Abstract
Predatory snails in the marine gastropod genus Conus stun prey by injecting a complex mixture of peptide neurotoxins. These conotoxins are associated with trophic diversification and block a diverse array of ion channels and neuronal receptors in prey species, but the evolutionary genesis of this functional diversity is unknown. Here we show that conotoxins with little amino acid similarity are in fact products of recently diverged loci that are rapidly evolving by strong positive selection in the vermivorous cone, Conus abbreviatus, and that the rate of conotoxin evolution is higher than that of most other known proteins. Gene duplication and diversifying selection result in the formation of functionally variable conotoxins that are linked to ecological diversification and evolutionary success of this genus.
Efforts to understand the molecular evolution of genes fundamental to complex adaptations have focused on key morphological or developmental features [e.g., Hox genes (1–2) and MADS-box genes (3–4)]. Genes that contribute to ecological diversification and the nature of evolutionary forces acting during this process are much more poorly known, partly because genes directly involved in ecological attributes are hard to identify.
Among the 500 species of the genus Conus, some hunt fishes, whereas others consume gastropods, polychaetes, or hemichordates (5). Venom is injected to stun prey and contains a wide variety of neurotoxins (“conotoxins”) that block a diversity of ion channels and neuronal receptors (6–16) in prey species. Different conotoxins are maximally effective on different prey species (6–8, 17–18); the ability to prey on these taxa among Conus species is thus linked to conotoxin evolution. To illuminate the genetic basis of ecological diversification in cone snails, we have begun to investigate the molecular evolution of conotoxins.
Members of the δ and ω conotoxin classes (“four-loop” conotoxins) have been sequenced (10, 19–20), are known to target a variety of sodium and calcium channels, respectively (10–16, 21), and have a “-C-C-CC-C-C-” cysteine “backbone” (8, 21–22) (C, cysteine; dashes refer to one to seven intercatenated amino acids of various types). These peptides are between 25 and 35 aa in length but are translated as precursor peptides of between 70 and 80 aa in which the N-terminal part of the propeptide (the “prepro” region) has been suggested to possess a signaling or processing function and that is cleaved from the toxin region during processing (19).
We examined the expression of four-loop conotoxin genes among two distantly related (unpublished data) vermivorous cone snails, Conus abbreviatus and Conus lividus to elucidate the evolution of these genes in Conus. These taxa have distinct diets; C. abbreviatus feeds primarily on eunicid and nereid polychaetes, whereas C. lividus feeds on a diversity of prey, including hemichordates and capitellid, nereid, spionid, and terebellid polychaetes (5, 23–25) Results show that the toxins from these species, particularly C. abbreviatus, comprise a rapidly duplicating multigene family that is diversifying by strong positive selection.
MATERIALS AND METHODS
Specimens of C. abbreviatus, Conus ebraeus, and C. lividus were collected from various sites around Oahu, Hawaii and kept in tanks until processing. C. abbreviatus and C. lividus are fairly distantly related, probably diverging during the Miocene, whereas C. abbreviatus and C. ebraeus are more closely related, probably diverging during the last 1.7–4.5 million years (unpublished data).
We extracted mRNA and synthesized cDNA from a portion of the venom duct of these species with variations on methods of Jakobsen et al. (26) and Lee and Vacquier (27). We amplified putative four-loop conotoxins from the cDNA of C. abbreviatus and C. ebraeus with a 5′ primer (CATCGTCAAGATGAAACTGACGTG) designed within the N terminus of the prepro region of four-loop conotoxins from published sequences (19–20) and an oligo[dT] for 40 cycles under the following conditions: 94°C for 30 seconds, 45°C for 30 seconds, 1-minute ramp to 68°C, and 68°C for 1 min. Amplification products were ligated into t-tailed pBluescript II KS(−) (28) that were then transformed into competent Escherichia coli. After growing the colonies overnight, we screened the white/positive colonies with either M13 (CATTTTGCTGCCGGTCA) or T3 (ATTAACCCTCACTAAAGGGAAC) vector primers and the 5′ conotoxin primer. Amplification products of expected sizes (200–400 bp) were sequenced. A primer (CACAGGTATGGATGACTCAGG) was then designed within the 3′ untranslated region from four-loop conotoxin sequences obtained from C. abbreviatus and C. ebraeus.
