Abstract
Insufficient blood supply during acute infarction and chronic ischemia leads to tissue hypoxia which can significantly alter gene expression patterns in the heart. In contrast to most mammals, some teleost fishes are able to adapt to extremely low oxygen levels. We describe here that chronic constant hypoxia (CCH) leads to a smaller ventricular outflow tract, reduced lacunae within the central ventricular cavity and around the trabeculae and an increase in the number of cardiac myocyte nuclei per area in the hearts of two teleost species, zebrafish (Danio rerio) and cichlids (Haplochromis piceatus). In order to identify the molecular basis for the adaptations to CCH, we profiled the gene expression changes in the hearts of adult zebrafish. We have analyzed over 15,000 different transcripts and found 376 differentially regulated genes, of which 260 genes showed increased and 116 genes decreased expression levels. Two notch receptors (notch-2 and notch-3) as well as regulatory genes linked to cell proliferation were transcriptionally upregulated in hypoxic hearts. We observed a simultaneous increase in expression of IGF-2 and IGFbp1 and upregulation of several genes important for the protection against reactive oxygen species (ROS). We have identified here many novel genes involved in the response to CCH in the heart, which may have potential clinical implications in the future.
Electronic supplementary material
The online version of this article (doi:10.1007/s00360-007-0201-4) contains supplementary material, which is available to authorized users.
Keywords: Hypoxia, Zebrafish, Heart, Hyperplasia, Gene expression
Introduction
Low oxygen levels (hypoxia) play important roles in clinical conditions such as stroke and heart failure. Insufficient blood supply leads to tissue hypoxia in the heart during acute infarction and chronic ischemia (Semenza 2001).
Effective protection of the heart against ischemia/reperfusion injury is one of the most important goals of experimental and clinical research in cardiology. Besides ischemic preconditioning as a powerful temporal protective phenomenon, adaptation to chronic hypoxia also increases cardiac tolerance to all major deleterious consequences of acute oxygen deprivation such as myocardial infarction, contractile dysfunction and ventricular arrhythmias (Kolar and Ostadal 2003). Although many factors have been proposed to play potential roles, the detailed mechanism of this long-term protection remains poorly understood. Some of the molecular mechanisms of cardiac protection by adaptation to chronic hypoxia and chronic high-altitude hypoxia have recently been reviewed (Kolar and Ostadal 2003; Ostadal and Kolar 2007). KATP channels, PKCδ as well as the different MAPK pathways were shown to be involved in the mechanism of increased tolerance of chronically hypoxic hearts and further the controversial role of ROS in hypoxia tolerance is discussed (Kolar and Ostadal 2003). Furthermore, a recent study has profiled the gene expression changes induced by chronic constant hypoxia (CCH) and chronic intermittent hypoxia (CIH) in newborn mice (Fan et al. 2005).
In contrast to most mammals (with the exception of some marine mammals), some teleosts, have developed the ability to withstand extreme chronic hypoxia (Stecyk et al. 2004). It is well assumed that these vertebrate species possess unique adaptations in order to survive short and long term oxygen deprivation. However, the molecular basis of these adaptations in fish has so far not been extensively investigated.
Several studies have profiled gene expression changes in teleosts exposed to hypoxia. Gracey et al. showed in adults of the euryoxic gobiid fish Gillichthys mirabilis (the long-jawed mudsucker), that 5 days of hypoxia induced a complex transcriptional response, including a shut down of energy requiring pathways like protein synthesis and locomotion, and an induction of genes needed for anaerobic ATP production in different tissues (Gracey et al. 2001). Recently, we described phenotypic and behavioral adaptations to long-term hypoxia and described the gene expression changes induced by chronic constant hypoxia in the gills of adult zebrafish (van der Meer et al. 2005).
Ton et al. identified global gene expression changes in zebrafish embryos. Zebrafish embryos at 48 h post fertilization were exposed to water with 5% oxygen content for 24 h. The authors identified 138 genes responsive to short-term hypoxia and could also show that transcriptional changes indicated metabolic depression, a switch from aerobic to anaerobic metabolism and energy conservation (Ton et al. 2003).
In this study, we have identified CCH-induced gene expression changes in the zebrafish heart by looking at over half of the zebrafish genome. We have compared several of these novel changes described in other species and tissues. We have here identified the heart-specific molecular adaptations to CCH. Future functional experiments are warranted to determine whether some of the findings can be used to better adapt mammalian hearts to CCH.
Material and methods
Animal handling
Adult wild-type zebrafish (Danio rerio) around 3 month of age, were obtained from a local pet store. Cichlids (Haplochromis piceatus) have been collected in the Mwanza Gulf of Lake Victoria in 1984 and were bred in our laboratory for about 20 generations. All animals were handled in compliance with animal care regulations. Our animal protocols were approved by the review board of Leiden University in accordance with the requirements of the Dutch government. Zebrafish were kept at 25°C in aquaria with day/night light cycles (12 h dark vs. 12 h light). Cichlids were kept at 25°C with the same day/night light cycles.
Hypoxia treatment
For gradual hypoxia treatment, oxygen levels were gradually decreased in 4 days from 100% air saturated water to 40% (day 1), 30% (day 2), 20% (day 3) and the final 10% air saturation (day 4). After day 4, the fish were kept for an additional 21 days at 10% air saturation. (at 100% air saturation and 28°C the O2 concentration is 8 mg/l and pO2 is 15 Torr). In parallel, a control group was kept at 100% air saturated water. Both groups were kept in identical aquaria of 100 l. The oxygen level on the hypoxia group was kept constant by a controller (Applikon Biotechnology, The Netherlands) connected to an O2-electrode and solenoid valve in line with an air diffuser. The oxygen level in the tank was kept constant by adding oxygen via the diffuser and thereby compensating the oxygen consumption of the fish. In case of immediate hypoxia exposure, tanks were pre-equilibrated to the respective pO2 concentration and fish were then directly set in the equilibrated aquaria.
Perfusion of cichlid hearts
In order to minimize blood clotting, a perfusion protocol was developed in which the whole blood volume of clinically dead animals was initially replaced with isotonic buffer and with a fixative solution secondarily.
Heart dissection
The fish were killed with an overdose of anesthetic (MS-222; Tricaine Methanesulfonate from Argent Chemical Laboratories, USA). Hearts were dissected from the fish immediately after the anesthetic worked. For RNA preparation the hearts were immediately shock-freezed in liquid nitrogen. For histology and microscopy, the hearts were left intact and fixed immediately in Karnovsky fixative (4% paraformaldehyde (PFA) and 2.5% glutaraldehyde in 0.1 M phosphate buffer, pH 7.2) for 4 h at 4°C. After three washes in 0.1 M phosphate buffer, pH 7.2, they were transferred to 70% ethanol.
Histology of adult fish hearts, statistical analysis and scanning electron microscopy
Hearts from zebrafish and cichlids raised either under normoxic or hypoxic conditions, were dissected from the fish and fixed for 24 h in 4% PFA in PBS. After fixation, hearts were washed with 1 × PBS, cut in halfs (sagittal through the midline) and then dehydrated through ethanol series starting at 70% ethanol, followed by 80, 90, 96 and 100% ethanol, each step was done once for 1 h except the last one which was done twice. After dehydration the hearts were embedded in increasing gradients of Historesin (Technovit 7100, Heraus Kulzer, Germany) (25, 50, 75 and 100% Historesin in ethanol, for 2 h at room temperature; 100% Historesin, 24 h at 4°C). Afterwards the plastic with the hearts was polymerized at 40°C (overnight). A 5 μm sagittal sections were made using the Ultramicrotome (Reichter-Jung) and a glass knife. Approximately 500 sections on each side of the midline were visually analyzed per heart (1,000 sections in total per heart). Sections were left to dry and later stained with hematoxylin–eosin staining. Pictures were taken with Axioplan 2 imaging, (Carl Zeiss, Jena). Azan staining of histological sections were done as follows. Paraffin sections of the hearts were prepared as described before (van der Meer et al. 2006). Sections were incubated with Azokarmine solution, for 30 min at 60°C. Afterwards they were washed in water and differentiated in 0.2% Anilin alcohol. They were then rinsed in 1% acetic acid in 95% alcohol, followed by 45 min incubation period, in 5% phosphotungstic acid. After that, sections were rinsed in distilled water and incubated for 45 min with aniline blue. Finally, they were rinsed with distilled water, differentiated and dehydrated in 95% alcohol followed by absolute, cleared in xylene and mounted in Entallan. The protocols used here for SEM had been described before (van der Meer et al. 2005). The statistical analysis of cardiac myocyte nuclei per section was done using Statistica by performing an independent t test. A P value of less than 0.05 was considered significant.
