Abstract
DNA replication in higher eukaryotic cells occurs at a large number of discrete sites called replication foci. We have previously purified a protein, focus-forming activity 1 (FFA-1), which is involved in the assembly of putative prereplication foci in Xenopus egg extracts. FFA-1 is the orthologue of the Werner syndrome gene product (WRN), a member of the RecQ helicase family. In this paper we show that FFA-1 colocalizes with sites of DNA synthesis and the single-stranded DNA binding protein, replication protein A (RPA), in nuclei reconstituted in the egg extract. In addition, we show that two glutathione S-transferase FFA-1 fusion proteins can inhibit DNA replication in a dominant negative manner. The dominant negative effect correlates with the incorporation of the fusion proteins into replication foci to form “hybrid foci,” which are unable to engage in DNA replication. At the biochemical level, RPA can interact with FFA-1 and specifically stimulates its DNA helicase activity. However, in the presence of the dominant negative mutant proteins, the stimulation is prevented. These results provide the first direct biochemical evidence of an important role for FFA-1 in DNA replication.
Keywords: FFA-1, Werner syndrome, Rec Q helicase, DNA replication, Xenopus laevis
Introduction
DNA replication in eukaryotic cells is initiated from a large number of origins distributed along chromosomes. In Saccharomyces cerevisiae, origins are composed of small pieces of DNA with specific sequence information. However, in other eukaryotes the relationship between origins and specific DNA sequences remains elusive (DePamphilis 1996). Some studies suggest that metazoan cells utilize specific DNA sequences as origins, whereas other studies suggest that replication can initiate from random DNA sequences. A careful analysis of these conflicting results has led to the model that metazoan chromosomes carry many potential origin sequences, but only a subset of them is actually selected for usage (DePamphilis 1993). As to the mechanism that further selects for an active origin among the potential ones, it appears that some kind of subnuclear structure may be involved. One striking feature about metazoan DNA replication is that it occurs at a large number of discrete sites called replication foci (or centers, factories) (Newport and Yan 1996). The existence of replication foci was first suggested by fiber autoradiography studies showing that replication bubbles were almost always observed in clusters of multiple similar-sized bubbles rather than single bubbles (Hand 1978). Such a pattern could be best explained if multiple consecutive replication origins were organized into a single unit and then initiated synchronously. Replication foci were later directly visualized by labeling somatic cells with bromodeoxyuridine or biotin-dUTP (Nakamura et al. 1986; Nakayasu and Berezney 1989). The replication-dependent incorporation of bromodeoxyuridine and biotin-dUTP clearly occurred at a large number of discrete foci within the nucleus. In addition, many proteins involved in DNA replication, such as proliferating cell nuclear antigen (Bravo and Macdonald-Bravo 1987; Leonhardt et al. 2000), DNA ligase I (Montecucco et al. 1995, Montecucco et al. 1998), CAF-1 (Krude 1995), DNA methyltransferase (Leonhardt et al. 1992; Liu et al. 1998), Cdk2/cyclin A, and replication protein A (RPA) (Cardoso et al. 1993), were found to be associated with these replication foci during DNA synthesis. Together, these observations have led to the suggestion that replication foci are formed by the clustering of multiple replicons. In this way, a real origin not only may contain certain DNA sequence information, but also have to be attached to replication foci (Newport and Yan 1996).
Despite the consistent observation of replication foci, little progress has been made toward understanding how these foci are assembled and whether they are important for DNA replication. It is very difficult to address these questions in somatic cells using visual observation methods. Extracts derived from unfertilized eggs, on the other hand, offer a good model system which allows a biochemical approach in addition to visual observation. In this system, a newly introduced DNA substrate, usually demembranated sperm chromatin, will induce nuclear envelope formation around itself, and the DNA is then replicated once semiconservatively (Lohka and Masui 1983; Blow and Laskey 1986; Newport 1987). As in somatic cells, replication in these reconstituted nuclei occurs at a large number of discrete foci (Mills et al. 1989; Cox and Laskey 1991). Moreover, structures similar to replication foci can also form on chromatin, even in the absence of membrane and consequently DNA replication (Adachi and Laemmli 1992, Adachi and Laemmli 1994; Yan and Newport 1995a). One component of these foci is the single-stranded DNA binding protein RPA, which is essential for eukaryotic DNA replication (Wold 1997). By fractionating the cytosol we have found that RPA associates with foci on chromatin only in the presence of another protein, focus forming activity 1 (FFA-1; Yan and Newport 1995b). Sequence analysis of the gene encoding FFA-1 has revealed an extensive homology between FFA-1 and the human Werner syndrome gene product (WRN) throughout the open reading frame, suggesting that FFA-1 is the true homologue (orthologue) of human WRN, the only one known outside mammals (Yan et al. 1998).
Werner syndrome (WS) is associated with the premature development of a variety of age-related diseases (Schellenberg et al. 1998). WRN gene encodes a member of the RecQ DNA helicase family (Yu et al. 1996), but unlike other members, it also contains an exonuclease domain similar to the proofreading exonuclease domain of Escherichia coli DNA polymerase I (Mushegian et al. 1997). The protein appears to concentrate in the nucleolus in human cells, but a nucleoplasmic localization has also been observed (Gray et al. 1998; Marciniak et al. 1998; Shiratori et al. 1999). However, upon replication arrest WRN migrates from nucleolus to discrete subnuclear foci characteristic of replication foci (Constantinou et al. 2000). Biochemical fractionation of mouse WRN suggests that at least some of it is associated with a large complex consisting of many replication proteins (Lebel et al. 1999). In vitro biochemical studies with recombinant WRN have confirmed that it possesses both helicase and exonuclease activities (Gray et al. 1997; Suzuki et al. 1997; Huang et al. 1998; Shen et al. 1998a,Shen et al. 1998b). In addition, WRN can stimulate the activity of DNA polymerase δ (Kamath-Loeb et al. 2000). At the cellular level, WS cells exhibit elevated rates of chromosomal rearrangements (Hoehn et al. 1975; Salk et al. 1981; Scappaticci et al. 1982). The rate of somatic mutations is also increased (Fukuchi et al. 1989, Fukuchi et al. 1990) and most of the mutations appear to be deletions of large segments of DNA (>20 kb) (Fukuchi et al. 1989). The genomic instability of WS does not appear to be due to a defect in DNA repair (Stefanini et al. 1989). Instead, DNA replication appears to be altered in WS cells: there are fewer initiation events for replication (Takeuchi et al. 1982a; Hanaoka et al. 1983), misfiring of origins (Fujiwara et al. 1985), reduced DNA chain elongation rates (Fujiwara et al. 1977), and a prolonged S phase (Takeuchi et al. 1982b). Although these phenotypes are consistent with a replication defect, WRN is not an essential gene in either humans or mice and as such the exact function of WRN remains a mystery.
With the cloning of FFA-1, we are able to directly test the function of FFA-1 and replication focus structure in DNA replication. In this paper we show that FFA-1 colocalizes with RPA and sites of DNA synthesis in nuclei reconstituted in interphase Xenopus egg extracts. We also demonstrate that two glutathione S-transferase (GST)–FFA-1 fusion proteins can act in a dominant negative way to inhibit DNA replication. Interestingly, these two fusion proteins are incorporated into replication foci to form “hybrid foci” that also contain RPA and the endogenous FFA-1 but are unable to engage in DNA replication. Finally, we show that RPA can interact with FFA-1 and strongly stimulate its helicase activity, but this stimulation is blocked by the two dominant negative fusion proteins. These results provide the first direct evidence for an important function of both FFA-1 and replication focus structure in DNA replication. They also suggest that WS is caused by defects in the DNA replication process.
Materials and Methods
Extract Preparation and Nuclei Reconstitution
Cytosol and membrane components of Xenopus egg extracts and demembranated sperm chromatin were prepared according to the procedure described by Smythe and Newport 1991. Nuclei were reconstituted at room temperature (22–24°C) by mixing cytosol (1/3 vol), membrane (1/10 vol), and ATP-regeneration system/sperm chromatin cocktail (1/10 vol; final concentration, 2 mM ATP, 20 mM phosphocreatine, 50 μg/ml creatine kinase, and 1,000 μl sperm chromatids). Egg lysis buffer (ELB: 10 mM Hepes, pH 7.5, 250 mM sucrose, 2.5 mM MgCl2, 50 mM KCl, 1 mM DTT) was used to make up the volume.
