Abstract
Prokaryotes and prokaryote-derived thylakoid membranes of chloroplasts share multiple, evolutionarily conserved pathways for protein export. These include the Sec, signal recognition particle (SRP), and Delta pH/Tat systems. Little is known regarding the thylakoid membrane components involved in these pathways. We isolated a cDNA clone to a novel component of the Delta pH pathway, Tha4, and prepared antibodies against pea Tha4, against maize Hcf106, a protein implicated in Delta pH pathway transport by genetic studies, and against cpSecY, the thylakoid homologue of the bacterial SecY translocon protein. These components were localized to the nonappressed thylakoid membranes. Tha4 and Hcf106 were present in ∼10-fold excess over active translocation sites. Antibodies to either Tha4 or Hcf106 inhibited translocation of four known Delta pH pathway substrate proteins, but not of Sec pathway or SRP pathway substrates. This suggests that Tha4 and Hcf106 operate either in series or as subunits of a heteromultimeric complex. cpSecY antibodies inhibited translocation of Sec pathway substrates but not of Delta pH or SRP pathway substrates. These studies provide the first biochemical evidence that Tha4 and Hcf106 are specific components of the Delta pH pathway and provide one line of evidence that cpSecY is used specifically by the Sec pathway.
Keywords: chloroplast protein transport, twin arginine, SecY, Hcf106, Tha4
Protein translocation across and into membranes is a fundamental cellular process, responsible for localization of ∼50% of the proteins in a eukaryotic cell. Schatz and Dobberstein 1996 classified eukaryotic protein translocation into import-type systems and export-type systems. Import systems translocate proteins from the cytosol into organelles and appear to have evolved after endosymbiosis. Export systems are derived from prokaryotic systems for exporting proteins across the cytoplasmic membrane and are present in the ER, the mitochondrial inner membrane, and chloroplast thylakoid membranes. Some proteins such as nucleus-encoded thylakoid proteins are localized by sequential action of an import system and an export system.
Recently, it has been shown that there are several different export systems present in thylakoid membranes and Escherichia coli plasma membranes (for reviews see Settles and Martienssen 1998; Dalbey and Robinson 1999; Keegstra and Cline 1999). In the case of thylakoids, these systems (or pathways) translocate distinct subgroups of proteins and can be distinguished by energy and stromal protein requirements, by competition with overexpressed precursors, and by identified components of the translocation machinery (Keegstra and Cline 1999). One pathway, termed the thylakoid Sec pathway, is responsible for translocation of plastocyanin (PC),1 OE33, PSI-F, and the plastid-encoded cytochrome F. The thylakoid Sec pathway appears to be homologous to the well-studied bacterial Sec pathway (Economou 1998) because it requires ATP and cpSecA, a homologue of the bacterial SecA protein, and is stimulated by the thylakoidal pH gradient (Keegstra and Cline 1999). The Sec pathway also employs membrane-bound machinery (Robinson et al. 1996), which is assumed to consist of cpSecY and cpSecE, homologues of the bacterial translocon proteins, SecY and SecE.
A second pathway, the chloroplast signal recognition particle (SRP) pathway, is responsible for targeting a family of integral thylakoid proteins, the LHCPs (Li et al. 1995), and probably also the plastid-encoded D1 protein (Nilsson et al. 1999). The SRP pathway appears to be homologous to the bacterial and ER SRP pathways because it employs a chloroplast homologue of the SRP54 protein, forms a soluble complex with LHCP substrates in the stroma, and requires GTP for integration (Keegstra and Cline 1999). The chloroplast SRP system possesses some unique features. It can operate posttranslationally, it lacks an associated RNA, and it possesses a novel protein component, cpSRP43, which apparently confers cpSRP54 with its posttranslational capability (Schuenemann et al. 1998). The chloroplast SRP system employs membrane protein(s) (Robinson et al. 1996) that have not yet been identified. One possibility is that the chloroplast SRP docks with a Sec-like translocon, similar to the ER SRP (Walter and Johnson 1994) and the bacterial SRP (Valent et al. 1998).
A third thylakoid pathway is termed the Delta pH pathway because it employs the thylakoidal pH gradient as sole energy source for transport of lumenal proteins (Cline et al. 1992). The Delta pH pathway is responsible for transport of OE23, OE17, PSI-N, and PSII-T and exhibits several distinctive and unusual features. It requires neither NTPs nor soluble protein factors. Its substrates possess conserved twin arginines in their signal peptide that, where examined, are essential for transport (Chaddock et al. 1995; Henry et al. 1997). Also, it seems capable of translocating tightly folded proteins (Clark and Theg 1997; Hynds et al. 1998).
The Delta pH system was initially thought to be a eukaryotic innovation because of its unique properties (Robinson and Klösgen 1994; Cline and Henry 1996). However, identification of a component of the machinery argues that it is of prokaryotic origin. Specifically, a maize mutant, hcf106, is selectively defective in the Delta pH pathway (Voelker and Barkan 1995). Hcf106 encodes a membrane protein that contains an amino-proximal transmembrane domain, a predicted amphipathic helix, and an acidic COOH-terminal domain (Settles et al. 1997). Hcf106 is present in thylakoids and is oriented with its amphipathic helix and COOH-terminal domain in the stroma (Settles et al. 1997). Database searches showed Hcf106 to be homologous to predicted proteins of unknown function in a wide range of bacteria (Settles et al. 1997). Recent studies in E. coli verify the existence of a prokaryotic Delta pH–like pathway. Mutation of genes, encoding E. coli Hcf106 homologues tatA, tatE (Sargent et al. 1998), or mttA (tatB) (Weiner et al. 1998), causes defects in export of a range of periplasmic redox proteins, all of which possess a twin arginine motif in their signal peptides (Berks 1996). The E. coli tat operon contains a gene (tatC) for a multispanning membrane protein that, when disrupted, also impairs export of periplasmic redox proteins (Bogsch et al. 1998). Homologues of TatC are encoded in chloroplast genomes of algae and a putative Arabidopsis chloroplast TatC homologue has been identified recently (Summer, E.J., and K. Cline, unpublished observations). A further correlation between the E. coli and thylakoid systems comes from the finding that bacterial Tat signal peptides direct efficient and exclusive transport by the thylakoid Delta pH system (Mori and Cline 1998; Wexler et al. 1998). These and other considerations argue that the bacterial Tat system is related in components and possibly mechanism to the Delta pH system (Settles and Martienssen 1998; Dalbey and Robinson 1999; Keegstra and Cline 1999).
These observations raise several important questions that are relevant for thylakoids and other prokaryote or prokaryote-derived membranes. First, are Hcf106 and its homologues directly involved in transport on the Delta pH/Tat systems and, if so, what roles do they play in the process? Currently, the only evidence for their involvement in protein transport comes from mutant analysis. Second, do the Delta pH/Tat systems and the cpSRP system employ distinct translocons or do they converge with the Sec-type translocon? Mutant and depletion analyses have addressed this question, but the results are uncertain. With regards to the Delta pH/Tat system, the E. coli Tat substrate trimethylamine N-oxide (TMAO) reductase is exported at normal levels in a conditional SecY mutant strain at the nonpermissive temperature or in a strain depleted of SecE (Santini et al. 1998), suggesting that the Tat system is Sec-independent. On the other hand, in chloroplasts, the cpSecY null mutant (Roy and Barkan 1998) is defective in transport on the Sec, SRP, and Delta pH pathways. Furthermore, the cpSecY null exhibits a more severe phenotype (virtual loss of thylakoids) than either the cpSecA or Hcf106 nulls individually, or even a double mutant in both pathways, suggesting interdependency of systems.
Here we have taken a biochemical approach to these questions. Antibodies were raised to the maize Hcf106 protein and a related pea protein, psTha4. These antibodies specifically inhibited transport of Delta pH pathway substrates with the respective maize and pea thylakoid membranes. Similarly, we have isolated a cDNA for pea cpSecY and produced an antibody to its COOH terminus. Antibody to cpSecY exclusively inhibited precursors that employ cpSecA for their transport. This provides direct evidence that cpSecY functions on the thylakoidal Sec pathway. The fact that neither the cpSRP pathway nor especially the Delta pH pathway was affected by antibody to cpSecY argues that the three kinds of thylakoid protein translocation systems are distinct at the level of the translocon.
Materials and Methods
cDNA Cloning of Pea cpSecY and Pea Tha4
The full coding regions of pea cpSecY and pea Tha4 were obtained in two steps. Nearly full-length cDNA clones were isolated by screening a pea lambda ZAP II cDNA library (a generous gift of Dr. Ken Keegstra, Michigan State University, East Lansing, MI) using standard protocols. For cDNA cloning of pea cpSecY, the hybridization probe was a DNA fragment of Arabidopsis cpSecY (accession number AF144684). For cDNA cloning of pea Tha4, the probe was a DNA fragment of maize Hcf106 (accession number AF027808) and Arabidopsis EST clone 182P20T7 (accession number H37534), which was identified by BLAST search as encoding a protein homologous to maize Hcf106. Both of the cDNA clones isolated were nearly complete but lacked initiation methionines. The missing 5′ sequences were isolated by 5′-RACE cloning (GIBCO BRL) following the manufacturer's recommendations. Clones containing the full-length coding regions for in vitro transcription and translation were engineered into pGem 4Z (Promega) in the SP6 orientation. A cDNA clone for atHcf106 was isolated by RT-PCR using primers based on genomic sequence (accession number ABO19226) and was cloned into pGem T (Promega) in the SP6 orientation. Sequencing of all clones on both strands was performed by the University of Florida Interdisciplinary Center for Biotechnology Research (ICBR) DNA Sequencing Core Facility.