Immunohistochemistry and statistical analysis
Zebrafish, raised either under normoxic or hypoxic conditions, were killed with an overdose of anesthetic MS-222 and frozen in liquid nitrogen. Subsequently, transversal cross-sections (10 μm thick) of the body, were cut using a cryostat at −20°C and mounted on glass slides coated with Vectabond (Vector Laboratories, Burlingame, USA). Sections were fixed in 4% formaldehyde in Tris-buffered saline (TBS; 50 mM Tris and 150 mM NaCl, pH 7.5) for 10 min and subsequently washed in TBS with 0.05% Tween-20 (TBST) (Sigma-Aldrich, Zwijndrecht, The Netherlands). Subsequently, sections were incubated for 10 min with 10% normal swine serum (Vector laboratories) in TBST after which sections were incubated for 24 h at 4°C with anti-phospho Akt polyclonal antibody (Santa-Cruz Biotechnology, USA) diluted 1:50 in TBST. After incubation with primary antibody, the slides were washed in TBST and subsequently placed in 0.25% (v/v) acetic anhydride in 0.1 M triethanolamine buffer (pH 8) for 10 min followed by rinsing in TBST. After this, sections were incubated for 60 min at 20°C with secondary anti-rabbit immunoglobulin G (IgG) antibody covalently coupled to alkaline phosphatase (Vector Laboaratories) diluted 1:100 and washed in TBST. After this, sections were incubated for 5 min with alkaline phosphatase (AP) buffer (0.1 M NaCl, 0.1 M Tris, 50 mM MgCl2 and 0.1% Tween-20, pH 9.5) followed by incubation with BM Purple AP substrate (Roche Applied Sciences, Almere, The Netherlands) for 30, 45 or 60 min which was followed by rinsing in TBST. All sections were mounted in glycerine-gelatin and stored at 4°C in the dark until staining intensity was measured. The absorbance values of the BM Purple in the sections were determined using a Leica DMRB microscope (Wetzlar, Germany) fitted with calibrated gray filters using different interference filters. Absorbances for BM Purple were determined at 550 nm. Images were recorded with a ×20 objective and a Sony XC-77CE camera (Towada, Japan) connected to a LG-3 frame grabber (Scion; Frederick, MD) in an Apple Power Macintosh computer. Recorded images were analysed with the public domain program NIH-Image V1.61 (US National Institutes of Health, available at http://rsb.info.nih.gov/nih-image/). Gray values were converted to absorbance values per pixel using the gray filters and a third-degree polynomial fit in the calibrate option of NIH-image programme. Morphometry was calibrated using a slide micrometer and the set scale option in NIH-image, taking the pixel-aspect ration into account. An independent t test was used to test for differences in phospho-Akt levels in cardiac myocytes of normoxic and hypoxic fish. A P value of less than 0.05 was considered significant. Values are means ± S.E.M.
RNA preparation and biological sampling
After dissection hearts were homogenized in a Dounce homogenizer using 1 ml Trizol solution (GibcoBrl, Life technologies). The whole heart was used and for each biological sample hearts were pooled from five different animals. After Trizol extraction, total RNA was further purified using RNAeasy columns (Qiagen). RNA samples were analyzed for quality control by Lab-on-a-chip analysis (Agilent) and on agarose gels. For the array experiment five arrays were done for normoxic and 5 arrays for the hypoxic condition. Biological samples (BS) came from two independent experiments and one technical replicate (TR) was included (2BS + 2BS + TR for normoxia and 2BS + 2BS + TR for hypoxia). As mentioned above for each BS, hearts from five different animals were pooled.
Microarray analysis
The Affymetrix GeneChip® Zebrafish Genome Arrays containing 15,509 Danio rerio gene transcripts were used. Probe sets on the arrays were designed with 16 oligonucleotide pairs to detect each transcript and procedures were in full support of MIAME standards. Labeling and microarray hybridization were performed by ServiceXS (Leiden, The Netherlands), including prior a standard round of RNA amplification according to standard Affymetrix protocols. The criteria used for differential expression were greater or equal than 2-fold induced or reduced and P ≤ 0.02. Data analysis was done using Rosetta Resolver. All expression data was submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo). The series entry number is GSE4989 and the following 10 accession numbers were assigned: GSM112796 and GSM112798-GSM112806. A complete list of differential regulated genes is shown in Supplemental Table 1.
Table 1.
Statistical analysis of histological sections (5 μm) from zebrafish (Danio rerio) and cichlid (Haplochromis piceatus) hearts
| Number of cardiomyocyte nuclei per section | ||||
|---|---|---|---|---|
| Zebrafish (nuclei per 900 μm2) | Cichlid (nuclei per 10,000 μm2) | |||
| Normoxia | Hypoxia | Normoxia | Hypoxia | |
| Mean | 9.81 | 13.67 | 14.7 | 24 |
| SD | 0.30 | 0.39 | 0.50 | 0.68 |
| P value | 9.9 × 10−17 | 7.6 × 10−12 | ||
Midline sections of zebrafish and cichlids raised under normoxic and hypoxic conditions were chosen and subdivided in smaller areas. In the case of D. rerio, each subarea of the section was 900 μm2 whereas in the case of the H. piceatus it was 10,000 μm2. Then, subareas were randomly picked and the amount of cardiac myocyte nuclei were counted. The vast majority of cardiac myocytes were mononucleated cells. Hundred subareas per section were counted per specimen. Three sections per heart were visually analyzed (with a magnification of 20×). In total 6 different zebrafish hearts (from three independent experiments) were investigated for hypoxic conditions and 6 for normoxic condition. In total 1,800 subareas were quantified per heart and condition used. The same was done for the cichlids (for which bigger subareas were counted). A two-tailed t test was applied and a significant difference (P < 0.001) in the amount of nuclei present in the hearts of normoxia versus hypoxia groups was observed for both species
Gene ontology analysis
The Gene ontology analysis was performed using eGOn and the database of the Norwegian Microarray Consortium (http://www.genetools.microarray.ntnu.no/egon/) and the most recent updated Unigene numbers for zebrafish (May 2007). 13414 Unigene numbers were annotated to the Affymetrix gene chip used. eGOn Enrichment analysis was used for the entire Affymetrix set versus the identified 376 gene set based on the new Unigene numbers. A master target test was performed (using Fishers exact test) and the cut off for positives was set to P < 0.05. The available three categories, molecular function, biological process and cellular component were all tested and are shown in this dataset (Supplemental Data 1). The fold enrichment was based on the percentage found within the differentially expressed genes divided by the percentage on the total chip. A list of all Unigene numbers found in the different categories is given in Supplemental Data 2.
Real-time quantitative RT-PCR
For verification of gene expression data, we used quantitative real-time RT-PCR. Biological RNA samples were obtained from a third independent experiment and as mentioned above for the microarrays, for each BS hearts from five different animals were pooled. The Roche Master SYBR Green kit was used for the RT-PCR reactions. The annealing and synthesis temperature was 55°C alternating with 96°C for 45 cycles. Dissociation protocols were used to measure melting curves and control for unspecific signals from the primers. A measure of 100 ng of total RNA was used per reaction. A standard curve for β-actin using 1, 5, 10, 100 and 500 ng of total RNA was used for normalization. Samples were measured in the Roche LightCycler. The primer3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) was used to design primers for short amplicons between 50 and 100 bases. The primers used are shown in Supplemental Table 2.
Results
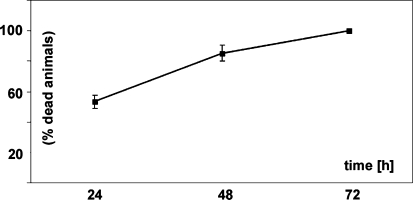
We describe here the survival rates of adult zebrafish upon immediate and gradual exposure to different pO2 concentrations. Immediate exposure to pO2 of 15 Torr (O2 concentration of 0.8 mg/l or 10% air saturated water) was lethal for the adult zebrafish and none survived for longer than 72 h (Fig. 1). If lowered gradually (see experimental procedures for regimen in Sect. “Hypoxia treatment”) zebrafish were able to grow and gain weight at O2 levels of 10% air saturation. The zebrafish were able to survive for longer than 6 months and no mortality was observed, demonstrating that they can well adapt to these conditions (data not shown). For the experiments with zebrafish and cichlids, the pO2 concentration was gradually lowered to 10% air saturation (0.8 mg/l) (see “Material and methods”) and fishes were kept under 10% air saturation for 3 weeks. Control groups were always kept in parallel under normoxic conditions.
Fig. 1.