We used both conotoxin primers to amplify putative four-loop conotoxins from two individuals of C. abbreviatus and C. lividus for 40 cycles under the following conditions: 94°C for 30 seconds, 50°C for 30 seconds, and 72°C for 30 seconds. The amplification products were cloned as above, and white/positive colonies were screened with amplifications with vector primers. We sequenced at least 40 screen amplifications of expected insert size from each individual.
Sequences were aligned manually first by species and then compiled and aligned together. The conserved arrangement of the cysteine backbone aided in alignments of the diverse sequences recovered. Molecular phylograms were constructed with neighbor-joining from Kimura two-parameter distances among the sequences with mega (29). Identical sequences were represented only once. Bootstrap values were also estimated with mega from 100 replicates. Sequence groups were denoted based on existence of distinct clades and similarity of predicted amino acid sequences.
The corrected proportions of nonsynonymous substitutions per nonsynonymous site (Dn) and synonymous substitutions per synonymous site (Ds) were estimated from the sequences representative of the diversity of the predicted amino acid sequences within each sequence group. Dn and Ds were estimated from the beginning of the prepro region to the last codon before the stop codon with method 1 of Ina (ref. 30; program by T.F.D.). Significance of differences between Dn and Ds was estimated by using a one-tailed t test with infinite degrees of freedom (29). We used Hochberg’s (31) Bonferroni technique to correct significance for multiple tests. Dn and Ds were also estimated with method 1 of Ina (30) over sliding/overlapping windows of 14 codons by using a “slide” of 7 codons for all sequence comparisons (program by T.F.D.).
With sequence data from a calmodulin locus and the fossil and biogeographic record of Conus, we have estimated the rate of synonymous substitutions within the nuclear genome of this genus to be 0.63–1.8% per million years (unpublished data). We used this rate and the amount of synonymous divergence between the most similar sequences from C. abbreviatus and C. lividus to estimate the times of divergence of conotoxin genes. Rates of nonsynonymous substitutions among the toxin regions of these genes were then calculated from the times of divergence and Dn values among sequences.
RESULTS AND DISCUSSION
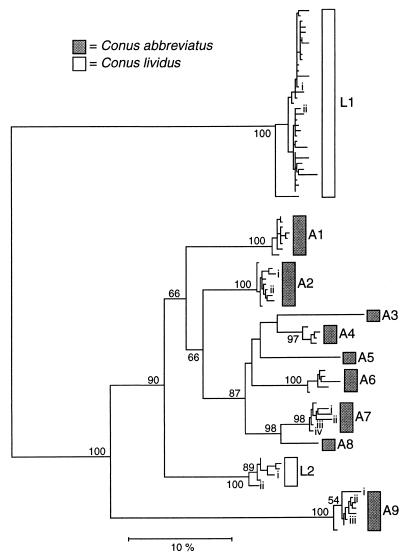
We report 180 sequences of four-loop conotoxins recovered from cDNA preparations (GenBank accession nos. AF089901–AF090080) that increase the known nucleotide sequence diversity of four-loop conotoxins by 30-fold and that assemble into 11 divergent groups in these species (Fig. 1). Considerably more diverse sequences were obtained from C. abbreviatus than from C. lividus. This may be because of the design of the 3′ conotoxin primer or because C. lividus has less diverse conotoxin gene families. Because of the possible bias in acquiring conotoxins from these taxa and the extreme divergence of sequences from C. lividus, we focused our analyses primarily on the evolution of four-loop conotoxins from C. abbreviatus.
Figure 1.
Neighbor-joining tree reconstructed from Kimura two-parameter distances computed from comparisons of entire conotoxin sequences (including 3′ untranslated regions). Bootstrap values >50% are indicated on branches. Roman numerals labeling branches within groups identify the representative sequences used in Dn and Ds analyses and Tables 1 and 2. Total numbers of sequences in each group from each individual: A1, n1 = 6, n2 = 9; A2, n1 = 19, n2 = 14; A3, n1 = 1, n2 = 0; A4, n1 = 5, n2 = 5; A5, n1 = 1, n2 = 0; A6, n1 = 5, n2 = 5; A7, n1 = 9, n2 = 2; A8, n1 = 1, n2 = 0; A9, n1 = 4, n2 = 14; L1, n1 = 26, n2 = 43; L2, n1 = 11, n2 = 0.
