chromatin. Absent the LIM domain, NLI is a dominant negative inhibitor of globin gene expression. Knock down of NLI/Ldb1 using shRNA in mouse erythroleukemia cells results in failure to activate β-globin transcription after induction. The NLI/Ldb1 complex forms at the β-globin LCR in these cells and after induction of β-globin transcription it is detected at the promoter of the gene: these new associations coincide with increased proximity, or loop formation, between the LCR and gene. Furthermore, chromatin immunoprecipitation-combined loop assays show that NLI/Ldb1 forms the base of a loop between the LCR and gene. Thus, we uncover a positive role for the E box/GATA complex in β-globin gene regulation and provide evidence that NLI may be a facilitator of long range gene activation in mammals.
Results and Discussion
The human β-globin LCR DNase I hypersensitive sites are targets of E box/GATA binding proteins
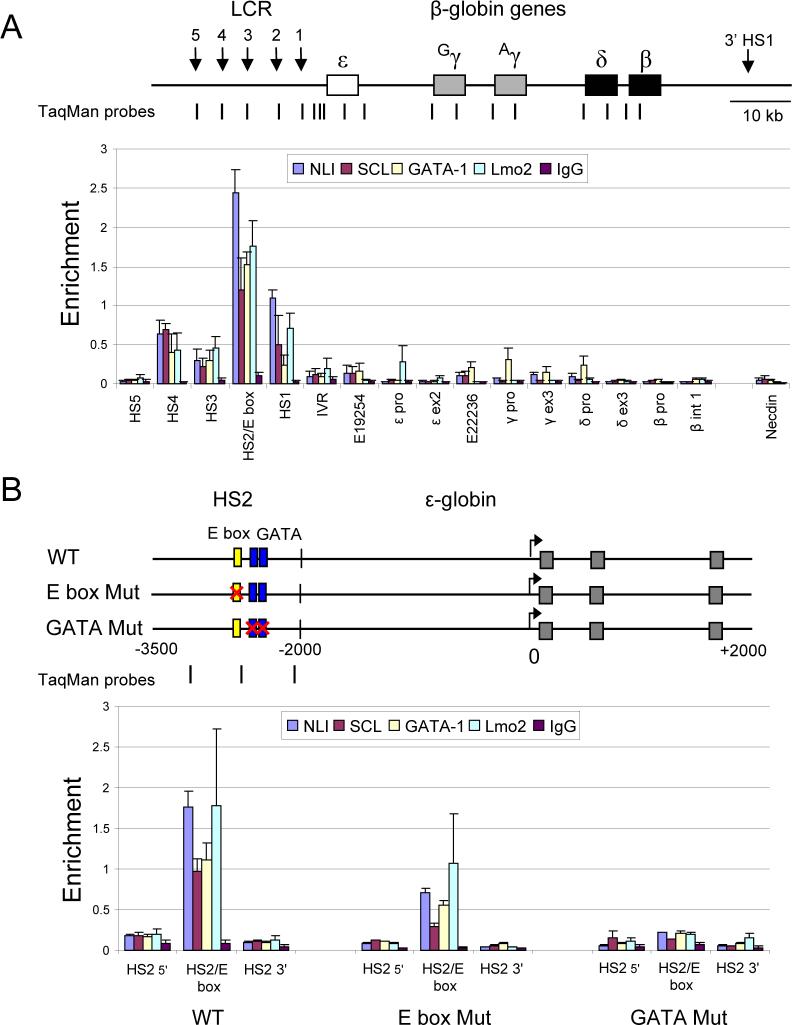
The large human β-globin locus contains five genes, embryonic ε, fetal Gγ and Aγ, and adult δ and β expressed sequentially during development under the influence of the upstream LCR (Figure 1A) which consists of a series of DNase I hypersensitive sites (HSS). A Transfac database search for compound E box/GATA motifs across the human β-globin locus revealed numerous candidate sites in which the motifs were separated by varying numbers of nucleotides (not shown). However, it is known that only a subset of GATA-1 sites are occupied in vivo in erythroid cells (Im et al., 2005). Thus, we performed chromatin immunoprecipitation (ChIP) assays to analyze the molecular pattern of the E box/GATA binding protein complex within the human β-globin locus in vivo (Figure 1A). Chromatin cross linked with 1% formaldehyde was prepared from human K562 cells that have a fetal-embryonic phenotype and express modest levels (compared to normal erythroid cells) of γ-globin transcripts, about 10-fold lower levels of ε-globin transcripts and trace amounts of δ-globin transcripts.
Figure 1. The human β-globin LCR is a natural target of E box/GATA binding proteins.
(A) The human β-globin locus is illustrated. Vertical arrows denote hypersensitive sites. The positions of TaqMan probes used for real-time qPCR are indicated below and named on the graph. Primer HS2/E box amplifies the HS2 E box/GATA motif (see Experimental Procedures). E19254 and E22236 are E box/GATA motifs upstream and downstream of the ε-globin gene, respectively. IVR denotes a position between the LCR and the ε-globin gene. Chromatin was prepared from 1% formaldehyde cross-linked K562 cells, digested with MNase and sonicated and then subjected to ChIP using antibodies to NLI, SCL, GATA-1, Lmo2 or control IgG. The results for at least three chromatin preparations are shown ± SEM. (B) A 1.46-kb HS2 fragment containing the HS2 core with 5' and 3' flanking sequences was linked to a complete genomic ε-globin gene on minichromosomes in the wild type (WT) construct. E box mutant and GATA mutant were identical except the E box or tandem GATA sites in the HS2 core were mutated by clustered point mutations. Gray boxes, exons of the gene. The positions of TaqMan primers used for real time PCR are indicated beneath the constructs. ChIP assays were performed using K562 cells stably carrying wild-type, E box mutant, or GATA mutant construct as described above. The results for at least three chromatin preparations are shown ± SEM.
Immunoprecipitation was carried out with antibodies to NLI, SCL, Lmo2 and GATA-1 and input and immunoprecipitated DNA were amplified by quantitative real-time PCR using TaqMan probes (Kim et al., 2007). As shown in Figure 1A, NLI binding was observed within the LCR HSS, especially in the HS2 core region that contains an E box and double GATA-1 motif. Prominent binding of GATA-1, Lmo2 and SCL co-localized with NLI, suggesting that the E box/GATA binding complex resides at this region. Very weak interaction of NLI, GATA-1 and SCL at two potential E box/GATA motifs 3' and 5' of the ε-globin gene and of GATA-1 at the γ and δ-globin promoters was observed. These data show robust binding of the E box/GATA complex is primarily detected in the human β-globin LCR in K562 cells.
To confirm that the E box/GATA complex proteins interact together on the same chromatin fragment, double ChIP experiments were performed. First, chromatin from K562 cells was immunoprecipitated with an anti-GATA-1 antibody. Bound complexes were eluted with 10mM DTT, followed by immunoprecipitation with anti-NLI, anti-Lmo2 or control IgG. Notably, enrichment of NLI and Lmo2 was detected along with GATA-1 at HS2, indicating that the E box/GATA complex forms at least at HS2 (Figure S1). Failure to detect the complex at other locations using sequential ChIP assays may be the result of the significantly lower levels of GATA-1, the first protein immunoprecipitated, present at those sites.