Survival of zebrafish embryos after immediate exposure to hypoxia. Zebrafish were immediately exposed to hypoxia (15 Torr; 0.8 mg/ml; 10% air saturated water). Results are derived from three independent experiments with n = 30 in each experiment (adding to a total of 90 fish tested). After 24, 48 and 72 h, dead and alive fish were counted. Shown here is the percentage of dead fish at the respective time points and the standard deviation. At 24 h 53.3%(±4.4) of fish were dead, at 48 hours 85.3% (±5.1) and at 72 hours 100% (±0). Control groups (n = 30), which were in parallel exposed to normoxic conditions showed no mortality (not shown). None of the zebrafish survived the immediate exposure to an O2 concentration of 0.8 mg/l (10% air saturation) over the 3-day period. In contrast, fishes gradually exposed to hypoxia showed no induced mortality for even the 25 day time periods used in our experiments
Phenotypic changes in the heart of adult teleosts under chronic constant hypoxia (CCH)
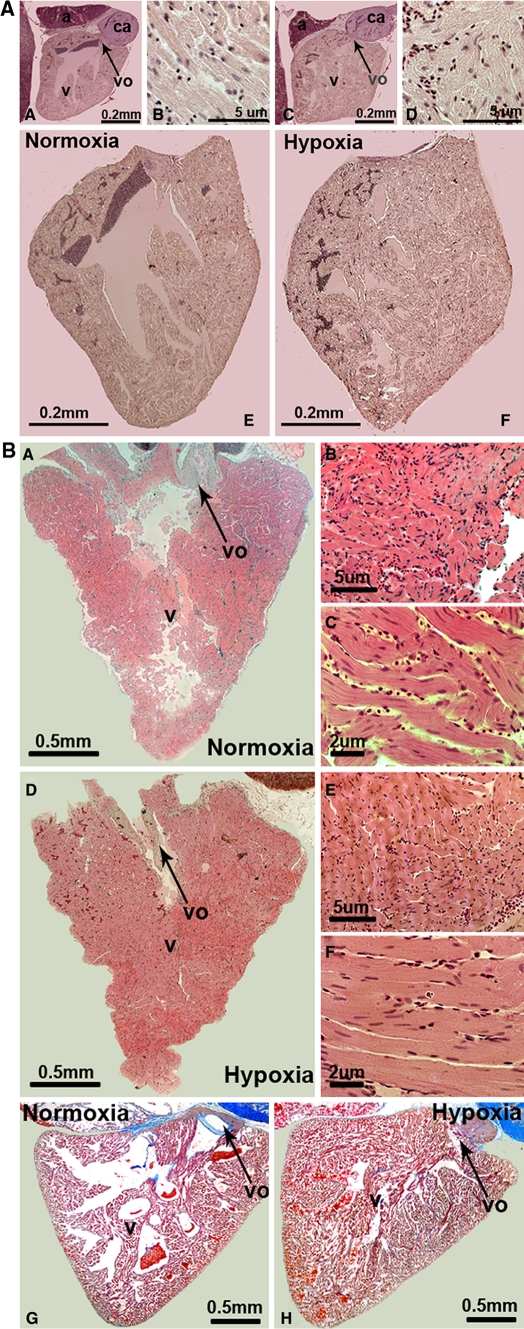
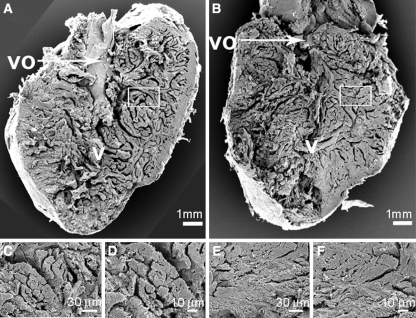
In comparison to the normoxic control groups, we observed a significantly smaller ventricular outflow tract and reduced lacunae within the central ventricular cavity and reduced lacunae around the trabeculae in midline sections of hearts of both zebrafish (Danio rerio) (Fig. 2a) and cichlids (Haplochromis piceatus) (Fig. 2b) exposed to CCH. In addition to the midline sections, none of the sections investigated from hypoxia treated fishes showed a ventricular outflow tract in comparable size to the normoxic controls (data not shown). In addition to the midline sections, none of the lateral sections (to both sides of the midline) investigated from hypoxia treated fishes showed a ventricular outflow tract as well as lacunae in a comparable size to the normoxic controls (data not shown). The larger cichlid hearts were also perfused and midline sectioned and showed similar results with a smaller ventricular outflow tract and reduced lacunae (Fig. 2b-H). This might represent ventricular hypertrophy or hyperplasia in both walls and trabeculae, which could lead to the observed cavity obliteration in these sections. We quantified the number of cardiac myocyte nuclei per area in the midline sections of both zebrafish and cichlids under normoxic, as well as hypoxic conditions. A significant difference in both species was observed and showed that hypoxia led to a 1.4- and 1.6-fold increase in the number of cardiac myocyte nuclei per area in zebrafish and cichlid hearts, respectively (Table 1). Cardiac myocyte nuclei in sections were clearly distinguishable from nuclei of other cells like erythrocytes and fibroblasts and only centralized nuclei in cardiac myocytes (which are more elongated than nuclei from erythrocytes) were counted. Furthermore, scanning electron microscopy (SEM) was used to confirm these findings in the smaller zebrafish hearts (Fig. 3). Future research is warranted in order to assess further how the cardiac myocytes adapt to CCH in the fish heart.
Fig. 2.
Histological changes of zebrafish and cichlid hearts after exposure to chronic constant hypoxia. a shows zebrafish hearts that were dissected, sectioned and stained with a hematoxylin–eosin staining with A, B and E representing normoxic and C, D and F hypoxic conditions. Cell nuclei are seen in dark (dark blue in online version) and cell cytoplasm in light (pink in online version). Pictures A, C, E and F have the same magnification (10×). Images B and D represent a 20× magnification of cardiac muscle (D). Abbreviations used are: a atrium; v ventricle; vo ventricular outflow tract and ca conus arteriosus. b (A–F) corresponds to sections of cichlid hearts, which were treated the same way as the ones above from zebrafish and G and H show cichlid hearts which have been perfused prior to dissection and were stained with either hematoxylin–eosin (A–F) or Azan blue (G, H). In A, B, C and G pictures of normoxic conditions are shown and D, E, F and H represent the corresponding hypoxic conditions. Similar results for both the zebrafish and the cichlid hearts were observed in three independent experiments
Fig. 3.
Morphological changes of zebrafish and cichlid hearts after exposure to chronic constant hypoxia. Scanning electron microscopy pictures of hearts from normoxia control zebrafish (a, c and d) and hypoxia-treated zebrafish (b, e and f). The bigger images show half of a heart ventricle, sectioned longitudinally. The smaller images represent a higher magnification of cardiac muscle. The scale is given at the bottom of each picture. Abbreviations used are: v ventricle and vo ventricular outflow tract
Gene expression changes in the heart of adult zebrafish under chronic constant hypoxia (CCH)
In this study, we used microarrays for the transcriptional profiling of up and downregulated genes in response to hypoxia in the zebrafish heart. We identified 376 genes that were differentially expressed under hypoxic conditions, out of which 116 genes showed a decrease in gene expression (30.9%) in comparison to 260 genes which showed increased expression levels (69.1%).
All 376 differentially expressed genes, including the ones with oligo sequences which could not be annotated so far (referred therein either as transcribed locus or zebrafish clone) are shown in the complete file (Supplemental Table 1). Functional groups are color coded and if possible, gene functions are briefly summarized and OMIM links given.
Functional groups of differentially expressed genes in the heart
We have clustered the differential expressed genes according to known functions (Table 2). Genes can have more than one particular function assigned, so some genes can appear in more than one group. In addition, a gene ontology analysis was performed using eGOn to determine gene enrichment and overrepresentation in the three categories of molecular function, biological processes and cellular components (Supplemental Data 1 and 2).
Table 2.