Expression and Purification of Stroma-exposed Domains of Maize Hcf106 and Pea Tha4
The stroma-exposed domain of maize Hcf106 (hcf106sd) and the stroma-exposed domain of psTha4 (tha4sd) were expressed in E. coli as His-tagged fusion proteins. The coding regions for amino acid residues 90–243 of maize Hcf106 and residues 78–137 of psTha4 were amplified by PCR with forward primers containing NdeI restriction sites and an in-frame sequence for six histidine residues. A cDNA clone for Hcf106 in pGem 4Z was used as template and the reverse primer was the pUC/M13 forward primer corresponding to the pGem 4Z plasmid. The cDNA clone for psTha4 was used as template and the reverse primer contained a SacI restriction site. The amplified DNA fragments were digested with NdeI and HindIII (maize hcf106sd), or NdeI and SacI (pea tha4sd), and cloned into pETH3c (McCarty et al. 1991). The resulting plasmids were introduced into BL21 (λDE3) and the expression was induced with IPTG. Both recombinant proteins accumulated in the soluble fraction of E. coli cells and were purified by nickel-nitrilotriacetic acid agarose chromatography according to the Novagen protocol. Purified protein was dialyzed against 20 mM Hepes-KOH, pH 8.0, and concentrated with a Centricon YM10.
Production of Antibodies
For cpSecY antibody production, the peptide NH2-CRAEIISQKYKNIELYDFDKY-COOH, equivalent to the COOH terminus of pea cpSecY plus an NH2-terminal cysteine, was synthesized and cross-linked to keyhole limpet hemocyanin by Genosys Biotechnologies. The keyhole limpet hemocyanin–linked peptide was used as antigen. E. coli–expressed maize hcf106sd and pea tha4sd were used an antigens. Antibodies to the proteins were prepared in rabbits by Cocalico Biologicals. IgG was purified from the serum with protein A–Sepharose as described (Harlow and Lane 1988). IgG was digested with immobilized papain (Pierce) at 37°C for 8 h to produce Fab fragments. Undigested IgG and Fc fragments were removed by passing the reaction mixture through protein A–Sepharose several times and the flow through was collected as Fab fragments. Purified IgG and Fab fragments were dialyzed against 20 mM Hepes-KOH, pH 8.0, concentrated with Centricon YM10 or YM50 devices (Millipore), and stored at −20°C. Immunodetection with all sera was performed with enhanced chemiluminescence (ECL; Amersham) according to the manufacturer's manual.
Preparation of Chloroplasts, Lysates, Thylakoids, Nonappressed, and Appressed Thylakoids
Intact pea chloroplasts were isolated from 9–10-d-old pea (Laxton's Progress 9) seedlings by a combination of differential and Percoll density gradient centrifugation as described (Cline 1986). Maize seeds were imbibed with running tap water for 2 d and seedlings were grown in vermiculite for 6–7 d under 14 h of light (1,000 μE/m2 per second) and 10 h of dark at 26°C. Maize leaf blades were harvested, chopped to 0.5–1-cm pieces, and then homogenized in GR medium (Cline 1986) lacking MgCl2 and MnCl2 (7–8 ml GR medium/g of leaves). Intact chloroplasts were isolated by the procedure used for isolating pea chloroplasts (Cline 1986), except that Percoll gradients lacked MgCl2 and MnCl2. Intact chloroplasts were resuspended in import buffer (330 mM sorbitol, 50 mM Hepes-KOH, pH 8.0) at a concentration of 1 mg chlorophyll/ml. Washed thylakoids and stromal extract were prepared from isolated chloroplasts (Cline et al. 1993). Nonappressed thylakoid subfractions were prepared from thylakoids with digitonin according to Leto et al. 1985. Appressed thylakoid subfractions were prepared by solubilizing thylakoids with Triton X-100 (Berthold et al. 1981), except that thylakoids were resuspended in buffer containing 0.4 M sucrose, 10 mM NaCl, 5 mM MgCl2, 40 mM MES-KOH, pH 6.5, with a final ratio of Triton X-100 to chlorophyll of 14 (wt/wt). In addition, the second Triton X-100 treatment was omitted.
Preparation of Labeled Precursor Proteins
In vitro transcription plasmids for pLHCP and iOE23 from pea, iOE17 from maize, iOE33 from wheat, pPSI-N from Arabidopsis, and pPSII-T from cotton have been described elsewhere (Cline et al. 1993; Henry et al. 1994, Henry et al. 1997; Hulford et al. 1994). Capped RNA for authentic and intermediate precursors was produced in vitro with SP6 polymerase (Cline 1988). RNA was translated in a wheat germ system in the presence of [3H]leucine or [35S]methionine (Cline 1988), or in rabbit reticulocyte lysate (Promega) in the presence of [3H]leucine following the manufacturer's guidelines. Translation products were diluted 3–12-fold and adjusted to import buffer containing 30 mM unlabeled leucine or 30 mM unlabeled methionine.
Assays for Chloroplast Protein Import and Thylakoid Protein Transport
Import of radiolabeled proteins into chloroplasts was conducted as previously described (Cline et al. 1993). Chloroplasts recovered from assays were repurified or posttreated with thermolysin and then repurified. Repurified chloroplasts were analyzed directly or subfractionated into soluble and membrane fractions. Transport of radiolabeled proteins into isolated thylakoids was conducted with washed thylakoids supplied with stromal extract as previously described (Cline et al. 1993). For antibody inhibition of protein transport, washed thylakoids were suspended in import buffer containing 10 mM MgCl2 plus 3% BSA at 1 mg of chlorophyll/ml and combined with Fab fragments or IgG. The suspension was adjusted with 20 mM Hepes/KOH, pH 8, to a final chlorophyll concentration of 0.33 mg/ml and antibody concentrations as indicated in figure legends. After 1 h on ice, thylakoids were recovered by centrifugation at 3,200 g for 8 min and washed with import buffer containing 10 mM MgCl2. Aliquots of pretreated thylakoids (equivalent to 25 μg chlorophyll) were supplemented with stromal extract equivalent to 50 μg chlorophyll of intact chloroplasts and Mg-ATP (5 mM final concentration) and then incubated with radiolabeled proteins in a final volume of 75 μl. Reactions were conducted at 25°C for 30 min in the light (70 μE/m2 per second) and terminated by transfer to ice. Thylakoids, recovered by centrifugation, were posttreated with thermolysin, washed with import buffer containing 5 mM EDTA, and then dissolved in SDS-PAGE sample buffer.
Immunoprecipitation
Radiolabeled cpSecY translation products or chloroplasts repurified after protein import reactions were dissolved in 0.05 M Tris-HCl, pH 6.8, 2% SDS, 4% glycerol, 2% β-mercaptoethanol, 10 mM EDTA, 0.008% bromophenol blue and were heated at 100°C for 2 min. Samples were then diluted 12-fold in 10 mM Tris-HCl, pH 7.5, 5 mM EDTA, 140 mM NaCl, 1 mM PMSF, 1% Triton X-100. 10 μl of antibody to pea cpSecY or pea Tha4 (as irrelevant antibody) was added and the samples were incubated with shaking for 1.5 h at 4°C. 40 μl of protein A–Sepharose (packed volume of beads washed in 10 mM Hepes-KOH, pH 8.0) was then added and the samples shaken for an additional 30 min at 4°C. The protein A–Sepharose/antibody/antigen complexes were pelleted (500 g, 2 min) and washed three times in 10 mM Tris-HCl, pH 7.5, 5 mM EDTA, 140 mM NaCl, 0.2% Triton X-100. The final pellets were resuspended in 40 μl SDS sample buffer, heated 2 min at 100°C, and the supernatant analyzed by SDS-PAGE and fluorography.
Miscellaneous
Chlorophyll concentrations were determined according to Arnon 1949. Protein concentrations were determined by the BCA method with BSA as a standard (Pierce). Digitonin was purified by dissolving the commercially obtained material (Calbiochem) in distilled water to a 10% aqueous solution, stirring overnight, and removing insoluble matter by centrifugation at 35,000 g for 10 min. The supernatant was transferred to a new tube, centrifuged as above, and the resulting supernatant lyophilized.
Results
cDNA Cloning of Pea cpSecY and Pea Tha4
Maize Hcf106 was shown to be a component required for the Delta pH pathway in vivo (Voelker and Barkan 1995). To examine its role in thylakoid protein transport, the stromal domain of Hcf106 was expressed in E. coli and antibodies were prepared to the purified recombinant protein. Because our objective was to examine the involvement of translocation components with pea chloroplasts, where most biochemical analysis of thylakoid protein transport has been carried out, we attempted to isolate cDNA clones for pea Hcf106 and pea cpSecY.
Screening for a pea Hcf106 homologue employed a mixed probe consisting of the maize Hcf106 cDNA and an Arabidopsis cDNA obtained from the EST program (Fig. 1). A nearly full-length pea cDNA was isolated and extended by 5′-RACE. Similar to the Arabidopsis EST, the pea cDNA encodes a protein that is related to Hcf106 in the transmembrane domain and amphipathic helix, but lacks the extended COOH-terminal acidic domain (Fig. 1). Based on sequence comparison, the predicted pea protein is more similar to Tha4, a newly identified maize protein that is related to Hcf106 in sequence and function (Walker, M.B., L.M. Roy, E. Coleman, R. Voelker, and A. Barkan, manuscript submitted for publication), than it is to Hcf106. Accordingly, the pea protein has been designated psTha4. We have now isolated an Arabidopsis cDNA, based on genomic sequence (accession number AB019226), that appears to encode the authentic Hcf106 orthologue (Fig. 1).
