Abstract
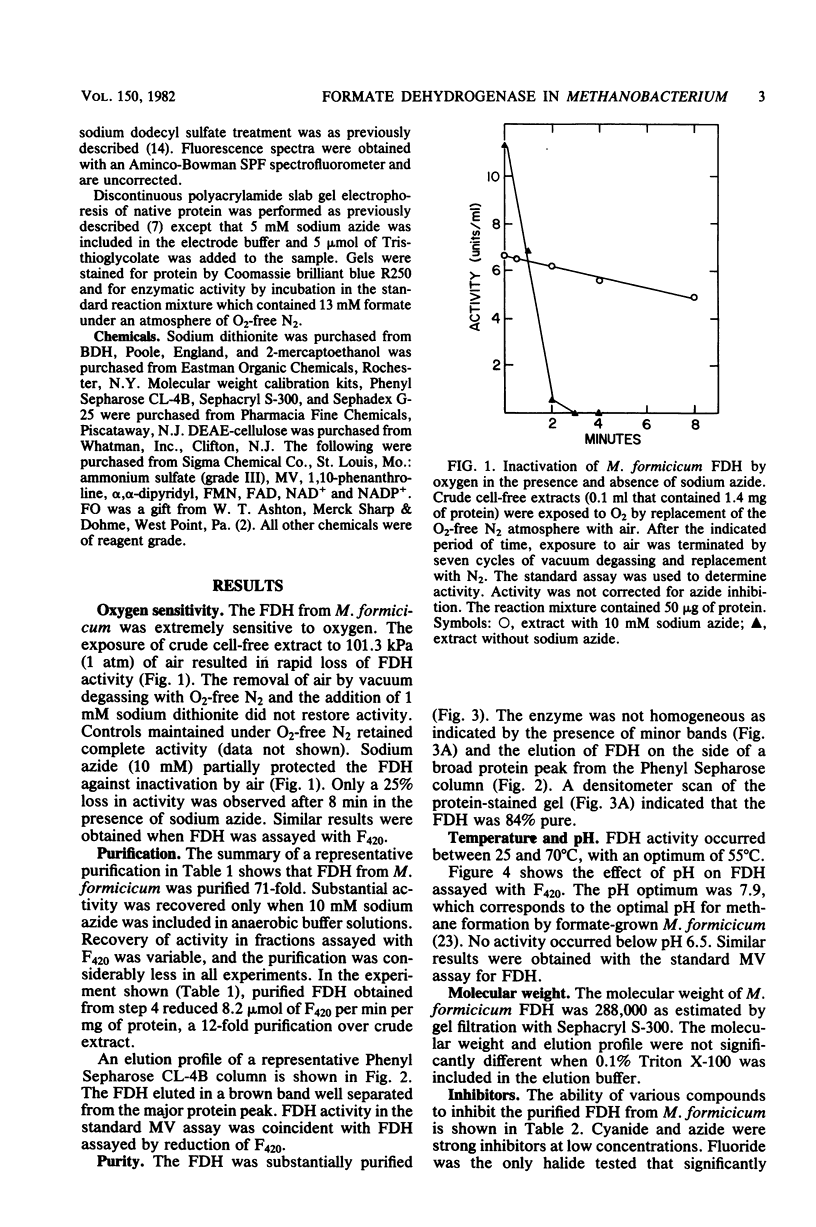
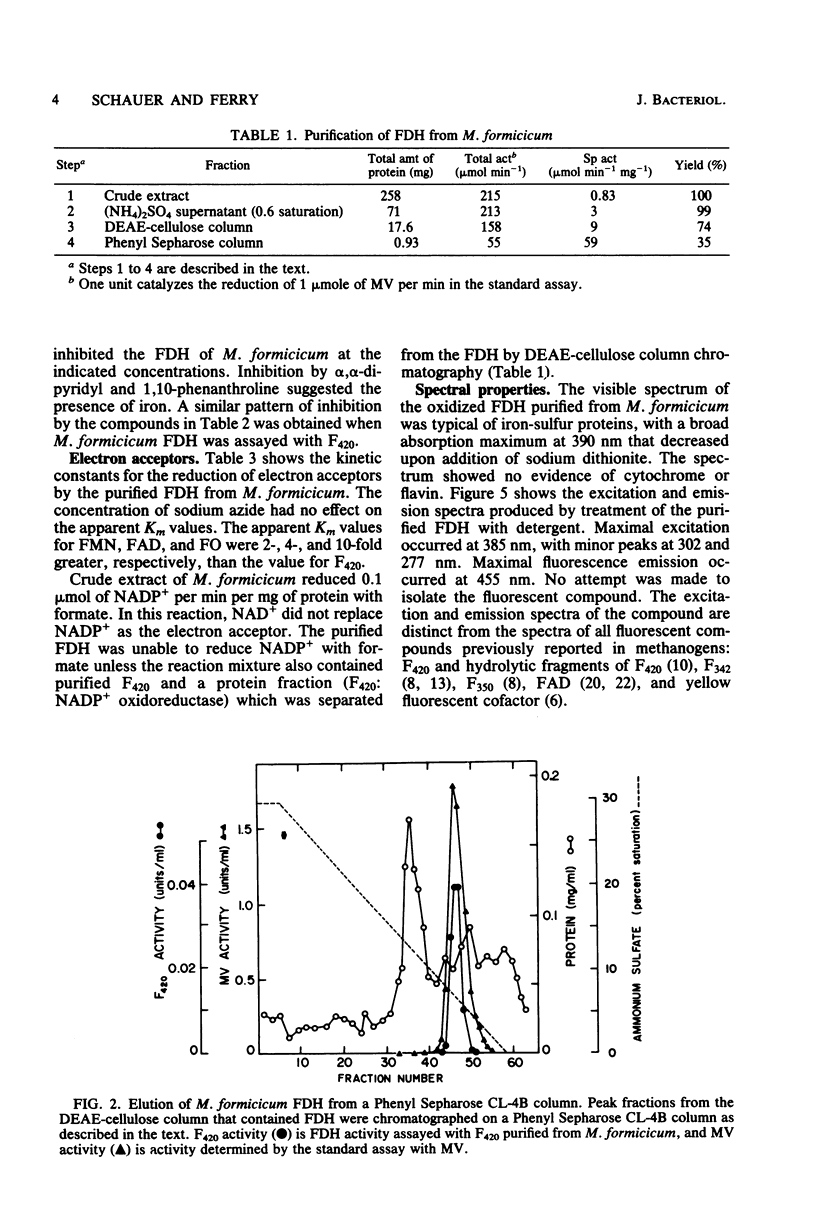
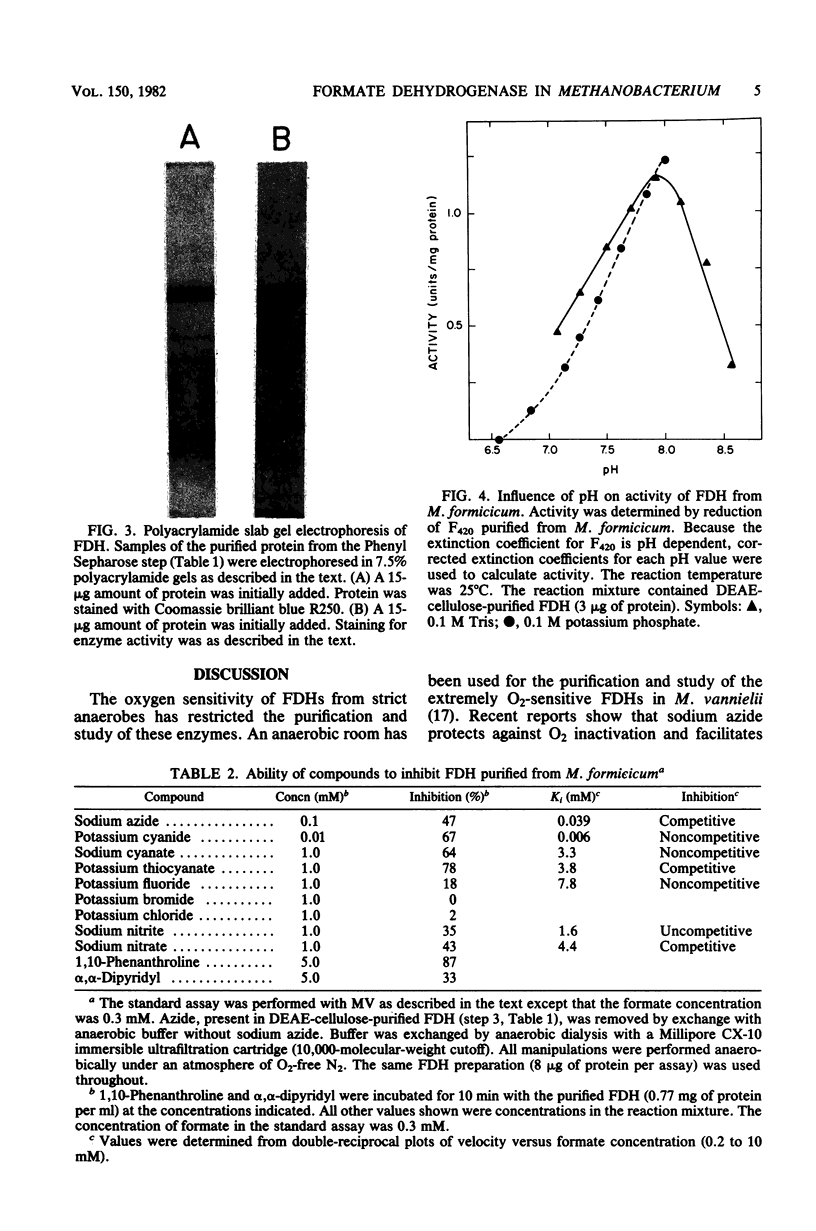
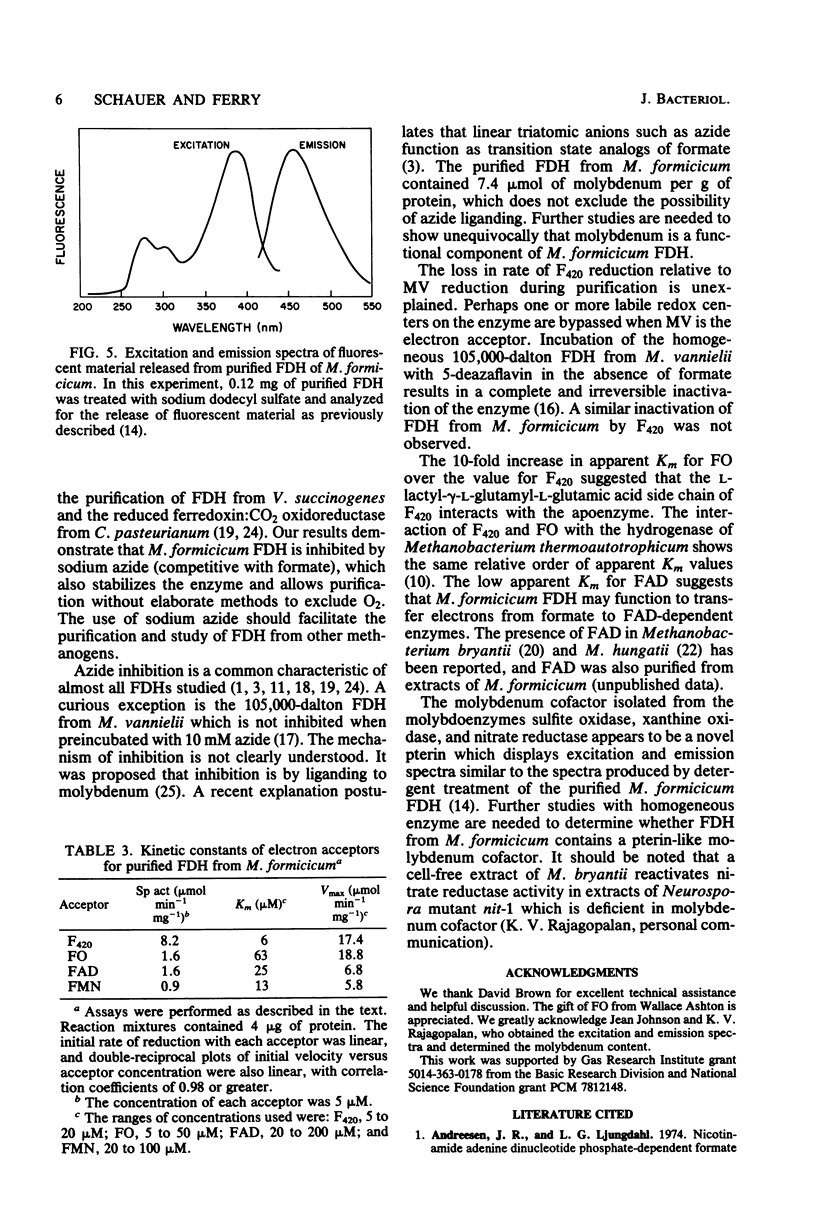
Soluble formate dehydrogenase from Methanobacterium formicicum was purified 71-fold with a yield of 35%. Purification was performed anaerobically in the presence of 10 mM sodium azide which stabilized the enzyme. The purified enzyme reduced, with formate, 50 mumol of methyl viologen per min per mg of protein and 8.2 mumol of coenzyme F420 per min per mg of protein. The apparent Km for 7,8-didemethyl-8-hydroxy-5-deazariboflavin, a hydrolytic derivative of coenzyme F420, was 10-fold greater (63 microM) than for coenzyme F420 (6 microM). The purified enzyme also reduced flavin mononucleotide (Km = 13 microM) and flavin adenine dinucleotide (Km = 25 microM) with