Functional groups of differentially expressed genes
| UniGene | GeneBank | Fold | Gene name |
|---|---|---|---|
| Upregulated genes | |||
| Angiogenesis | |||
| Dr.11575 | NM_173244 | 2.3 | T-cell acute lymphocytic leukemia 1; TAL1 |
| Dr.845 | BG729013 | 2.8 | Fibrinogen alpha/alpha-E chain |
| Dr.4907 | BC045868 | 4.2 | Fibrinogen, gamma polypeptide |
| Apoptosis | |||
| Dr.15862 | AF493987 | 2.1 | BCL2adenovirus E1b 19 interacting protein3 |
| DrAffx.1.39 | AF302789 | 2.3 | Death receptor |
| Dr.20106 | AI722277 | 2.8 | Apoptosis inhibitor 5 |
| Dr.4039 | BQ480688 | 21.8 | BAX inhibitor 1 |
| Cell adhesion | |||
| Dr.6007 | NM_131820 | 2.9 | Cadherin 1 |
| Dr.25140 | BQ262802 | 3.3 | Tumor-associated calcium signal transducer |
| Dr.4409 | BC049036 | 4.4 | CD9 antigen |
| Dr.25140 | BQ262802 | 7.7 | Tumor-associated calcium signal transducer glycoprotein |
| Development | |||
| Dr.11575 | NM_1732 | 2.3 | T-cell acute lymphocyte leukemia 1 (tal 1) |
| Dr.23348 | BE201653 | 2.6 | Bone morphogenetic protein 3b; (bmp 3) |
| Dr25405 | BC013923 | 2.8 | SOX2 SRY-box 2 |
| Dr.6382 | AW165053 | 2.9 | Hedgehog-interacting protein |
| Dr.10879 | U97669 | 3.0 | NOTCH3 Notch homolog 3 (Drosophila) |
| Dr.15055 | BC050172 | 3.6 | Chemokine receptor 4a |
| Dr.6787 | BI533426 | 5.5 | Noelin |
| Dr.16720 | BI980847 | 6.3 | notch 2 |
| Disease related | |||
| Dr.6349 | AW116668 | 2.4 | Eparin cofactor II |
| Dr.21064 | BC046075 | 4.5 | 4hydroxyphenylpyruvate dioxygenase HPD |
| Dr.12584 | NM_131211 | 5.4 | Gata binding protein 3 (GATA3) |
| Dr.3530 | AI497545 | 79.3 | Prion protein (prp) gene |
| Growth regulation | |||
| Dr.8145 | NM_13143 | 2.2 | Insulin like growth factor 2 (IGF-2) |
| Dr.7609 | BI475857 | 2.4 | Prolactin receptor |
| Dr.8285 | NM_13136 | 2.4 | Mad homolog 2 |
| Dr.8947 | CD594735 | 2.5 | Spint 2 |
| Dr.822 | BM184127 | 2.5 | Spint 2 |
| Dr.3563 | CD014488 | 2.8 | Tetraspan membrane protein IL-TMP |
| Dr.8587 | NM_17328 | 2.9 | Insulin-like growth factor binding protein 1 |
| Dr.2596 | BM342901 | 3.2 | Cyclin I |
| Dr.8587 | AL910822 | 3.4 | Insulin-like growth factor binding protein 1 |
| Dr.26458 | BC053206 | 5.6 | m-ras |
| Heart related | |||
| Dr.15088 | BM181749 | 4.3 | Lectin galactoside-binding soluble 1; (galectin10-like 3) |
| Dr.4867 | AI496840 | 5.5 | Haptoglobin |
| Dr.3585 | AY049731 | 6.6 | Angiotensinogen |
| Dr.2452 | BQ284848 | 4.3 | Complement component C9 |
| Dr.18453 | BC044525 | 4.8 | Uridine phosphorylase |
| Dr.3025 | BG738204 | 2.7 | Alpha-2 macro-globulin; A2MG |
| Inflammation | |||
| Dr.12491 | BI672168 | 2.1 | Complement C4–2 |
| Dr.4047 | NM_131627 | 2.3 | Small inducible cytokine A (scyba) |
| Dr.5053 | NM_131723 | 2.3 | Kruppel-like factor 4 |
| Dr25207 | X06465 | 2.5 | Complement component 8, gamma polypeptide |
| Dr.6845 | K02765 | 2.9 | C3 complement component 3 |
| Dr.5741 | BU710482 | 3.2 | Complement component b fb |
| Dr.7722 | BI878414 | 3.5 | Complement C3-H1 |
| Dr.22244 | AW019781 | 3.6 | Complement C1s |
| Dr.22133 | AW076768 | 3.7 | c1rs-A and clrs-B |
| Dr.5528 | AI497212 | 4.2 | Complement component C9 |
| Dr.2452 | BQ284848 | 4.3 | Complement component C9 |
| Dr.1730 | AI721528 | 4.8 | cfI-B complement control protein factor I-B |
| Dr.2452 | BM778002 | 5.8 | Complement component C9 |
| Dr.20291 | BM036389 | 6.5 | Complement C3-S |
| Dr.190 | NM_131338 | 7.9 | Complement component factor B |
| Dr.1192 | AB071601 | 2. | Lipocalin-type prostaglandin D synthase-like protein |
| Metabolism | |||
| Dr.9492 | BI882244 | 2.0 | Sulfide dehydrogenase like |
| Dr.15574 | BM571467 | 2.1 | Hypoxanthine _hosphor-ribosyltransferase 1 |
| Dr.3332 | AI943053 | 2.2 | Angiopoietin 5 |
| Dr.16130 | CD014898 | 2.3 | Alcohol dehydrogenase 8 b |
| Dr.3959 | BI43001 | 2.5 | 5′-nucleotidase |
| Dr.22205 | AW019477 | 2.6 | Oxidoreductase |
| Dr.1699 | AI667249 | 2.7 | Pyruvate kinase |
| Dr.5504 | BI879550 | 3.2 | Cystathionine-beta-synthase |
| Dr.1202 | AJ245491 | 3.9 | Apolipoprotein A-I |
| Dr.4111 | BC053267 | 4.2 | Fructose-1,6-bisphosphatase 1 |
| Dr.18834 | AW019321 | 4.2 | Urate oxidase |
| Dr.19224 | BC050167 | 4.3 | Aldolase b |
| Dr.4938 | NM_131645 | 4.4 | Fatty acid desaturase 2 |
| Dr.12654 | BC046901 | 14.8 | ELOVL family member 6, |
| Dr.5488 | AI545593 | 17.3 | Apolipoprotein A-IV |
| Muscle related | |||
| Dr.3585 | AY049731 | 6.6 | Angiotensinogen |
| Dr.2452 | BQ284848 | 4.3 | Complement component C9 |
| Proteolysis | |||
| Dr.20934 | AF541952 | 2.6 | Trypsin precursor |
| Dr.3025 | BG738204 | 2.7 | Alpha-2-macroglobulin |
| Dr.22139 | AW018965 | 3.0 | Alpha-1-antitrypsin |
| Dr.25331 | AI658072 | 4.1 | Alpha-2-macroglobulin-2 |
| Dr.12602 | NM_139180 | 4.3 | Lysozyme |
| Dr.1605 | BM185388 | 4.4 | Protease inhibitor 1 |
| Dr.17459 | CD586837 | 4.8 | Inter-alpha-trypsin inhibitor heavy chain H3 |
| Dr.3073 | AI585030 | 5.0 | Serine protease inhibitor alpha 1 |
| Dr.26371 | AI667676 | 5.4 | Prostasin |
| Dr.3025 | BM530427 | 5.6 | Alpha-2-macroglobulin-1 |
| Dr.3025 | BM316867 | 6.5 | Alpha-2-macroglobulin-2 |
| Dr.2960 | X67055 | 3.5 | ITIH3 pre-alpha (globulin) inhibitor, H3 polypeptide |
| Dr.25379 | BI326783 | 6.7 | Alpha-2-macroglobulin |
| Dr.4797 | AI959534 | 7.8 | 26–29 kD-Proteinase protein |
| ROS protection | |||
| Dr.20068 | NM_131075 | 2.1 | Metallothionein (mt) |
| Dr.5399 | AI957765 | 2.3 | Biliverdin I Beta Reductase |
| Dr.14058 | CD015351 | 3.5 | Glutathione S-transferase theta 1 |
| Dr.25160 | BC049475 | 5.9 | Metallothionein 2 |
| Dr.3613 | BC048037 | 6.0 | Cerulopasmin |
| Dr.4905.1 | BC045464 | 6.5 | Uncoupling protein 4 |
| Signal transduction | |||
| Dr.9852 | AW826425 | 2.1 | CAM kinase 1 |
| Dr.8591 | BM186508 | 2.9 | Rho guanine nucleotide exchange factor 10 |
| Dr.6236 | AW115973 | 3.1 | Rho guanine nucleotide exchange factor 5 |
| Dr.1267 | BC051157 | 3.4 | Phospholipase C delta |
| Dr.22129 | BC016668 | 3.9 | RRAGC Rag C (Ras-related GTP binding C) |
| Dr.7255 | AW116479 | 4.4 | Protein phosphatase 1, |
| Dr.4453 | BC044421 | 5.8 | Phosphoprotein phosphatase |
| Translation | |||
| Dr.13234 | BM036471 | 2.0 | Ribonuclease P |
| Dr.382 | CB363830 | 2.1 | Nucleolin |
| Dr.6949 | AW078116 | 2.1 | RNA 3′-terminal phosphate cyclase-like protein (HSPC338) |
| Dr.13563 | BI890729 | 2.3 | Methionyl aminopeptidase 2 |
| Dr.26328 | AL723696 | 2.3 | Eukaryotic translation initiation factor 4A, |
| Dr.17693 | BQ078285 | 3.7 | 40 S ribosomal protein S6 |
| Dr.20270 | BI674050 | 5.9 | Ribosomal protein L12 |
| Dr.25224 | CD015330 | 20.4 | Ribosomal protein L12 |
| Dr.12439 | BM533848 | 17.5 | Heterogeneous nuclear ribonucleoprotein K |