Figure 1.
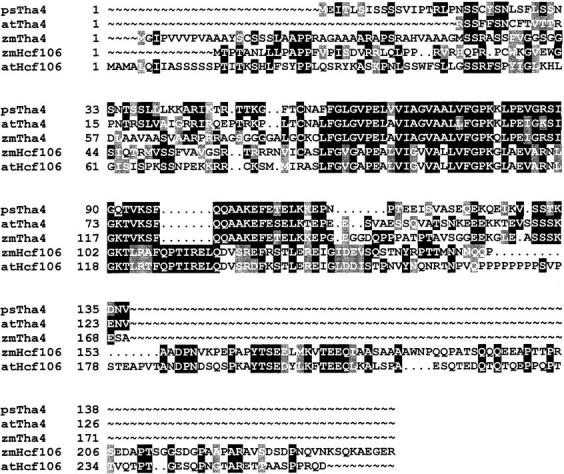
Pea Tha4 is a novel protein related to maize Tha4 and Hcf106. Tha4 is a distinct Hcf106 paralogue with a truncated COOH terminus. The deduced amino acid sequences of Tha4 from pea (psTha4, accession number AF144708), maize (zmTha4, accession number AF145755), and the predicted Arabidopsis Tha4 EST (atTha4, accession number H37534), as well as Hcf106 from maize (zmHcf106, accession number AF027808), and Arabidopsis (atHcf106, accession number AF139188) were aligned by PILEUP (Genetics Computer Group) and the alignment was enhanced by BOXSHADE (http://www.isrec.isb-sib.ch/software/BOX_form.html), which shows identical amino acids boxed in black and related amino acids boxed in gray.
The psTha4 cDNA encodes a protein with 137 residues. The amino terminus has characteristics of a chloroplast transit peptide. This was experimentally verified with an in vitro chloroplast import assay. The psTha4 translation product migrated at 19 kD (Fig. 2 A, lane 1). Upon incubation with chloroplasts, a faster migrating 16-kD band was produced that copurified with intact chloroplasts and was protected from exogenous protease (Fig. 2 A, lanes 2 and 3). This indicates that the precursor was imported into chloroplasts and processed to mature size. The imported psTha4 protein fractionated predominantly with the thylakoid membranes (Fig. 2 A, lane 5) and was integrated into the membrane as assessed by resistance to alkaline extraction (Fig. 2 A, lane 7). Similar to Hcf106, the hydrophilic domain of imported psTha4 was exposed to the stromal compartment as it was degraded by exogenous protease (Fig. 2 A, lane 6).
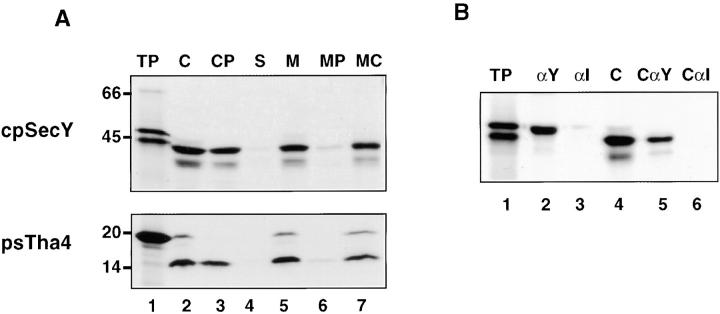
Figure 2.
cDNAs for pea cpSecY and psTha4 encode precursor proteins that are imported into chloroplasts and localized to thylakoids. (A) In vitro synthesized pea cpSecY and psTha4 were incubated with intact chloroplasts in the presence of ATP for 10 min. Chloroplasts were repurified without (C, lane 2) or with (CP, lane 3) posttreatment with thermolysin. Repurified chloroplasts were also fractionated into stroma (S, lane 4) and total membranes (M, lane 5) and the membranes treated with thermolysin (MP, lane 6), or extracted with 0.2 M Na2CO3 (MC, lane 7). Lane 1 contains the equivalent of 1 μl of translation product and lanes 2–7 contain samples representing 5 μl of the amount of translation product added to the import reaction. SDS-PAGE/fluorograms are depicted. (B) Radiolabeled pea cpSecY translation product (lane 1) or chloroplasts recovered from an import reaction with precursor to cpSecY (lane 4) were immunoprecipitated with antibody directed against the COOH terminus of pea cpSecY (see Materials and Methods) (lanes 2 and 5) or an irrelevant antibody (anti-psTha4; lanes 3 and 6) and the samples were analyzed by SDS-PAGE/fluorography.
A partial pea cpSecY cDNA was isolated and then extended by 5′-RACE (accession number AF144684). The predicted protein is highly homologous to other plant cpSecY proteins, with notable sequence divergence only in the amino-terminal transit peptide and the extreme COOH terminus. Upon in vitro translation, two bands were produced, one at 47 kD (Fig. 2 A, lane 1) and one migrating slightly faster. Immunoprecipitation with an antibody to the cpSecY COOH terminus showed that the larger translation product is the full-length precursor (Fig. 2 B, lanes 2 and 3). Incubation with chloroplasts produced a major band of processed cpSecY protein at 42 kD and a minor band migrating slightly faster (Fig. 2 A, lanes 2 and 3). The imported cpSecY fractionated with the thylakoid membranes (Fig. 2 A, lane 5) was resistant to alkaline extraction (Fig. 2 A, lane 7), but degraded by protease (Fig. 2 A, lane 6) to produce a 20-kD degradation product (data not shown).
Antibodies against Maize Hcf106, Pea Tha4, and Pea cpSecY
The stromal domains of Hcf106 and psTha4 were expressed in E. coli with NH2-terminal His tags for purification. Both proteins accumulated in the soluble fraction of E. coli, presumably in a native conformation. Purified pea tha4sd and maize hcf106sd migrated on SDS-PAGE with molecular masses of ∼16 kD and ∼35 kD, respectively (Fig. 3 A, lanes 1 and 2). Both proteins were used as antigens without further treatment.
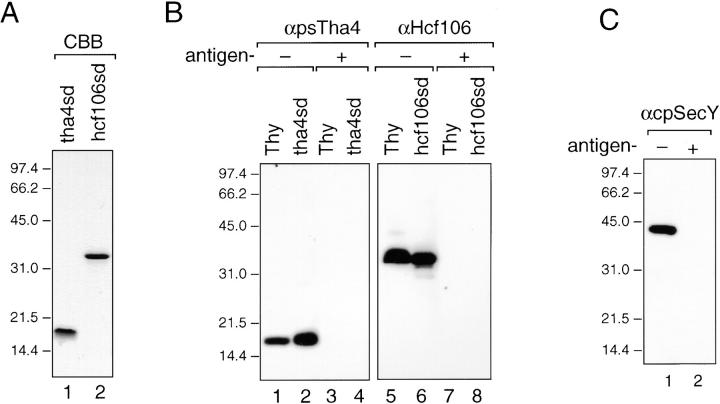
Figure 3.
Antibodies against pea cpSecY, psTha4, and maize Hcf106. (A) Coomassie brilliant blue–stained gel of E. coli–expressed and purified pea tha4sd (500 ng, lane 1) and maize hcf106sd (500 ng, lane 2). (B) Immunodetection of psTha4 and maize Hcf106. Pea thylakoid proteins (equivalent to 5 μg chlorophyll, lanes 1 and 3), pea tha4sd (10 ng, lanes 2 and 4), maize thylakoid proteins (equivalent to 2 μg chlorophyll, lanes 5 and 7), and maize hcf106sd (10 ng, lanes 6 and 8) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Blots were probed with anti-Hcf106 or anti-psTha4. Antibodies were preincubated without or with ∼1 μg/ml of each antigen protein for 1 h on ice before immunodecoration. (C) Immunodetection of pea cpSecY. Pea thylakoid protein (equivalent to 5 μg chlorophyll) was separated by SDS-PAGE and blotted to nitrocellulose membrane. The blots were immunodetected with anti-cpSecY that was preincubated without (lane 1) or with 1 μg/μl antigen peptide (lane 2) for 1 h on ice.
Anti-psTha4 reacted on immunoblots with an ∼16-kD pea thylakoid polypeptide (Fig. 3 B, lanes 1 and 2). The specificity of the reaction was verified by conducting the antibody incubation in the presence of the tha4sd antigen (Fig. 3 B, lanes 3 and 4). Similarly anti-Hcf106 recognized a ∼35-kD band in maize thylakoids in an antigen-reversible manner (Fig. 3 B, lanes 5–8). Anti-Hcf106 did not react with pea thylakoids on immunoblots and anti-psTha4 did not react with maize thylakoids (data not shown). However, as will be seen below, anti-Hcf106 binds to a pea Hcf106 orthologue in its native form.
Three cpSecY synthetic peptides were used as antigens in rabbits. These peptides correspond to several different stroma-facing hydrophilic segments based on the topology of E. coli SecY (Akiyama and Ito 1987). Although all of the resulting antibodies immunoprecipitated the cpSecY translation product, only an antibody to the COOH-terminal peptide reacted with immunoblots of thylakoid proteins and inhibited translocation (data not shown). Antibody to the COOH-terminal 20 residues of pea cpSecY immunodecorated a 42-kD band in pea thylakoids (Fig. 3 C, lane 1) in an antigen-reversible manner (Fig. 3 C, lane 2). As seen above, the antibody also immunoprecipitated the cpSecY translation product and imported protein (Fig. 2 B). Anti-pea cpSecY did not cross-react with maize cpSecY (data not shown).
