Abstract
Lipooligosaccharide (LOS), a predominant surface-exposed component of the outer membrane, has been implicated as a virulence factor in the pathogenesis of Moraxella catarrhalis infections. However, the critical steps involved in the biosynthesis and assembly of M. catarrhalis LOS currently remain undefined. In this study, we used random transposon mutagenesis to identify a 3-deoxy-d-manno-octulosonic acid (KDO) biosynthetic operon in M. catarrhalis with the gene order pyrG-kdsA-eno. The lipid A-KDO molecule serves as the acceptor onto which a variety of glycosyl transferases sequentially add the core and branch oligosaccharide extensions for the LOS molecule. KdsA, the KDO-8-phosphate synthase, catalyzes the first step of KDO biosynthesis and is an essential enzyme in gram-negative enteric bacteria for maintenance of bacterial viability. We report the construction of an isogenic M. catarrhalis kdsA mutant in strain 7169 by allelic exchange. Our data indicate that an LOS molecule consisting only of lipid A and lacking KDO glycosylation is sufficient to sustain M. catarrhalis survival in vitro. In addition, comparative growth and susceptibility assays were performed to assess the sensitivity of 7169kdsA11 compared to that of the parental strain. The results of these studies demonstrate that the native LOS molecule is an important factor in maintaining the integrity of the outer membrane and suggest that LOS is a critical component involved in the ability of M. catarrhalis to resist the bactericidal activity of human sera.
Moraxella catarrhalis is a gram-negative aerobic diplococcus that is frequently identified as part of the nasopharyngeal floras, particularly in pediatric populations (4). This bacterium is an important mucosal pathogen of the upper and lower respiratory tracts in humans. In particular, the organism is a leading cause of otitis media and sinusitis in young children and is associated with pulmonary exacerbations in adults with chronic lung disease or compromised immune function (26, 27). Research over the past decade has focused on the identification and characterization of M. catarrhalis surface antigens, including lipooligosaccharides (LOS), as potential vaccine candidates (for recent reviews, see references 17, 22, 23, and 42).
LOS, a predominant surface-exposed component of the outer membrane, has been implicated as a virulence factor in the pathogenesis of M. catarrhalis infections. The LOS of M. catarrhalis is similar to that of other mucosal pathogens in that it lacks a repeating O-antigen attached to the core oligosaccharide, which is characteristic of the lipopolysaccharide (LPS) molecule. Instead, the LOS molecule contains a lipid A-proximal conserved inner core and one or more structurally diverse oligosaccharide branch extensions that determine serologic specificity. Although the LOS of M. catarrhalis appears to be more antigenically conserved than the LOS of other bacteria, three LOS serotypes (termed A, B, and C) have been identified on the basis of structural and immunologic analyses of the terminal oligosaccharide branches (15, 41). Interest in the evaluation of this glycolipid as an effective vaccine candidate has been strengthened by preliminary studies identifying a strong humoral immune response to the conserved inner core of the LOS molecule following M. catarrhalis infections (9, 32). Despite these data, the critical steps involved in the biosynthesis and assembly of M. catarrhalis LOS currently remain undefined. Thus, to begin to understand the role of LOS in the pathogenesis of M. catarrhalis infections and in the human immune response to this molecule, additional studies focused on the enzymology and molecular genetics involved in the biosynthesis of this important glycolipid are warranted.
The lipid A moiety of the LOS molecule is connected to the oligosaccharide chain via 3-deoxy-d-manno-octulosonic acid (KDO). The first step in the biosynthesis of KDO is catalyzed by the KDO-8-phosphate synthase, KdsA, and involves the aldol-type condensation of phosphoenolpyruvate (PEP) and arabinose-5-phosphate. KDO is dephosphorylated by KDO-8-phosphate phosphatase and is subsequently incorporated into lipid A by KDO transferase(s) following activation by a CMP-KDO synthetase, with CTP used as the nucleotide donor. The lipid A-KDO molecule serves as the acceptor onto which a variety of glycosyl transferases sequentially add the core and branch oligosaccharide extensions for the LOS and LPS molecules. In this report, we describe the identification and characterization of the M. catarrhalis kdsA homologue. Our studies indicate that kdsA is a component of a KDO biosynthetic operon in M. catarrhalis with the gene order pyrG-kdsA-eno. PyrG is a CTP synthase, and Eno (enolase) catalyzes the formation of PEP.
We constructed and analyzed an isogenic M. catarrhalis kdsA mutant, and our data indicate that an LOS molecule consisting only of lipid A and lacking KDO glycosylation is sufficient to sustain M. catarrhalis survival in vitro. In addition, the findings presented in this paper demonstrate that the native LOS molecule is an important factor in maintaining the integrity of the outer membrane and suggest that it is a critical component involved in the ability of M. catarrhalis to resist the bactericidal activity of normal human sera.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The pediatric middle-ear isolate M. catarrhalis 7169 (previously described) (20) was used to construct the kdsA-deficient mutant 7169kdsA11. M. catarrhalis O35E, kindly provided by Eric Hansen (University of Texas Southwestern Medical Center, Dallas, Tex.), was used to construct the EZ::TN transposon (TN) mutants. M. catarrhalis strains were routinely cultured on brain heart infusion (BHI) agar plates at 35.5°C in 5% CO2; mutant strains were grown on BHI agar supplemented with kanamycin at 30 μg per ml. For broth cultures, bacteria were inoculated to an optical density at 600 nm (OD600) of 0.08 in BHI or GC broth (minimal salts medium without supplementation) and grown at 37°C with rotary shaking at 225 rpm. The bacterial cultures were monitored spectrophotometrically (OD600) at 1.5-h intervals for growth curve analysis. All data shown for growth experiments represent averages of the results of three independent assays. Additional studies comparing the growth of strain 7169kdsA11 to that of 7169 in media at various levels of nutritional repleteness were performed as previously described (19). Escherichia coli XL1-Blue was used as the host strain for plasmid DNA manipulations. E. coli strain cultures were grown using Luria-Bertani agar plates and broth with antibiotic supplementation (with ampicillin [100 μg per ml] and kanamycin [40 μg per ml]) as required.
General DNA manipulations.
Restriction endonucleases and standard molecular biology reagents were obtained from New England Biolabs, Inc. (Beverly, Mass.). Platinum TaqDNA high-fidelity polymerase was purchased from Invitrogen (Carlsbad, Calif.), and a pGEM-T Easy vector system was acquired from Promega (Madison, Wis.). Plasmid isolation, purification of PCR products, and gel purification of electrophoretically separated DNA fragments were performed using kits manufactured by Qiagen (Santa Clarita, Calif.). Restriction enzyme digestions, ligations, and transformations by electroporation were performed using standard methods. Chromosomal DNA was isolated as previously described (33). PCR amplifications of chromosomal DNA were performed using a GeneAMP PCR System 9700 (P.E. Applied Biosystems, Foster City, Calif.) for 25 cycles; annealing temperatures and extension lengths were primer set dependent. DNA nucleotide sequences of all constructs were obtained via automated DNA sequencing (Roswell Park Cancer Institute Biopolymer Facility, Roswell Park Cancer Institute, Buffalo, N.Y.) and analyzed with MacVector 6.5.3 software and a Wisconsin Sequence Analysis package (Genetics Computer Group, Madison, Wis.).