To identify the cis-elements through which the E box/GATA complex interacts at HS2, we compared the binding properties of wild type HS2 and HS2 mutated at specific sites. These studies could be conveniently carried out by using minichromosomes which link LCR-HS2 to ε-globin and are replicated and physiologically assembled into chromatin in K562 cells (Kim and Dean, 2003). NLI, SCL, GATA-1 and Lmo2 binding to HS2 was strongly detected when the ChIP assay was performed with cells carrying wild type HS2 minichromosomes, as expected (Figure 1B). To examine the requirements for detection of NLI on chromatin, we mutated the E box or double GATA-1 binding sites within HS2. The E box mutation reduced binding of the factors to between 30% and 62% of the WT level. However, binding of NLI, SCL, GATA-1, and Lmo2 were all strongly decreased to about 10∼20% of WT when the two tandem GATA sites were mutated, suggesting that although both motifs are important for formation of the E box/GATA complex on HS2 in vivo, GATA-1 binding is of critical importance.
Taken together, these data indicate that the human β-globin locus LCR is a natural target of the E box/GATA protein complex and that the complex interacts with chromatin through both E box and GATA-1 sites in HS2. This result contrasts with the observation that the complex interacts solely through the GATA-1 motif at an auto-regulatory site upstream of the GATA-1 gene (Vyas et al., 1999).
The E box/GATA protein complex has an important role in HS2 enhancer function
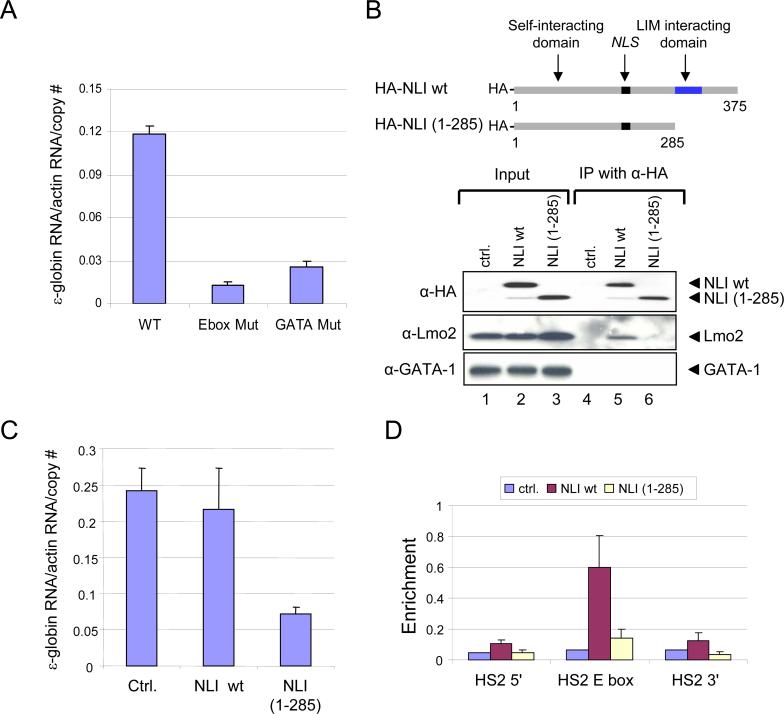
The β-globin LCR HSS subsume the enhancer activity of the LCR and collectively activate high levels of expression of the globin genes. HS2, independently, has strong activity in classical enhancer assays and contains well known cis-regulatory elements for NF-E2, GATA-1 and SCL that influence its activity (Stamatoyannopoulos and Grosveld, 2001). To investigate the importance of the E box/GATA protein complex to enhancer activity of HS2, we used RT-PCR to measure ε-globin transcription from minichromosomes where the gene was linked to WT HS2 or to HS2 containing the E box or GATA mutations (Figure 2A). Similar to the HS2 NF-E2 mutation, which completely eliminates enhancer activity (Gong et al., 1996), disruption of the E box or double GATA-1 sites essentially abolished ε-globin transcription, indicating that the interaction of the E box/GATA complex with both sites in chromatin is critical for the enhancer activity of HS2.
Figure 2. Enhancer activity of the E box/GATA complex on ε-globin expression.
(A) RT-qPCR analysis of ε-globin transcription in K562 cells carrying HS2ε minichromosomes with WT HS2 or with HS2 mutated to destroy the E box or GATA sites (see Figure 1B). Graphed are the averages of three RNA preparations ± SEM. Data for the different clones were normalized to the actin signal. (B) Wild-type (WT) or C terminal deleted NLI mutant (1−285) proteins are depicted. K562 cells were stably co-transfected with HS2ε minichromosomes (Kim and Dean, 2003) and HA-tagged-wild-type (WT) or C terminal deleted NLI mutant constructs (1−285). NLS, nuclear localization signal. Nuclear extract from stable cell lines were immunoprecipitated by anti-HA agarose antibody, and input proteins or precipitates were analyzed by western blot analysis with antibodies shown on the left. Ctrl., control plasmid without insert. (C) RNA was isolated from stable cell lines and minichromosomal ε-globin mRNA was detected by RNase protection assay. The mean ε-globin mRNA normalized to actin RNA (± SEM) for several clones of each type is illustrated graphically. (D) ChIP assays were carried out using stable cell lines expressing either HA-tagged WT or truncated (1−285) NLI with anti-HA antibody as described for Figure 1. The results for at least three chromatin preparations are presented (± SEM).
Next, we examined the function of NLI in globin gene expression. The C-terminal LIM interaction domain of NLI is required for binding to LIM domain proteins such as Lmo2 and through self-interaction NLI may bridge between LMO2 complexes bound to chromatin through GATA-1 and the SCL/E2A heterodimer (Visvader et al., 1997; Jurata and Gill, 1997). To address the functional importance of the C-terminal region of NLI to globin gene expression, minichromosomes expressing HA-tagged wild-type (WT) or C-terminal truncated NLI (1−285) were stably transfected into K562 cells. HA-tagged WT or mutant (1−285) NLI were expressed equivalently as determined by western blot analysis with anti-HA antibody (Figure 2B, lanes 1−3, upper panel). Using these stable cell lines, expression of ε-globin RNA was examined by RNase protection assay. We found that HA-NLI (1−285) exerts a dominant negative effect reducing ε-globin expression (Figure 2C).