The nine sequence groups of C. abbreviatus, in which seven include sequences from both individuals, must represent at least five loci in this species. Because of the levels of divergence among these groups (>7% nucleotide divergence) and because each individual has sequences belonging to at least seven groups, it is likely that each represents a distinct locus.
Predicted amino acid sequences of the sequences are highly variable (Table 1). Even among the closely related monophyletic set encompassing groups A3–A8, nearly every amino acid in the toxin region (excluding the structure-defining cysteine residues) has been substituted at least once. Most of the amino acid variation is nonconservative; that is, substitutions involve amino acids of different chemical classes. Moreover, there is considerable length variation among the toxin peptides, which range from 25 to 36 aa. Despite this variation, phylogenetic reconstruction of the untranslated and coding regions shows the distinct signal of gene duplication and subsequent divergence (Fig. 1). Without the evidence of similarity provided by the more slowly evolving prepro region, the recent divergence of these radically different peptides and their evolutionary relationships would never have been apparent.
Table 1.
Predicted amino acid sequences representative of the diversity of conotoxins from each sequence group
| Conotoxin | Prepro region Toxin region |
|---|---|
| A1 | VLIIAVLFLTACQLTTAE-TSSRGEQKHRALRSTDKKFKVALLCSPPGSYCFGPAACCSNF-CSTLSDVCQESWSG |
| A2i | .I......................K...........NS.LTRG.T...GA..G.A....QS..DI.AST.NA---- |
| A2ii | ........................K...........NS.LTRG.T...GA..G.A....QS..NI.AST.NA---- |
| A3 | ..........................E.L.......NSRMTKR.T.RHGV..YSYF...KA..NPS.KR.L----- |
| A4 | ........................K...........YSRMTKH.T..EVG.LFAYE...KI..WRPR--.YP.--- |
| A5 | ............................L.......NSRMSKR.T...G..YH.DP...QV..NFPRKH.L----- |
| A6 | .............H..........K...........NSRMTKR.TA..GA.YAAYT....A..NLNTKK.VL.--- |
| A7i | ........................K...........DSRMTKR.T.A.DA.DATTK..IP...NLATKK..VPTFP |
| A7ii | ...........R............K...........NSRMTKR.T.A.DA.DATTE..IL...NLATKK..VPTFP |
| A7iii | .........A..............K...........NSRMTKR.T.A.DA.DATTE..IL...NLATKE..VPTFP |
| A7iv | ........................K...........NSRMTKR.T.A.GA.DATTE..IL...NLATKK..VPTFP |
| A8 | ..............................P.....NSRMTKR.T.G.EA.DATTN..FLT..NLATNK.RSPNFP |
| A9i | .I.F..........IATA.SYA.S.R..PD..LSSRNS.LSKR.LGS.EL.VRDTS...MS..TNNI--.F----- |
| A9ii | .I............IATA.SYA.S.R..PD..LSSRNS.LSKR.LGSREQ.VRDTS...MS..TNNI--.F----- |
| A9iii | .I.I..........IATA.SYA.S.R..PD..LSSRNS.LSKR.LGSREL.VRDTS...MS..TNNI--.F----- |
| L1i | ...........SE.V..DY.RDKWQYRAAS..DAMRN.RDTRT...A.EV.TSKSP..TG.L..HIGGM.HH---- |
| L1ii | .......S...SE.V..DY.RDKWQYRAAS..DAMRN.RDTR-...G.EV.TRHSP..TG.L.NHIGGM.HH---- |
| L2i | ....................YP..Q.R.H.......NS.LTRG.T.RNGF.RYHSH.......H.WAIM.L----- |
| L2ii | ....................YP..Q.R.H.......NS.LTRG.T.RNGA.GYHSH.......H.WAN..L----- |
Single-letter amino acid codes are used. Underlined residues denote presumed proteolytic cleavage sites (22). Names of sequences were used to facilitate discussion of these sequences; proper nomenclature for conotoxins (14) is A1, ABVIA; A2i, ABVIB; A2ii, ABVIC; A3, ABVID; A4, ABVIE; A5, ABVIF; A6, ABVIG; A7i, ABVIH; A7ii, ABVII; A7iii, ABVIJ; A7iv, ABVIK; A8, ABVIL; A9i, ABVIM; A9ii, ABVIN; A9iii, ABVIO; L1i, LVVIA; L1ii, LVVIB; L2i, LVVIC; and L2ii, LVVID.