Isolation and analysis of M. catarrhalis LOS.
For analysis of the TN mutants, LOS was prepared from proteinase-K-treated whole-cell lysates (PKL), resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 16% acrylamide separating gel, and visualized by silver staining (3, 37). For structural analysis, LOS from strains 7169 and 7169kdsA11 was isolated using the following modified PKL protocol. Plate-grown organisms were resuspended in 15 ml of phosphate-buffered saline (PBS) to an OD600 of 1.0 and washed twice with PBS. The resulting cell pellet was incubated for 2 h at room temperature in 20 mM Tris-HCl (pH 7.4) containing 4 mM MgCl2 and 0.5 mg of lysozyme per ml. Following the addition of RNase and DNase at 2 μg each per ml, the suspension was incubated with agitation overnight. An equivalent volume of 60 mM Tris-HCl (pH 6.8), containing 10 mM EDTA and 2% SDS, was added and the samples were incubated at 100°C for 5 min. Proteinase-K (10 μg per ml) was added, and the samples were incubated for 2 h at 65°C and 6 h at 37°C. The samples were sonicated for 15 min and then placed at −20°C overnight after the addition of a 1/10 volume of 3 M NaOAc (pH 5.2) and 2 volumes of ice-cold ethanol. Following centrifugation at 14,000 rpm in a microcentrifuge (Eppendorf model 5415) the pellets were washed twice in 70% EtOH and resuspended in 200 μl of distilled water. An equivalent amount of a 2:1 solution of chloroform:methanol was added to the rehydrated samples, and after centrifugation, the aqueous layer was reextracted. A total of 8 μl of each sample was analyzed using the bilayer-stacker SDS-PAGE urea gel protocol described by Inzana and Apicella (16) followed by either silver staining or Western blot analysis with the monoclonal antibody (MAb) 4G5 (20). MAb 4G5-immunoprobed colony blot and immunodot assays were performed using standard methods as previously described (18, 20).
MS analysis.
LOS from M. catarrhalis 7169 was analyzed by mass spectrometry (MS) to determine its molecular mass, the generic compositions of the oligosaccharide, and the phosphorylation states of the lipid A moiety. LOS was O-deacylated by treatment with anhydrous hydrazine for 35 min at 37°C (10). The conversion of LOS to di-N-acyl LOS makes it more water soluble and amenable to MS analysis (7). The O-deacylated LOS was then taken up in water and desalted by drop dialysis using a 0.025-μm-pore-size nitrocellulose membrane (Millipore, Bedford, Mass.). Approximately 25% of the sample was dried in a Speed-Vac (Savant Instruments Inc., Holbrook, N.Y.), reconstituted in 1 μl of Milli-Q water, and desalted with cation-exchange resin beads (DOWEX) (50×; NH4+) (28). Prior to spotting onto the stainless steel target, the sample was mixed with 1 μl of 160 mM 2,5-dihydroxybenzoic acid-87.5 mM 1-hydroxyisoquinoline solution in 4:1 acetone-water and left to air dry on the target (25). These O-deacylated LOS samples were then analyzed by matrix-assisted laser desorption (MALDI)-MS using an Applied Biosystems Voyager DESTR+ apparatus (Framingham, Mass.) with a nitrogen laser (337 nm) in linear negative-ion mode under delayed extraction conditions. The delay time was 165 ns, and the grid voltage was 94% of the full acceleration voltage (20 kV). Spectra were acquired, averaged, and mass calibrated with an external calibrant consisting of an equimolar mixture of angiotensin I, adrenocorticotropin 18-39, and adrenocorticotropin 7-38 (Bachem, Torrance, Calif.).
Isolation and MS analysis of lipid A from the 7169kdsA11 mutant.
Lipid A from the M. catarrhalis 7169kdsA11 was isolated by The Complex Carbohydrate Research Center (University of Georgia, Athens, Georgia) using the chloroform-methanol-petroleum ether method as previously described by Tzeng et al. (39). Reflectron negative-ion MALDI-MS analysis of lipid A was carried out by The Complex Carbohydrate Research Center using an Applied Biosystems 4700 Proteomics analyzer equipped with a Nd:YAG laser (3,550-nm wavelength).
TN mutagenesis and identification of kdsA.
Competent M. catarrhalis cultures were prepared and subjected to electroporation as previously described (11). Random TN mutagenesis of M. catarrhalis was performed using an EZ::TN <KAN-2> Tnp Transposome kit (Epicentre, Madison, Wis.) per the manufacturer's specifications. Transformants mutagenized by the EZ::TN TN were selected on the basis of resistance to kanamycin and screened for loss of reactivity to MAb 4G5 by colony blot assay to identify mutants specifically defective in the production of the full-length LOS molecule. The kanamycin-resistant MAb 4G5-negative transformants were subsequently evaluated by examining the LOS profiles of these isolates by SDS-PAGE analysis. One of the clones (termed MCTN38) had no discernible LOS and was selected for further study. The TN insertion site within the MCTN38 genome was identified by rescue cloning the TN and flanking M. catarrhalis chromosomal DNA into pUC18 and subjecting the recombinant clones to nucleotide sequence analysis. Preceding construction of an isogenic kdsA mutant in strain 7169, primers 316 and 317 were used to amplify a 1,300-nucleotide product from strain 7169 chromosomal DNA and the amplicon was subjected to nucleotide sequence analysis.
Construction of a kdsA isogenic mutant.
The nonpolar mutagenesis cassette aphA-3 from pUC18K has previously been used to successfully construct isogenic mutants of M. catarrhalis (18-20, 24), and this resistance determinant was used to insertionally inactivate the kdsA in strain 7169. In summary, plasmid pk3-B was constructed by cloning a 395-bp fragment containing the 3′ portion of the 7169 kdsA coding region and downstream flanking sequence (generated with the primers 334 and 335) (Table 1) into the TA-cloning vector pGEM-T Easy. aphA-3 was PCR amplified (using primers 341 and 342) from pUC18K and directionally cloned into pkd3B, following enzymatic digestion of both the plasmid and the amplicon with SacII and BamHI such that the ATG codon 3′ of the resistance determinant was placed in frame with the kdsA stop codon. This construct was termed pkd3BnpS. A 480-bp NcoI/SacII fragment containing the 5′ portion of kdsA, including the ATG start codon, was amplified using primers 343 and 344 and subsequently cloned into NcoI/SacII-restricted pkd3BnpS.
TABLE 1.