To explore the importance of the NLI LIM domain for formation of the E box/GATA complex on HS2 chromatin, a ChIP assay was performed with anti-HA antibodies using chromatin from cells stably expressing HA-NLI-WT or HA-NLI (1−285). Interestingly, only NLI wild-type, but not NLI (1−285), was detected at HS2 in vivo (Figure 2D). Next, to examine why C-terminal truncated NLI cannot bind HS2 in vivo, we performed a co-immunoprecipitation assay with anti-HA antibody in wild type or mutant NLI expressing K562 cells. As shown in Fig. 2B (lane 5), HA-tagged wild type NLI was able to immunoprecipitate endogenous Lmo2. However, C-terminal truncated NLI failed to interact with Lmo2 (Figure 2B, lane 6), indicating a necessary role for the C-terminal region of NLI in complex formation with Lmo2. We did not observe direct interaction between NLI and GATA-1 by immunoprecipitation (Figure 2B, lanes 5) although biochemical evidence supports this interaction in Drosophila (Ramain et al., 2000). Taken together, the NLI LIM interaction domain is necessary for Lmo2 interaction and to co-localize NLI protein with other E box/GATA complex proteins at HS2.
These results indicate that both E box and GATA-1 sites in HS2 are important for the formation and activation function of the Ebox/GATA protein complex and that the LIM domain of NLI is required for interaction with Lmo2 and inclusion in the complex on chromatin.
Knock-down of NLI/Ldb1 by shRNA results in failure of MEL cells to activate β-globin expression
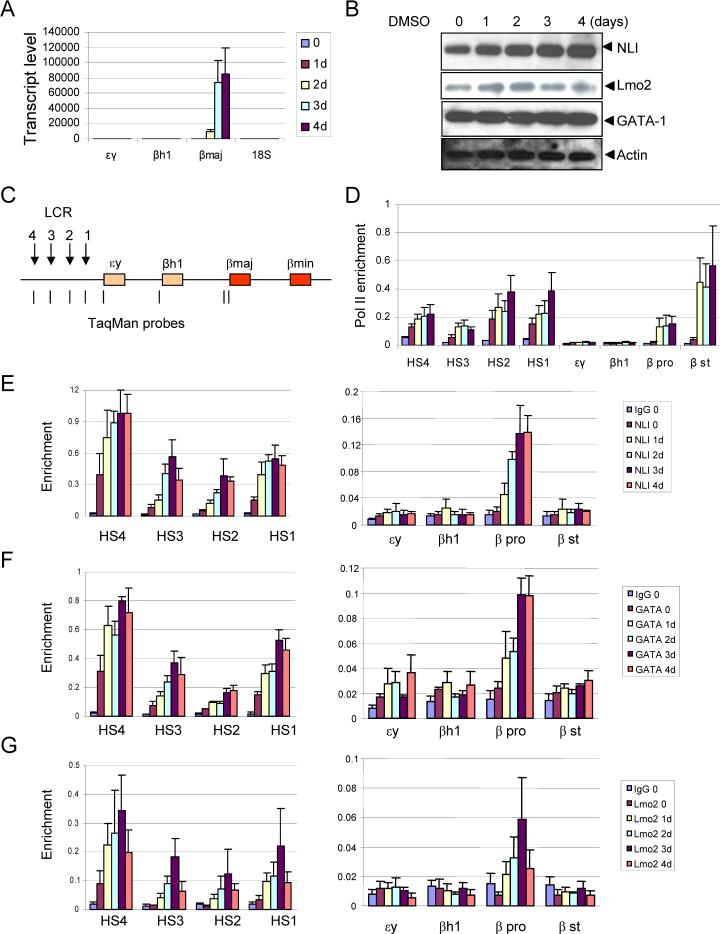
Similar to the human locus, the mouse β-globin locus has an upstream LCR (HS1-HS4) and four developmentally regulated genes, embryonic εy and βh1, and adult βmaj and βmin . MEL cells are erythroid precursors that are blocked at the pro-erythroblast stage. When induced to differentiate by treatment with DMSO, MEL cells undergo phenotypic changes that resemble the final stages of normal erythropoiesis and β-globin mRNA levels increase dramatically. During DMSO-induced differentiation, MEL cells continue to proliferate while accumulating hemoglobin and other erythroid specific proteins and then they arrest. We adopted this system in an effort to obtain a more robust and physiological picture of NLI function in β-globin regulation than offered by the K562 system.
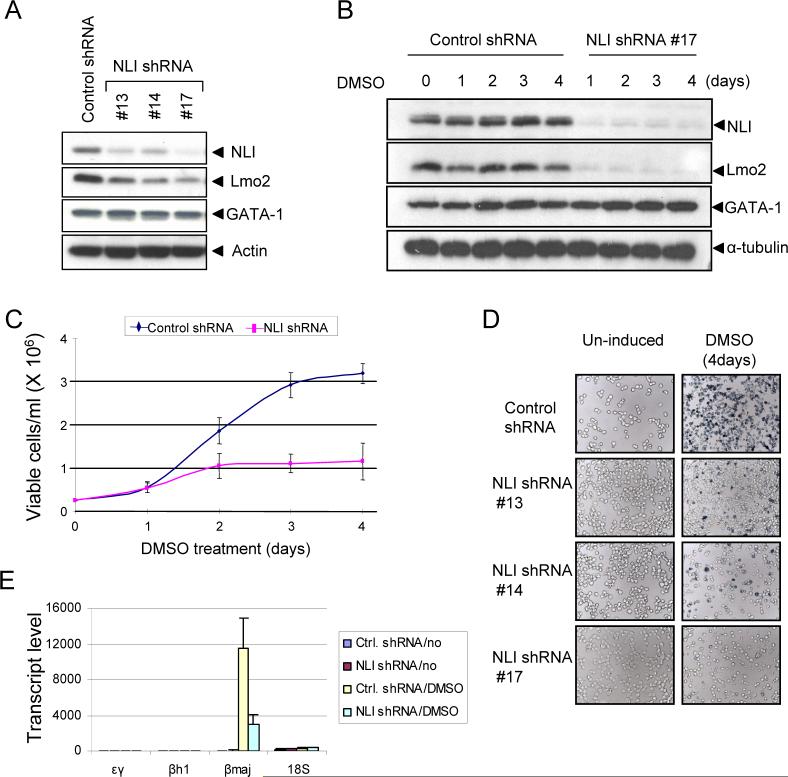
First, we reduced the expression of NLI/Ldb1 by RNA interference in MEL cells. MEL cells were stably transduced by a lentivirus expressing control or NLI-directed shRNA. Western blot analysis demonstrated decreased NLI expression in NLI knockdown cells compared with control shRNA expressing cells (Figure 3A). Next, stable clones expressing either control or NLI shRNA were differentiated in the presence of DMSO. NLI levels in cells expressing control shRNA were unaffected during induction and there was no detection of NLI in NLI knockdown cells (Figure 3B). Decreased NLI expression was associated with reduced proliferation of MEL cells (Figure 3C).
Figure 3. NLI knock-down inhibits MEL cell differentiation.