Preparation of Recombinant GST Fusion Proteins
Various fragments of FFA-1 cDNA were subcloned into the appropriate pGEX expression vectors downstream of the GST gene to create in frame fusions. The plasmids were transformed into E. coli BL21 (DE3) strain and the cells were grown at 37°C to OD600 = 0.3 and then induced with 0.5 mM IPTG for 3 h. The fusion proteins were purified on a glutathione affinity column following standard procedures (GST Gene Fusion System [1997]; Amersham Pharmacia Biotech). They were dialyzed three times against 100 vol of ELB at 4°C, frozen in liquid nitrogen in 5-μl aliquots, and stored at −80°C.
Antibody Preparation
The following antibodies were used in this study: rabbit anti-RPA (all three subunits), rat anti-RPA (large subunit), rabbit and rat anti–FFA-1 C (amino acids 1030–1436), and rabbit anti-GST. The rabbit anti-RPA (three subunits) was used directly, without further purification, as described previously (Fang and Newport 1993). All the other antibodies were raised against the gel-purified recombinant GST fusion proteins according to the standard procedure (Goding 1986). The antibodies were purified by first passing the sera through the affinity columns containing the corresponding fusion proteins (the affinity columns were prepared as described previously [Yan et al. 1993]). The bound antibodies were eluted with 50 mM glycine (pH 2.5) and then rapidly renatured with 1/10 vol of 1 M Tris.HCl (pH 8). Contaminating antibodies against GST and other E. coli proteins were then removed by another column containing the total proteins of E. coli cells overexpressing GST. Pure anti-GST antibody was finally obtained by purifying the contaminating antibodies eluted from the previous column again on a column containing purified GST.
Indirect Immunofluorescence Staining
Indirect immunofluorescence staining was performed as described previously (Yan and Newport 1995a). In brief, nuclei were fixed with an equal volume of 3% formaldehyde (Sigma-Aldrich)/2% sucrose/PBS. After 10 min, they were spun through 1 M sucrose/PBS onto coverslips and then treated with 0.1% Triton X-100/PBS for 5 min. The coverslips were then blocked with 10% FCS for 20 min and stained with the appropriate primary and secondary antibodies. For visual assays of DNA replication, 20 μM biotin-dCTP (GIBCO BRL) was added to the reaction and detected by Streptavidin–Texas red (Calbiochem). On average, two focal planes were required to observe all the foci in one nucleus. Therefore, the total number of foci was calculated by doubling the number of foci on the focal plane that cut across the middle of the nucleus. Images were collected with a monochrome DAGE-MTI cooled CCD-300-RT camera run under Image v1.6.1 (Scion) and processed in Photoshop® v5.5 (Adobe Systems).
DNA Helicase Assay
The substrate for DNA helicase assays is a DNA sequencing reaction product. In brief, the −40 sequencing primer was hybridized to ss-M13mp18 DNA and then extended by Sequenase (USB Biochemicals) in the presence of dNTP, [32P]dATP (NEN Life Science Products), and dilute ddTTP. The DNA was then purified on microspin S-300 HR columns (Amersham Pharmacia Biotech) and diluted to 5 ng/μl in TES buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA, 100 mM NaCl). Each reaction contains 5 ng of DNA substrate, 2 mM ATP/MgCl2, and various proteins at the indicated final concentrations. For the helicase inhibition experiments, the fusion proteins were preincubated with FFA-1 and RPA at room temperature for 20 min before the addition of DNA and ATP. The helicase reactions were terminated after a 30-min incubation at room temperature by mixing with equal volumes of stop solution (0.5% SDS, 20 mM EDTA) and then separated on 8% polyacrylamide/TAE gels.
Replication Assays
Radioactive assays of DNA replication were carried out by including 1 μCi [32P]dATP in each replication reaction. Samples were taken at various time points, processed with proteinase K, and separated on 1% agarose TAE gels as described previously (Smythe and Newport 1991). For replication run-on assays, nuclei were assembled in the presence of 50 μg/μl of aphidicolin (Sigma-Aldrich) for 80 min, diluted two times with PBS, and spun through 1 M sucrose/PBS onto coverslips. The coverslips were then placed over 100 μl of dNTP solution (5 mM dATP, dGTP, TTP, and biotin-dCTP) on parafilm. After 20 s of incubation, they were fixed and stained for FFA-1 and biotin-dCTP following the same immunofluorescence staining procedure as described above.
Protein Interaction Assays and Immunoprecipitation
For immunoprecipitation, 20 μl of cytosol was diluted to 50 μl with ELB and then incubated with 10 μl of Affi-gel protein A beads (Bio-Rad Laboratories) precoated with 20 μl of the various antibodies (purified rabbit anti–FFA-1 C and rabbit IgG, rabbit anti-RPA serum, and preimmune) at 4°C for 1 h. For the mapping of the interaction domain, 5 μl of the fusion proteins (1 μg) was incubated with 10 μl of cytosol (ELB was added to make the final volume 30 μl) at room temperature for 1 h. For the interaction between the fusion proteins and the purified RPA, 5 μl of the fusion proteins (1 μg) was pretreated with 1 μl of DNase I (40 U; Sigma-Aldrich) or ELB at room temperature for 10 min and then mixed with 5 μl of Xenopus RPA (0.1 μg; Fang and Newport 1993) and 19 μl of ELB. After 1 h at room temperature, the reactions were mixed with 10 μl of glutathione-agarose beads (Sigma-Aldrich) at 4°C for another 1 h. The beads (Affi-gel protein A or glutathione-agarose) were brought down by low speed centrifugation and washed four times with 0.5 ml of ELB with 0.02% NP-40. The proteins bound to the beads were separated on 8% SDS-PAGE, transferred to PVDF, probed with the purified rat anti–FFA-1 C or anti-RPAp70, and detected by SuperSignal chemiluminescence (Pierce Chemical Co.).
Results
Colocalization of FFA-1 with RPA and Replication Foci
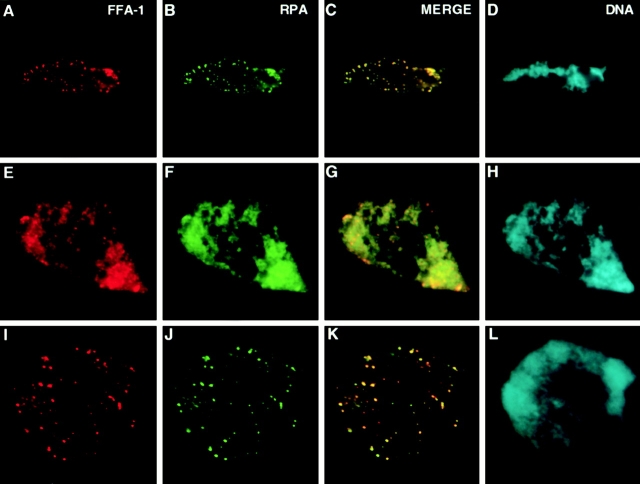
We first used indirect immunofluorescence staining to establish the relationship among FFA-1, RPA, and sites of DNA replication in nuclei reconstituted in Xenopus egg extracts. To do this, nuclei were reconstituted by mixing demembranated sperm chromatin and egg extracts. After various lengths of time they were fixed and then stained for FFA-1, RPA, and DNA synthesis. Replication began asynchronously, but generally occurred between 45 and 85 min in most nuclei. It was monitored by the incorporation of biotin-dCTP which was added to the reaction 5 min before fixation. As shown in Fig. 1, FFA-1 and RPA displayed an almost identical spatial distribution throughout DNA replication. During the early stage (Fig. 1, A–D; 40–45 min), they formed discrete foci on chromatin. Replication was not detectable in most nuclei, which is consistent with the observation that focus structure is formed before DNA synthesis. But when replication was detected, it occurred at a subset of FFA-1 foci (Fig. 2, A–C), suggesting that FFA-1 foci are indeed where replication is initiated. During the middle stage, both FFA-1 and RPA showed a more diffuse staining colocalizing with chromatin (Fig. 1, E–H; 45–85 min). Replication proceeded rapidly and was also detected throughout chromatin (Fig. 2, D–F). The diffuse FFA-1 and RPA staining subsided by the end of this stage and FFA-1 and RPA could again be detected as foci which often showed incorporation of biotin-dCTP (data not shown). During the late stage (after 85 min), replication was no longer detectable (data not shown), the diffuse staining of FFA-1 and RPA further decreased, but more discrete foci containing FFA-1 and RPA appeared again (Fig. 1, I–L) and persisted up to at least 150 min (the maximum time assayed).