Nucleotide sequence of oligonucleotide primers used for PCR-based cloning procedures and RT-PCR analyses
| Primer | Sequence (5′-3′) |
|---|---|
| 316 | ATCCTTGGTTTGTGGCGGTG |
| 317 | AACATCTGCTTGAATGGTTGG |
| 334 | GGATCCGTTACGCACGCACTTCAAGAGCa |
| 335 | TTGGGTTGCCACGAGAATCC |
| 341 | CCGCGGGGGTGACTAACTAGGAGGAATAAATGGCTAb |
| 342 | GGATCCGTCGACTCTAGAGGATCCCCGGGTCATTAa |
| 343 | CCATGGTCACCCAGAGTTTACCAGCc |
| 344 | CCGCGGGCCAAGTCAAGCCATTCTCb |
| 381 | CGATTTTGGTTCCAGGTGGC |
| 382 | CATCAGAGCGGGTTTGAAGTTC |
| 383 | TGATAGTGATTTGGGCGGCAC |
| 384 | AAGGCTTGGACCACGAAAAGAG |
| 385 | GGCGGTATGAATGTGCTTGAAAG |
| 386 | GCTCTTGAAGTGCGTGCGTAAC |
| 387 | TTACGCACGCACTTCAAGAGC |
| 388 | TTGGGTTGCCACGAGAATCC |
| 389 | AACGCCAATAGCCAAATCCG |
| 390 | CCGACAAATCAAAACCCTTACCAC |
Engineered BamHI site is underlined.
Engineered SacII site is underlined.
Engineered NcoI site is underlined.
After verification by sequence analysis, this mutagenesis construct (containing a nonpolar insertion of the aphA-3 resistance determinant within an internal deletion of the kdsA coding region) was amplified by PCR using primers 343 and 335 and the PCR-generated DNA product was purified and used to naturally transform M. catarrhalis 7169. In brief, a 100-μl aliquot of an early log-phase 7169 culture (OD600 of 0.2) was plated onto BHI agar and 20 ng of the purified amplicon DNA was spotted onto a portion of the bacterial lawn. After incubation for 5 h under standard growth conditions, the area of the bacterial lawn that had been inoculated with the mutagenesis construct was swabbed onto selective plates containing kanamycin.
After 48 h of incubation, the resulting kanamycin-resistant colonies were screened for loss of reactivity to MAb 4G5 and one of these transformants (termed 7169kdsA11) was selected for further study. Insertional inactivation of kdsA by the aphA-3 mutagenesis construct was verified by sequence analysis of amplicons obtained (using primers 316 and 317) from PCR analysis of strain 7169kdsA11 chromosomal DNA. LOS was purified from 7169kdsA11 and 7169 and analyzed as described above. Outer-membrane protein (OMP) profiles were examined by fractionating Zwittergent-extracted OMP preparations on SDS-PAGE gels as previously described (2).
RT-PCR.
Total RNA was isolated (using an RNeasy Mini kit [Qiagen] according to the manufacturer's recommendations) from log-phase bacterial cells. To remove any residual contaminating chromosomal DNA from the purified RNA, the samples were subjected to DNase treatment using an on-column RNase-Free DNase set (Qiagen). Primer sets for the reverse-transcription (RT)-PCRs (designed internally to each open reading frame and spanning the intergenic regions between adjacent genes) are listed in Table 1 and depicted in Fig. 1. RT-PCR was performed using a Qiagen One-Step RT-PCR kit and following the manufacturer's instructions. In parallel, RT-PCRs were performed with DNA as the nucleic acid template. To ensure there was no residual or contaminating DNA in the RNA samples, reactions were also performed without activation of the RT. Amplicons were resolved on 1.5% agarose gels and visualized by ethidium bromide staining.
FIG. 1.
Genetic organization of the M. catarrhalis LOS biosynthesis gene cluster containing pyrG-kdsA-eno. The large arrows represent the direction of transcription, and the site of the TN insertion identified in MCTN38 is denoted (EZ::TN IS). The short, numbered arrows indicate the relative annealing positions of the oligonucleotide primers used in PCR (A) and RT-PCR (B) analyses.
In vitro susceptibility assays.
Susceptibility assays to assess the sensitivity of strain 7169kdsA11 to a panel of hydrophobic agents compared to that of strain 7169 were performed using standard disk-diffusion assays as described previously, with several modifications (29, 47). Bacteria were resuspended to an OD600 of 0.2, and 100-μl aliquots of the bacterial suspension were spread onto BHI plates. Sterile blank paper disks (Becton Dickinson, Cockeysville, Md.) saturated with the various agents were placed on the lawn in triplicate, and plates were incubated at 35.5°C in 5% CO2. Sensitivity to each hydrophobic agent was assessed by measuring the diameter of the zone of growth inhibition in two axes, and the mean value (in millimeters) was recorded. The data represent the averages of the results of three separate experiments (each performed in triplicate).
Bactericidal activity of NHS.
A 200-μl microscale serum bactericidal assay was performed in a 24-well plate essentially as described but with the following modifications (48). Briefly, normal human serum (NHS) (pooled from 10 healthy adult donors) was diluted to 1.0, 2.5, 5.0, and 10.0% in phosphate-buffered saline solution (PBSS). Bacteria (10 μl of 106 cells) were inoculated into 190-μl reaction wells containing the diluted NHS or control wells containing heat-inactivated NHS or PBSS alone (0.0% NHS) and incubated at 37°C with shaking for 30 min. Serial dilutions (1:10) of each reaction well were plated onto BHI agar, and following 48 h of incubation, the resulting colonies were counted. The percentage of survival was calculated by comparing the number of CFU after the 30-min incubation in serum with the number of CFU of the inoculum incubated in PBSS alone. The data represent the averages of the results of three independent and highly reproducible assays (see Fig. 7).
FIG. 7.
Bactericidal effects of NHS on strains 7169 (black bars) and 7169kdsA11 (gray bars). The comparative serum sensitivity microscale assay was performed as described in Materials and Methods. Bacterial recovery is expressed as log CFU per milliliter, and the data represent the averages of the results of three independent assays.
Nucleotide sequence accession numbers.
The nucleotide sequences of the M. catarrhalis O35E pyrG-kdsA-eno gene cluster and of the 7169 kdsA strain have been deposited at GenBank under accession numbers AY174100 and AY174101, respectively.
RESULTS
Identification of the M. catarrhalis kdsA.
An M. catarrhalis mutant defective in LOS biosynthesis was identified using random TN mutagenesis as described in Materials and Methods. The resulting kanamycin-resistant colonies were screened for loss of reactivity to the M. catarrhalis LOS-specific MAb 4G5. MAb 4G5 reacts to a terminal LOS epitope that is conserved in all strains of M. catarrhalis LOS analyzed to date. The growth of one of the MAb 4G5-nonreactive TN mutants (termed MCTN38) was greatly diminished on agar plates compared to that of the wild type, requiring 48 h instead of 16 h for visible single-colony formation. LOS (prepared from PKL) of the mutant and parental strains was resolved by SDS-PAGE and visualized by silver staining. This analysis indicated that in contrast to the findings for the wild-type LOS expressed by the parent strain, the mutant did not produce any discernible LOS (data not shown).