| Dr.12439 | BM533848 | 24.2 | Heterogeneous nuclear ribonucleoprotein K |
| Dr.14821 | BM071714 | 33.8 | Heterogeneous nuclear ribonucleoprotein K |
| Dr.12502 | BQ284686 | 40.7 | Heterogeneous nuclear ribonucleoprotein K |
| Dr.12439. | BM534432 | 40.9 | Heterogeneous nuclear ribonucleoprotein K |
| Dr.12439 | BQ616930 | 45.4 | Heterogeneous nuclear ribonucleoprotein K |
| Transport | |||
| Dr.1084 | BQ109772 | 3.0 | Clathrin coat assembly protein AP19 |
| Dr.5562 | X04506 | 3.0 | APOB apolipoprotein B (including Ag(x) antigen) |
| Dr.13231 | BM778646 | 4.2 | Solute carrier family 22 |
| Dr.30444 | AY329629 | 4.3 | Embryonic globin beta e2 |
| Dr.24250 | AF489105 | 2.0 | Uroporphyrinogen III synthase |
| Dr.10343 | NM_131687 | 4.7 | Na+K+ transporting, alpha 1a.2 polypeptide |
| Dr.7634 | AW115757 | 11.3 | Hemopexin |
| Downregulated genes | |||
| Angiogenesis | |||
| Dr.26411 | BQ783571 | −8.9 | Fast muscle troponin I |
| Dr.15501 | BM316040 | −2.1 | Similar to CYR6 HUMAN CYR61 protein precursor, Insulin-like growth factor-binding protein 10 |
| Cell adhesion | |||
| Dr.251 | BQ285646 | −2.3 | Cadherin 11 |
| Disease related | |||
| Dr.22774 | AW280206 | −5.7 | ras-like GTP-binding protein RAB27A |
| Dr.1816 | AL720262 | −4.4 | Ataxin 2-binding protein |
| Dr.9893 | BM036473 | −2.3 | Fibrillarin |
| Dr.16726 | BI429372 | −2.0 | netrin G1 |
| Growth regulation | |||
| Dr.12986 | CA787334 | −5.3 | v-fos |
| Dr.12986 | BI881979 | −5.0 | v-fos |
| Dr.12986 | BM957279 | −4.5 | v-fos |
| Dr.1221 | AW510198 | −4.3 | Pmx-1b (PHOX-1) |
| Dr.12986 | BI881979 | −4.2 | v-fos |
| Dr.12410 | NM_131826 | −2.4 | Sprouty homolog 4 |
| Dr.6431 | BC049326 | −2.3 | Suppressors of cytokine signaling 3 |
| Dr.6511 | NM_130922 | −2.2 | B-cell translocation gene 2 |
| Dr.5365 | AI601685 | −2.2 | Dual specificity phosphatase 5 |
| Dr.12062 | BC047814 | −2.1 | Epidermal growth factor receptor kinase substrate EPS8 |
| Dr.17286 | BM777144 | −2.0 | Hormone-regulated proliferation-associated 20 kDa protein |
| Dr.9448 | BM156058 | −2.0 | TGF-beta-inducible early growth response protein 2 |
| Heart related | |||
| Dr.20010 | BQ826502 | −7.0 | ATPase, Ca++ transporting, cardiac muscle (ATP2A1) |
| Dr.1448 | AL717344 | −3.5 | Fast skeletal myosin light chain 1a |
| Dr.20990 | AY033829 AY081167 | −2.4–2.1 | Titin |
| Metabolism | |||
| Dr.24950 | BC053305 | −4.1 | Creatine kinase CKM3 |
| Dr.9528 | BC045993 | −3.5 | Pyruvate dehydrogenase kinase |
| Dr.146 | AI477401 | −2.9 | Carnitine O-palmitoyltransferase II |
| Dr.21501 | AI667180 | −2.4 | Short-chain acyl-CoA dehydrogenase |
| Dr.19643 | AL918850 | −2.4 | FabG beta-ketoacyl -reductase |
| Dr.15059 | BM530407 | −2.2 | Elongation of very long chain fatty acids (Cig30) |
| Dr.21040 | BC045479 | −2.1 | Glucose-6-phosphatase, transport protein 1 |
| Dr.988 | AW154697 | −2.1 | Dodecenoyl-coenzyme A delta isomerase |
| Dr.11971 | BG727588 | −2.0 | Carnitine O-acetyl-transferase |
| Dr.4777 | AW420997 | −2.0 | Succinate-CoA ligase |
| Dr.11252 | BC047826 | −2.0 | Creatine kinase, mitochondrial 1 |
| Muscle related | |||
| Dr.21800 | AI883923 | −5.0 | Myosin binding protein C |
| Dr.5066 | AF524840 | −3.4 | Alpha-actinin 3 |
| Dr.24260 | NM_131619 | −3.0 | Myosin, light polypeptide 3 |
| Dr.2914 | BC045520 | −2.5 | Myosin light polypeptide 2; mylz2 |
| Dr.20990 | AY033829 AY081167 | −2.4–2.1 | Titin |
| Dr.1435 | AI353817 | −2.0 | Caveolin 3 |
| Dr.18657 | BQ479700 | −2.1 | Carbonic anhydrase II |
| Dr.26411 | BQ783571 | −8.9 | Troponin I |
| Proteolysis | |||
| Dr.3581 | BM101561 | −8.3 | Chymotrypsinogen B1 |
| Dr.3581 | BM101561 | −7.5 | Chymotrypsinogen B1 |
| Signal transduction | |||
| Dr.22841 | AI641080 | −2.4 | Serum deprivation response protein (SDPR) |
| Translation | |||
| Dr.7939 | AW281840 | −2.7 | Mitochondrial elongation factor G1 |
| Dr.1286 | BM036808 | −2.2 | Mitochondrial ribosomal protein L48 |
| Dr.18218 | AL909921 | −2.1 | Mitochondrial 28 S ribosomal protein S12 |
| Transport | |||
| Dr.676 | BC050956 | −4.4 | ADT2, ADP,ATP carrier protein |
| Dr.25199 | CD014403 | −2.1 | Calcium-binding mitochondrial carrier protein Aralar2 (Citrin) |
| Dr.2784 | AI942949 | −2.0 | Solute carrier family 25 |
| Dr.11127 | BG306498 | −3.7 | Synaptotagmin I |
| Dr.11127 | AW826278 | −3.2 | Synaptotagmin I |
| Dr.13273 | BI885460 | −2.3 | GTP-binding protein rab15 |
| Dr.22748 | AW280026 | −2.7 | trpn1 |
| Dr.11302 | BG306530 | −2.1 | ATPase (Ca++ transporting plasma membrane 2) |
Proteinbiosynthesis (Translation): In the group linked to proteinbiosynthesis, only three genes showed decreased expression under hypoxic conditions. All three were found to be part of the mitochondrial translational machinery. In contrast, 9 non mitochondrial genes linked to proteinbiosynthesis showed all increased expression (Table 2).
Metabolism: The group with metabolic genes contains 11 repressed and 28 genes with enhanced expression (Table 2). The repression majorly involves metabolic genes linked to β-oxidation and lipid metabolism. Among the metabolic genes with enhanced expression, are pyruvate kinase and aldolase which both are key enzymes for glycolysis indicating a shift from aerobic to anaerobic metabolism induced by hypoxia.
Protection against reactive oxygen species (ROS): The group of genes important for protection against ROS contains 6 genes which are all enhanced under hypoxia.
Apoptosis: We found four genes linked to programmed cell death to be enhanced by hypoxia. Two of these, the death receptor 5 (DR5) and the BNIP3 homologue, are considered to be pro-apoptotic, whereas apoptosis inhibitor 5 and Bax inhibitor have been shown to have anti-apoptotic properties (Tewari et al. 1997; Xu and Reed 1998).
Growth regulation: In the group of genes linked to growth regulation, we found 9 genes to be repressed and 8 genes with enhanced expression. Within the group of 8 repressed genes, we found 5 antiproliferative genes: spry4 and dual specificity phosphatase 5 both inhibit mitogen-activated kinases (MAPK), SOCS3 binds and inhibits Janus kinases (JAK) and thereby prevents STAT3 activation. BTG2 which is important in the G1/S transition and TIEG2 is a transcriptional repressor with antiproliferative functions. Although some genes involved in cell proliferation, like the transcription factor c-fos were repressed by hypoxia (see also “Discussion”), the regulation of the vast majority (13 of 17) of identified genes in this group suggests stimulation of proliferation (Table 2).
Inflammation: 14 genes involved in the inflammatory response were identified to be differentially regulated and all showed increased expression upon hypoxia treatment.