Figure 1.
Colocalization of FFA-1 and RPA in nuclei reconstituted around sperm chromatin in the interphase egg extract. Representative nuclei from early (40–45 min; A–D), middle (45–85 min; E–H), and late (85–150 min; I–L) stages are shown. (A, E, and I) FFA-1 staining. (B, F, and J) RPA staining. (C, G, and K) Merge of FFA-1 staining and RPA staining. (D, H, and L) DNA staining by Hoechst.
Figure 2.
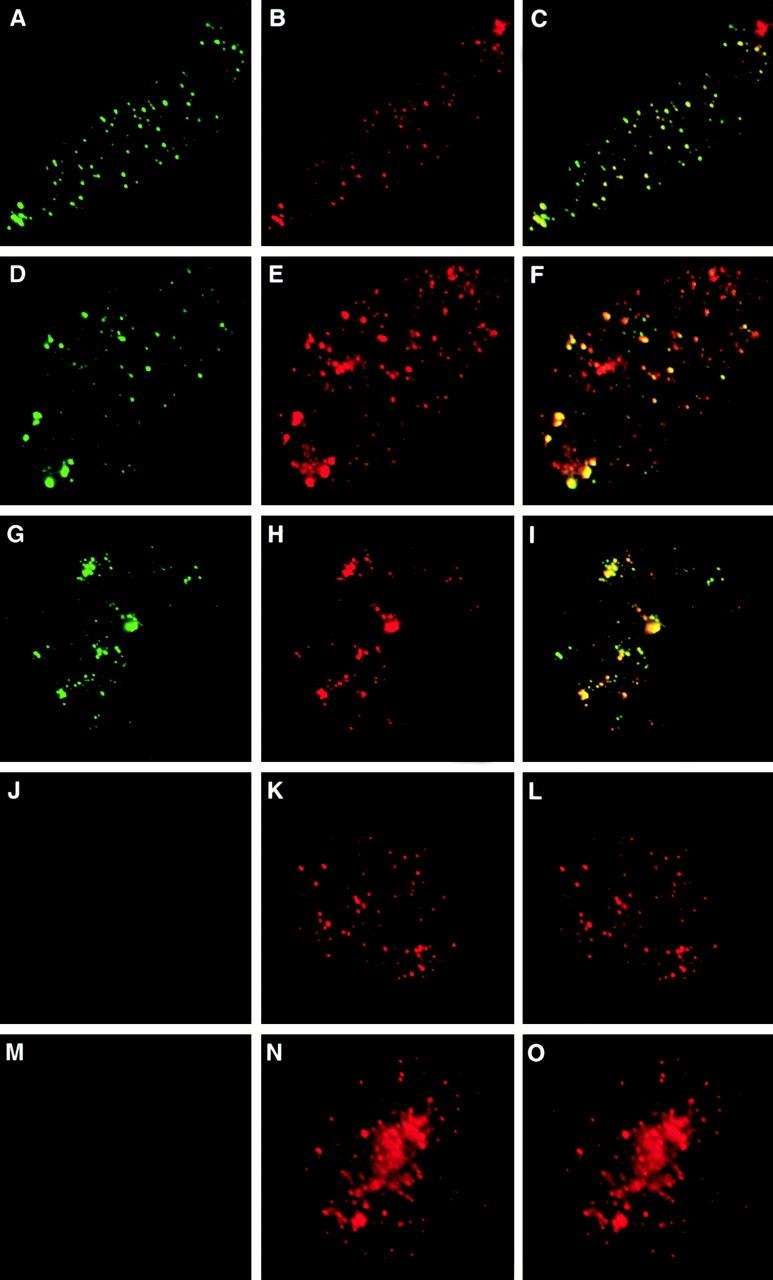
Colocalization of FFA-1 and sites of DNA synthesis. (A–F) In normal reconstituted nuclei. Biotin-dCTP was added 5 min before fixation. (A–C) 45 min. (D–F) 60 min. (G–L) In reconstituted nuclei formed in the presence of aphidicolin. (G–I) An aphidicolin-arrested nucleus directly fixed and stained. Biotin-dCTP was added to the reaction at the beginning. (H–L) An aphidicolin-arrested nucleus pelleted through 1 M sucrose cushion and then labeled with dATP, dGTP, TTP, and biotin-dCTP. (A, D, G, and J) FFA-1 staining. (B, E, H, and K) Biotin staining. (C, F, I, and L) Merge of FFA-1 staining and biotin staining.
The more diffuse staining of FFA-1 and RPA was always observed in the nuclei that displayed extensive DNA synthesis, suggesting that the two phenomena might be mechanistically linked. A reasonable explanation is that during DNA synthesis more single-stranded DNA is generated, which in turn attracts more RPA and FFA-1 and the foci consequently appear expanded in size. To test this hypothesis, we blocked DNA synthesis at the priming step with aphidicolin, which inhibits DNA polymerases α and δ (Melendy and Stillman 1991). As shown in Fig. 2G–I, aphidicolin completely blocked the incorporation of biotin-dCTP, yet FFA-1 foci still formed and their number increased to >1,000 after 80 min of incubation. To determine whether these foci were where replication would have occurred, we designed a replication run-on experiment based on the observation that replication elongation resumes synchronously after removal of aphidicolin (Strausfeld et al. 1994). In brief, the aphidicolin-arrested nuclei were spun onto a coverslip through a 1 M sucrose cushion. The nuclei were then incubated with a mixture of dATP, dGTP, TTP, and biotin-dCTP to allow replication elongation to resume for a short period of time (usually 20 s). As shown in Fig. 2J–L, the sites of biotin-dCTP incorporation overlapped with FFA-1 foci, suggesting that FFA-1 foci are indeed sites for DNA replication even in the middle stage nuclei.
Immunodepletion of FFA-1 on DNA Replication
We then directly tested the role of FFA-1 in DNA replication by immunodepleting it from egg extracts. As shown in Fig. 3 A, the anti–FFA-1 antibody but not the control antibody removed FFA-1 to a level below detection (>98% depletion). Yet when assayed for DNA replication in the nuclei reconstituted with the depleted extracts, no significant difference was observed between the two, suggesting that FFA-1 is not essential for DNA replication (Fig. 3 B). However, we also failed to notice a significant reduction in the ability of the FFA-1–depleted extract to form RPA foci (Fig. 3C and Fig. D). Although it remains formally possible that our depletion may have been incomplete, in the context that neither human WRN nor its mouse homologue is an essential gene, this experiment suggests that there is at least one more focus-forming activity in the egg extract. It should be mentioned that this result does not contradict our previous report that depletion of FFA-1 blocks RPA focus formation (Yan et al. 1998). In those experiments we used partially purified column fractions of RPA and FFA-1 as the starting material for depletion and presumably the redundant activity had already been removed.
Figure 3.

The effect of FFA-1 depletion on DNA replication and RPA focus formation. (A) Western blot for FFA-1 in the extracts depleted with either anti–FFA-1 antibody or control antibody. Arrowhead indicates the position of FFA-1. (B) DNA synthesis in nuclei reconstituted around sperm chromatin with the depleted extracts. Samples were taken at the times indicated. (C and D) Immunofluorescence staining for RPA on sperm chromatin incubated in either FFA-1–depleted (C) or control-depleted cytosol (D).