To identify the site of TN insertion within MCTN38, purified genomic DNA was digested with PvuII and rescue cloned into the HincII site of pUC18. Several ampicillin- and kanamycin-resistant clones were analyzed by restriction enzyme digestion, and all were found to contain an approximately 3.6-kb insert. The entire 3.6-kb genomic fragment was subjected to sequence analysis and was found to contain one complete open reading frame (ORF) (ORF2) and two flanking incomplete ORFs (ORF1 and ORF3), all in the same orientation; the precise insertion site of the 1.2-kb TN was identified within the coding region of the ORF2. Database searches with the deduced polypeptide sequences of the three potential coding regions revealed high homology of ORF1 to PyrG (63% identity with the PyrG of Pseudomonas aeruginosa), of ORF2 to KdsA (69% identity with the KdsA of P. aeruginosa), and of ORF3 to Eno (67% identity with the Eno of P. aeruginosa) (Fig. 1).
The biosynthesis of KDO and its subsequent activation prior to transfer to lipid A involve the condensation of PEP and arabinose-5-P catalyzed by the KDO-8-phosphate synthase encoded by kdsA. Enolase (the gene product of eno) catalyzes the formation of PEP, one of the precursors of KDO, from phosphoglycerate during glycolysis. pyrG encodes a CTP synthase, which catalyzes the transfer of ammonia to UTP to form the nucleotide CTP. Preceding transfer to lipid A, KDO is activated to CMP-KDO using CTP as the nucleotide donor. It is interesting that pyrG, kdsA, and eno were reported to be genetically linked in an identical arrangement in only one previous study that identified a uniquely large KDO biosynthetic operon comprised of these genes in P. aeruginosa (45). The initial analysis of the genomic region surrounding the TN-insertion in the LOS mutant MCTN38 suggested that the M. catarrhalis pyrG, kdsA, and eno homologues might also be cotranscribed in an operon. In addition, it has been reported that KdsA, the KDO-8-phosphate synthase, is an essential enzyme in gram-negative enteric bacteria and that a minimal LPS structure of lipid A-KDO is required to maintain bacterial viability (8, 31, 34). Therefore, we constructed an isogenic kdsA mutant in M. catarrhalis strain 7169 to confirm that the altered LOS phenotype of MCTN38 was due to the complete functional inactivation of kdsA alone and not due to contributing downstream polar effects resulting from the TN insertion.
Analysis of an isogenic kdsA mutant.
The M. catarrhalis 7169kdsA11 isogenic mutant was constructed by integrating the nonpolar kanamycin resistance determinant aphA-3 into a sequence deleted from the kdsA coding region as described above. DNA sequence analysis of amplicons generated using primers flanking the predicted site of aphA-3 insertion confirmed that the nonpolar mutagenesis cassette had inserted into the kdsA of 7169kdsA11 at the predicted chromosomal location. Furthermore, total RNA isolated from both the parental 7169 and mutant 7169kdsA11 strains was subjected to RT-PCR analysis using oligonucleotide primer pairs designed to investigate whether pyrG, kdsA, and eno were cotranscribed. As shown in Fig. 2 (top panel), these analyses confirmed that this LOS biosynthetic gene cluster constitutes a polycistronic operon in M. catarrhalis. Transcription through kdsA in 7169kdsA11 combined with the detection of mRNA transcripts of the downstream eno confirmed the nonpolar effects of the insertional mutation (bottom panel).
FIG. 2.
Detection of the pyrG-kdsA-eno operon by RT-PCR analysis using total RNA isolated from strain 7169 (top panel) or 7169kdsA11 (bottom panel). Refer to Fig. 1 and its legend for the annealing position and polarity of each gene-specific primer. The RT-PCRs were performed using the following nucleic acid templates: lanes 1, strain 7169 chromosomal DNA; lanes 2, total RNA isolated from 7169; lanes 6, strain 7169kdsA11 chromosomal DNA; lanes 7, total RNA purified from 7169kdsA11. Reaction sets contained the following primers: a, 381 and 382; b, 383 and 384; c, 385 and 386; d, 387 and 388; e, 389 and 390. The results for one representative set of control reactions (performed using primers 385 and 386) are depicted on the agarose gels as follows: lanes 3 and 8, RT-PCRs using purified total RNA as the nucleic acid template but without activation of the RT; lanes 4 and 9, RT-PCR with no added nucleic acid template; lanes 5 and 10, RT-PCR using chromosomal DNA as the template. Molecular size standards (lanes M) are depicted in 100-bp increments.
LOS isolated from proteinase-K lysates of 7169 and 7169kdsA11 were analyzed by SDS-PAGE and Western blotting (Fig. 3). In contrast to the LOS profile of the parental strain (panel A, lane 1), there was no detectable LOS produced by the 7169kdsA11 isogenic mutant (panel A, lane 2). In addition, the results of the Western blot analysis shown in panel B demonstrate that the M. catarrhalis LOS-specific MAb 4G5 only reacted to the LOS of the parental 7169 strain, further confirming the mutated LOS phenotype of 7169kdsA11. These results were identical to those of the analysis of the LOS isolated from the TN mutant MCTN38.
FIG. 3.
A silver-stained SDS-PAGE gel (A) and an MAb4G5- probed immunoblot (B) depicting the LOS profiles of M. catarrhalis 7169 (lanes 1) and 7169kdsA11 (lanes 2). Molecular size standards are shown in kilodaltons.
Structural analysis of LOS composition.
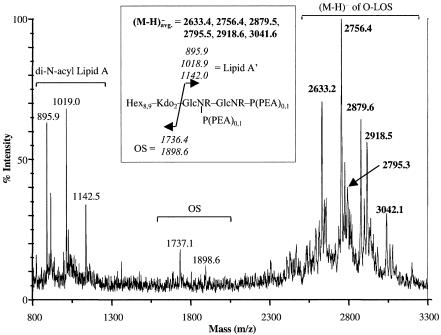
Taken together, these results suggest that consistent with the expected phenotype of a KdsA-deficient mutant, strain 7169kdsA11 expresses a deeply truncated LOS molecule consisting solely of a lipid A molecule devoid of KDO. This conclusion is supported by the MALDI-MS analysis of the LOS prepared from both the wild-type and mutant strains. These studies demonstrated that in contrast to findings obtained with a similar sample prepared from the kdsA isogenic mutant, spectral data were only obtained from the O-deacylated LOS of M. catarrhalis wild-type strain 7169. The lack of any signal from the LOS extracted from the kdsA mutant 7169kdsA11 is consistent with the expectation that due to its increased hydrophobicity, truncation of sugars distal to the KDO moiety would result in a LOS molecule that would no longer partition into the aqueous phase of the chloroform-methanol-water partition used as the final stage of the proteinase-K purification of the LOS. In contrast, the wild-type LOS preparation partitioned into the aqueous phase and, after hydrazine treatment, the O-deacylated LOS yielded a set of six molecular ions by negative-ion linear MALDI-MS, as seen in Fig. 4. The findings for these six deprotonated (M-H)− ions at m/z 2633.2, 2756.4, 2795.3, 2879.6, 2918.5, and 3042.1 are consistent with a generic oligosaccharide composition for a serotype B strain that contains eight to nine hexoses and two KDO sugars but lacks HexNAc. Previous studies have indicated that the LOS from M. catarrhalis serotypes A and C contain a GlcNAc residue in the oligosaccharide branch which clearly was not detected in investigations of the experimental masses of wild-type 7169 LOS determined by MALDI-MS.