Heart-related function: Several genes linked to cardiac hypertrophy, cardiomyopathy (disease of the heart muscle) and cardiac infarction were identified in this study (see Table 2 and "Discussion").
Muscle-related function: Several sarcomeric genes linked to hypertrophy showed decreased expression under hypoxic conditions (Table 2 and “Discussion”).
Development: The genes in this group were all found to be upregulated by hypoxia among them the gene for notch-2 and notch-3. Notch receptors are transmembrane receptors with essential roles in development including heart development.
Transport (cellular and vascular): A heterologous group containing the gene for embryonic hemoglobin beta e2, important for oxygen transport and hemopexin which is important for heme and iron transport and was found to be upregulated.
Angiogenesis: It is well known that hypoxia via the hypoxia inducible factor 1α (HIF1α) pathway leads to angiogenesis. In the zebrafish heart under CCH, we observed increased expression of HIF1α as well as fibrinogen-α and -γ. Fibrinogen-α has been shown to stimulate HIF1α and VEGF expression and thereby induces angiogenesis (Shiose et al. 2004).
Expression changes of known hypoxia responsive genes
Examples for regulation of known hypoxia responsive genes are HIF1α, insulin growth factor-2 (IGF-2), insulin growth factor binding protein 1 (IGFbp1) and caveolin 3. The transcription factor Hif1α is pivotal in the cellular response to hypoxic stress (Semenza 1999). We further observed increased expression of the egl nine homolog, a gene which was shown to be induced by hypoxia through the Hif1α pathway (Pescador et al. 2005). IGF-2 gene- and protein expression had been shown to be upregulated by hypoxia (Beilharz et al. 1995). It was shown that IGFbp1 also is a hypoxia-inducible gene in zebrafish embryos and it mediates hypoxia-induced embryonic growth-inhibition and developmental-retardation (Kajimura et al. 2005a). Both IGF-2 and IGFbp1 were found upregulated in our study. We observed decreased expression of caveolin 3 in the hearts of zebrafish exposed to CCH, earlier findings showed that chronic myocardial hypoxia led to decreased caveolin-3 protein expression in rabbit hearts (Shi et al. 2000). These findings indicate that the hypoxic conditions used lead to hypoxic stress in the fish heart.
Evaluation of microarray results by quantitative real-time RT-PCR
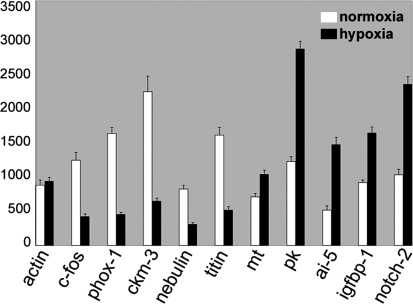
To further verify our results, we used quantitative real-time PCR for 10 of the transcripts. We confirmed the gene expression changes found in the microarray studies for these 5 up- and 5 downregulated transcripts by this independent method (Fig. 4). The downregulated zebrafish genes tested were: c-fos, phox1, creatine kinase (ckm3), nebulin, titin, and the upregulated genes tested were: metallothionein, pyruvate kinase, apoptosis inhibitor 5, igfbp1 and notch-2. The fold induction values were not always directly comparable to the array data but in all cases induction or reduction was confirmed. Quantitative differences between array data and qPCR results have been reported before (Meijer et al. 2005; Ton et al. 2003; van der Meer et al. 2005).
Fig. 4.
Verification of gene expression changes by quantitative real-time PCR. 10. selected genes, which were found to be differentially expressed on the microarrays, were further analyzed by quantitative real-time RT-PCR. Relative expression is given based on normalization to β-actin. A standard curve for β-actin was included in each experiment and data represents three independent experiments each done in triplicates. The primers used are given in Supplemental Table 2
Assessment of microarray results for the IGF/PI3K/Akt pathway by comparing phospho-Akt levels in cardiac myocytes of normoxic versus hypoxic zebrafish hearts
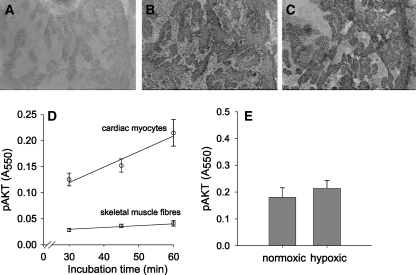
The IGF/PI3K/Akt pathway is activated by IGFs, which are antagonized by the IGFbp1. To test the effects of the upregulation of both IGF-2 and IGFbp1, we assayed phospo-Akt levels in cardiac myocytes and showed that phospho-Akt levels were not different between normoxic and hypoxic cells. Figure 5 shows cytoplasmic immunohistochemical staining of phospho-Akt. The antibody recognizes phosphorylated and detects the phospho-Akt1/2/3 forms. The incubation times for the primary and secondary antibody as well as the BM Purple were optimized to obtain a good signal to noise ratio. Absorbances were linearly related to the time of incubation with the primary and secondary antibodies as well as that of BM Purple. Figure 5d shows for normoxic fish the absorbance values of the phospho-Akt staining in cardiac myocytes as well as skeletal muscle fibers from the tail as a function of the incubation time with BM Purple AP substrate. The absorbance values for the heart are considerably higher than for the skeletal muscle fibers. However, for both cardiac and skeletal muscle the absorbances are linearly related with the incubation time with BM Purple AP substrate and increase at the same relative rate. This implicates that the absorbance of BM Purple after 45 min incubation with BM Purple AP substrate did not reach saturation and therefore provides a semi-quantitative estimate of the phospho-Akt content in the cardiac myocytes. Absorbances of staining for phospho-Akt in normoxic and hypoxic cardiac myocytes were not shown to be significantly different (Fig. 5E, P < 0.48), which indicates that hypoxia did not change the activation of the Akt pathway in the cardiac myocytes.
Fig. 5.
Effects of chronic constant hypoxia of the activation of Akt in zebrafish cardiac myocytes. Immunohistochemical staining of phospho-Akt in cardiac myocytes of zebrafish raised under hypoxic a or normoxic b conditions. Specificity is shown by the control sections obtained from normoxic fish which were not incubated with primary antibody against phospho-Akt c. For both cardiac myocytes and skeletal muscle fibers, the absorbance of the BM Purple is linearly increasing at the same relative rate with the incubation time with BM Purple AP substrate at the same relative rate d. Mean absorbances of phospho-Akt staining (+S.E.M.) from normoxic and hypoxic cardiac myocytes was not different e)
Discussion
In the aquatic environment, oxygen concentrations can often vary, and being able to adapt to changes in oxygen levels can be advantageous for the survival of aquatic animals. This might be in part the reason why some teleosts have developed the ability to withstand extreme hypoxic conditions.
In this study, we have focused on the long-term response to hypoxia in the fish heart. The hypothesis is that the zebrafish heart, in contrast to most mammalian hearts, which are characterized by relative intolerance to injury or the lack of oxygen, are able to adapt to extreme hypoxic conditions.
We showed that chronic hypoxia of zebrafish caused a smaller ventricular outflow tract, reduced lacunae and increased cardiac myocyte densities in the heart. These findings suggest that hypoxia induced an increase of the cardiac myocyte volume or at least did not result in a loss of cardiac myocytes. This is in contrast to mammals where tissue hypoxia in chronic heart failure leads to apoptosis and considerable losses of cardiac myocytes (see for review, Sabbah et al. 2000a). In mammals, compensation for this loss of cardiac myocytes occurs mainly by hypertrophy of the remaining cardiac myocytes (Ostadal and Kolar 2007), although regeneration of myocardium by proliferation of cardiac myocytes may occur also but to a limited extend (Beltrami et al. 2001). Our assay for phosho-Akt did not show any enhancement of Akt activity in response to the CCH, suggesting a lack of hypertrophic signaling via the phosphotadilinositol 3 kinase pathway. However, the density of cardiac myocyte nuclei increased by 50% during CCH, which indicates substantial proliferation of cardiac myocytes and/or nuclear hyperplasia. Recently, it has been shown that the zebrafish heart has the ability to regenerate from mechanical cardiac injury by proliferation of cardiac myocytes (Poss et al. 2002). If the zebrafish heart responds to chronic hypoxia in a similar way as to mechanical dissection, this will be beneficial in preventing apoptosis of cardiac myocytes as the diffusion distance for oxygen are not increased as during hypertrophy, which will help to prevent the development of anoxic cores in the cardiac myocytes (Des Tombe et al. 2002; van der Laarse et al. 2005). In the case that the increase in cardiac myocyte nuclear density during chronic hypoxia was due to nuclear hyperplasia and nuclear ploidy, the observed smaller ventricular outflow tract and reduced lacunae may have been the result of hypertrophy of the existing cardiac myocytes. The histological assay in this study does not clearly distinguish between hyperplasia and hypertrophy of cardiac myocytes. Future investigation of the mechanisms underlying the general morphological adaptations of teleost fish heart in response to CCH requires a nuclear stain in combination with a clear membrane stain, which allows determination of cell sizes in addition to counts of cardiac myocyte nuclei.