Inhibitory Effect of Dominant Negative FFA-1 Mutants on DNA Replication in Reconstituted Nuclei
Therefore, we tried an alternative approach by using dominant negative mutants. FFA-1's involvement in focus formation suggests that it should interact with itself and/or other proteins. Therefore, it should be possible to make mutants that are functionally deficient, but still capable of protein–protein interaction. Adding such mutant proteins to the extract may interfere with the function of not only the endogenous FFA-1 protein but also the redundant protein.
We tested several GST fusion proteins containing various regions of FFA-1 (Fig. 4B and Fig. C) for inhibitory effects on DNA replication. Nuclei were reconstituted in the extract in the presence of the fusion proteins and [32P]dATP for various lengths of time. The incorporation of [32P]dATP into DNA was assayed by running the replication products on agarose gels. As shown in Fig. 4 A, GST-Xho (containing FFA-1 amino acids 1–587, just upstream of the helicase domain) efficiently inhibited DNA synthesis (5–10-fold) (Fig. 4 A). GST alone did not exert any effect on DNA synthesis. The inhibition was most effective when GST-Xho was added at the beginning of the reaction and was leaky as DNA synthesis did occur later. The minimal concentration of GST-Xho to inhibit DNA synthesis was ∼400 nM (data not shown), about 80 times that of the endogenous FFA-1 (estimated at 5 nM in the reaction). This is most likely an overestimate, because not all the fusion protein is in active conformation and the concentration of the putative redundant protein is not taken into account.
Figure 4.

Effect of GST–FFA-1 fusion protein on DNA replication. (A) Replication of nuclei reconstituted around sperm chromatin in the presence of various fusion proteins (500 nM final concentration for GST-Xho and GST-Stu/Xho; 1 μM for GST-Stu and GST) or buffer. The fusion proteins and [32P]dATP were added at the beginning of the reactions. Samples were taken at the indicated times and separated on 0.8% agarose/TAE gel. The top band is the DNA that was too large to enter the gel and the bottom band is the sheared DNA (>20 kb) introduced during sample preparation. (B) Diagram of the GST–FFA-1 fusion proteins used in this study. Numbers indicate the positions of the amino acids that demarcate the various domains in FFA-1. (C) The fusion proteins used in this study separated on a 12% SDS polyacrylamide gel and stained by Coomassie blue. Molecular weight markers are labeled on the right in kilodaltons.
Although these characteristics are consistent with a dominant negative effect, there are two potentially trivial explanations. The first is that GST-Xho nonspecifically inactivates the egg extract. This is not the case, however, since nuclear reformation was not significantly affected (see below; Fig. 5, A–F). The second is that since GST-Xho contains the putative 3′→5′ exonuclease domain, it may inhibit replication simply by degrading DNA. To rule out the this possibility, we expressed and tested a smaller fusion protein, GST-Stu (containing FFA-1 amino acids 1–296), which still contains the nuclease domain. In vitro assays indicated that it possessed good 3′→5′ exonuclease activity (data not shown). However, it did not exert any significant inhibitory effect on DNA replication in reconstituted nuclei even at twice the molar concentration of GST-Xho (Fig. 4 A). This result suggests that the inhibitory domain is located in the region spanning amino acids 297–587, which lies between the nuclease domain and the helicase domain. When the GST fusion protein containing this region (GST-Stu/Xho) was expressed and tested, it indeed effectively inhibited DNA replication at about the same concentration as that of GST-Xho (Fig. 4 A). Together these experiments, in combination with the immunofluorescence studies, strongly suggest that FFA-1 participates in DNA replication in reconstituted nuclei.
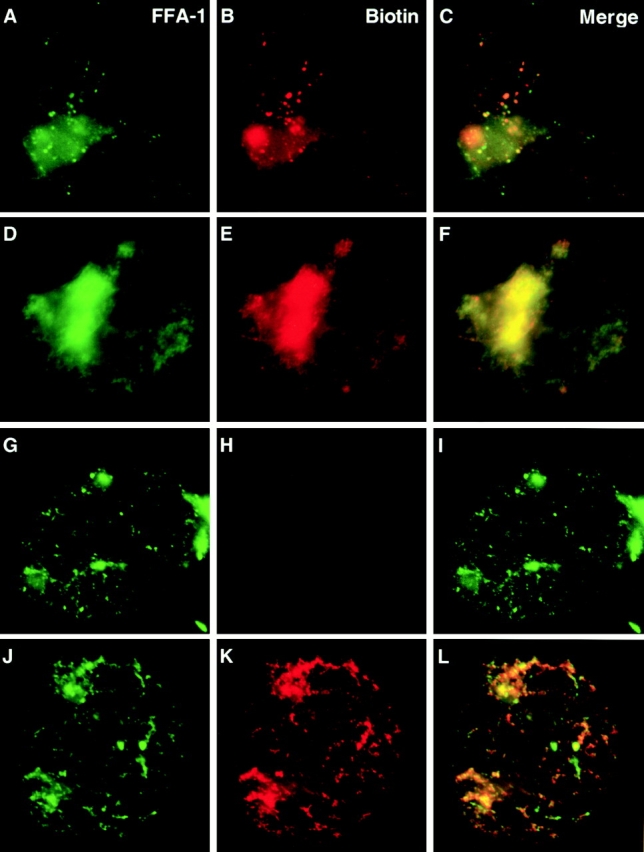
Figure 5.
Formation of hybrid foci. (A–F) Nuclei were reconstituted around sperm chromatin for 52 min in the presence of GST-Xho and stained with the affinity-purified anti-GST (A and D), RPA (B), or FFA-1 (E). (C) Merge of GST staining and RPA staining. (F) Merge of GST staining and FFA-1 staining. The anti–FFA-1 antibody is against the COOH-terminal end of FFA-1 and does not react with GST-Xho. (G–O) Nuclei reconstituted in the presence of GST-Stu/Xho (G–I), GST-Stu (J–L), and GST (M–O) for 52 min. (G, J, and M) GST staining (green). (H, K, and N) RPA staining (red). (I, L, and O) Merge of GST and RPA staining.
Formation of Hybrid Replication Foci
The presence of FFA-1 at replication foci raises the question of whether the inhibitory fusion proteins block DNA replication by interfering with the assembly, or the activity of replication foci. To address this issue, we determined the localization of the GST-Xho fusion protein by immunofluorescence staining. Interestingly, GST-Xho was itself incorporated into 300–500 discrete foci (Fig. 5, A–F). The GST-Xho foci were mostly hybrid in nature because they also contained RPA (Fig. 5 B) and the endogenous FFA-1 (Fig. 5 E). They were well formed by 52 min, but did not show incorporation of biotin-dCTP, whereas extensive incorporation of biotin-dCTP had already occurred by this time in the control nuclei. In addition, whereas normal foci tend to expand in size during DNA synthesis, the GST foci changed very little in size upon further incubation (data not shown).
In support of the correlation between the formation of “hybrid foci” and the inhibitory effect on replication, we found that the number of foci containing GST-Xho per nucleus decreased dramatically from 300–500 at 400 nM down to <50 at 100 nM, which correlates well with the precipitous drop in replication inhibition by GST-Xho at the lower concentration (data not shown). Moreover, we found that GST-Stu/Xho, which can inhibit DNA replication, also induced the formation of hybrid foci within reconstituted nuclei (Fig. 5, G–I). In contrast, GST and GST-Stu, which cannot inhibit DNA replication, could not form any foci detectable by the anti-GST antibody (Fig. 5, J–O). Together, these observations strongly suggest that the Stu/Xho domain inhibits DNA replication by forming “hybrid foci” that are incapable of executing DNA synthesis.