FIG. 4.
Linear negative-ion MALDI-MS spectrum of O-deacylated LOS from wild-type strain 7169. All ion values are represented as average (avg.) masses. In the high-mass region above m/z 2500, six deprotonated molecular ion species, (M-H)−, were identified at m/z 2633.2, 2756.4, 2795.3, 2879.6, 2918.5, and 3042.1. At lower mass levels, several prompt fragments were observed that were generated by cleavage at the labile KDO-lipid A glycosidic linkage yielding two oligosaccharide (OS) fragments at m/z 1737.1 and 1898.6 and three di-N-acyl lipid A (lipid A′) fragments at m/z 895.9, 1019.0, and 1142.5, respectively. The two OS fragments differed by the mass of a single hexose (Hex; ΔM = 162 Da) and the three lipid A′ species differed by the mass of one or two PEAs (ΔM = 123 Da). The heterogeneity of the OS (two isoforms) and lipid A′ (three isoforms) was consistent with the presence of the six LOS glycoforms differing in the total number of hexose residues (8 or 9) and/or the number of PEAs (0, 1, or 2) that modified one or more of the two phosphates (P) on lipid A′. The calculated average masses for the intact molecular ions and their OS and lipid A′ fragments are shown in the boxed inset structure.
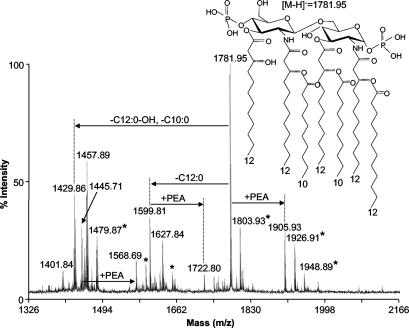
To further investigate the phenotype of this mutant, a phenol-chloroform-petroleum ether extraction (39) was performed on strain 7169kdsA11. Negative-ion reflectron MALDI-MS of this extract demonstrated the presence of a number of deprotonated molecular ions (Fig. 5). The major species with (M-H)− at m/z 1781.95 corresponds to a diphosphoryl, heptaacyl-lipid A, lacking any oligosaccharide (see inset in Fig. 5), in similarity to findings for the structure described by Masoud et al. for the acid-liberated lipid A of M. catarrhalis serotype A (21). Judging on the basis of analysis of the previously published structure, it is likely that the acyl groups attached to the lipid A dihexosamine core consist of a 3-hydroxydodecanoic acid [C12:0(3-OH)], a 3-dodecanoyloxydodecanoic acid [C12:0(3-O-)C12:0], and two 3-decanoyloxydodecanoic acids [C12:0(3-O-)C10:0]. The other deprotonated molecular ions in this spectrum correspond to various lipid A forms (lacking acyl substitutions or with various acyl chain lengths), as summarized in Table 2. The m/z 1904.93 ion corresponds to the predominant lipid A species with an additional phosphoethanolamine (PEA) group (m/z 1,904 = m/z 1781 + 123), and ions m/z 1722.80 and 1568.69 represent PEA-containing species lacking a dodecanoyl (C12:0) acyl chain (m/z 1,722 = m/z 1,904 − 182) or a C12:0 acyl chain and a decanoyl acyl chain (C10:0) (m/z 1,568 = m/z 1,904 − 336), respectively. These data confirm that in the absence of a functional kdsA gene, the LOS becomes truncated and devoid of KDO, leading to the expression of an unsubstituted lipid A molecule by the 7169kdsA11 mutant.
FIG. 5.
Reflectron negative-ion MALDI-MS of lipid A isolated from the M. catarrhalis 7169kdsA11 mutant. Results for all ions are represented as deprotonated (M-H)− exact masses. The upper right portion of the figure shows the structure of the major species at m/z 1781.95. Abbreviations: C12:0-OH, 3-hydroxy-dodecanoyl acyl chain; C12:0, dodecanoyl acyl chain; C10:0, decanoyl acyl chain. *, sodiated species.
TABLE 2.
Mr values observed for lipid A structures of the 7169kdsA11 mutant as detected by negative reflectron MALDI-MS spectra
| Mra | Predicted lipid A structureb | |
|---|---|---|
| Observed | Calculated | |
| 1905.94 | 1906.18 | 4× C12:0(3-OH), C12:0, 2× C10:0, PEA |
| 1782.95 | 1783.17 | 4× C12:0(3-OH), C12:0, 2× C10:0 |
| 1723.81 | 1724.01 | 4× C12:0(3-OH), 2× C10:0, PEA |
| 1628.85 | 1629.03 | 4× C12:0(3-OH), C12:0, C10:0 |
| 1600.82 | 1601.00 | 4× C12:0(3-OH), 2× C10:0 |
| 1569.70 | 1569.87 | 4× C12:0(3-OH), C10:0, PEA |
| 1458.89 | 1458.90 | 3× C12:0(3-OH), C12:0, C10:0, + 28 Da (2× CH2) |
| 1446.72 | 1446.86 | 4× C12:0(3-OH), C10:0 |
| 1430.87 | 1430.87 | 3× C12:0(3-OH), C12:0, C10:0 |
| 1402.85 | 1402.84 | 3× C12:0(3-OH), 2× C10:0 |
All mass values represent exact monoisotopic masses.
All structures are listed on the basis of a core containing two glucosamine and two phosphate moieties with additional groups as listed.
Comparative growth studies.
In vitro growth studies were used to compare the growth rate of the KdsA-deficient mutant 7169kdsA11 to that of the parental 7169 strain. In comparison to 7169, 7169kdsA11 exhibited a reduced growth rate for development of single colonies on agar plates (as previously noted for the initial TN mutant MCTN38). The growth of the wild-type and mutant strains was monitored spectrophotometrically in vigorously shaken broth cultures. As shown in Fig. 6A, the growth rate of the mutant was severely reduced in comparison to that of the wild type. Additional studies using several different growth media with various degrees of nutritional richness indicated that the growth deficit of 7169kdsA11 was consistently observed under all culture conditions evaluated (data not shown). Interestingly, comparative analysis of OMP preparations isolated from both the wild-type and mutant strains indicated that the loss of a functional KdsA had no major effect on the OMP profile (Fig. 6B).
FIG. 6.
(A) In vitro growth of the parental 7169 (▪) and the mutant 7169kdsA11 (○) strains was monitored spectrophotometrically at 1.5-h intervals. (B) The corresponding OMP profiles of 7169 (lane 1) and 7169kdsA11 (lane 2) were evaluated by SDS-PAGE analysis and Coomassie brilliant blue staining. Molecular size standards are shown in kilodaltons.