Sequences from groups A1–A8 of C. abbreviatus cluster together monophyletically and are most closely related to sequences from group L2 from C. lividus; sequences from group A9 in C. abbreviatus are more distantly related to this cluster. These relationships show that some conotoxin loci in C. abbreviatus have persisted since the separation of C. abbreviatus and C. lividus in the Miocene (unpublished data), whereas others—perhaps the bulk of four-loop conotoxins—have duplicated and diversified subsequently.
Conotoxin sequence divergence is driven by strong selection, especially the recently duplicated loci A1–A8. Dn is significantly greater than Ds in the majority of between-group pairwise comparisons (43 of 59; Table 2). All comparisons with P values ≤ 0.001 (n = 9) remain significant after correcting for multiple tests. The average Dn and Ds values across overlapping windows for all sequence comparisons between representative sequences from groups A1 toA8 (Fig. 2) show that the amino acid variation and signature of diversifying selection (Dn > Ds) is mostly restricted to the toxin part of these sequences.
Table 2.
Dn:Ds ratios, significance levels, and Dn and Ds values among conotoxin sequences
| Conotoxin | Conotoxin
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2i | A2ii | A3 | A4 | A5 | A6 | A7i | A7ii | A7iii | A7iv | A8 | |
| A1 | 3.56 | 4.16 | 6.65 | 2.57 | 3.36 | 4.34 | 2.55 | 2.80 | 2.77 | 2.65 | 5.19 | |
| ∗∗∗∗ | ∗∗∗∗ | ∗∗∗∗ | ∗ | ∗∗∗ | ∗∗∗∗ | ∗∗ | ∗∗∗ | ∗∗∗ | ∗∗ | ∗∗∗∗ | ||
| A2i | 28.16 | 0.40 | 4.63 | 1.63 | 3.04 | 2.31 | 2.12 | 2.37 | 2.54 | 2.01 | 4.34 | |
| 7.91 | ∗∗∗∗ | ∗∗∗ | ∗ | ∗ | ∗ | ∗ | ∗ | ∗∗∗∗ | ||||
| A2ii | 24.55 | 1.52 | 5.80 | 1.44 | 3.24 | 2.63 | 2.37 | 2.70 | 2.90 | 2.18 | 5.95 | |
| 5.90 | 3.80 | ∗∗∗∗ | ∗∗∗ | ∗ | ∗ | ∗ | ∗∗ | ∗ | ∗∗∗∗ | |||
| A3 | 31.84 | 30.92 | 26.64 | 2.52 | 2.60 | 1.97 | 2.17 | 2.49 | 2.91 | 2.14 | 2.60 | |
| 4.79 | 6.68 | 4.59 | ∗ | ∗ | ∗ | ∗ | ∗∗∗ | ∗ | ∗ | |||
| A4 | 24.57 | 21.89 | 19.27 | 15.81 | 1.27 | 2.23 | 1.64 | 1.77 | 1.84 | 1.49 | 2.59 | |
| 9.55 | 13.42 | 13.37 | 6.28 | ∗ | ||||||||
| A5 | 27.71 | 25.14 | 23.25 | 18.68 | 18.77 | 1.37 | 2.82 | 3.52 | 3.67 | 3.58 | 3.67 | |
| 8.25 | 8.27 | 7.17 | 7.19 | 14.81 | ∗ | ∗∗ | ∗∗∗ | ∗∗ | ∗∗∗ | |||
| A6 | 28.64 | 23.77 | 20.78 | 15.93 | 14.65 | 19.05 | 1.53 | 1.90 | 2.08 | 1.55 | 2.61 | |
| 6.60 | 10.28 | 7.91 | 8.10 | 6.58 | 13.91 | ∗ | ||||||
| A7i | 30.10 | 22.60 | 19.67 | 24.54 | 18.07 | 19.69 | 14.61 | 0.79 | 1.32 | 1.08 | 2.68 | |
| 11.82 | 10.66 | 8.31 | 11.31 | 11.04 | 6.99 | 9.53 | ||||||
| A7ii | 27.66 | 21.30 | 18.25 | 23.35 | 16.87 | 18.23 | 14.42 | 1.87 | UND | UND | 6.14 | |
| 9.89 | 8.99 | 6.75 | 9.39 | 9.50 | 5.18 | 7.60 | 2.35 | ∗ | ||||
| A7iii | 27.58 | 23.01 | 19.79 | 24.68 | 17.60 | 19.02 | 15.96 | 3.15 | 1.23 | UND | 7.18 | |
| 9.97 | 9.05 | 6.82 | 8.47 | 9.58 | 5.18 | 7.66 | 2.38 | 0.00 | ∗ | ∗∗ | ||
| A7iv | 27.51 | 20.06 | 17.12 | 22.05 | 15.74 | 18.30 | 13.29 | 2.52 | 0.61 | 1.86 | 3.24 | |
| 10.38 | 9.98 | 7.87 | 10.32 | 10.55 | 5.11 | 8.59 | 2.33 | 0.00 | 0.00 | |||
| A8 | 31.80 | 25.05 | 21.45 | 21.71 | 20.41 | 23.65 | 15.58 | 8.38 | 7.33 | 8.67 | 7.70 | |
| 6.13 | 5.74 | 3.60 | 8.35 | 7.89 | 6.44 | 5.97 | 3.12 | 1.19 | 1.21 | 2.38 | ||