Pea cpSecY and psTha4 Are Integral Thylakoid Proteins Located in the Nonappressed Membranes
Antibodies were used to verify the expected properties of endogenous pea cpSecY and psTha4 (Fig. 4). As with the imported proteins, endogenous psTha4 and cpSecY were recovered primarily in the thylakoid membrane fraction (data not shown). They were resistant to carbonate extraction, but digested by added thermolysin (Fig. 4, lanes 2 and 3). A band corresponding to the 20-kD protease-protected fragment of imported cpSecY was not immunodecorated with anti-cpSecY (data not shown), indicating that the protease protected fragment corresponds to an NH2-terminal or internal fragment. These results verify that cpSecY and psTha4 are integral membrane proteins exposed to the stroma. Thylakoid membranes consist of two structurally and functionally distinct domains, the nonappressed membranes and the appressed membranes. Upon subfractionation of thylakoids with digitonin (see Materials and Methods), cpSecY and psTha4 were recovered with the nonappressed membranes (Fig. 4, lane 5) rather than the appressed membranes (Fig. 4, lane 4). cpSecA, which is peripherally associated with thylakoids (Fig. 4, lanes 1 and 3), was also recovered in the nonappressed membranes (Fig. 4, lane 5). Maize Hcf106 was localized in the nonappressed membranes of maize thylakoids (data not shown). These results are consistent with other studies implicating the nonappressed membranes as the site of protein transport/integration (Kirwin et al. 1988; Kohorn and Yakir 1990; Hashimoto et al. 1997).
Figure 4.
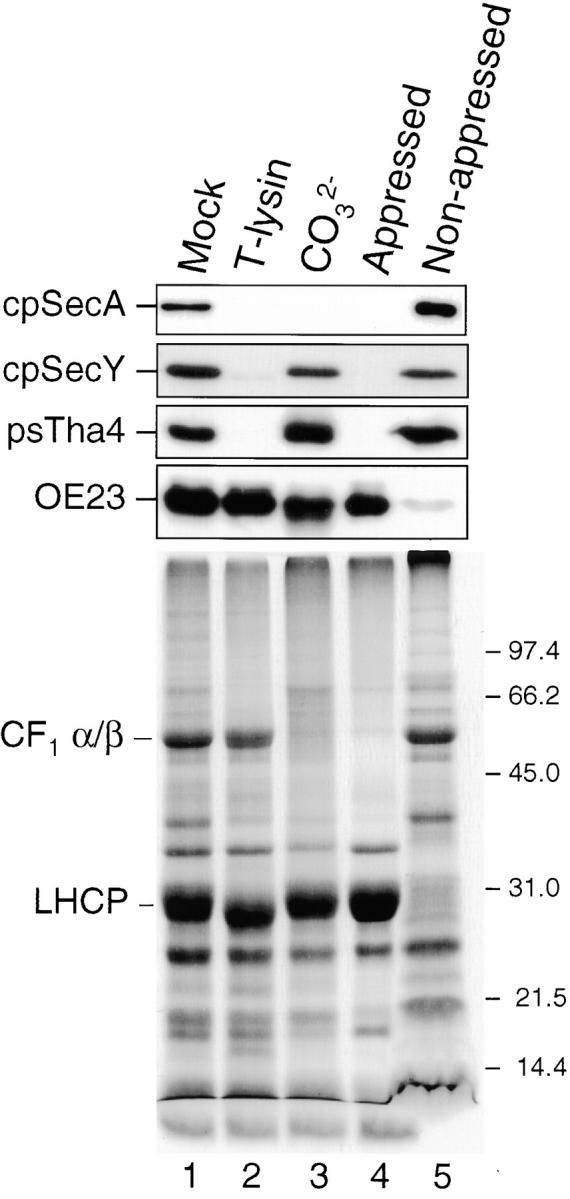
cpSecY and psTha4 are integral membrane proteins of the nonappressed membrane regions of thylakoids. Pea thylakoids were treated with import buffer (Mock, lane 1), 0.1 mg/ml thermolysin (lane 2), or 0.2 M Na2CO3 (lane 3). Appressed membranes (lane 4) and nonappressed membranes (lane 5) were prepared from pea thylakoids (see Materials and Methods). Samples, equivalent to 5 μg chlorophyll of thylakoids, were fractionated by SDS-PAGE, blotted to nitrocellulose, and proteins were immunodetected by the ECL method with the antibodies indicated to the left of the panel (top). A Coomassie-stained gel of the samples is shown in the bottom panel. CF1 α/β, the two large subunits of the chloroplasts coupling factor, is a marker for nonappressed membranes. LHCP is a marker for appressed membranes.
Quantification of psTha4 on immunoblots by comparison to a standard dilution series of tha4sd showed that psTha4 is present at ∼150 fmol/μg chlorophyll of thylakoid, which corresponds to ∼90,000 molecules per chloroplast. A similar quantification showed that Hcf106 is present at ∼100,000 molecules per maize chloroplast. This is interesting because our estimate of the number of active Delta pH translocation sites in pea thylakoids is ∼8,000 per chloroplast. This was estimated from the Vmax for transport of iOE23 in an intact chloroplast (8,000/chloroplast per minute; Cline et al. 1993) and from the minimum time required to observe a fully translocated protein on the Delta pH pathway (1 min; Cline, K., unpublished results; Teter and Theg 1998). This observation suggests that Hcf106 and Tha4 are present in a great excess over the functional translocation sites. cpSecY could not be reliably quantified with immunoblots.
Antibodies to Hcf106 Specifically Inhibit the Delta pH Pathway of Maize Thylakoids
Genetic disruption of the maize Hcf106 gene and genes for homologous proteins in E. coli results in mislocalization of precursor proteins (Voelker and Barkan 1995; Sargent et al. 1998; Weiner et al. 1998). Fig. 5 provides biochemical evidence that Hcf106 is directly involved in Delta pH pathway transport. Maize thylakoids were preincubated with or without anti-Hcf106 IgG or with preimmune IgG. The thylakoids were then washed with buffer and used for in vitro transport/integration assays. Transport of Delta pH pathway substrates, iOE17 and iOE23, was inhibited ∼95% by anti-Hcf106 IgG (Fig. 5 A, lane 4), but not by preimmune IgG (Fig. 5 A, lane 6). Anti-Hcf106 had no effect on transport of Sec pathway substrates, iOE33 and pPC, or on integration of the SRP substrate pLHCP.
Figure 5.
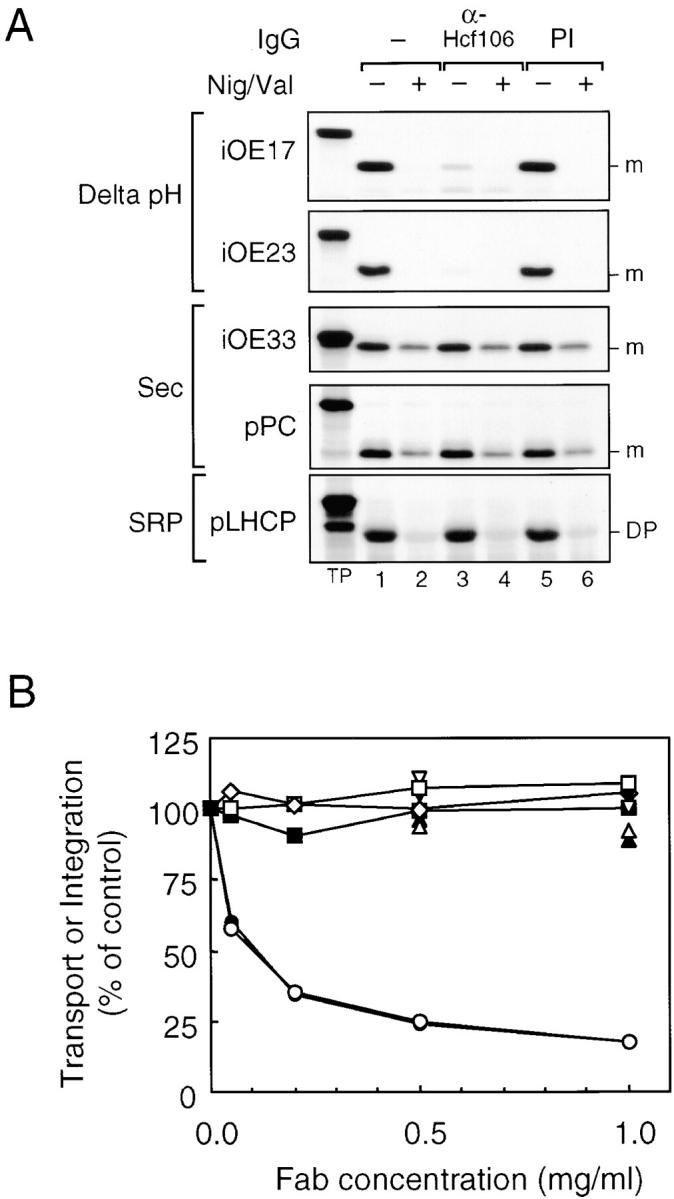
Inhibition of Delta pH pathway protein transport by antibodies to Hcf106. (A) Maize thylakoids were preincubated with 0.1 mg/ml anti-Hcf106 or preimmune (PI) IgGs. Thylakoids were then washed and assayed for transport or integration of various precursors in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of 0.5 μM nigericin and 1.0 μM valinomycin (Nig/Val). The radiolabeled precursors used in each assay are indicated next to the panel. (B) Maize thylakoids were preincubated with increasing amounts of anti-Hcf106 Fab fragments in the absence or presence of 20 μM hcf106sd or with PI-Fab. After washing, the thylakoids were assayed for protein transport or integration. iOE17 (•, anti-Hcf106; ▴, plus hcf106sd; ▾, PI-Fab), iOE23 (○, anti-Hfc106; ▵, plus hcf106sd; ▿, PI-Fab), iOE33 (□), pPC (⋄), and pLHCP (▪). Radiolabeled bands were extracted from excised gel bands and quantified by scintillation counting.