Effects of kdsA disruption on susceptibility to hydrophobic agents and human sera.
The mutagenesis of genes involved in the synthesis of the inner-core structure of LPS in enteric bacteria was previously shown to cause alterations in the integrity of the hydrophobic barrier of the outer membrane (5, 6, 36). To investigate whether the inactivation of the M. catarrhalis kdsA and the resulting defects in the LOS molecule expressed by the mutant had an effect on the permeability of the outer membrane of this strain, we examined the sensitivity profiles of strains 7169kdsA11 and 7169 to a panel of hydrophobic agents. As shown in Table 3, the LOS mutant 7169kdsA11 showed increased susceptibility to all of the various hydrophobic agents examined compared to that exhibited by 7169. These studies confirmed that the M. catarrhalis LOS inner-core molecule is essential to the integrity of the outer-membrane barrier function.
TABLE 3.
Susceptibilities of strains 7169 and 7169kdsA11 to a panel of hydrophobic agents
| Compound | Zone of growth inhibition (mm) for strain:
|
|
|---|---|---|
| 7169 | 7169kdsA11 | |
| Novobiocin (10 mg/ml) | 41.3 | 52.3 |
| Polymyxin B (10 mg/ml) | 24.7 | 36.8 |
| Deoxycholate (100 mg/ml) | 33 | 43 |
| SDS (100 mg/ml) | 25.7 | 38.3 |
| Tween 20 (5% [wt/vol]) | 14.2 | 18.3 |
| Triton X-100 (5% [wt/vol]) | 19.5 | 36 |
To evaluate whether the loss of a functional KdsA and the production of a deeply truncated LOS molecule by strain 7169kdsA11 affected serum sensitivity, bactericidal assays using NHS were performed. The results of these studies (summarized in Fig. 7) indicated that 7169kdsA11 was exceedingly serum sensitive compared to the parental strain. Although 7169 was resistant to the bactericidal activity of the sera at every concentration evaluated, at 1.0% NHS only 6.5% of the 7169kdsA11 inoculum population survived and at 10.0% NHS the entire 7169kdsA11 population was killed.
DISCUSSION
In this report, we describe the use of random TN mutagenesis for identification of a KDO biosynthetic locus in M. catarrhalis comprised of pyrG-kdsA-eno. It is interesting that prior to this study, the only other KDO biosynthetic operon containing kdsA (encoding the KDO-8-P synthase) genetically linked to pyrG and eno was described for P. aeruginosa, an organism expressing LPS (46). In both LPS and LOS, the hydrophobic lipid A component of the molecule is linked to the hydrophilic core oligosaccharide by one to four molecules of KDO. The biosynthesis of KDO has been extensively studied and well characterized in the Enterobacteriaceae, for which it has been demonstrated that KDO is an essential component of LPS. In these organisms, a minimal LPS molecule consisting of KDO-lipid A is essential for the maintenance of cell viability and only conditionally lethal mutants defective in KDO biosynthesis have been identified (for recent reviews, see references 8, 31, and 34).
In contrast to the findings for enteric bacteria, the results presented in this report indicate that an M. catarrhalis kdsA mutant defective in KDO biosynthesis was viable for in vitro growth. The observation that a KdsA-deficient phenotype was not lethal to M. catarrhalis was first demonstrated by the initial isolation of an LOS-deficient TN mutant that contained a disrupted kdsA coding sequence and then confirmed via the construction and analysis of a deletion-insertion isogenic kdsA mutant termed 7169kdsA11. As expected due to the defined genetic defect of the mutated strain, 7169kdsA11 did not produce a discernible LOS profile by SDS-PAGE analysis consistent with an extremely truncated LOS phenotype containing only lipid A and lacking KDO glycosylation. This observation was supported by the loss of MAb 4G5 reactivity to purified LOS preparations from 7169kdsA11 (as analyzed by immunoblot analysis) and to whole-bacterium suspensions (as evaluated by immunodot analysis) (data not shown). In addition, the data obtained from MS analysis indicated that in contrast to findings for the wild-type strain, the LOS samples prepared from the mutant were highly hydrophobic and consisted of an LOS molecule truncated at the KDO moiety, leaving a diphosphoryl-heptacyl-lipid A as the major species. These data indicated that a kdsA mutation is not lethal to M. catarrhalis, supporting the conclusion that M. catarrhalis is viable with a minimal LOS molecule consisting of lipid A alone. Interestingly, these results are consistent with recent reports of Neisseria meningitidis mutants that are viable in the absence of endotoxin or KDO-glycosylated lipid A (35, 39, 40). These data support the hypothesis that the biosynthesis of the KDO-lipid A backbone of the oligosaccharide inner core is not conserved between LPS-expressing enteric organisms and LOS-producing nonenteric bacteria (39).
Although M. catarrhalis does not require KDO glycosylation of the lipid A for maintenance of viability, growth analyses demonstrated that the KdsA-deficient isogenic mutant 7169kdsA11 strain was consistently impeded under all standard growth conditions and that this growth deficiency could not be overcome by enhancing the nutritional repleteness of the medium. Consistent with previous studies that reported that mutants exhibiting a deep rough LPS phenotype are hypersensitive to hydrophobic compounds and display altered outer-membrane integrity and stability (30, 47), the inactivation of kdsA and the resulting defects in the LOS molecule produced by the mutated strain had a direct and drastic effect on the permeability barrier of the outer membrane. These data offer additional support for the evaluation of KdsA as a target for the development of novel chemotherapeutic agents against gram-negative human pathogens, as KDO is found in prokaryotes expressing LOS-LPS and deficiencies in KDO biosynthesis result in either bacterial death or severely compromised bacterial growth and outer-membrane function (36). Although the M. catarrhalis kdsA mutant was viable under in vitro conditions, these data suggest that KdsA-deficient mutants may be attenuated for virulence in vivo. Furthermore, there is interest in the identification and development of new antimicrobial therapies against M. catarrhalis infections, as recent studies have identified an increasing prevalence of antibiotic resistance among clinical isolates (1, 4, 12, 38).
Only one gene involved in the biosynthesis of M. catarrhalis LOS, a galE homologue, has been identified to date (48). It was reported that the truncated LOS molecule expressed by the serotype A GalE-deficient mutant lacks the two terminal hexose residues and that this mutant is sensitive to the bactericidal activity of human sera at 25% (48). In comparison, the KdsA-deficient mutant 7169kdsA11 strain exhibited exquisite serum sensitivity, as over 90% of the organisms were killed after incubation with 1% NHS and no viable bacteria were detected after exposure to 10% NHS. In contrast, the wild-type 7169 strain was completely resistant under these experimental conditions. Several previous studies demonstrated that the ability of M. catarrhalis to resist complement-mediated killing is an important virulence factor in the pathogenesis of this organism (13, 14, 43, 44, 48). Given the serum concentrations used in these studies, the results suggest that the loss of the full-length LOS molecule might facilitate the binding of existing human antibodies to bacterial surface antigens normally occluded on wild-type strains, thus resulting in highly efficient classical complement-mediated lysis. These findings strengthen the hypothesis that an intact LOS molecule expressed on the surface of M. catarrhalis is critical for protection against the bactericidal effects of NHS.