Above diagonal: Dn/Ds ratios among representative sequences of combined prepro and toxin regions from groups A1 to A8 (see Fig. 1). Significance of pairwise comparisons for the difference between Dn and Ds is indicated below their ratios. ∗, P < 0.005; ∗∗, P < 0.01; ∗∗∗, P < 0.005; ∗∗∗∗, P < 0.001. UND, undefined (Ds = 0). Below diagonal: Dn (above) and Ds (below) values (%). Within-group comparisons are denoted by boxes.
Figure 2.
Sliding window analysis of average Dn and Ds estimates for all toxin sequence comparisons. Codons 1–42 primarily include the prepro region; codons 43–63 only contain toxin sequences terminating before the stop codon; because the presumed cleavage site varies in position, window position 36–49 includes about a 1:1 ratio of prepro/toxin codons (see Table 1; gaps in the alignment are excluded from this codon-numbering scheme).
Moreover, the average Dn among the toxin regions of sequence groups A1–A8 (0.47, SE = 0.20) is significantly greater than that among the prepro regions (0.053, SE = 0.04). Interestingly, the average Ds among toxin regions (0.18, SE = 0.10) is also significantly greater than the average Ds among prepro regions (0.024, SE = 0.020) (see Fig. 2). This pattern is also observed in abalone lysin genes under diversifying selection (32) and may reflect the shift of nucleotide positions from synonymous to nonsynonymous sites during protein evolution. Alternatively, if most changes among conotoxin loci involve nonsynonymous substitutions, Ina’s (30) method might overestimate Ds while underestimating Dn in regions of low sequence identity because it equally weights all possible pathways for multiple step changes of a codon. In either case, toxin regions appear to be accumulating both synonymous and nonsynonymous substitutions at a greater rate than prepro regions.
Sequences from C. abbreviatus in the A1–A8 cluster and those from C. lividus in the L2 cluster show a divergence at synonymous sites of ≈12.0% in the prepro and 3′ untranslated regions. This is similar to the 12.4% divergence of calmodulin introns from these species (unpublished data) and suggests that the A1–A8 and L2 clusters diverged when C. abbreviatus and C. lividus lineages diverged. Prior rate calibrations for calmodulin show that sequences diverge at synonymous sites at a rate of 0.63–1.8% per million years in Conus—about average for eukaryotic genes (33). If we use these rates to calculate the average divergence time of the A1–A8 and L2 clusters, the result is about 6.7–19 million years. Likewise, the average time of divergence among the A1–A8 clusters is about 4.9–14 million years. From these times and the average Dn, we estimate that the rates of nonsynonymous substitutions among the toxin regions of the A1–A8 clusters average 1.7–4.8% per million years. The lower rate of 1.7% per million years (= 17 nonsynonymous substitutions per site per 109 years) is five times greater than the highest nonsynonymous rate reported by Li (33) for mammals (interferon γ nonsynonymous rate = 3.1 substitutions per site per 109 years) and nearly three times greater than the highest nonsynonymous rates reported for Drosophila. Conotoxins are thus diversifying at an extraordinarily high rate.