Physical Interaction between FFA-1 and RPA
We then studied the biochemical mechanism by which the dominant negative fusion proteins inhibit DNA synthesis at “hybrid foci.” The simplest explanation is that the fusion proteins displace the endogenous FFA-1 and the redundant protein from “hybrid foci.” However, since the endogenous FFA-1 is still present at “hybrid foci,” this explanation is unlikely to be the case. The second explanation is that, since helicases usually act as oligomers, the dominant negative fusion proteins, which do not contain the helicase domain, might interact with the endogenous FFA-1, leading to the formation of inactive helicase oligomers. We have tested this idea, but found no interaction between the fusion proteins and FFA-1 (data not shown); as such it is also unlikely to be true. The third explanation is based on the report that the helicase activity of human WRN can be stimulated by the human RPA protein, probably by protein–protein interaction (Shen et al. 1998b; Brosh et al. 1999). Conceivably, the helicase activity of FFA-1 may also be stimulated by RPA through protein–protein interaction. If the dominant negative fusion proteins can also interact with RPA, they may then interfere with this stimulation. To test this idea, we first determined whether FFA-1 could indeed interact with RPA by coimmunoprecipitation. Protein A beads were coated with different antibodies and then incubated with the cytosol. The proteins bound to the beads were then separated on SDS-PAGE, transferred to PVDF membranes, and probed with either anti–FFA-1 or anti-RPA antibodies. As shown in Fig. 6 A, the anti–FFA-1 antibody brought down not only FFA-1, but also a small amount of RPA. Conversely, the anti-RPA antibody brought down RPA and a small amount of FFA-1. In neither case did the control antibody bring down FFA-1 and RPA. This experiment suggests that FFA-1 and RPA can physically interact with each other.
Figure 6.

Interaction between FFA-1 and RPA. (A) Coimmunoprecipitation of FFA-1 and RPA. Western blot analysis of the proteins brought down from the cytosol by the Affi-gel protein A beads precoated with the indicated antibodies. Blots were probed with the rabbit anti–FFA-1C (top) and rabbit anti-RPA (bottom). (B) Mapping of the RPA interaction domain in FFA-1. The various GST–FFA-1 fusion proteins were incubated with the cytosol and then brought down by glutathione beads. The bound proteins were then subject to Western blot analysis with the purified rat anti-RPA antibody. (C) Interaction between GST-Xho and the purified Xenopus RPA in the presence or absence of DNase I. The proteins brought down by glutathione beads were analyzed by Western blot with the purified rat anti-RPA antibody. (D) Amount of RPA bound to GST-Stu/Xho. GST-Stu/Xho (500 nM) was incubated with 5 μl of cytosol in a 15-μl reaction and then brought down by glutathione beads. Indicated amounts (as percentages of cytosol) of the beads and supernatant fractions were analyzed by Western blot with the purified rat anti-RPA antibody.
We then determined which region of FFA-1 mediates its interaction with RPA. Various GST–FFA-1 fusion proteins were incubated in the cytosol and then brought down by glutathione-agarose beads. Proteins bound to the beads were analyzed by Western blot for the presence of RPA. As shown in Fig. 6 B, RPA was efficiently brought down with GST-Xho and GST-Stu/Xho, but not with GST or GST-Stu. Two other fusions containing the middle and COOH-terminal regions of FFA-1 also failed to bring down RPA (data not shown). Therefore, the Stu/Xho domain, which can inhibit DNA replication and induce the formation of “hybrid foci,” also mediates FFA-1's interaction with RPA. Further characterization revealed that the interaction can still be observed between the fusion proteins and the purified RPA, and in the presence of DNase I (Fig. 6 C), suggesting that it is by direct contact, rather than indirectly mediated by other proteins or DNA. These results are consistent with the colocalization of FFA-1, RPA, and the GST fusions containing the Stu/Xho domain at “hybrid foci.”
The direct interaction between the Stu/Xho domain and RPA raised the possibility that replication inhibition is due to the titration of RPA, rather than the interference with FFA-1 activity. To test this possibility, we determined whether RPA was completely bound to the GST-Stu/Xho fusion protein in the pull-down assay. As shown in Fig. 6 D, at the concentration effective for replication inhibition, the fusion protein bound to only a small fraction of RPA (∼10%). This result suggests that FFA-1 interacts with only a subset of RPA and the dominant negative effect is not due to the titration of RPA.
Functional Effect of FFA-1–RPA Interaction on FFA-1 Helicase Activity
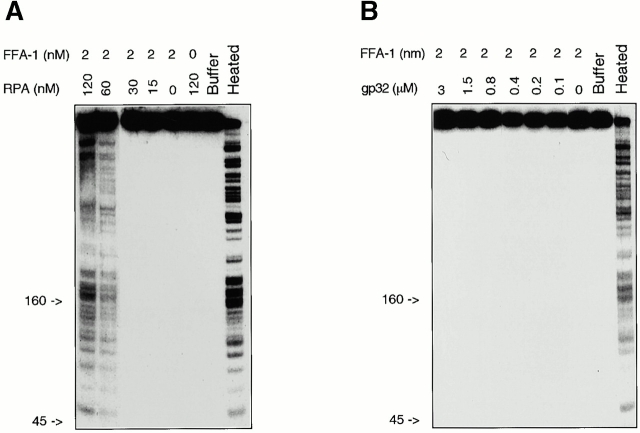
We then determined whether the interaction between FFA-1 and RPA could augment the DNA helicase activity of FFA-1. FFA-1 was incubated with a radioactively labeled DNA substrate and various amounts of RPA in the presence of ATP. The substrate was made by extending a primer hybridized to single-stranded m13 DNA in the presence of [32P]dATP, dNTPs, and dilute ddTTP. The rationale for using this type of substrate is that RPA may preferentially stimulate the unwinding of long DNA strands. After incubation, the reactions were terminated and separated on 8% polyacrylamide gels. The undissociated fragments could not enter the gel due to the large size of m13 DNA, whereas the dissociated fragments would run as a series of bands of different sizes. As shown in Fig. 7 A, FFA-1 alone showed very weak unwinding activity at the concentration used in this experiment (∼2 nM). However, the unwinding of both short and long fragments was greatly stimulated by RPA at a concentration as low as 60 nM. RPA alone, even at the highest concentration (120 nM), did not cause any significant dissociation of the substrate.
Figure 7.
Effect of RPA and gp32 on the helicase activity of FFA-1. (A) Helicase reactions containing the indicated amounts of FFA-1 and RPA (in nM). (B) Helicase reactions containing the indicated amounts of FFA-1 (in nM) and gp32 (in μM). In both A and B, the second lanes from the right contain buffer only, whereas the rightmost lanes contain the heated DNA substrate. All the reactions also contain 2.5 μM (in nucleotides) DNA and 2 mM ATP. Indicated on the left of each gel are the migration positions of double-stranded DNA markers.
There are two likely mechanisms to explain how RPA might stimulate FFA-1 helicase activity. The more trivial explanation is that RPA simply prevents the dissociated DNA from reassociation. Alternatively, the effect may be due to the specific interaction between the two proteins. To differentiate between these two possibilities, we tested the effect on FFA-1 helicase activity by another single-stranded DNA binding protein, the gene 32 protein of E. coli phage T4 (gp32). As shown in Fig. 7 B, this protein had no significant stimulatory effect on the unwinding reaction over a wide range of concentrations (from 37.5 nM to 3 μM). The size of binding site of gp32 is seven nucleotides (Ferrari et al. 1994), so 3 μM of protein is in ∼10-fold excess over the number of binding sites. In contrast, assuming that the size of binding site of Xenopus RPA is similar to that of human RPA (30 nucleotides; Wold 1997), 60 nM of RPA occupies at most 75% of the binding sites. These results suggest that the stimulation on FFA-1 helicase activity by RPA depends on the direct interaction between the two proteins.