Using an in vivo transposome-based approach, we identified a M. catarrhalis kdsA homologue and localized this gene within a KDO biosynthetic operon. In addition, a viable isogenic kdsA mutant that produces an LOS molecule lacking KDO glycosylation was constructed and analyzed, which we believe represents the only successful construction of a stable KdsA-deficient organism to date. Taken together, our studies of the isogenic KdsA-null mutant indicated that the native M. catarrhalis LOS molecule is critical for maintenance of outer-membrane stability and the ability of this organism to resist the bactericidal effects of human sera.
Acknowledgments
This research was supported by Public Health Service Research grants AI46422, DC005837 (A.A.C.), and AI31254 (B.W.G.). N.R.L. is supported as a postdoctoral fellow by NIAID training grant AI07614. This work was supported in part by the Department of Energy-funded (DE-FGOZ-93ER-20097) Center for Plant and Microbial Complex Carbohydrates.
We thank the Center for Complex Carbohydrate Research at the University of Georgia, in particular Biswa Choudhury and Parastoo Azadi, for the preparation and analysis of the data involving the lipid A isolated from the kdsA mutant.
Editor: D. L. Burns
REFERENCES
- 1.Bandak, S. I., M. R. Turnak, B. S. Allen, L. D. Bolzon, D. A. Preston, S. K. Bouchillon, and D. J. Hoban. 2001. Antibiotic susceptibilities among recent clinical isolates of Haemophilus influenzae and Moraxella catarrhalis from fifteen countries. Eur. J. Clin. Microbiol. Infect. Dis. 20:55-60. [DOI] [PubMed] [Google Scholar]
- 2.Campagnari, A. A., T. F. Ducey, and C. A. Rebmann. 1996. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrane of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect. Immun. 64:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campagnari, A. A., L. M. Wild, G. E. Griffiths, R. J. Karalus, M. A. Wirth, and S. M. Spinola. 1991. Role of lipooligosaccharides in experimental dermal lesions caused by Haemophilus ducreyi. Infect. Immun. 59:2601-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright, M. C., and H. McKenzie. 1997. Moraxella (Branhamella) catarrhalis—clinical and molecular aspects of a rediscovered pathogen. J. Med. Microbiol. 46:360-371. [DOI] [PubMed] [Google Scholar]
- 5.Fralick, J. A., and L. L. Burns-Keliher. 1994. Additive effect of tolC and rfa mutations on the hydrophobic barrier of the outer membrane of Escherichia coli K-12. J. Bacteriol. 176:6404-6406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujishima, H., A. Nishimura, M. Wachi, H. Takagi, T. Hirasawa, H. Teraoka, K. Nishimori, T. Kawabata, K. Nishikawa, and K. Nagai. 2002. kdsA mutations affect FtsZ-ring formation in Escherichia coli K-12. Microbiology 148:103-112. [DOI] [PubMed] [Google Scholar]
- 7.Gibson, B. W., A. A. Campagnari, W. Melaugh, N. J. Phillips, M. A. Apicella, S. Grass, J. Wang, K. L. Palmer, and R. S. Munson, Jr. 1997. Characterization of a transposon Tn916-generated mutant of Haemophilus ducreyi 35000 defective in lipooligosaccharide biosynthesis. J. Bacteriol. 179:5062-5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gronow, S., and H. Brade. 2001. Lipopolysaccharide biosynthesis: which steps do bacteria need to survive? J. Endotoxin Res. 7:3-23. [PubMed] [Google Scholar]
- 9.Gu, X. X., J. Chen, S. J. Barenkamp, J. B. Robbins, C. M. Tsai, D. J. Lim, and J. Battey. 1998. Synthesis and characterization of lipooligosaccharide-based conjugates as vaccine candidates for Moraxella (Branhamella) catarrhalis. Infect. Immun. 66:1891-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helander, I. M., K. Nummila, I. Kilpelainen, and M. Vaara. 1995. Increased substitution of phosphate groups in lipopolysaccharides and lipid A of polymyxin-resistant mutants of Salmonella typhimurium and Escherichia coli. Prog. Clin. Biol. Res. 392:15-23. [PubMed] [Google Scholar]
- 11.Helminen, M. E., I. Maciver, M. Paris, J. L. Latimer, S. L. Lumbley, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A mutation affecting expression of a major outer membrane protein of Moraxella catarrhalis alters serum resistance and survival in vivo. J. Infect. Dis. 168:1194-1201. [DOI] [PubMed] [Google Scholar]
- 12.Hoban, D. J., G. V. Doern, A. C. Fluit, M. Roussel-Delvallez, and R. N. Jones. 2001. Worldwide prevalence of antimicrobial resistance in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis in the SENTRY Antimicrobial Surveillance Program, 1997-1999. Clin. Infect. Dis. 32(Suppl. 2):S81-S93. [DOI] [PubMed] [Google Scholar]
- 13.Hol, C., C. M. Verduin, E. van Dijke, J. Verhoef, and H. van Dijk. 1993. Complement resistance in Branhamella (Moraxella) catarrhalis. Lancet 341:1281. [DOI] [PubMed] [Google Scholar]
- 14.Hol, C., C. M. Verduin, E. E. Van Dijke, J. Verhoef, A. Fleer, and H. van Dijk. 1995. Complement resistance is a virulence factor of Branhamella (Moraxella) catarrhalis. FEMS Immunol. Med. Microbiol. 11:207-211. [DOI] [PubMed] [Google Scholar]
- 15.Holme, T., M. Rahman, P. E. Jansson, and G. Widmalm. 1999. The lipopolysaccharide of Moraxella catarrhalis structural relationships and antigenic properties. Eur. J. Biochem. 265:524-529. [DOI] [PubMed] [Google Scholar]
- 16.Inzana, T. J., and M. A. Apicella. 1999. Use of a bilayer stacking gel to improve resolution of lipopolysaccharides and lipooligosaccharides in polyacrylamide gels. Electrophoresis 20:462-465. [DOI] [PubMed] [Google Scholar]
- 17.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 18.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 67:5815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luke, N. R., R. J. Karalus, and A. A. Campagnari. 2002. Inactivation of the Moraxella catarrhalis superoxide dismutase SodA induces constitutive expression of iron-repressible outer membrane proteins. Infect. Immun. 70:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luke, N. R., T. A. Russo, N. Luther, and A. A. Campagnari. 1999. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane protein B1 defined by monoclonal antibody 11C6. Infect. Immun. 67:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masoud, H., M. B. Perry, and J. C. Richards. 1994. Characterization of the lipopolysaccharide of Moraxella catarrhalis. Structural analysis of the lipid A from M. catarrhalis serotype A lipopolysaccharide. Eur. J. Biochem. 220:209-216. [DOI] [PubMed] [Google Scholar]