We were interested in the underlying gene expression changes of these adaptations. We found gene regulations in a transcriptional network of the serum response element (SRE), which are opposed to the ones described in mammals. In the zebrafish heart hypoxia repressed c-fos and phox1 expression. Both genes are important in the same transcriptional network, phox1 can transduce serum-responsive transcriptional activity to the c-fos (SRE) by interacting with serum response factor (SRF) (Simon et al. 1997). In mammals many studies showed increased expression of c-fos by hypoxia, e.g. in rats hypoxia induces c-fos expression in the LV and RV (Deindl et al. 2003) and the same held true in tissue culture cells (Webster et al. 1993). Interestingly, repression of c-fos by hypoxia has also been shown for the short-term response to anoxia in anoxia tolerant turtles (Greenway and Storey 2000). Is it possible that repression of c-fos and phox-1 are important adaptations in hypoxia tolerant animals?
Several novel gene expression changes induced by CCH have been identified in this study (Table 2 and Supplemental Table 1). An example is the two Notch receptors, notch-2 and notch-3, whose expression were induced by CCH. Notch receptors have been shown to be important for heart patterning and differentiation (Armstrong and Bischoff 2004), cell fate determination and self renewal of stem cells (Androutsellis-Theotokis et al. 2006; Silvia Bianchi 2006) but so far have not been linked to the hypoxic response.
Several genes with links to human heart pathology were found to be upregulated in this study. Among these were two markers for myocardial infarction, complement component C9 and haptoglobin. C9 is used as a marker for myocardial infarction (Doran et al. 1996) and a polymorphism for haptoglobin predicts 30-day mortality and heart failure in patients with diabetes and acute myocardial infarction (Levy et al. 2002). We also observed increased expression of fetuin-α, a circulating calcium-regulatory glycoprotein that inhibits vascular calcification. Low fetuin-α levels have been associated with heart failure in mice (Merx et al. 2005). Upregulation of fetuin-α in the zebrafish heart could help to better tolerate CCH. Selenoprotein P (SEPP1) is a heparin-binding protein that appears to be associated with endothelial cells and has been implicated as an oxidant defense in the extracellular space. Human populations that are selenium deficient are susceptible to the development of Keshan disease, a potentially fatal form of cardiomyopathy (Nezelof et al. 2002). SEPP1 expression was found to be increased in our study indicating a potential protective mechanism against oxidative stress in the heart. This was further supported by the increased expression observed for several genes important for the protection against reactive oxygen species (ROS) (Table 2). Among them were metallothionein and glutathione S-transferase, which are both known ROS scavengers. Metallothionein has further been described to be protective against hypoxia-induced apoptosis when overexpressed (Wang et al. 2001). Our findings suggest ROS protection as an important adaptation to CCH in the fish heart.
Furthermore, our data show for the first time that CCH simultaneously induced upregulation of IGF-2 and the insulin-like growth factor binding protein 1 (IGFbp1). In isolated cardiomyoblasts, angiotensin II stimulates IGF-2 expression which is involved in the induction of apoptotic signaling in rat hearts (Lee et al. 2006). We have observed here both angiotensin and IGF-2 upregulation (Table 2). The upregulation of IGFbp1 seems to be a general response to hypoxia in zebrafish embryos, where it mediates hypoxia-induced embryonic growth retardation and developmental delay (Kajimura et al. 2005b). IGFbp1 is a secreted protein, which binds to IGFs in the extracellular environment and prevents receptor activation (Florini et al. 1996; Stewart and Rotwein 1996). Here, IGFbp1 by binding IGF-2, may have prevented both cardioprotective as well as apoptotic effects of enhanced IGF-2 expression. To test the effects of the upregulation of both IGF-2 and IGFbp1, we assayed phospo-Akt levels in cardiac myocytes and showed that phospho-Akt levels were not different between normoxic and hypoxic cells. This suggests that either hypoxia-induced changes in mRNA expression did not occur at the protein level or the effects of increased IGF-2 expression on the IGF-1 receptor (which can be activated by IGF-2) were blunted by the upregulation of the IGFbp1. The fact that we did not observe increased phospho-Akt levels suggests a lack of hypertrophic signaling as enhanced phospho-Akt is required for cardiac hypertrophy (DeBosch et al. 2006). This may be beneficial for the heart as hypertrophy of cardiac myocytes implicates an increase in the diffusion distance for oxygen which may cause the development of anoxic cores (Des Tombe et al. 2002), release of cytochrome c (van Beek-Harmsen and van der Laarse 2005) and production of ROS (Lee et al. 2006; Powers et al. 2007) and eventually causing apoptosis of cardiac myocytes. The importance of the IGF-2/IGFbp1 signaling in the protection of the zebrafish heart and its underlying mechanisms remain to be determined.
The gene expression changes we observed for the specific response to CCH in the fish heart were very different from the responses we observed in an earlier study in the gills (van der Meer et al. 2005). Under the same criteria as used in this study, we found that the majority of differentially regulated genes in the gills showed a decrease in gene expression (68.1% or 250 genes in total) in comparison to genes, which showed increased expression levels (31.9% or 117 genes in total). This is opposed to the heart where 69.1% of differentially regulated genes (260 genes in total) showed increased and 30.9% (116 in total) decreased expression levels. Many genes linked to protein synthesis showed a similar trend, and were downregulated in the gills (van der Meer et al. 2005) and upregulated in the heart (Table 2). The major differences observed in gene regulation between heart and gills point to very tissue specific responses to CCH. A list of genes identified in both studies, as identified by a direct comparison based on Accession numbers and old Unigenes numbers are shown in Supplemental Data 3.
Teleosts have developed specific phenotypic adaptations to low oxygen, due to the natural occurrence of hypoxia in the water environment. Here, we identified the changes in gene expression as well as associated morphological changes of the fish heart to hypoxia. We observed repression of c-fos, which differs compared to the described increase in expression in mammals (Deindl et al. 2003). Other changes found like upregulation of the two Notch receptors have not been described before to the best of our knowledge. Similarly, the simultaneous increase in expression of IGF-2 and IGFbp1 has not been shown. The changes identified here can contribute to the ability of teleosts to adapt to severe hypoxia (for example, the upregulation of fetuin-α and sepp1 levels). Future functional studies are warranted to validate the role of the identified genes in cardiac protection to hypoxia.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We would like to thank Guido van den Thillart for essential help with the hypoxia set up, Gerda Lamers for intensive help with EM microscopy. Maximilian Corredor for his help with data analysis and software, Carlo Rutjes and Patrick Niemantsverdriet for animal care. Dr. Willem van der Laarse and Dr. Siona Slob are acknowledged for their assistance in the optimization of immunohistochemistry. This work was in part supported by a European Commission 6th Framework Programme grant (contract LSHG-CT-2003-503496, ZF-Models) and a grant from the Portuguese foundation for Science and Technology (SFRH/BD/27262/2006). All experiments comply with the current laws of the Netherlands.
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1007/s00360-007-0201-4) contains supplementary material, which is available to authorized users.