(A) Reduced NLI expression in stable NLI shRNA clones. MEL cells were stably transfected with control or NLI shRNA. Western blot analysis was performed with the indicated antibodies. Actin served as an internal control. #13, #14, and #17 represent individually derived NLI shRNA stable clones. (B) Western blot analysis of NLI expression during differentiation. Stable clones were treated with 1.5 % of DMSO and whole cell extracts were prepared at the indicated times. α-tubulin served as a loading control. Similar results were obtained from independent experiments with other clones. (C) Cell viability of stable clones during differentiation. Cells were cultured with 1.5 % DMSO, and cell viabilities were assessed daily by the trypan blue dye exclusion method (averages of 3 clones ± SEM). (D) Cells were treated with 1.5 % DMSO for 4 days and then hemoglobinization was assayed by staining with benzidine. (E) Stable clones with control or NLI shRNA were treated with or without 1.5% DMSO. Total RNA was isolated at day 4, and εy, βh1, and β-major mRNA expression was analyzed by quantitative real-time PCR (averages of 3 clones ± SEM). 18S ribosomal RNA served as a control.
We then determined whether reduced NLI expression affects terminal differentiation of MEL cells by measuring benzidine positivity, reflecting accumulation of hemoglobin in the cells. As shown in Figure 3D, DMSO treatment resulted in strong benzidine positivity, reflecting hemoglobin accumulation in control cells. However, DMSO-induced differentiation was abrogated in NLI knockdown cells which showed few or no benzidene positive cells. This reduction was confirmed by RT-PCR analysis indicating reduced induction of the β-globin mRNA by DMSO (Figure 3E). Together, these data indicate that NLI is essential for optimal proliferation and hemoglobin synthesis during MEL cell differentiation and this finding is consistent with results of targeted deletion of the Ldb1 gene in mice (Mukhopadhyay et al., 2003).
NLI but not GATA-1 stabilizes Lmo2 through the LIM domain
Lmo2 is an oncogenic protein involved in T cell leukemia (Larson et al., 1995; Grutz et al., 1998; Ferrando et al., 2002) and is necessary for hematopoietic development (Yamada et al., 1998). In addition, Lmo proteins down regulate LIM homeodomain (LIM-HD) transcription factor activity by competing for Ldb1 (Shoresh et al., 1998; Milan and Cohen, 1999). Thus, complex formation by NLI and Lmo2 is thought to be important to regulate the expression of numerous genes. Interestingly, Lmo2 detection was strongly associated with NLI expression. Most stable clones expressing NLI shRNA had reduced Lmo2 protein (Figure 3A). In addition, when induced to differentiate by DMSO, reduced Lmo2 expression was not restored in cells with stable NLI silencing (Figure 3B).
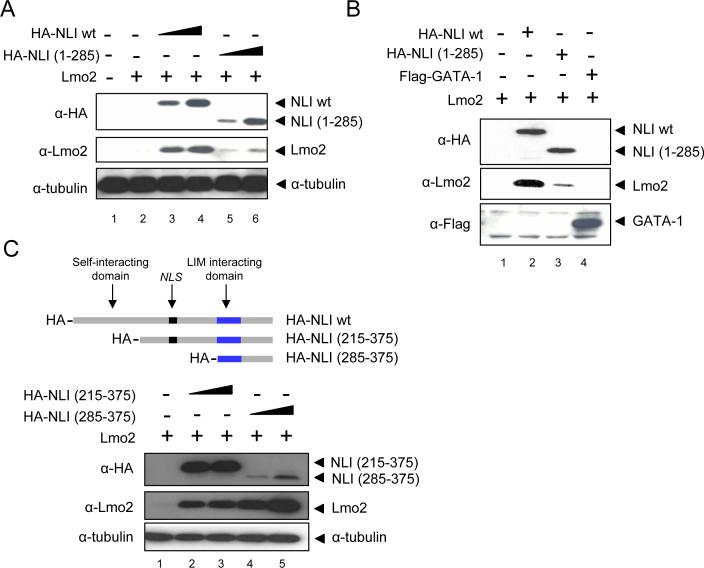
To further investigate the mechanism underlying the decrease in Lmo2 expression under conditions of reduced levels of NLI, we performed transient transfection assays in 293T cells which lack endogenous Lmo2 and GATA-1 expression. When an Lmo2-expressing plasmid alone was transfected, the protein could not be visualized (Figure 4A, lane 2). However, Lmo2 was robustly detected in a dose dependent manner after co-expression of wild-type NLI (Figure 4A, lane 3 and 4), indicating that Lmo2 protein might be stabilized by NLI. We next asked whether the C-terminal NLI LIM interacting domain is important for Lmo2 stabilization. Co-expression of truncated NLI lacking the LIM interacting domain failed to substantially stabilize Lmo2 (Figure 4A, lanes 5 and 6). Previously, it had been shown that Lmo2 can directly bind with GATA-1 in vitro (Osada et al., 1997). To elucidate whether GATA-1 also stabilizes Lmo2 protein, GATA-1 was co-expressed with Lmo2 and nuclear extracts were prepared. GATA-1 expression had no effect on Lmo2 stability (Figure 4B, lane 4). However, expression of NLI along with Lmo2 and GATA-1 permitted Lmo2 to co-immunoprecipitate GATA-1 (Figure S2).
Figure 4. Stabilization of Lmo2 protein through the LIM domain of NLI.
(A) Increasing amounts of wild type or mutant NLI (5ug and 10ug) expression vectors were co-transfected in to 293 cells with a fixed amount of Lmo2 construct (5ug). After 48hr, whole cell extracts were prepared. Western blotting was performed with the indicated antibodies. (B) Nuclear extracts prepared from 293T cells transfected with the indicated constructs for 48 hr. The immunoblot was incubated with antibodies shown on the left. (C) 293T cells were transfected with the indicated constructs for 48hr. Whole cell extracts were prepared and western blots were incubated with the indicated antibodies
To further define the region of NLI required to stabilize Lmo2, two HA-tagged versions of NLI with N-terminal truncations of different sizes were co-expressed with Lmo2 (Figure 4C). We found that C-terminal NLI (285−375) containing the LIM domain was sufficient to stabilize NLI. There was a relatively low signal for this peptide using HA antibodies and the level of peptide expressed by the vector could not be independently determined with the NLI antibody which is directed against the N-terminus. Since we do not suspect a difference in stoichiometry of interaction between Lmo2 and full length NLI versus C-terminal peptide NLI, the low signal may reflect inherent difficulties in immunoblotting the peptide or decreased reactivity of the HA antibodies.
Together, our results show that NLI forms a complex with Lmo2 through the C-terminal LIM domain containing region of NLI. The binding is important for Lmo2 stabilization and for E box/GATA binding complex formation to regulate β-globin gene expression.
Recruitment of the Ebox/GATA complex to the β-globin promoter after MEL cell induction is associated with physical proximity to the LCR
The formation of a chromatin loop has been reported that brings the LCR and globin target genes into proximity in actively transcribing erythroid cells (Tolhuis et al., 2002). GATA-1 is involved in this physical interaction (Vakoc et al., 2005). However, the molecular mechanism underlying this proximity, in particular the role of GATA-1 is not understood.