Protein transport on the Delta pH pathway absolutely depends on the thylakoidal ΔpH (Cline et al. 1992). To verify that the ΔpH was not impaired by the antibody treatment, the assays were conducted in the absence and presence of the ionophores nigericin and valinomycin, which completely dissipate the proton motive force (Fig. 5 A, lanes 2, 4, and 6). Transport of OE33 and PC and integration of LHCP are stimulated by the ΔpH, and translocation levels are reduced 60–70% in its absence (Cline et al. 1992; Yuan and Cline 1994). As can be seen in Fig. 5, the ΔpH-mediated stimulation of Sec pathway transport and LHCP integration was similarly eliminated by ionophores regardless of antibody treatment. Therefore, we conclude that the binding of anti-Hcf106 IgG to maize thylakoids did not compromise the thylakoidal ΔpH.
As preincubation of anti-Hcf106 IgG with maize thylakoids may induce aggregation of Hcf106 in the plane of thylakoids due to the divalent binding sites of IgG, we prepared monovalent Fab fragments from anti-Hcf106 IgG and tested the Fab fragments with in vitro thylakoid protein transport assays (Fig. 5 B). Increasing concentrations of anti-Hcf106 Fab fragments inhibited transport of Delta pH pathway substrates, iOE17 and iOE23, nearly as well as IgGs. Preimmune Fab fragments had no effect on transport, and the inhibition by anti-Hcf106 Fab fragments was suppressed by inclusion of hcf106sd during the antibody incubation step. Anti-Hcf106 Fab fragments had no effect on integration of pLHCP or on transport of iOE33 and pPC. Thus, inhibition of Delta pH pathway transport resulted directly from binding of the antibody, rather than as a secondary effect of aggregation.
Antibodies to psTha4 Specifically Inhibit the Delta pH Pathway of Pea Thylakoids
A similar analysis was conducted with pea thylakoid membranes and antibodies to psTha4 (Fig. 6). Preincubation of pea thylakoids with increasing amounts of anti-psTha4 IgG inhibited protein transport of the Delta pH pathway substrates iOE17 and iOE23. Preimmune IgG had no effect on transport, and the inhibition by anti-Tha4 was suppressed by including antigen tha4sd during the preincubation step. Transport of the Sec pathway substrate iOE33 and integration of the SRP pathway substrate pLHCP were unaffected by anti-psTha4 IgG. Assays conducted in the absence and presence of ionophores confirmed that anti-psTha4 IgG binding did not compromise the thylakoidal ΔpH (data not shown). These results indicate that psTha4 also directly functions in protein transport on the Delta pH pathway.
Figure 6.
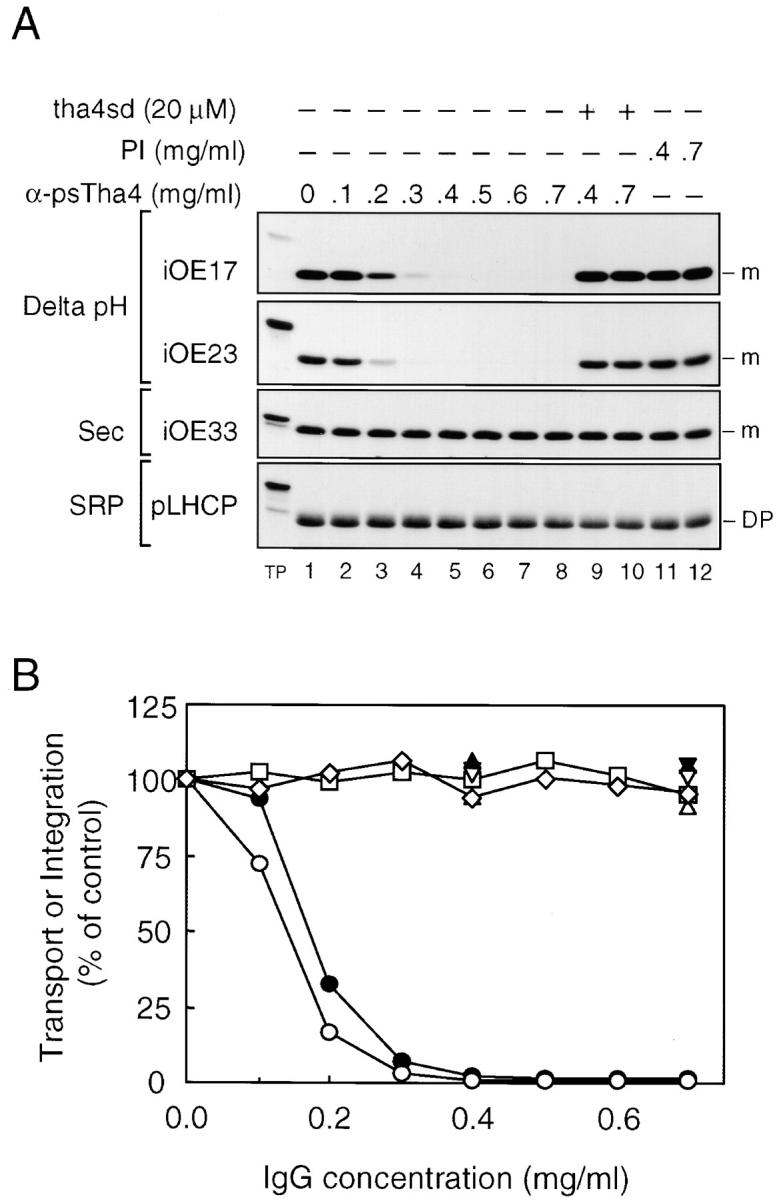
Inhibition of Delta pH pathway protein transport by antibodies to psTha4. Pea thylakoids were preincubated with increasing amounts of anti-psTha4 IgG in the absence or presence of 20 μM tha4sd or with preimmune (PI) IgG. After washing, the thylakoids were assayed for protein transport or integration. (A) Fluorogram of transport assays. Assay conditions were as shown in the figure. (B) Quantification of transport assays shown in A. iOE17 (•, anti-psTha4; ▴, plus tha4sd; ▾, PI-IgG), iOE23 (○, anti-psTha4; ▵, plus tha4sd; ▿, PI-IgG), iOE33 (□), and pLHCP (⋄). Radiolabeled bands were extracted from excised gel bands and quantified by scintillation counting.
Anti-Hcf106 or Anti-psTha4 Inhibits Transport of All Known Delta pH Pathway Substrates
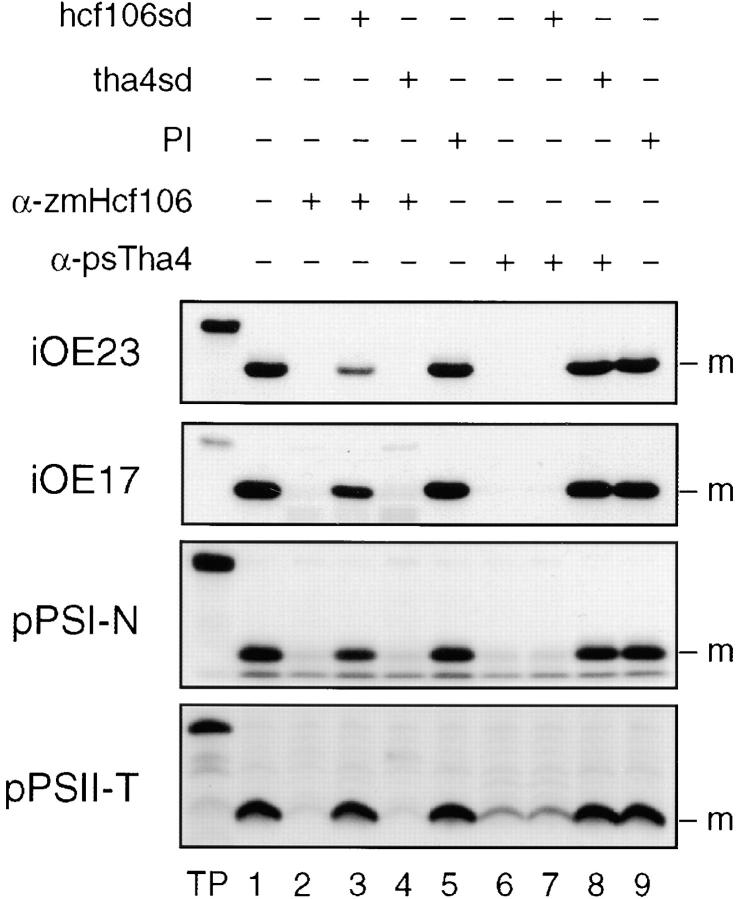
E. coli has three genes that are related to Hcf106: tatA, tatB, and tatE. Disruption of each of these genes singly causes partial inhibition of the translocation of overlapping sets of substrates (Sargent et al. 1998; Weiner et al. 1998), whereas double mutations result in a more complete inhibition (Sargent et al. 1998). One interpretation of these genetic results is that individual Hcf106 homologues have preferences for certain substrates. To assess whether Hcf106 or Tha4 show any selectivity for substrate, antibody inhibition experiments were conducted with four well-documented Delta pH pathway substrates: OE23, OE17, PSI-N, and PSII-T. We observed that antibodies to maize Hcf106 inhibit the Delta pH pathway, but not the Sec or SRP pathways, of pea thylakoid membranes (data not shown). This allowed us to simultaneously test the effects of anti-Hcf106 and anti-Tha4 with one membrane system. As shown in Fig. 7, either anti-Hcf106 or anti-psTha4 IgGs virtually eliminated transport of all Delta pH pathway substrates with pea thylakoids. In the experiment shown in Fig. 7, inhibition of PSII-T transport was not complete. However, in other experiments anti-Tha4 inhibited PSII-T transport to the same extent as that of the other substrates. Importantly, the inhibition by anti-Hcf106 was suppressed by hcf106sd, but not tha4sd (Fig. 7, lanes 2–4), and the inhibition by anti-Tha4 was suppressed by tha4sd, but not by hcf106sd (Fig. 7, lanes 6–8). This suggests that anti-Hcf106 binds to the pea Hcf106 orthologue. Neither antibody inhibited the integration of LHCP (data not shown), which was conducted as a control. We interpret these results to mean that antibody binding to either Hcf106 or Tha4 in vitro can disable the entire pathway.