Significant differences between Dn and Ds among the nucleotide sequences of these peptides are signals of diversifying selection and adaptive evolution (34). These signals are mainly due to the divergence of sequences within the toxin and not the prepro region, implying that diversifying selection has involved toxin sequences and not the entire propeptide sequences. The considerable length variation among mature conotoxin peptides also suggests adaptive evolution for conotoxin size. Although we do not know whether these conotoxins have functional differences and it is unclear how conotoxin length affects functionality, results from other studies have demonstrated that four-loop conotoxins that differ in amino acid sequences have different specificities for particular cell channels (6–16). Thus, the rapid evolution we document here among conotoxin loci probably leads to substantial variation in venom effectiveness against particular prey types.
Our results show that conotoxin diversity is associated with an ongoing process of locus duplication and rapid divergence. Because conotoxins are intricately related to a species’ ability to paralyze its prey, the rapid adaptive evolution of these loci suggests that conotoxins are under strong selection in response to changes in the availability of or accessibility to particular prey species over time or because of a type of “arms race” between conotoxins and the cell channels and receptors of prey. Coevolution of predator and prey may generate evolutionary forces similar to those seen in host-pathogen evolution (34–38) and provide the means by which ecologically relevant genetic loci may rapidly diversify.
Acknowledgments
We thank G. C. Fiedler, D. Strang, P. S. Armstrong, and K. A. del Carmen for aid in collections of specimens. We also thank F. Cipriano, M. Hare, D. Hartl, S. Lavery, and two anonymous reviewers for input and reviews of earlier drafts of this manuscript. This work was supported by the Research Council of the University of Hawaii and Organismic and Evolutionary Biology Department of Harvard University and grants from the Conchologists of America, Hawaiian Malacological Society, Lerner-Gray Fund for Marine Research, Sigma Xi, the Western Society of Malacologists, and grants to S.R.P. from the National Science Foundation.
Footnotes
References
- 1.Sharman A C, Holland P W H. Neth J Zool. 1996;46:46–67. [Google Scholar]
- 2.Bailey W J, Kim J, Wagner G P, Ruddle F H. Mol Biol Evol. 1997;14:843–853. doi: 10.1093/oxfordjournals.molbev.a025825. [DOI] [PubMed] [Google Scholar]
- 3.Purugganan M D, Rounsley S D, Schmidt R J, Yanofsky M F. Genetics. 1995;140:345–356. doi: 10.1093/genetics/140.1.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theissen G, Kim J T, Saedler H. J Mol Evol. 1996;43:484–516. doi: 10.1007/BF02337521. [DOI] [PubMed] [Google Scholar]
- 5.Kohn A J. Ecol Monogr. 1959;29:47–90. [Google Scholar]
- 6.Olivera B M, Gray W R, Zeikus R, McIntosh J M, Varga J, Rivier J, de Santos V, Cruz L J. Science. 1985;230:1338–1343. doi: 10.1126/science.4071055. [DOI] [PubMed] [Google Scholar]
- 7.Olivera B M, Rivier J, Clark C, Ramilo C A, Corpuz G P, Abogadie F C, Mena E E, Woodward S R, Hillyard D R, Cruz L J. Science. 1990;249:257–263. doi: 10.1126/science.2165278. [DOI] [PubMed] [Google Scholar]
- 8.Olivera B M, Rivier J, Scott J K, Hillyard D R, Cruz L J. J Biol Chem. 1991;266:22067–22070. [PubMed] [Google Scholar]
- 9.Cruz L J, Gray W R, Yoshikami D, Olivera B. J Toxicol Toxin Rev. 1985;4:107–132. [Google Scholar]
- 10.Monje V D, Haack J A, Naisbitt S R, Miljanich G, Ramachandran J, Nasdasdi L, Olivera B M, Hillyard D R, Gray W R. Neuropharmacology. 1993;32:1141–1149. doi: 10.1016/0028-3908(93)90008-q. [DOI] [PubMed] [Google Scholar]