Since NLI has been proposed to mediate long range gene activation, we examined the molecular pattern of E box/GATA protein complex formation on the globin locus during induction of β-globin expression in MEL cells. β-globin mRNA levels increase dramatically over four days of induction with DMSO (Figure 5A). The detection of NLI by western blot analysis appears to increase modestly during induction of β-globin expression while the levels of Lmo2 and GATA-1 do not appear to be affected (Figure 5B). We also compared pol II recruitment in the mouse β-globin locus by ChIP and qPCR in un-induced and DMSO-induced MEL cells (Figure 5D). Similar to previous results (Johnson et al., 2001; Johnson et al., 2003), the recruitment of pol II to the LCR and βmaj promoter and start site is strongly increased after induction of terminal differentiation of MEL cells by DMSO treatment.
Figure 5. Enrichment of E box/GATA complex on β-globin promoter region during mouse MEL cell differentiation.
(A) MEL cells were treated with 2% DMSO and analyzed over a 4 day time course and globin mRNA was analyzed at each stage of differentiation by quantitative real-time PCR (± SEM). 18S ribosomal RNA served as a control. (B) Proteins were analyzed by western blot with specific antibodies described in the legend to Figure 3. Actin served as loading control. (C) The mouse β-globin locus is diagrammed and the positions of TaqMan probes used for real-time PCR in panels D-G are indicated below and named on the graphs. (D) Chromatin was prepared from MEL cell treated with 2% DMSO over 4 days. ChIP assays were performed as described in the legend for Figure 1 with antibodies against RNA pol II. The color key for each time point is the same as in (A). The results are averages of three chromatin preparations ± SEM. (E-G) The same chromatin preparations used for pol II ChIP were analyzed with antibodies against NLI (E), GATA-1 (F) or Lmo2 (G). The control IgG value is plotted for time 0 and was similar at all time points.
Consistent with our observations in the human β-globin locus, GATA-1 and Lmo2 co-localized with NLI at the LCR HSS and their detection increased as a function of time of induction (Figure 5E-G). This result contrasts with other studies in which NLI was not detected at HS2 until after DMSO differentiation (Brand et al., 2004). Importantly, upon induction of β-globin transcription by DMSO, the E box/GATA protein complex was also detected at the β-globin promoter, which contains an E box/GATA motif, at levels significantly above controls although below levels at the LCR (Figure 5E-G). Recruitment of the E box/GATA protein complex was not detected at the inactive embryonic εy and βh1 genes.
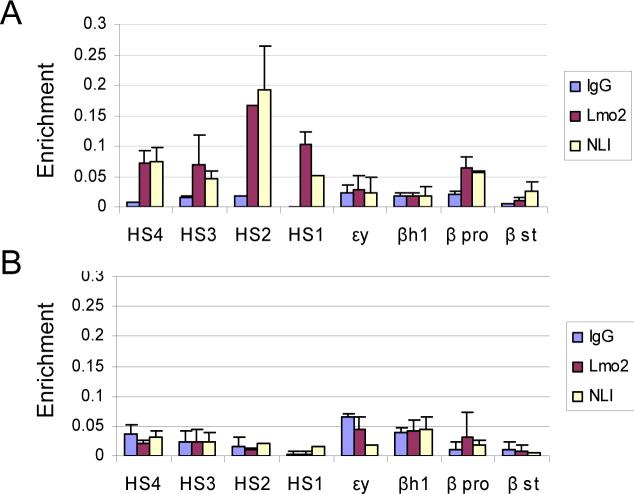
To corroborate these results in primary erythroid cells, we performed ChIP assays for NLI and Lmo2 using E14.5 mouse fetal liver and brain cells. The proteins are recruited to the LCR and active β-globin promoter in liver (Figure 6A) but not in brain cells (Figure 6B). In view of these results, our failure to detect the NLI complex proteins at active globin promoters in K562 cells likely results from infrequent or less stable interaction there, consistent with a much lower level of globin transcription.
Figure 6. NLI and Lmo2 occupy the LCR and β-globin promoter in E14.5 mouse fetal liver cells.
Single cell suspensions of E14.5 fetal liver and brain cells were prepared. Cross-linking and ChIP was then carried out as for MEL cells using antibodies to NLI and Lmo2 or IgG as control. The results are averages of two chromatin preparations ± SEM. (A) NLI and Lmo2 occupancy in E14.5 fetal liver. (B) NLI and Lmo2 occupancy in E14.5 fetal brain.
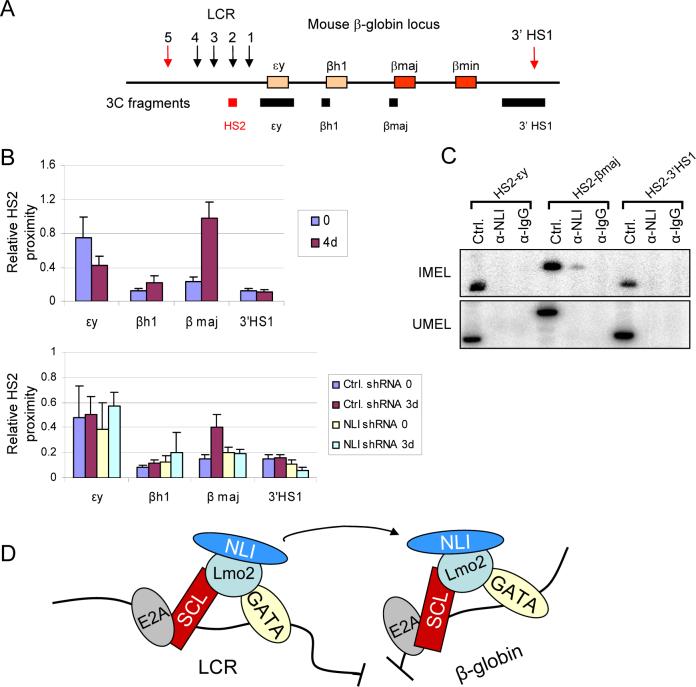
Our findings suggested to us that the E box/GATA protein complex might be directly involved in loop formation between the LCR and an active β-globin gene (Tolhuis et al., 2002; Vakoc et al., 2005). To assess this possibility directly, we performed a 3C or chromosome conformation capture experiment (Dekker et al., 2002). This assay depends on nuclear restriction enzyme digestion followed by re-ligation under dilute conditions and PCR quantitation of ligation products. Restriction sites quite distant from each other will be ligated to each other at a very low frequency which will be increased if the distant sequences are brought in to physical proximity by protein-protein interaction between complexes bound to DNA at both sites.