Figure 7.
Antibodies to Hcf106 and psTha4 inhibit the entire range of known Delta pH pathway substrates. Pea thylakoids were preincubated with anti-Hcf106 IgG (1 mg/ml) or anti-psTha4 IgG (0.5 mg/ml) in the absence or presence of 20 μM hcf106sd or tha4sd as shown above the panel. Thylakoids were incubated with the corresponding preimmune IgGs at the same concentrations. After washing, thylakoids were assayed for transport of Delta pH pathway substrates (depicted to the left of each panel). Assay conditions were as shown above the panels.
Antibodies to cpSecY Specifically Inhibit Sec Pathway Transport
To explore the function of cpSecY in thylakoid protein transport, the effect of anti-cpSecY IgG on the three pathways was examined (Fig. 8). Preincubation of pea thylakoids with increasing amounts of anti-cpSecY inhibited transport of Sec pathway precursors, iOE33 and pPC (Fig. 8 A, lanes 1–5). Inclusion of 20 μM antigen peptide during the antibody preincubation step suppressed the inhibition (Fig. 8 A, lanes 6 and 7). No inhibition was observed for control thylakoids preincubated with preimmune IgG (Fig. 8 A, lanes 8 and 9). These results demonstrate that cpSecY operates in conjunction with cpSecA for thylakoid Sec pathway transport. Anti-cpSecY IgG had no effect on transport of the Delta pH pathway precursors, iOE23 and iOE17, or on integration of the SRP pathway substrate pLHCP. Even at higher concentrations of cpSecY IgGs (2.0 mg/ml), there was no inhibition of OE23 transport or LHCP integration (data not shown). Of interest is that Fab fragments prepared from anti-cpSecY IgG were ineffective inhibitors of Sec pathway transport (data not shown). This contrasts with the situation of anti-Hcf106 inhibition and suggests either that aggregation of the Sec translocons is required for inhibition or that IgG inhibits Sec pathway transport by steric hindrance.
Figure 8.
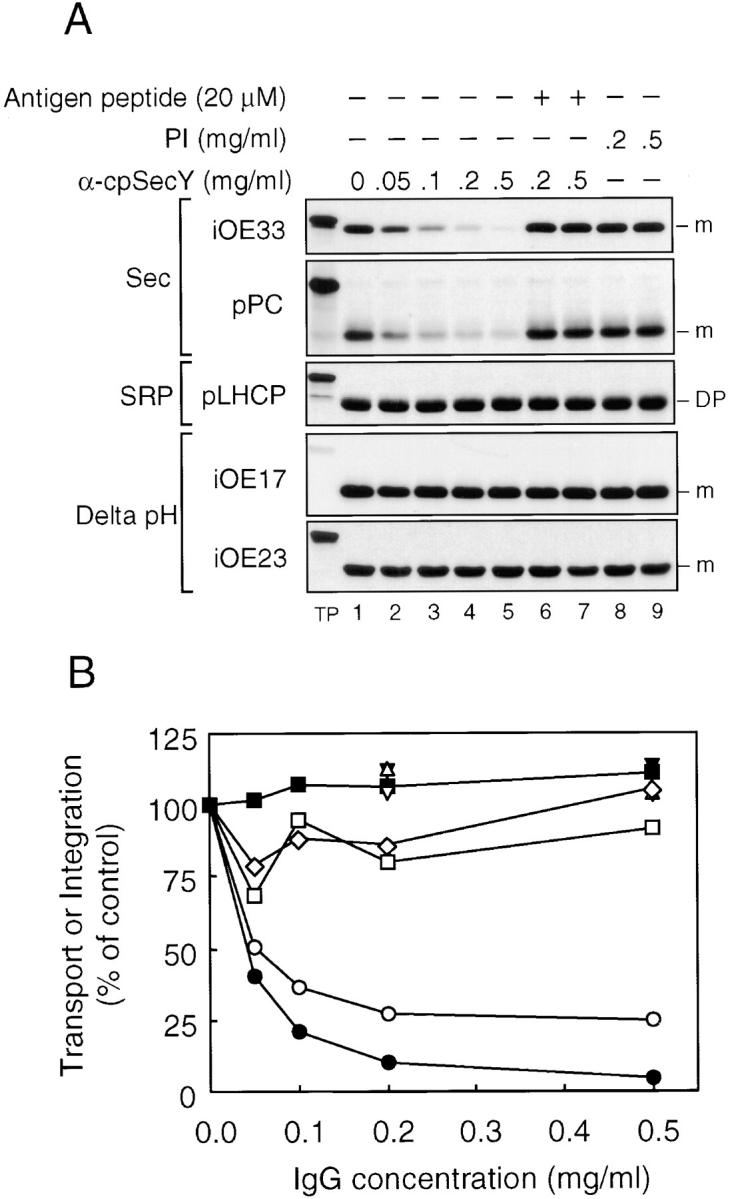
Antibodies to cpSecY inhibit protein transport of cpSecA-dependent substrates, but not those of the Delta pH or cpSRP pathways. (A) Pea thylakoids were preincubated and assayed for transport or integration as in Fig. 6 except that anti-cpSecY IgG and 20 μM antigen peptide were used. (A) A fluorogram of the transport assays. Precursors used and other conditions were as shown in the figure. (B) Quantification of transport assays shown in A. iOE33 (•, anti-cpSecY; ▴, plus antigen peptide; ▾, PI-IgG), pPC (○, anti-cpSecY; ▵, plus antigen peptide; ▿, PI-IgG), iOE17 (□, anti-cpSecY), iOE23 (⋄, anti-cpSecY), and pLHCP (▪, anti-cpSecY). Radiolabeled bands were extracted from excised gel bands and quantified by scintillation counting.
Discussion
Hcf106 was first identified as a component of the Delta pH transport pathway by genetic studies in maize, wherein mutations in Hcf106 resulted in defective transport of Delta pH pathway substrates in vivo (Voelker and Barkan 1995) and in vitro (Settles et al. 1997). Subsequently, point mutation or disruption of Hcf106 homologues in E. coli was shown to result in defective transport and localization of Tat substrates in vivo (Sargent et al. 1998; Weiner et al. 1998). Here we have used specific antibodies and in vitro assays to provide biochemical evidence that Hcf106 is directly involved in the targeting and/or translocation of proteins by the Delta pH pathway. Consistent with in vivo results, anti-Hcf106 inhibited transport of Delta pH substrates, but had no effect on transport by the Sec pathway or on integration of the SRP substrate LHCP.
We also identified a gene from pea that is related to, but not orthologous to, Hcf106. Our original intention was to isolate the pea Hcf106 orthologue. However, sequence comparisons indicate that the pea protein as well as the protein coded by the Arabidopsis EST are orthologues of maize Tha4 (Fig. 1). Our data also support the idea that the Delta pH pathway generally employs at least two Hcf106-like proteins. Genetic studies in maize (Settles et al. 1997; Roy and Barkan 1998; Walker, M.B., L.M. Roy, E. Coleman, R. Voelker, and A. Barkan, submittedmanuscript for publication) and the existence of sequences for two Hcf106 paralogues in Arabidopsis (Fig. 1) provide one line of evidence for this conclusion. Antibody inhibition studies shown in Fig. 7 provide biochemical evidence that there are two functional Hcf106-like proteins in pea thylakoids. Antibody to maize Hcf106 inhibited pea thylakoid transport of Delta pH pathway substrates (Fig. 7), but not Sec pathway substrates or LHCP (data not shown). Furthermore, anti-Hcf106 inhibition was suppressed by hcf106sd, the maize Hcf106 antigen, but not by tha4sd, the pea Tha4 antigen (Fig. 7). Likewise, anti-Tha4 inhibited the pea Delta pH pathway in a reaction that was only suppressed by tha4sd. This argues that anti-Hcf106 recognizes a protein distinct from psTha4, most likely the pea Hcf106 orthologue. The inability of anti-Hcf106 to immunoblot the pea orthologue may relate to the fact that the antibody was raised to native expressed protein and that antibody inhibition also involved binding to the native structure.