- 22.McMichael, J. C. 2000. Progress toward the development of a vaccine to prevent Moraxella (Branhamella) catarrhalis infections. Microbes Infect. 2:561-568. [DOI] [PubMed] [Google Scholar]
- 23.McMichael, J. C. 2000. Vaccines for Moraxella catarrhalis. Vaccine 19(Suppl. 1):S101-S107. [DOI] [PubMed] [Google Scholar]
- 24.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohr, M. D., K. O. Bornsen, and H. M. Widmer. 1995. Matrix-assisted laser desorption/ionization mass spectrometry: improved matrix for oligosaccharides. Rapid Commun. Mass Spectrom. 9:809-814. [DOI] [PubMed] [Google Scholar]
- 26.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, T. F. 1998. Lung infections. 2. Branhamella catarrhalis: epidemiological and clinical aspects of a human respiratory tract pathogen. Thorax 53:124-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordhoff, E., A. Ingendoh, R. Cramer, A. Overberg, B. Stahl, M. Karas, F. Hillenkamp, and P. F. Crain. 1992. Matrix-assisted laser desorption/ionization mass spectrometry of nucleic acids with wavelengths in the ultraviolet and infrared. Rapid Commun. Mass Spectrom. 6:771-776. [DOI] [PubMed] [Google Scholar]
- 29.Ochsner, U. A., A. I. Vasil, Z. Johnson, and M. L. Vasil. 1999. Pseudomonas aeruginosa fur overlaps with a gene encoding a novel outer membrane lipoprotein, OmlA. J. Bacteriol. 181:1099-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raetz, C. R. 1990. Biochemistry of endotoxins. Annu. Rev. Biochem. 59:129-170. [DOI] [PubMed] [Google Scholar]
- 31.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahman, M., T. Holme, I. Jonsson, and A. Krook. 1995. Lack of serotype-specific antibody response to lipopolysaccharide antigens of Moraxella catarrhalis during lower respiratory tract infection. Eur. J. Clin. Microbiol. Infect. Dis. 14:297-304. [DOI] [PubMed] [Google Scholar]
- 33.Russo, T. A., J. E. Guenther, S. Wenderoth, and M. M. Frank. 1993. Generation of isogenic K54 capsule-deficient Escherichia coli strains through TnphoA-mediated gene disruption. Mol. Microbiol. 9:357-364. [DOI] [PubMed] [Google Scholar]
- 34.Schnaitman, C. A., and J. D. Klena. 1993. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol. Rev. 57:655-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steeghs, L., R. den Hartog, A. den Boer, B. Zomer, P. Roholl, and P. van der Ley. 1998. Meningitis bacterium is viable without endotoxin. Nature 392:449-450. [DOI] [PubMed] [Google Scholar]
- 36.Taylor, W. P., G. Y. Sheflyan, and R. W. Woodard. 2000. A single point mutation in 3-deoxy-D-manno-octulosonate-8-phosphate synthase is responsible for temperature sensitivity in a mutant strain of Salmonella typhimurium. J. Biol. Chem. 275:32141-32146. [DOI] [PubMed] [Google Scholar]
- 37.Tsai, C. M., and C. E. Frasch. 1982. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119:115-119. [DOI] [PubMed] [Google Scholar]
- 38.Turnak, M. R., S. I. Bandak, S. K. Bouchillon, B. S. Allen, and D. J. Hoban. 2001. Antimicrobial susceptibilities of clinical isolates of Haemophilus influenzae and Moraxella catarrhalis collected during 1999-2000 from 13 countries. Clin. Microbiol. Infect. 7:671-677. [DOI] [PubMed] [Google Scholar]
- 39.Tzeng, Y. L., A. Datta, V. K. Kolli, R. W. Carlson, and D. S. Stephens. 2002. Endotoxin of Neisseria meningitidis composed only of intact lipid A: inactivation of the meningococcal 3-deoxy-d-manno-octulosonic acid transferase. J. Bacteriol. 184:2379-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tzeng, Y. L., A. Datta, C. Strole, V. S. Kolli, M. R. Birck, W. P. Taylor, R. W. Carlson, R. W. Woodard, and D. S. Stephens. 2002. KpsF is the arabinose-5-phosphate isomerase required for 3-deoxy-D-manno-octulosonic acid biosynthesis and for both lipooligosaccharide assembly and capsular polysaccharide expression in Neisseria meningitidis. J. Biol. Chem. 277:24103-24113. [DOI] [PubMed] [Google Scholar]
- 41.Vaneechoutte, M., G. Verschraegen, G. Claeys, and A. M. Van Den Abeele. 1990. Serological typing of Branhamella catarrhalis strains on the basis of lipopolysaccharide antigens. J. Clin. Microbiol. 28:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verduin, C. M., C. Hol, A. Fleer, H. van Dijk, and A. van Belkum. 2002. Moraxella catarrhalis: from emerging to established pathogen. Clin. Microbiol. Rev. 15:125-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verduin, C. M., M. Jansze, C. Hol, T. E. Mollnes, J. Verhoef, and H. van Dijk. 1994. Differences in complement activation between complement-resistant and complement-sensitive Moraxella (Branhamella) catarrhalis strains occur at the level of membrane attack complex formation. Infect. Immun. 62:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Verduin, C. M., M. Kools-Sijmons, J. van der Plas, J. Vlooswijk, M. Tromp, H. van Dijk, J. Banks, H. Verbrugh, and A. van Belkum. 2000. Complement-resistant Moraxella catarrhalis forms a genetically distinct lineage within the species. FEMS Microbiol. Lett. 184:1-8. [DOI] [PubMed] [Google Scholar]
- 45.Walsh, A. G., L. L. Burrows, and J. S. Lam. 1999. Genetic and biochemical characterization of an operon involved in the biosynthesis of 3-deoxy-D-manno-octulosonic acid in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 173:27-33. [DOI] [PubMed] [Google Scholar]
- 46.Walsh, A. G., M. J. Matewish, L. L. Burrows, M. A. Monteiro, M. B. Perry, and J. S. Lam. 2000. Lipopolysaccharide core phosphates are required for viability and intrinsic drug resistance in Pseudomonas aeruginosa. Mol. Microbiol. 35:718-727. [DOI] [PubMed] [Google Scholar]
- 47.West, N. P., H. Jungnitz, J. T. Fitter, J. D. McArthur, C. A. Guzman, and M. J. Walker. 2000. Role of phosphoglucomutase of Bordetella bronchiseptica in lipopolysaccharide biosynthesis and virulence. Infect. Immun. 68:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zaleski, A., N. K. Scheffler, P. Densen, F. K. Lee, A. A. Campagnari, B. W. Gibson, and M. A. Apicella. 2000. Lipooligosaccharide Pk (Galα1-4Galβ1-4Glc) epitope of Moraxella catarrhalis is a factor in resistance to bactericidal activity mediated by normal human serum. Infect. Immun. 68:5261-5268. [DOI] [PMC free article] [PubMed] [Google Scholar]