Effect of GST–FFA-1 Fusion Proteins on FFA-1/RPA Helicase Activity
Since GST-Xho and GST-Stu/Xho can interact with RPA, we then addressed whether they can inhibit the stimulation of FFA-1 helicase activity by RPA. To do this, various amounts of the fusion proteins were added to the helicase reaction containing FFA-1 and RPA. As shown in Fig. 8 A, GST-Xho and GST-Stu/Xho inhibited the unwinding reaction in a dose-dependent manner. In contrast, GST-Stu had no effect on unwinding, even at the highest concentration (2.4 μM). In the absence of RPA, FFA-1 had a low intrinsic helicase activity, particularly on small fragments. But as shown in Fig. 8 B, this activity was not significantly affected by the fusion proteins. In addition, the affinity of RPA for single-stranded DNA was not affected by the fusion proteins (data not shown). Collectively, these results support the interpretation that RPA stimulates FFA-1 through direct protein–protein interaction and this stimulation is blocked by the two fusion proteins containing the Stu/Xho domain.
Figure 8.
Effect of GST–FFA-1 fusion proteins on FFA-1 helicase activity. (A) In the presence of RPA. Lanes 1–10 contain 2 nM FFA-1 and 60 nM RPA, but lanes 1–9 also contain the various fusion proteins at the indicated final concentrations (in μM). Lane 11 contains the substrate incubated in buffer only and lane 12 contains the heated substrate. (B) In the absence of RPA. Lanes 1–3 contain 5 nM FFA-1 and GST-Xho (0.75 μM), GST-Stu/Xho (0.75 μM), and GST-Stu (1.5 μM). Lane 4 contains 5 nM FFA-1 only, lane 5 contains the substrate incubated in buffer only, and lane 6 contains the heated substrate. More FFA-1 was used in this experiment to better detect the weak helicase activity in the absence of RPA. The film was also exposed four times as long as that of A. All the reactions contain 2.5 μM DNA and 2 mM ATP.
It is worth mentioning that the concentrations of the proteins in the helicase inhibition experiment agree reasonably well with those in the replication inhibition experiment. In the helicase experiment, the concentrations for FFA-1, RPA, and the two dominant negative fusion proteins are 2, 60, and 300 nM (minimum), respectively. In the replication experiment, the corresponding concentrations are 5, 33, and 400 nM (minimum). Therefore, it is reasonable to believe that the FFA-1 helicase activity is also stimulated by RPA during normal DNA replication and that this stimulation is abolished by the two dominant negative fusion proteins at “hybrid foci.”
Discussion
Summary
In this paper, we have shown that FFA-1 colocalizes with RPA foci from which DNA replication then initiates in nuclei reconstituted in Xenopus egg extracts. In addition, we have found that two fusion proteins containing the Stu/Xho domain of FFA-1 can act in a dominant negative manner to block DNA replication in reconstituted nuclei. These results strongly suggest that FFA-1 participates in DNA replication. At the biochemical level, we have confirmed that RPA can physically interact with FFA-1 and greatly stimulates its helicase activity. As to the mechanism of the dominant negative effect by the two fusion proteins, we have observed that they can be incorporated into foci to form “hybrid foci” that are unable to engage in DNA synthesis. Interestingly, the same Stu/Xho domain that mediates the inhibition of DNA replication also mediates the interaction between FFA-1 and RPA. Moreover, the two inhibitory fusion proteins can block the stimulation of FFA-1 DNA helicase activity by RPA, presumably by competing against the endogenous FFA-1 protein for interaction with RPA.
FFA-1 Helicase and DNA Replication
The results with the dominant negative mutants and immunofluorescence staining strongly suggest that FFA-1 plays an important role in DNA replication. In support of this conclusion, human WS cells display many phenotypes that are consistent with defects in DNA replication (Schellenberg et al. 1998). In addition, biochemical fractionation of mouse WRN suggests that it is present in “replisome,” a large complex consisting of many replication proteins, including proliferating cell nuclear antigen and topoisomerase I (Lebel et al. 1999). It has also been demonstrated recently that human WRN can stimulate DNA polymerase δ (Kamath-Loeb et al. 2000). Therefore, it is reasonable to conclude that at least one function for FFA-1 and WRN is in DNA replication. The unsettled issue is whether this function is essential for or only peripheral to DNA replication. Our depletion data suggest that removal of FFA-1 from total interphase extracts yields only a minor effect on DNA replication. Similarly, neither human nor mouse WRN gene is essential for cell viability. Although these observations indicate that FFA-1 and WRN are not essential for DNA replication, they do not necessarily suggest that the function executed by FFA-1 and WRN is not essential. Considering that WRN is only one of five known RecQ family members in human cells, it is very likely that other members can partially compensate for the loss of WRN and FFA-1. In support of this hypothesis, the FFA-1 depletion experiment suggests that there is at least one additional focus-forming activity in Xenopus egg extracts. Similarly, in S. cerevisiae, two helicases, SGS1 and SRS2, are partially redundant to each other and disruption of both genes causes cells to arrest with an unreplicated genome (Lee et al. 1999) or a slow growth phenotype (Gangloff et al. 2000). Although SGS1 is a RecQ family member, SRS2 is not, suggesting that the redundant protein for FFA-1 and WRN does not have to be a RecQ family member. In higher eukaryotes, database searches have not detected any protein with extensive homology to Srs2 so far, yet a functional homologue of SRS2 may still exist.
Nature of RPA Foci in Egg Extracts
RPA foci can form on sperm chromatin in interphase cytosol in the absence of membranes (Adachi and Laemmli 1992). Because DNA replication does not occur under this condition, it is assumed that these RPA foci are formed “prereplicatively.” Later studies in mammalian cells detected RPA foci in S phase, but not G1 phase nuclei (Dimitrova et al. 1999). One simple explanation is that the RPA foci formed in the absence of membranes are qualitatively different from the replication foci formed inside reconstituted nuclei. However, there is another likely explanation. If replication forks are arrested with aphidicolin, a large number of FFA-1 (and RPA) containing foci are observed. The minimal number is 1,000 per nucleus, whereas in many nuclei the staining has a granular appearance, suggesting an even higher number of foci. This is much more than what can be formed on sperm chromatin decondensed in cytosol only (at most 200 per sperm chromatin; Adachi and Laemmli 1992; Yan and Newport 1995a). Therefore, the optimal condition for focus formation is inside reconstituted nuclei (which are equivalent to S phase nuclei in somatic cells), whereas the foci formed in cytosol alone most likely represent the basal level due to the large amount of replication proteins stockpiled in eggs. In somatic cells, replication proteins are not overproduced and therefore only S phase nuclei provide the right condition for efficient RPA focus formation.
Replication Foci and DNA Replication
Replication foci have been observed by many laboratories with various detection methods, but their importance in DNA replication has not been determined. The data in this paper suggest that replication foci do play an important role in DNA replication. Immunofluorescence staining suggests that replication does occur at foci containing RPA and FFA-1. Moreover, the two dominant negative mutant proteins do not block the assembly of replication foci, but instead are incorporated into the so-called “hybrid foci,” which also contain RPA and the endogenous FFA-1. At the biochemical level, our data suggest that the inhibition on DNA replication is due, at least in part, to the block of stimulation of FFA-1 helicase activity by RPA. Presumably, at normal foci the helicase activity of FFA-1 is greatly stimulated by the interaction with RPA. At “hybrid foci,” however, we infer that FFA-1 cannot interact with RPA due to competition from the fusion proteins and consequently cannot efficiently unwind DNA.