The antibodies against Hcf106 and Tha4 exerted a more complete block in Delta pH transport than would be expected from analyses of mutant plants. In the hcf106 null mutant, Delta pH substrates, OE23 and OE17, accumulate to varying degrees between 10 and 40% (see Roy and Barkan 1998). Tha4 mutants exhibit an even less severe phenotype than hcf106, but this might be explained by the existence of a second functional tha4 gene (Walker, M.B., L.M. Roy, E. Coleman, R. Voelker, and A. Barkan, manuscript submitted for publication). A similar situation was seen with mutations of the tat genes in E. coli. Single mutations of tatA or tatE result in partial disabling of Tat transport. Only double mutations or deletion of the multispanning membrane protein TatC result in a complete block in the export of the five precursors tested (Bogsch et al. 1998; Sargent et al. 1998). In contrast to these in vivo results, either anti-psTha4 or anti-Hcf106 virtually eliminated transport of four Delta pH transport substrates (Fig. 7).
There are several possible explanations for the difference between in vivo and in vitro data. One possibility is that in vivo, low transport levels may be compensated by reduced protein turnover and increased protein synthesis. In fact, pulse labeling of hcf106 maize leaves indicates that protein transport is more severely blocked than protein accumulation data suggest (Voelker and Barkan 1995). In addition, chloroplasts isolated from hcf106 plants were also severely defective in Delta pH transport in vitro (Settles et al. 1997).
Another possibility is that Hcf106 and Tha4 are part of the same protein complex. Antibody binding to any of the components of a critical multimeric complex might disable the entire complex, whereas another family member might replace a missing component in vivo. The structural similarity of Tha4 and Hcf106 suggests that they perform highly related functions in the targeting/translocation process. In this regard, it is notable that most bacterial species and chloroplasts from at least three plant species have two Hcf106 homologues, suggesting the possibility that they function in heteromultimeric complexes (Settles and Martienssen 1998).
SecY is an indispensable component for the bacterial Sec pathway (Economou 1998), where it functions as part of the translocon, i.e., the membrane machinery through which polypeptide chains are translocated. Chloroplast SecY genes have been identified in several plants and algae, but cpSecY function had not been experimentally demonstrated. Here we present evidence that cpSecY functions in the chloroplast Sec pathway. Antibodies to cpSecY inhibited transport of proteins shown previously to require cpSecA and ATP (Yuan et al. 1994). Thus, cpSecY functions similarly to the bacterial SecY, which cooperates with SecA for protein translocation.
There were at least two reasons to examine the involvement of the cpSecY in other thylakoid pathways. First is that SecYEG/Sec61 is considered to be a general conserved translocation channel (Matlack et al. 1998). When combined with a variety of other proteins that serve as motors, receptors, and channel gating mechanisms, several distinct translocation machineries result. One of these is the SRP-linked system. Second is the fact the cpSecY null mutant has defects in Sec, Delta pH, and SRP pathways and was more severe than either the cpSecA null or the Hcf106 null mutants (Roy and Barkan 1998). Thus, it was plausible that cpSecY/E could function as part of more than one pathway. However, antibody to cpSecY had no effect on the Delta pH pathway in our experiments, suggesting that cpSecY is not part of the Delta pH pathway translocon. During the preparation of this manuscript, Schuenemann et al. 1999 published a similar experiment showing that antibody to Arabidopsis cpSecY does not inhibit OE23 transport. Other explanations are possible for these results. For example, antibodies to E. coli SecY inhibit SecA and precursor binding to the membrane (Watanabe and Blobel 1989; Hartl et al. 1990). Thus the effect of cpSecY antibodies might be related to the function of cpSecA, which is not employed on the Delta pH pathway. Another possibility is that cpSecY exists in two different translocon complexes that differ in their accessibility to antibodies. In yeast, Sec61p, homologous to SecY, is a component of two different translocons, one of which is involved in posttranslational protein translocation, the other in cotranslational translocation. Antibodies to the COOH terminus of Sec61p immunoprecipitates the cotranslational Sec61 complex, but not the posttranslational complex, apparently because of masking by additional components present in the latter complex (Finke et al. 1996). Nevertheless, our biochemical data are consistent with genetic results that the E. coli Tat pathway is independent of SecY and E (Santini et al. 1998) and lends overall support to the idea that the Delta pH/Tat systems operate without a Sec translocon.
A similarly important question regards the integration of the SRP substrate LHCP. In E. coli, in vivo and in vitro data indicate that for some precursors, SRP and Sec pathways converge at the translocon level, i.e., the SRP pathway employs SecA and SecY/E for translocation (Valent et al. 1998). The data presented here clearly showed that integration of the chloroplast SRP substrate was unaffected by antibodies to cpSecY. In addition, LHCP integration is not inhibited by azide, a SecA inhibitor, indicating that cpSecA does not participate in its integration (Yuan et al. 1994). Thus, the chloroplast SRP substrate LHCP seems not to utilize the Sec machinery for its integration, despite the fact that membrane protein(s) are required for LHCP integration. Whether this will hold true for other chloroplast SRP substrates remains to be determined.
If the Sec translocon is not functional for the Delta pH and SRP pathways, it raises intriguing possibilities as to the identity and mode of action of such translocons. For the Delta pH pathway, Hcf106 and Tha4 could conceivably play a role in the translocation step. The topology of these proteins suggests that they serve as receptors for the pathway. However, evidence for that role is currently lacking. On the other hand, both proteins contain conserved acidic and proline residues in their transmembrane domains, suggesting that some of their function is conducted within the bilayer. A TatC homologue is another candidate for the Delta pH pathway translocon. TatC was identified as essential for Tat pathway transport (Bogsch et al. 1998). TatC as well as chloroplast homologues of TatC are predicted to be multispanning membrane proteins. As such, they invoke comparison with the multispanning SecY. Another thylakoid membrane protein that appears to play a role in thylakoid biogenesis is the chloroplast Oxa1p homologue, Albino3 (Sundberg et al. 1997; Hell et al. 1998). The Albino3 protein is a candidate for the translocation machinery used by the chloroplast SRP pathway. Determining the role of each of these proteins in the targeting and translocation step is certainly the next challenge for understanding thylakoid protein transport systems.
Acknowledgments
The authors thank Alice Barkan, Ralph Henry, and Mark Settles for critical review of this manuscript and helpful suggestions, and Drs. Hajime Tokuda and Ken-ichi Nishiyama for advice on immunodetection of cpSecY. The authors also thank Mike McCaffery for excellent technical assistance.
This work was supported in part by National Institutes of Health grant R01 GM46951 and National Science Foundation grant MCB-9419287 to K. Cline.
This manuscript is Florida Agricultural Experiment Station Journal series number R-06936.