References
- Androutsellis-Theotokis A, Leker RR, Soldner F, Hoeppner DJ, Ravin R, Poser SW, Rueger MA, Bae S-K, Kittappa R, McKay RDG (2006) Notch signalling regulates stem cell numbers in vitro and in vivo. Nature (advanced online publication) [DOI] [PubMed]
- Armstrong EJ, Bischoff J (2004) Heart valve development: endothelial cell signaling and differentiation. Circ Res 95:459–470 [DOI] [PMC free article] [PubMed]
- Beilharz EJ, Bassett NS, Sirimanne ES, Williams CE, Gluckman PD (1995) Insulin-like growth factor II is induced during wound repair following hypoxic-ischemic injury in the developing rat brain. Mol Brain Res 29:81–91 [DOI] [PubMed]
- Beltrami AP, Urbanek K, Kajstura J, Yan SM, Finato N, Bussani R, Nadal-Ginard B, Silvestri F, Leri A, Beltrami CA, Anversa P (2001) Evidence that human cardiac myocytes divide after myocardial infarction. N Engl J Med 344:1750–1757 [DOI] [PubMed]
- Bianchi S, Dotti MT, Federico A (2006) Physiology and pathology of notch signalling system. J Cell Physiol 207:300–308 [DOI] [PubMed]
- DeBosch B, Treskov I, Lupu TS, Weinheimer C, Kovacs A, Courtois M, Muslin AJ (2006) Akt1 is required for physiological cardiac growth. Circulation 113:2097–2104 [DOI] [PubMed]
- Deindl E, Kolar F, Neubauer E, Vogel S, Schaper W, Ostadal B (2003) Effect of intermittent high altitude hypoxia on gene expression in rat heart and lung. Physiological Res 52:47–57 [PubMed]
- Des Tombe AL, Van Beek-Harmsen BJ, Lee-De Groot MB, Van Der Laarse WJ (2002) Calibrated histochemistry applied to oxygen supply and demand in hypertrophied rat myocardium. Microsc Res Tech 58:412–420 [DOI] [PubMed]
- Doran JP, Howie AJ, Townend JN, Bonser RS (1996) Detection of myocardial infarction by immunohistological staining for C9 on formalin fixed, paraffin wax embedded sections. J Clin Pathol 49:34–37 [DOI] [PMC free article] [PubMed]
- Fan C, Iacobas DA, Zhou D, Chen Q, Lai JK, Gavrialov O, Haddad GG (2005) Gene expression and phenotypic characterization of mouse heart after chronic constant or intermittent hypoxia. Physiol Genomics 22:292–307 [DOI] [PMC free article] [PubMed]
- Florini JR, Ewton DZ, Coolican SA (1996) Growth hormone and the insulin-like growth factor system in myogenesis. Endocr Rev 17:481–517 [DOI] [PubMed]
- Gracey AY, Troll JV, Somero GN (2001) Hypoxia-induced gene expression profiling in the euryoxic fish Gillichthys mirabilis. PNAS 98:1993–1998 [DOI] [PMC free article] [PubMed]
- Greenway SC, Storey KB (2000) Mitogen-activated protein kinases and anoxia tolerance in turtles. J Exp Zool 287:477–484 [DOI] [PubMed]
- Kajimura S, Aida K, Duan C (2005a) From the Cover: Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. PNAS 102:1240–1245 [DOI] [PMC free article] [PubMed]
- Kajimura S, Aida K, Duan C (2005b) Insulin-like growth factor-binding protein-1 (IGFBP-1) mediates hypoxia-induced embryonic growth and developmental retardation. Proc Natl Acad Sci USA 102:1240–1245 [DOI] [PMC free article] [PubMed]
- Kolar F, Ostadal B (2003) Molecular mechanisms of cardiac protection by adaptation to chronic hypoxia. Physiol Res 53:S3–S13 [PubMed]
- Lee SD, Chu CH, Huang EJ, Lu MC, Liu JY, Liu CJ, Hsu HH, Lin JA, Kuo WW, Huang CY (2006) Roles of insulin-like growth factor II in cardiomyoblast apoptosis and in hypertensive rat heart with abdominal aorta ligation. Am J Physiol Endocrinol Metab 291:E306–E314 [DOI] [PubMed]
- Levy AP, Hochberg I, Jablonski K, Resnick HE, Lee ET, Best L, Howard BV (2002) Haptoglobin phenotype is an independent risk factor for cardiovascular disease in individuals with diabetes: the strong heart study. J Am Coll Cardiol 40:1984–1990 [DOI] [PubMed]
- Meijer AH, Verbeek FJ, Salas-Vidal E, Corredor-Adamez M, Bussman J, van der Sar AM, Otto GW, Geisler R, Spaink HP (2005) Transcriptome profiling of adult zebrafish at the late stage of chronic tuberculosis due to Mycobacterium marinum infection. Mol Immunol 42:1185–1203 [DOI] [PubMed]
- Merx MW, Schafer C, Westenfeld R, Brandenburg V, Hidajat S, Weber C, Ketteler M, Jahnen-Dechent W (2005) Myocardial stiffness, cardiac remodeling, and diastolic dysfunction in calcification-prone fetuin-a-deficient mice. J Am Soc Nephrol 16:3357–3364 [DOI] [PubMed]
- Nezelof C, Bouvier R, Dijoud F (2002) Multifocal myocardial necrosis: a distinctive cardiac lesion in cystic fibrosis, lipomatous pancreatic atrophy, and keshan disease. Fetal Pediatr Pathol 21:343–352 [DOI] [PubMed]
- Ostadal B, Kolar F (2007) Cardiac adaptation to chronic high-altitude hypoxia: Beneficial and adverse effects. Respir Physiol Neurobiol (in press) [DOI] [PubMed]
- Pescador N, Cuevas Y, Naranjo S, Alcaide M, Villar D, Landazuri MO, Del Peso L (2005) Identification of a functional hypoxia-responsive element that regulates the expression of the egl nine homologue 3 (egln3/phd3) gene. Biochem J 390:189–197 [DOI] [PMC free article] [PubMed]
- Poss KD, Wilson LG, Keating MT (2002) Heart regeneration in zebrafish. Science 298:2188–2190 [DOI] [PubMed]
- Powers SK, Kavazis AN, McClung JM (2007) Oxidative stress and disuse muscle atrophy. J Appl Physiol 102(6):2389–2397 [DOI] [PubMed]
- Sabbah HN, Sharov VG, Goldstein S (2000a) Cell death, tissue hypoxia and the progression of heart failure. Heart Fail Rev 5:131–138 [DOI] [PubMed]
- Semenza GL (1999) Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol 15:551–578 [DOI] [PubMed]
- Semenza GL (2001) Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med 7:345–350 [DOI] [PubMed]
- Shi Y, Pritchard Jr KA, Holman P, Rafiee P, Griffith OW, Kalyanaraman B, Baker JE (2000) Chronic myocardial hypoxia increases nitric oxide synthase and decreases caveolin-3. Free Radic Biol Med 29:695–703 [DOI] [PubMed]
- Shiose S, Hata Y, Noda Y, Sassa Y, Takeda A, Yoshikawa H, Fujisawa K, Kubota T, Ishibashi T (2004) Fibrinogen stimulates in vitro angiogenesis by choroidal endothelial cells via autocrine VEGF. Graefes Arch Clin Exp Ophthalmol 242:777–783 [DOI] [PubMed]
- Simon KJ, Grueneberg DA, Gilman M (1997) Protein and DNA contact surfaces that mediate the selective action of the Phox1 homeodomain at the c-fos serum response element. Mol Cell Biol 17:6653–6662 [DOI] [PMC free article] [PubMed]
- Stecyk JAW, Stenslokken K-O, Farrell AP, Nilsson GE (2004) Maintained cardiac pumping in anoxic crucian carp. Science 306:77 [DOI] [PubMed]
- Stewart CE, Rotwein P (1996) Growth, differentiation, and survival: multiple physiological functions for insulin-like growth factors. Physiol Rev 76:1005–1026 [DOI] [PubMed]
- Tewari M YM, Ross B, Dean C, Giordano A, Rubin R (1997) AAC-11, a novel cDNA that inhibits apoptosis after growth factor withdrawal. Cancer Res 57:4063–4069 [PubMed]
- Ton C, Stamatiou D, Liew C-C (2003) Gene expression profile of zebrafish exposed to hypoxia during development. Physiol Genom 13:97–106 [DOI] [PubMed]
- van Beek-Harmsen BJ, van der Laarse WJ (2005) Immunohistochemical determination of cytosolic cytochrome C concentration in cardiomyocytes. J Histochem Cytochem 53:803–807 [DOI] [PubMed]
- van der Laarse WJ, des Tombe AL, van Beek-Harmsen BJ, Lee-de Groot MB, Jaspers RT (2005) Krogh’s diffusion coefficient for oxygen in isolated Xenopus skeletal muscle fibers and rat myocardial trabeculae at maximum rates of oxygen consumption. J Appl Physiol 99:2173–2180 [DOI] [PubMed]
- van der Meer DL, van den Thillart GE, Witte F, de Bakker MA, Besser J, Richardson MK, Spaink HP, Leito JT, Bagowski CP (2005) Gene expression profiling of the long-term adaptive response to hypoxia in the gills of adult zebrafish. Am J Physiol Regul Integr Comp Physiol 289:R1512–R1519 [DOI] [PubMed]
- van der Meer DL, Marques IJ, Leito JT, Besser J, Bakkers J, Schoonheere E, Bagowski CP (2006) Zebrafish cypher is important for somite formation and heart development. Dev Biol 299:356–372 [DOI] [PubMed]
- Wang G-W, Zhou Z, Klein JB, Kang YJ (2001) Inhibition of hypoxia/reoxygenation-induced apoptosis in metallothionein-overexpressing cardiomyocytes. Am J Physiol Heart Circ Physiol 280:H2292–H2299 [DOI] [PubMed]
- Webster KA, Discher DJ, Bishopric NH (1993) Induction and nuclear accumulation of fos and jun proto-oncogenes in hypoxic cardiac myocytes. J Biol Chem 268:16852–16858 [PubMed]
- Xu Q, Reed JC (1998) Bax inhibitor-1, a mammalian apoptosis suppressor identified by functional screening in yeast. Mol Cell 1:337–346 [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.