FFA-1 and Other Replicative Helicases
As a DNA helicase, one role of FFA-1 in DNA replication is presumably to unwind DNA. What then is the relationship between FFA-1 and the other helicases, such as the mini-chromosome maintenance proteins which are also implicated in DNA replication (Tye 1999)? Although in vitro replication of small genomes like SV40 DNA involves only one replicative helicase, accumulating evidence suggests that in vivo replication of genomic DNA requires multiple helicases. In S. cerevisiae, in addition to the MCMs, SGS1, and SRS2, the DNA2 helicase is also required for DNA replication and appears to be involved in the maturation of Okazaki fragments (Budd et al. 1995; Budd and Campbell 1997). In addition, two other helicases, PIF1 and RRM3, participate in the replication of rDNA regions with opposing roles. Although RRM3 promotes replication fork progression throughout rDNA, PIF1 stops the leftward fork movement at the replication fork barrier located near the 3′ end of the 35S transcription unit (Ivessa et al. 2000). The multitude of helicases involved in cellular DNA replication is probably an adaptation to the enormous complexity of genomic DNA, both in size and in organization. Conceivably, in eukaryotic cells, the MCMs may facilitate in the initial unwinding of origins, but other helicases, like SGS1, SRS2, or DNA2, may take over at a later stage. In addition, certain regions like the rDNA repeat locus may be compacted differently from the rest of the genome and may require special helicases, like PIF1 and RRM3, to properly unwind them without irreversibly disrupting the local chromatin organization. As to FFA-1 and WRN, one likely role for them is to organize replication foci. But since FFA-1 associates with DNA throughout replication and human WRN interacts with other replication proteins and stimulates DNA polymerase δ, FFA-1 and WRN may also participate in the elongation stage. As to the potential mechanistic role in elongation, Constantinou et al. 2000 have recently reported that WRN has an activity to translocate Holliday junction and, upon replication arrest, it migrates from nucleolus to discrete RPA-containing subnuclear foci characteristic of replication foci. In addition, genetic analyses in E. coli, budding yeast, and fission yeast suggest that RecQ type of helicases is involved in the stabilization of replication forks (Stewart et al. 1997; Courcelle and Hanawalt 1999; Frei and Gasser 2000). In particular, the slow growth phenotype of Sgs1 Srs2 double mutant can be rescued by the inactivation of the homologous recombination pathway (Gangloff et al. 2000). Together, these observations suggest that FFA-1 and WRN may protect stalled replication forks from aberrant recombination events. These events are probably rare in normal somatic cells, but more prevalent in nuclei reconstituted in egg extracts owing to the much larger number of replication forks in the latter system. Further study is required to uncover the details of how FFA-1/WRN and other RecQ helicases coordinate the replication machinery and the recombination machinery to ensure the accurate duplication of cellular DNA.
Acknowledgments
We thank Drs. John Burch and Ken Zaret for critically reading the manuscript.
Supported by grants from the V Foundation, Ellison Medical Foundation, and the National Institutes of Health (R01-GM57962-02).
Footnotes
Abbreviations used in this paper: ELB, egg lysis buffer; FFA-1, focus-forming activity 1; GST, glutathione S-transferase; RPA, replication protein A; WRN, WS gene product; WS, Werner syndrome.
References
- Adachi Y., U.K. Laemmli Identification of nuclear pre-replication centers poised for DNA synthesis in Xenopus egg extractsimmunolocalization study of replication protein A. J. Cell Biol. 1992;119:1–15. doi: 10.1083/jcb.119.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi Y., U.K. Laemmli Study of the cell cycle-dependent assembly of the DNA pre-replication centres in Xenopus egg extracts. EMBO (Eur. Mol. Biol. Organ.) J. 1994;13:4153–4164. doi: 10.1002/j.1460-2075.1994.tb06733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow J.J., Laskey R.A. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Bravo R., Macdonald-Bravo H. Existence of two populations of cyclin/proliferating cell nuclear antigen during the cell cycleassociation with DNA replication sites. J. Cell Biol. 1987;105:1549–1554. doi: 10.1083/jcb.105.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh R.M., Jr., Orren D.K., Nehlin J.O., Ravn P.H., Kenny M.K., Machwe A., Bohr V.A. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- Budd M.E., Campbell J.L. A yeast replicative helicase, Dna2 helicase, interacts with yeast FEN-1 nuclease in carrying out its essential function. Mol. Cell. Biol. 1997;17:2136–2142. doi: 10.1128/mcb.17.4.2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M.E., Choe W.C., Campbell J.L. DNA2 encodes a DNA helicase essential for replication of eukaryotic chromosomes. J. Biol. Chem. 1995;270:26766–26769. doi: 10.1074/jbc.270.45.26766. [DOI] [PubMed] [Google Scholar]
- Cardoso M.C., Leonhardt H., Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replicationcyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- Constantinou A., Tarsounas M., Karow J.K., Brosh R.M., Bohr V.A., Hickson I., West S.C. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO (Eur. Mol. Biol. Organ.) J. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Hanawalt P.C. RecQ and RecJ process blocked replication forks prior to the resumption of replication in UV-irradiated Escherichia coli . Mol. Gen. Genet. 1999;262:543–551. doi: 10.1007/s004380051116. [DOI] [PubMed] [Google Scholar]
- Cox L.S., Laskey R.A. DNA replication occurs at discrete sites in pseudonuclei assembled from purified DNA in vitro. Cell. 1991;66:271–275. doi: 10.1016/0092-8674(91)90617-8. [DOI] [PubMed] [Google Scholar]
- DePamphilis M.L. Origins of DNA replication in metazoan chromosomes. J. Biol. Chem. 1993;268:1–4. [PubMed] [Google Scholar]
- DePamphilis M.L. Origins of DNA replication. In: DePamphilis M.L., editor. DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. pp. 45–86. [Google Scholar]
- Dimitrova D.S., Todorov I.T., Melendy T., Gilbert D.M. Mcm2, but not RPA, is a component of the mammalian early G1-phase prereplication complex. J. Cell Biol. 1999;146:709–722. doi: 10.1083/jcb.146.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F., Newport J.W. Distinct roles of cdk2 and cdc2 in RP-A phosphorylation during the cell cycle. J. Cell Sci. 1993;106:983–994. doi: 10.1242/jcs.106.3.983. [DOI] [PubMed] [Google Scholar]
- Ferrari M.E., Bujalowski W., Lohman T.M. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J. Mol. Biol. 1994;236:106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- Frei C., Gasser S.M. The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 2000;14:81–96. [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., Higashikawa T., Tatsumi M. A retarded rate of DNA replication and normal level of DNA repair in Werner's syndrome fibroblasts in culture. J. Cell. Physiol. 1977;92:365–374. doi: 10.1002/jcp.1040920305. [DOI] [PubMed] [Google Scholar]
- Fujiwara Y., Kano Y., Ichihashi M., Nakao Y., Matsumura T. Abnormal fibroblast aging and DNA replication in the Werner syndrome. Adv. Exp. Med. Biol. 1985;190:459–477. doi: 10.1007/978-1-4684-7853-2_23. [DOI] [PubMed] [Google Scholar]
- Fukuchi K., Martin G.M., Monnat R.J., Jr. Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl. Acad. Sci. USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi K., Tanaka K., Kumahara Y., Marumo K., Pride M.B., Martin G.M., Monnat R.J., Jr. Increased frequency of 6-thioguanine-resistant peripheral blood lymphocytes in Werner syndrome patients. Hum. Genet. 1990;84:249–252. doi: 10.1007/BF00200569. [DOI] [PubMed] [Google Scholar]
- Gangloff S., Soustelle C., Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 2000;25:192–194. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- Goding G.W. Monoclonal AntibodiesPrinciples and Practice 1986. Academic Press; San Diego: pp. 315 pp [Google Scholar]
- Gray M.D., Shen J.C., Kamath-Loeb A.S., Blank A., Sopher B.L., Martin G.M., Oshima J., Loeb L.A. The Werner syndrome protein is a DNA helicase. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- Gray M.D., Wang L., Youssoufian H., Martin G.M., Oshima J. Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Exp. Cell Res. 1998;242:487–494. doi: 10.1006/excr.1998.4124. [DOI] [PubMed] [Google Scholar]