Footnotes
1.used in this paper: i, intermediate precursor form; LHCP, light-harvesting chlorophyll a/b protein; m, mature form; OE33, OE23, and OE17, 33-, 23-, and 17-kD subunits of the photosystem II oxygen-evolving complex; p, precursor form; PC, plastocyanin; PSI-N, N subunit of the photosystem I complex; PSII-T, T subunit of the photosystem II complex; SRP, signal recognition particle
References
- Akiyama Y., Ito K. Topology analysis of the SecY protein, an integral membrane protein involved in protein export in Escherichia coli . EMBO (Eur. Mol. Biol. Organ.) J. 1987;6:3465–3470. doi: 10.1002/j.1460-2075.1987.tb02670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D.I. Copper enzymes in isolated chloroplasts. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks B.C. A common export pathway for proteins binding complex redox cofactors. Mol. Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- Berthold D.A., Babcock G.T., Yocum C.F. A highly resolved oxygen evolving photosystem II preparation from spinach thylakoid membraneselectron paramagnetic resonance and electron transport properties. FEBS Lett. 1981;134:231–234. [Google Scholar]
- Bogsch E.G., Sargent F., Stanley N.R., Berks B.C., Robinson C., Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- Chaddock A.M., Mant A., Karnauchov I., Brink S., Herrmann R.G., Klösgen R.B., Robinson C. A new type of signal peptidecentral role of a twin-arginine motif in transfer signals for the Delta pH-dependent thylakoidal protein translocase. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:2715–2722. doi: 10.1002/j.1460-2075.1995.tb07272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.A., Theg S.M. A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol. Biol. Cell. 1997;8:923–934. doi: 10.1091/mbc.8.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K. Import of proteins into chloroplasts. Membrane integration of a thylakoid precursor protein reconstituted in chloroplast lysates. J. Biol. Chem. 1986;261:14804–14810. [PubMed] [Google Scholar]
- Cline K. Light-harvesting chlorophyll a/b protein-membrane insertion, proteolytic processing, assembly into LHC-II, and localization to appressed membranes occurs in chloroplast lysates. Plant Physiol. 1988;86:1120–1126. doi: 10.1104/pp.86.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K., Henry R. Import and routing of nucleus-encoded chloroplast proteins. Annu. Rev. Cell Dev. Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- Cline K., Ettinger W.F., Theg S.M. Protein-specific energy requirements for protein transport across or into thylakoid membranes. Two lumenal proteins are transported in the absence of ATP. J. Biol. Chem. 1992;26:2688–2696. [PubMed] [Google Scholar]
- Cline K., Henry R., Li C., Yuan J. Multiple pathways for protein transport into or across the thylakoid membrane. EMBO (Eur. Mol. Biol. Organ.) J. 1993;12:4105–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey R.E., Robinson C. Protein translocation into and across the bacterial plasma membrane and the plant thylakoid membrane. Trends Biochem. Sci. 1999;24:17–22. doi: 10.1016/s0968-0004(98)01333-4. [DOI] [PubMed] [Google Scholar]
- Economou A. Bacterial preprotein translocasemechanism and conformational dynamics of a processive enzyme. Mol. Microbiol. 1998;27:511–518. doi: 10.1046/j.1365-2958.1998.00713.x. [DOI] [PubMed] [Google Scholar]
- Finke K., Plath K., Panzner S., Prehn S., Rapoport T.A., Hartmann E., Sommer T. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae . EMBO (Eur. Mol. Biol. Organ.) J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D. AntibodiesA Laboratory Manual 1988. Cold Spring Harbor Laboratory, ; Cold Spring Harbor, NY: pp. 726 pp [Google Scholar]
- Hartl F.-U., Lecker S., Schiebel E., Hendrick J.P., Wicker W. The binding cascade of SecB to SecA to SecY/E mediates preprotein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- Hashimoto A., Ettinger W.F., Yamamoto Y., Theg S.M. Assembly of newly imported oxygen evolving complex subunits in isolated chloroplastssites of assembly and mechanism of binding. Plant Cell. 1997;9:441–452. doi: 10.1105/tpc.9.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K., Herrmann J.M., Pratje E., Neupert W., Stuart R.A. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc. Natl. Acad. Sci. USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry R., Kapazoglou A., McCaffery M., Cline K. Differences between lumen targeting domains of chloroplast transit peptides determine pathway specificity for thylakoid transport. J. Biol. Chem. 1994;269:10189–10192. [PubMed] [Google Scholar]
- Henry R., Carrigan M., McCaffery M., Ma X., Cline K. Targeting determinants and proposed evolutionary basis for the Sec and Delta pH protein transport systems in chloroplast thylakoid membranes. J. Cell Biol. 1997;136:823–832. doi: 10.1083/jcb.136.4.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulford A., Hazell L., Mould R.M., Robinson C. Two distinct mechanisms for the translocation of proteins across the thylakoid membrane, one requiring the presence of a stromal protein factor and nucleotide triphosphates. J. Biol. Chem. 1994;269:3251–3256. [PubMed] [Google Scholar]
- Hynds P.J., Robinson D., Robinson C. The sec-independent twin-arginine translocation system can transport both tightly folded and malfolded proteins across the thylakoid membrane. J. Biol. Chem. 1998;273:34868–34874. doi: 10.1074/jbc.273.52.34868. [DOI] [PubMed] [Google Scholar]
- Keegstra K., Cline K. Protein import and routing systems of chloroplasts. Plant Cell. 1999;11:557–570. doi: 10.1105/tpc.11.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwin P.M., Elderfield P.D., Williams R.S., Robinson C. Transport of proteins into chloroplastsorganization, orientation, and lateral distribution of the plastocyanin processing peptidase in the thylakoid network. J. Biol. Chem. 1988;263:18128–18132. [PubMed] [Google Scholar]
- Kohorn B.D., Yakir D. Movement of newly imported light-harvesting chlorophyll-binding protein from unstacked to stacked thylakoid membranes is not affected by light treatment or absence of amino-terminal threonines. J. Biol. Chem. 1990;265:2118–2123. [PubMed] [Google Scholar]
- Leto K.J., Bell E., McIntosh L. Nuclear mutation leads to an accelerated turnover of chloroplast-encoded 48 kD and 34.5 kD polypeptides in thylakoids lacking photosystem II. EMBO (Eur. Mol. Biol. Organ.) J. 1985;4:1645–1653. doi: 10.1002/j.1460-2075.1985.tb03832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Henry R., Yuan J., Cline K., Hoffman N.E. A chloroplast homologue of the signal recognition particle subunit SRP54 is involved in the posttranslational integration of a protein into thylakoid membranes. Proc. Natl. Acad. Sci. USA. 1995;92:3789–3793. doi: 10.1073/pnas.92.9.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlack K.E., Mothes W., Rapoport T.A. Protein translocationtunnel vision. Cell. 1998;92:381–390. doi: 10.1016/s0092-8674(00)80930-7. [DOI] [PubMed] [Google Scholar]
- McCarty D.R., Hattori T., Carson C.B., Vasil V., Lazar M., Vasil I.K. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- Mori H., Cline K. A signal peptide that directs non-Sec transport in bacteria also directs efficient and exclusive transport on the thylakoid Delta pH pathway. J. Biol. Chem. 1998;273:11405–11408. doi: 10.1074/jbc.273.19.11405. [DOI] [PubMed] [Google Scholar]
- Nilsson R., Brunner J., Hoffman N.E., van Wijk K.J. Interactions of ribosome nascent chain complexes of the chloroplast-encoded D1 thylakoid membrane protein with cpSRP54. EMBO (Eur. Mol. Biol. Organ.) J. 1999;18:733–742. doi: 10.1093/emboj/18.3.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C., Klösgen R.B. Targeting of proteins into and across the thylakoid membranea multitude of mechanisms. Plant Mol. Biol. 1994;26:15–24. doi: 10.1007/BF00039516. [DOI] [PubMed] [Google Scholar]
- Robinson D., Karnauchov I., Herrmann R.G., Klösgen R.B., Robinson C. Protease-sensitive thylakoidal import machinery for the Sec-, ΔpH- and signal recognition particle-dependent protein targeting pathways, but not for CfoII integration. Plant J. 1996;10:149–155. [Google Scholar]
- Roy L.M., Barkan A. A SecY homologue is required for the elaboration of the chloroplast thylakoid membrane and for normal chloroplast gene expression. J. Cell Biol. 1998;141:385–395. doi: 10.1083/jcb.141.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini C.-L., Ize B., Chanal A., Müller M., Giordano G., Wu L.-F. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli . EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent F., Bogsch E.G., Stanley N.R., Wexler M., Robinson C., Berks B.C., Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO (Eur. Mol. Biol. Organ.) J. 1998;13:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz G., Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- Schuenemann D., Gupta S., Persello-Cartieaux F., Klimyuk V.I., Jones J.D.G., Nussaume L., Hoffman N.E. A novel signal recognition particle targets light harvesting proteins to the thylakoid membranes. Proc. Natl. Acad. Sci. USA. 1998;95:10312–10316. doi: 10.1073/pnas.95.17.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuenemann D., Amin P., Hartmann E., Hoffman N.E. Chloroplast secY is complexed to secE and involved in the translocation of the 33-kD but not the 23-kD subunit of the oxygen-evolving complex. J. Biol. Chem. 1999;274:12177–12182. doi: 10.1074/jbc.274.17.12177. [DOI] [PubMed] [Google Scholar]
- Settles A.M., Martienssen R. Old and new pathways of protein export in chloroplasts and bacteria. Trends Cell Biol. 1998;8:494–501. doi: 10.1016/s0962-8924(98)01387-7. [DOI] [PubMed] [Google Scholar]
- Settles A.M., Yonetani A., Baron A., Bush D.R., Cline K., Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- Sundberg E., Slagter J.G., Fridborg I., Cleary S.P., Robinson C., Coupland G. ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell. 1997;9:717–730. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teter S.A., Theg S.M. Energy-transducing thylakoid membranes remain highly impermeable to ions during protein translocation. Proc. Natl. Acad. Sci. USA. 1998;95:1590–1594. doi: 10.1073/pnas.95.4.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent Q.A., Scotti P.A., High S., de Gier J.W.L., von Heijne G., Lentzen G., Wintermeyer W., Oudega B., Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO (Eur. Mol. Biol. Organ.) J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelker R., Barkan A. Two nuclear mutations disrupt distinct pathways for targeting proteins to the chloroplast thylakoid. EMBO (Eur. Mol. Biol. Organ.) J. 1995;14:3905–3914. doi: 10.1002/j.1460-2075.1995.tb00062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Johnson A.E. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Watanabe M., Blobel G. Site-specific antibodies against the PrlA (secY) protein of Escherichia coli inhibit protein export by interfering with plasma membrane binding of preproteins. Proc. Natl. Acad. Sci. USA. 1989;86:1895–1899. doi: 10.1073/pnas.86.6.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner J.H., Bilous P.T., Shaw G.M., Lubitz S.P., Frost L., Thomas G.H., Cole J.A., Turner R.J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- Wexler M., Bogsch E.G., Klosgen R.B., Palmer T., Robinson C., Berks B.C. Targeting signals for a bacterial Sec-independent export system direct plant thylakoid import by the delta pH pathway. FEBS Lett. 1998;431:339–342. doi: 10.1016/s0014-5793(98)00790-x. [DOI] [PubMed] [Google Scholar]
- Yuan J., Cline K. Plastocyanin and the 33-kD subunit of the oxygen-evolving complex are transported into thylakoids with similar requirements as predicted from pathway specificity. J. Biol. Chem. 1994;269:18463–18467. [PubMed] [Google Scholar]
- Yuan J., Henry R., McCaffery M., Cline K. SecA homolog in protein transport within chloroplastsevidence for endosymbiont-derived sorting. Science. 1994;266:796–798. doi: 10.1126/science.7973633. [DOI] [PubMed] [Google Scholar]