MEL cells were treated with or without DMSO to induce β-globin transcription. Cross-linked chromatin was digested with Hind III and re-ligated. After reversal of cross-links, real-time qPCR was performed using one primer in HS2 and a second primer in the β-globin gene (Figure 7A). As shown in Figure 7B, after DMSO treatment the proximity of HS2 and the β-globin gene increased 4 fold , a magnitude similar to that seen comparing non-expressing and expressing erythroid tissues and G1E cells (Tolhuis et al., 2002; Vakoc et al., 2005). There was no increase in HS2 proximity when the second primer was within the εy and βh1 genes which do not become activated after DMSO treatment indicating that looping occurs only between the LCR and the active β-globin gene. Furthermore, when NLI was knocked down by shRNA, there was no increase in interaction between HS2 and the β-globin after DMSO induction (Figure 7B). Thus, over the time course of differentiation during which the E box/GATA complex became detectible at the β-globin promoter as well as at the LCR, proximity of these elements is established in an NLI dependent fashion.
Figure 7. Spatial communication between the β-globin LCR and gene depends on E box/GATA complex occupancy.
(A) The murine β-globin locus and the Hind III fragments used in this study. (B) Relative proximity of HS2 with different globin genes in MEL cells (top) before DMSO (0) and after 4 days of DMSO treatment (n=3, ± SEM). All PCR products were identified as the expected ligation dependent products by sequencing. Quantitation of the data was by real-time qPCR. Signals were normalized to a control template and an interaction in the ERCC locus to adjust for template amount and quality. In the lower graph, proximity of HS2 and globin genes in MEL cells stably expressing either control shRNA or NLI shRNA are compared. The 3C experiment was carried out for three days to assure survival of NLI shRNA cells. (C) The ChIP-loop assay was performed by digesting cells with Hind III after 4 days of DMSO induction (IMEL, upper panel) or with no induction (UMEL, lower panel). Chromatin was immunoprecipitated with antibodies against NLI or control IgG followed by ligation and PCR amplification with 3C primers. Ctrl. represents the PCR control template of the size expected for amplification between the HS2 primer and the β-maj primer or other globin genes where NLI interaction with chromatin was not observed (Figure 5 and data not shown). (D) A model of long range communication between the β-globin LCR and gene facilitated by E box/GATA complex formation through the self-interacting domain of NLI/Ldb1.
In order to investigate whether NLI is central to the formation of a loop between the LCR and the β-globin gene, we performed a ChIP-loop experiment in which chromatin was first immunoprecipitated using an antibody to NLI and the resultant material was subjected to 3C analysis (Horike et al., 2005). The HS2 and β-globin primers uniquely amplified a band of the expected size and sequence in MEL cells after induction with DMSO that is not amplified in uninduced MEL cells (Figure 7C). We conclude that NLI/Ldb1, and likely its protein partners, anchors a chromatin loop in the active β-globin locus that brings together the LCR and β-globin gene.
In summary, we have shown that long range interaction of the β-globin LCR and the active β-globin gene is facilitated by NLI through formation of a loop structure between the LCR and the β-globin gene. NLI is not a DNA binding protein and its detection at the LCR and β-globin gene is the result of formation of a multi-protein complex requiring the NLI LIM interacting domain. It is through this domain that Lmo2 connects NLI to the DNA binding complex components GATA-1 and SCL. We propose that the NLI self-interacting domain underlies the establishment of proximity between the β-globin LCR and gene (Figure 7D). This kind of complex interaction was envisioned to exist between two proximal promoter elements in the P4.2 gene (Xu et al., 2003). These results fulfill certain predictions made for the function of the NLI homologue, Drosophila Chip (Dorsett, 1999). However, at least using current ChIP technology, the interaction of the complex with chromatin was specifically detected at the LCR and target β-globin gene and not at multiple sites across the locus as originally envisioned. Given the wide spread expression pattern of NLI, this versatile protein may be a participant in long range enhancer interactions in diverse cell types through participation in different protein complexes that integrate its interaction with chromatin.
Experimental Procedures
Cell lines and Benzidine Staining
K562 cells were cultured as described previously (Kim and Dean, 2004). MEL cells were obtained from Dr. Gerd Blobel (University of Pennsylvania). MEL, 293T and 293FT cells were cultured in DMEM supplemented with 10% fetal bovine serum. For MEL differentiation, cells at a concentration of 2.5 × 105 cells per ml were incubated with 1.5∼2 % DMSO for the indicated times. Benzidine (Sigma, St. Louis, MO) was dissolved in 12% acetic acid to a final concentration of 0.5% (w/v). Just before the cells were stained for hemoglobin, 40 ul of 30% H2O2 was added to 1 ml 0.5% benzidine solution. 100 ul of this mixture was added to the cells. The reaction was allowed to proceed for 15 min at room temperature. Cells were then prepared on slides using a Cytospin centrifuge prior to photography.
Plasmids and Minichromosomes
pCI-HA-NLI(1−375) and pCI-HA-NLI(1−285) were kindly provided by Dr. Dale Dorsett (Saint Louis University) and HA-NLI(215−375) and HA-NLI(285−375) were prepared from full length NLI. pcDNA3-Flag-GATA-1 was a kind gift from Dr. Gerd Blobel, and pEFIRES-Lmo2 was from Dr. Stephen Brandt (Vanderbilt University) (Xu et al., 2003). Minichromosomes carrying the ε-globin gene (3.7-kb Eco RI fragment) and HS2 (1.46-kb Kpn I to Bgl II fragment) have been described (Kim and Dean, 2003). The E box and GATA-1 binding motifs in HS2 were mutated by site-directed mutagenesis (Gong et al., 1996). Minichromosomes were stably transfected into K562 cells, and multiple individual clones were selected and maintained as described previously (Gong et al., 1996). Motifs are underlined and changed nucleotides are bolded.
Wild type sequence: GCCCAGATGTTCTCAGCCTAGAGTGATGACTCCTATCTTGGG.
GATA mutant: GCCCAGATGTTCTCAGCCTAGAGTTCATACTCCGCGATTGGG.
E box mutant: GCCGAGATTTTCTCAGCCTAGAGTGATGACTCCTATCTTGGG.
Immunoprecipitation and Immunoblotting analysis
Nuclear extract (Gong et al., 1996) were precipitated with anti-HA agarose (Sigma) overnight at 4°C, and bound proteins were eluted with HA peptides (Sigma). To prepare whole cell extract, cells were suspended in lysis buffer [50 mM Tris pH8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM Na3VO4, 25 mM NaF, 0.5 mM AEBSF (4-(2-Aminoethyl)-benzenesulfonyl fluoride hydrochloride), 0.1 mM pepstatin A, 0.2 mM leupeptin, and 10 μg/ml aprotinin]. Lysates were cleared by centrifugation at 13,000 rpm for 20 min. Protein extracts were separated by SDS-PAGE and Immunoblotting was performed as described previously (Song et al., 2001). Immunoblots were developed using the ECL Plus detection system (Amersham).