- Hanaoka F., Takeuchi F., Matsumura T., Goto M., Miyamoto T., Yamada M. Decrease in the average size of replicons in a Werner syndrome cell line by Simian virus 40 infection. Exp. Cell Res. 1983;144:463–467. doi: 10.1016/0014-4827(83)90425-1. [DOI] [PubMed] [Google Scholar]
- Hand R. Eucaryotic DNAorganization of the genome for replication. Cell. 1978;15:317–325. doi: 10.1016/0092-8674(78)90001-6. [DOI] [PubMed] [Google Scholar]
- Hoehn H., Bryant E.M., Au K., Norwood T.H., Boman H., Martin G.M. Variegated translocation mosaicism in human skin fibroblast cultures. Cytogenet. Cell Genet. 1975;15:282–298. doi: 10.1159/000130526. [DOI] [PubMed] [Google Scholar]
- Huang S., Li B., Gray M.D., Oshima J., Mian I.S., Campisi J. The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa A.S., Zhou J.Q., Zakian V.A. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- Kamath-Loeb A.S., Johansson E., Burgers P.M., Loeb L.A. Functional interaction between the Werner syndrome protein and DNA polymerase delta. Proc. Natl. Acad. Sci. USA. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp. Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Lebel M., Spillare E.A., Harris C.C., Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Johnson R.E., Yu S.L., Prakash L., Prakash S. Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science. 1999;286:2339–2342. doi: 10.1126/science.286.5448.2339. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Page A.W., Weier H.U., Bestor T.H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Leonhardt H., Rahn H.P., Weinzierl P., Sporbert A., Cremer T., Zink D., Cardoso M.C. Dynamics of DNA replication factories in living cells. J. Cell Biol. 2000;149:271–280. doi: 10.1083/jcb.149.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Oakeley E.J., Sun L., Jost J.P. Multiple domains are involved in the targeting of the mouse DNA methyltransferase to the DNA replication foci. Nucleic Acids Res. 1998;26:1038–1045. doi: 10.1093/nar/26.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohka M.J., Masui Y. Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science. 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- Marciniak R.A., Lombard D.B., Johnson F.B., Guarente L. Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl. Acad. Sci. USA. 1998;95:6887–6892. doi: 10.1073/pnas.95.12.6887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melendy T., Stillman B. Purification of DNA polymerase delta as an essential simian virus 40 DNA replication factor. J. Biol. Chem. 1991;266:1942–1949. [PubMed] [Google Scholar]
- Mills A.D., Blow J.J., White J.G., Amos W.B., Wilcock D., Laskey R.A. Replication occurs at discrete foci spaced throughout nuclei replicating in vitro. J. Cell Sci. 1989;94:471–477. doi: 10.1242/jcs.94.3.471. [DOI] [PubMed] [Google Scholar]
- Montecucco A., Savini E., Weighardt F., Rossi R., Ciarrocchi G., Villa A., Biamonti G. The N-terminal domain of human DNA ligase I contains the nuclear localization signal and directs the enzyme to sites of DNA replication. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:5379–5386. doi: 10.1002/j.1460-2075.1995.tb00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecucco A., Rossi R., Levin D.S., Gary R., Park M.S., Motycka T.A., Ciarrocchi G., Villa A., Biamonti G., Tomkinson A.E. DNA ligase I is recruited to sites of DNA replication by an interaction with proliferating cell nuclear antigenidentification of a common targeting mechanism for the assembly of replication factories. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:3786–3795. doi: 10.1093/emboj/17.13.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mushegian A.R., Bassett D.E., Jr., Boguski M.S., Bork P., Koonin E.V. Positionally cloned human disease genespatterns of evolutionary conservation and functional motifs. Proc. Natl. Acad. Sci. USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Morita T., Sato C. Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp. Cell Res. 1986;165:291–297. doi: 10.1016/0014-4827(86)90583-5. [DOI] [PubMed] [Google Scholar]
- Nakayasu H., Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitrostages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Newport J., Yan H. Organization of DNA into foci during replication. Curr. Opin. Cell Biol. 1996;8:365–368. doi: 10.1016/s0955-0674(96)80011-1. [DOI] [PubMed] [Google Scholar]
- Salk D., Au K., Hoehn H., Martin G.M. Cytogenetics of Werner's syndrome cultured skin fibroblastsvariegated translocation mosaicism. Cytogenet. Cell Genet. 1981;30:92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- Scappaticci S., Cerimele D., Fraccaro M. Clonal structural chromosomal rearrangements in primary fibroblast cultures and in lymphocytes of patients with Werner's Syndrome. Hum. Genet. 1982;62:16–24. doi: 10.1007/BF00295599. [DOI] [PubMed] [Google Scholar]
- Schellenberg G.D., Miki T., Yu C.E., Nakura J. Werner Syndrome. In: Vogelstein B., Kinzler K.W., editors. The Genetic Basis of Human Cancer. Vol. 18. McGraw-Hill; New York: 1998. pp. 347–359. [Google Scholar]
- Shen J.C., Gray M.D., Oshima J., Kamath-Loeb A.S., Fry M., Loeb L.A. Werner syndrome protein. I. DNA helicase and DNA exonuclease reside on the same polypeptide J. Biol. Chem 273 1998. 34139 34144a [DOI] [PubMed] [Google Scholar]
- Shen J.C., Gray M.D., Oshima J., Loeb L.A. Characterization of Werner syndrome protein DNA helicase activitydirectionality, substrate dependence and stimulation by replication protein A Nucleic Acids Res 26 1998. 2879 2885b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori M., Sakamoto S., Suzuki N., Tokutake Y., Kawabe Y., Enomoto T., Sugimoto M., Goto M., Matsumoto T., Furuichi Y. Detection by epitope-defined monoclonal antibodies of Werner DNA helicases in the nucleoplasm and their upregulation by cell transformation and immortalization. J. Cell Biol. 1999;144:1–9. doi: 10.1083/jcb.144.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C., Newport J.W. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 1991;35:449–468. doi: 10.1016/s0091-679x(08)60583-x. [DOI] [PubMed] [Google Scholar]
- Stefanini M., Scappaticci S., Lagomarsini P., Borroni G., Berardesca E., Nuzzo F. Chromosome instability in lymphocytes from a patient with Werner's syndrome is not associated with DNA repair defects. Mutat. Res. 1989;219:179–185. doi: 10.1016/0921-8734(89)90013-1. [DOI] [PubMed] [Google Scholar]
- Stewart E., Chapman C.R., Al-Khodairy F., Carr A.M., Enoch T. rqh1+, a fission yeast gene related to the Bloom's and Werner's syndrome genes, is required for reversible S phase arrest. EMBO (Eur. Mol. Biol. Organ.) J. 1997;16:2682–2692. doi: 10.1093/emboj/16.10.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld U.P., Howell M., Rempel R., Maller J.L., Hunt T., Blow J.J. Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases. Curr. Biol. 1994;4:876–883. doi: 10.1016/s0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- Suzuki N., Shimamoto A., Imamura O., Kuromitsu J., Kitao S., Goto M., Furuichi Y. DNA helicase activity in Werner's syndrome gene product synthesized in a baculovirus system. Nucleic Acids Res. 1997;25:2973–2978. doi: 10.1093/nar/25.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi F., Hanaoka F., Goto M., Akaoka I., Hori T., Yamada M., Miyamoto T. Altered frequency of initiation sites of DNA replication in Werner's syndrome cells Hum. Genet 60 1982. 365 368a [DOI] [PubMed] [Google Scholar]
- Takeuchi F., Hanaoka F., Goto M., Yamada M., Miyamoto T. Prolongation of S phase and whole cell cycle in Werner's syndrome fibroblasts Exp. Gerontol 17 1982. 473 480b [DOI] [PubMed] [Google Scholar]
- Tye B.K. MCM proteins in DNA replication. Annu. Rev. Biochem. 1999;68:649–686. doi: 10.1146/annurev.biochem.68.1.649. [DOI] [PubMed] [Google Scholar]
- Wold M.S. Replication protein Aa heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Yan H., Newport J. An analysis of the regulation of DNA synthesis by cdk2, Cip1, and licensing factor J. Cell Biol 129 1995. 1 15a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Newport J. FFA-1, a protein that promotes the formation of replication centers within nuclei Science 269 1995. 1883 1885b [DOI] [PubMed] [Google Scholar]
- Yan H., Merchant A.M., Tye B.K. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes Dev. 1993;7:2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]
- Yan H., Chen C.Y., Kobayashi R., Newport J. Replication focus-forming activity 1 and the Werner syndrome gene product. Nat. Genet. 1998;19:375–378. doi: 10.1038/1263. [DOI] [PubMed] [Google Scholar]
- Yu C.E., Oshima J., Fu Y.H., Wijsman E.M., Hisama F., Alisch R., Matthews S., Nakura J., Miki T., Ouais S. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]