- 11.Fainzilber M, Vanderschors R, Lodder J C, Li K W, Geraerts W P M, Kits K S. Biochemistry. 1995;34:5364–5371. doi: 10.1021/bi00016a007. [DOI] [PubMed] [Google Scholar]
- 12.Fainzilber M, Kofman O, Zlotkin E, Gordon D. J Biol Chem. 1994;269:2574–2580. [PubMed] [Google Scholar]
- 13.Kristipati R, Nadasdi L, Tarczyhornoch K, Lau K, Miljanich G P, Ramachandran J, Bell J R. Mol Cell Neurosci. 1994;5:219–228. doi: 10.1006/mcne.1994.1026. [DOI] [PubMed] [Google Scholar]
- 14.Hasson A, Shon K J, Olivera B M, Spira M E. J Neurophysiol. 1995;73:1295–1302. doi: 10.1152/jn.1995.73.3.1295. [DOI] [PubMed] [Google Scholar]
- 15.Kits K S, Lodder J C, Vanderschors R C, Li K W, Geraerts W P M, Fainzilber M. J Neurochem. 1996;67:2155–2163. doi: 10.1046/j.1471-4159.1996.67052155.x. [DOI] [PubMed] [Google Scholar]
- 16.Gandia L, Lara B, Imperial J S, Villarroya M, Albillos A, Maroto R, Garcia A G, Olivera B M. Eur J Physiol. 1997;435:55–64. doi: 10.1007/s004240050483. [DOI] [PubMed] [Google Scholar]
- 17.Endean R, Rudkin C. Toxicon. 1963;1:49–64. doi: 10.1016/0041-0101(65)90021-8. [DOI] [PubMed] [Google Scholar]
- 18.Endean R, Rudkin C. Toxicon. 1965;2:225–249. doi: 10.1016/0041-0101(65)90021-8. [DOI] [PubMed] [Google Scholar]
- 19.Woodward S R, Cruz L J, Olivera B M, Hillyard D R. EMBO J. 1990;9:1015–1020. doi: 10.1002/j.1460-2075.1990.tb08204.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colledge C J, Hunsperger J P, Imperial J S, Hillyard D R. Toxicon. 1992;30:1111–1116. doi: 10.1016/0041-0101(92)90056-b. [DOI] [PubMed] [Google Scholar]
- 21.Cruz L J, Ramilo C A, Corpuz G P, Olivera B M. Biol Bull. 1992;183:159–164. doi: 10.2307/1542418. [DOI] [PubMed] [Google Scholar]
- 22.Hillyard D R, Olivera B M, Woodward S, Corpuz G P, Gray W R, Ramilo C A, Cruz L J. Biochemistry. 1989;28:358–361. doi: 10.1021/bi00427a049. [DOI] [PubMed] [Google Scholar]
- 23.Kohn A J. Pac Sci. 1980;34:359–369. [Google Scholar]
- 24.Kohn A J, Nybakken J W. Mar Biol. 1975;29:211–234. [Google Scholar]
- 25.Reichelt R E, Kohn A J. Proc Fifth Int Coral Reef Congr. 1985;5:191–196. [Google Scholar]
- 26.Jakobsen K S, Breivold E, Hornes E. Nucleic Acids Res. 1990;18:3669. doi: 10.1093/nar/18.12.3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee Y H, Vacquier V D. Anal Biochem. 1992;206:206–207. doi: 10.1016/s0003-2697(05)80035-1. [DOI] [PubMed] [Google Scholar]
- 28.Marchuk D, Drumm M, Saulino A, Collins F S. Nucleic Acids Res. 1991;19:1154. doi: 10.1093/nar/19.5.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar S, Tamura K, Nei M. mega: Molecular Evolutionary Genetics Analysis. University Park, PA: Pennsylvania State Univ.; 1993. , Version 1.01. [Google Scholar]
- 30.Ina Y. J Mol Evol. 1995;40:190–226. doi: 10.1007/BF00167113. [DOI] [PubMed] [Google Scholar]
- 31.Hochberg Y. Biometrika. 1988;75:800–802. [Google Scholar]
- 32.Metz E M, Robles-Sikisaka R, Vacquier V D. Proc Natl Acad Sci USA. 1998;95:10676–10681. doi: 10.1073/pnas.95.18.10676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer; 1997. [Google Scholar]
- 34.Hughes A L, Nei M. Nature (London) 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 35.Hughes A L. Mol Biol Evol. 1992;9:381–393. doi: 10.1093/oxfordjournals.molbev.a040730. [DOI] [PubMed] [Google Scholar]
- 36.Hill R E, Hastie N D. Nature (London) 1987;326:96–99. doi: 10.1038/326096a0. [DOI] [PubMed] [Google Scholar]
- 37.Riley M A. Mol Biol Evol. 1993;10:1048–1059. doi: 10.1093/oxfordjournals.molbev.a040054. [DOI] [PubMed] [Google Scholar]
- 38.Zhang J, Rosenberg H F, Nei M. Proc Natl Acad Sci USA. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]