Chromatin Immunoprecipitation (ChIP) and Quantitative real-time PCR analysis
ChIP assays were carried out essentially as described (Zhao et al., 2006) with some modification. Briefly, 2∼5 × 107 cells were cross-linked with 1% formaldehyde for 10 min at room temperature. The reaction was terminated by the addition of glycine to 0.125 M for 5 min at room temperature. Nuclei were prepared and digested with 200U of MNase at 37 °C for 15 min, and then sheared by sonication to an average chromatin fragment size of 200 to 400 bp. Chromatin was pre-cleared and reacted with antibodies overnight at 4 °C and immunoprecipitated by protein A/G plus agarose (Santa Cruz) or protein A agarose (Upstate) which was pre-equilibrated with sonicated salmon sperm DNA and BSA. Immunoprecipitated material was then washed extensively and the cross-links reversed. DNA from eluted chromatin was purified by phenol extraction and ethanol precipitation. For ChIP assays with E14.5 mouse fetal liver and brain cells, single cell suspensions were prepared from the dissected tissue and then cross-linking and subsequent steps were carried out as above.
Differences in DNA enrichment for ChIP samples were determined by real time PCR using the ABI Prism 7900 (PE Applied Biosystems). The threshold was set to cross a point at which PCR amplification was linear, and the number of cycles (Ct) required to reach the threshold was collected and analyzed using Microsoft Excel. The PCR was performed as described previously using 2.5% of the precipitated sample DNA and 0.02% of the input DNA (Zhao et al., 2006). Primer and TaqMan probe sequences are available on request. PCR product was measured by SYBR green fluorescence for ChIP assays with E14.5 liver and brain cells.
RNase protection assay
RNA was prepared from 5 × 106 K562 cells carrying minichromosomes by use of the PUREscript kit (Gentra). The episomal copy of the ε-globin gene is marked by a distinguishing mutation in the 5'-untranslated region (Gong et al., 1996). RNase digestion and gel analyses were performed according to the protocol of the manufacturer of the reagents (RPA II kit; Ambion). RNase protection assay results were normalized to the actin control signal and corrected for minichromosome copy number.
Reverse-Transcription Reaction
Two micrograms of RNA was treated with RNase-free DNase I for 15 min at 25°C, and then RNA was reverse transcribed by using Superscript III First Strand Synthesis Kit as suggested by the manufacturer (Invitrogen). cDNA was diluted to 50 ul, and 5 ul of cDNA was amplified in a 25 ul reaction volume by real-time PCR using TaqMan chemistry. Primer sequences are available on request.
Chromosome Conformation Capture (3C) assay
The 3C assay was performed as described previously (Tolhuis et al., 2002; Vakoc et al., 2005) with minor modification. Briefly, formaldehyde-crosslinked chromatin was digested with Hind III restriction enzyme overnight, followed by ligation with T4 DNA ligase at 16 °C for 3 hr. Crosslinks were reversed and then DNA was purified by phenol extraction and ethanol precipitation. DNA was further purified with a QIAquick PCR purification Kit (Qiagen). All PCR products were amplified, cloned and the correct sequence confirmed. To determine the PCR efficiency among different primer pairs, equimolar amounts of each plasmid were mixed, digested and ligated (control template) (Vakoc et al., 2005). Quantitation of the data was performed by real-time qPCR using SYBR green fluorescence. To compare the cross-linking frequency and ligation efficiency between two restriction fragments present on an unrelated locus, PCR reactions were normalized to ERCC (Palstra et al., 2003).
ChIP-Loop Assay
The ChIP-loop assay (Horike et al., 2005) was performed with some modifications. Crosslinked nuclei were purified on a 5−10% sucrose gradient (Gong et al., 1996), digested with Hind III, briefly sonicated and immunoprecipitated with antibodies to NLI or normal IgG as described above. Immunoprecipitated material was washed and diluted in ligation buffer containing 10 mMol DTT followed by incubation with T4 ligase overnight. After reversal of cross links the DNA was processed as for 3C (above).
Virus production and transduction
Control and NLI-directed TRC lentiviral shRNAs were purchased from Open Biosystems. Lentiviruses were made by transducing 293FT cells with shRNAs and Virapower™ packaging mix (Invitrogen) according to the manufacturer's instructions. After 2 days, complete media was added, and the culture was harvested on day 4. To collect virus, the media was collected and centrifuged at 3000 rpm for 15 min at 4 °C, followed by filtration and then aliquoted and stored at −70 °C. To make stable cells, MEL cells were incubated with viral particles in the presence of 6 ug/ml of polybrene. After 4 days, transfected cells were selected in 2 μg/ml of puromycin (Sigma). After 2 weeks of continuous selection in the indicated conditions, several single colonies were selected and further expanded. Silencing was confirmed by Western blot analysis.
Antibodies
NLI, GATA-1, NF-E2 and Pol II antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Flag M2, actin and α-tubulin antibodies were from Sigma. α-Lmo2 was from R&D Systems. α-HA was from Roche. α-SCL was a kind gift of Dr. C. Porcher (University of Oxford) (Porcher et al., 1999).
Acknowledgements
We thank Dale Dorsett for insightful discussions. We are grateful to Catherine Porscher for the gift of SCL antibodies, Dale Dorsett and Stephen Brandt for plasmids and Gerd Blobel for plasmids and MEL cells. We thank Ina Ifrim for expert technical assistance with some of these experiments. We also thank Hyo-Sung Jeon for generous help with virus production and members of our group for discussions. This work was supported by the Intramural Program of the NIDDK, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
References
- Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Interactions of the LIM-domain-binding factor Ldb1 with LIM homeodomain proteins. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- Anderson KP, Crable SC, Lingrel JB. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J. Biol. Chem. 1998;273:14347–14354. doi: 10.1074/jbc.273.23.14347. [DOI] [PubMed] [Google Scholar]
- Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol Biol. 2004;11:73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- Cohen-Kaminsky S, Maouche-Chretien L, Vitelli L, Vinit MA, Blanchard I, Yamamoto M, Peschle C, Romeo PH. Chromatin immunoselection defines a TAL-1 target gene. EMBO J. 1998;17:5151–5160. doi: 10.1093/emboj/17.17.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Dorsett D. Distant liaisons: long-range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. Dev. 1999;9:505–514. doi: 10.1016/s0959-437x(99)00002-7. [DOI] [PubMed] [Google Scholar]
- Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrando AA, Neuberg DS, Staunton J, Loh ML, Huard C, Raimondi SC, Behm FG, Pui CH, Downing JR, Gilliland DG, Lander ES, Golub TR, Look AT. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/s1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- Goardon N, Lambert JA, Rodriguez P, Nissaire P, Herblot S, Thibault P, Dumenil D, Strouboulis J, Romeo PH, Hoang T. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 2006;25:357–366. doi: 10.1038/sj.emboj.7600934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong QH, McDowell JC, Dean A. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the ε-globin gene in vivo by 5' hypersensitive site 2 of the β-globin locus control region. Mol. Cell. Biol. 1996;16:6055–6064. doi: 10.1128/mcb.16.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutz GG, Bucher K, Lavenir I, Larson T, Larson R, Rabbitts TH. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 1998;17:4594–4605. doi: 10.1093/emboj/17.16.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Loss of silent-chromatin looping and impaired imprinting of DLX5 in Rett syndrome. Nat. Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- Im H, Grass JA, Johnson KD, Kim SI, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. U. S. A. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue- specific locus control region and a downstream promoter. Mol. Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH. Highly Restricted Localization of RNA Polymerase II within a Locus Control Region of a Tissue-Specific Chromatin Domain. Mol. Cell. Biol. 2003;23:6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Gill GN. Functional analysis of the nuclear LIM domain interactor NLI. Mol. Cell. Biol. 1997;17:5688–5698. doi: 10.1128/mcb.17.10.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurata LW, Pfaff SL, Gill GN. The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J. Biol. Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- Kim A, Dean A. A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol. Cell. Biol. 2003;23:8099–8109. doi: 10.1128/MCB.23.22.8099-8109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Dean A. Developmental stage differences in chromatin sub-domains of the β-globin locus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7028–7033. doi: 10.1073/pnas.0307985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and noncoding human β-globin sequences. Mol. Cell. Biol. 2007;27:1271–1279. doi: 10.1128/MCB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahlil R, Lecuyer E, Herblot S, Hoang T. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol. Cell. Biol. 2004;24:1439–1452. doi: 10.1128/MCB.24.4.1439-1452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson RC, Osada H, Larson TA, Lavenir I, Rabbitts TH. The oncogenic LIM protein Rbtn2 causes thymic developmental aberrations that precede malignancy in transgenic mice. Oncogene. 1995;11:853–862. [PubMed] [Google Scholar]
- Lecuyer E, Herblot S, Saint-Denis M, Martin R, Begley CG, Porcher C, Orkin SH, Hoang T. The SCL complex regulates c-kit expression in hematopoietic cells through functional interaction with Sp1. Blood. 2002;100:2430–2440. doi: 10.1182/blood-2002-02-0568. [DOI] [PubMed] [Google Scholar]
- Liu Z, Garrard WT. Long-range interactions between three transcriptional enhancers, active Vkappa gene promoters, and a 3' boundary sequence spanning 46 kilobases. Mol. Cell. Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- Lopes S, Lewis A, Hajkova P, Dean W, Oswald J, Forne T, Murrell A, Constancia M, Bartolomei M, Walter J, Reik W. Epigenetic modifications in an imprinting cluster are controlled by a hierarchy of DMRs suggesting long-range chromatin interactions. Hum. Mol Genet. 2003;12:295–305. doi: 10.1093/hmg/ddg022. [DOI] [PubMed] [Google Scholar]
- Matthews JM, Visvader JE. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan M, Cohen SM. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol Cell. 1999;4:267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Baylies MK, Dorsett D. Chip, a widely expressed chromosomal protein required for segmentation and activity of a remote wing margin enhancer in Drosophila. Genes Dev. 1997;11:2729–2740. doi: 10.1101/gad.11.20.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcillo P, Rosen C, Dorsett D. Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics. 1996;144:1143–1154. doi: 10.1093/genetics/144.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D, Westphal H. Functional ablation of the mouse Ldb1 gene results in severe patterning defects during gastrulation. Development. 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- Osada H, Grutz GG, Axelson H, Forster A, Rabbitts TH. LIM-only protein Lmo2 forms a protein complex with erythroid transcription factor GATA-1. Leukemia. 1997;11(Suppl 3):307–312. [PubMed] [Google Scholar]
- Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The β-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- Porcher C, Liao EC, Fujiwara Y, Zon LI, Orkin SH. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development. 1999;126:4603–4615. doi: 10.1242/dev.126.20.4603. [DOI] [PubMed] [Google Scholar]
- Ramain P, Khechumian R, Khechumian K, Arbogast N, Ackermann C, Heitzler P. Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol. Cell. 2000;6:781–790. doi: 10.1016/s1097-2765(05)00079-1. [DOI] [PubMed] [Google Scholar]
- Shoresh M, Orgad S, Shmueli O, Werczberger R, Gelbaum D, Abiri S, Segal D. Overexpression Beadex mutations and loss-of-function heldup-a mutations in Drosophila affect the 3' regulatory and coding components, respectively, of the Dlmo gene. Genetics. 1998;150:283–299. doi: 10.1093/genetics/150.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SH, Jong HS, Choi HH, Inoue H, Tanabe T, Kim NK, Bang YJ. Transcriptional silencing of Cyclooxygenase-2 by hyper-methylation of the 5' CpG island in human gastric carcinoma cells. Cancer Res. 2001;61:4628–4635. [PubMed] [Google Scholar]
- Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatoyannopoulos G, Grosveld F. Hemoglobin Switching. In: Stamatoyannopoulos G, Majerus PW, Perlmutter RM, Varmus H, editors. The Molecular Basis of Blood Diseases. W.B.Saunders; Philadelphia: 2001. pp. 135–182. [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Torigoi E, Bennani-Baiti IM, Rosen C, Gonzalez K, Morcillo P, Ptashne M, Dorsett D. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc Natl. Acad. Sci. U. S. A. 2000;97:2686–2691. doi: 10.1073/pnas.050586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Valverde-Garduno V, Guyot B, Anguita E, Hamlett I, Porcher C, Vyas P. Differences in the chromatin structure and cis-element organization of the human and mouse GATA1 loci: implications for cis-element identification. Blood. 2004;104:3106–3116. doi: 10.1182/blood-2004-04-1333. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Mao X, Fujiwara Y, Hahm K, Orkin SH. The LIM-domain binding protein Ldb1 and its partner LMO2 act as negative regulators of erythroid differentiation. Proc Natl. Acad. Sci. U. S. A. 1997;94:13707–13712. doi: 10.1073/pnas.94.25.13707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli L, Condorelli G, Lulli V, Hoang T, Luchetti L, Croce CM, Peschle C. A pentamer transcriptional complex including tal-1 and retinoblastoma protein downmodulates c-kit expression in normal erythroblasts. Mol. Cell. Biol. 2000;20:5330–5342. doi: 10.1128/mcb.20.14.5330-5342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas P, McDevitt MA, Cantor AB, Katz SG, Fujiwara Y, Orkin SH. Different sequence requirements for expression in erythroid and megakaryocytic cells within a regulatory element upstream of the GATA-1 gene. Development. 1999;126:2799–2811. doi: 10.1242/dev.126.12.2799. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Xu Z, Huang S, Chang LS, Agulnick AD, Brandt SJ. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol Cell Biol. 2003;23:7585–7599. doi: 10.1128/MCB.23.21.7585-7599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Meng X, Cai Y, Koury MJ, Brandt SJ. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem. J. 2006;399:297–304. doi: 10.1042/BJ20060873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Warren AJ, Dobson C, Forster A, Pannell R, Rabbitts TH. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3890–3895. doi: 10.1073/pnas.95.7.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Kim A, Song SH, Dean A. Enhancer blocking by chicken beta -globin 5' HS4: Role of enhancer strength and insulator nucleosome depletion. J. Biol. Chem. 2006;281:30573–30580. doi: 10.1074/jbc.M606803200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.