formate, but did not reduce NAD+ or NADP+. The reduction of NADP+ with formate required formate dehydrogenase, coenzyme F420, and coenzyme F420:NADP+ oxidoreductase. The formate dehydrogenase had an optimal pH of 7.9 when assayed with the physiological electron acceptor coenzyme F420. The optimal reaction rate occurred at 55 degrees C. The molecular weight was 288,000 as determined by gel filtration. The purified formate dehydrogenase was strongly inhibited by cyanide (Ki = 6 microM), azide (Ki = 39 microM), alpha,alpha-dipyridyl, and 1,10-phenanthroline. Denaturation of the purified formate dehydrogenase with sodium dodecyl sulfate under aerobic conditions revealed a fluorescent compound. Maximal excitation occurred at 385 nm, with minor peaks at 277 and 302 nm. Maximal fluorescence emission occurred at 455 nm.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blanchard J. S., Cleland W. W. Kinetic and chemical mechanisms of yeast formate dehydrogenase. Biochemistry. 1980 Jul 22;19(15):3543–3550. doi: 10.1021/bi00556a020. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Coughlan M. P., Johnson J. L., Rajagopalan K. V. Mechanisms of inactivation of molybdoenzymes by cyanide. J Biol Chem. 1980 Apr 10;255(7):2694–2699. [PubMed] [Google Scholar]
- DAVIS B. J. DISC ELECTROPHORESIS. II. METHOD AND APPLICATION TO HUMAN SERUM PROTEINS. Ann N Y Acad Sci. 1964 Dec 28;121:404–427. doi: 10.1111/j.1749-6632.1964.tb14213.x. [DOI] [PubMed] [Google Scholar]
- Daniels L., Zeikus J. G. One-carbon metabolism in methanogenic bacteria: analysis of short-term fixation products of 14CO2 and 14CH3OH incorporated into whole cells. J Bacteriol. 1978 Oct;136(1):75–84. doi: 10.1128/jb.136.1.75-84.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doddema H. J., Vogels G. D. Improved identification of methanogenic bacteria by fluorescence microscopy. Appl Environ Microbiol. 1978 Nov;36(5):752–754. doi: 10.1128/aem.36.5.752-754.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirich L. D., Vogels G. D., Wolfe R. S. Distribution of coenzyme F420 and properties of its hydrolytic fragments. J Bacteriol. 1979 Oct;140(1):20–27. doi: 10.1128/jb.140.1.20-27.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirich L. D., Vogels G. D., Wolfe R. S. Proposed structure for coenzyme F420 from Methanobacterium. Biochemistry. 1978 Oct 31;17(22):4583–4593. doi: 10.1021/bi00615a002. [DOI] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- Ferry J. G., Wolfe R. S. Nutritional and biochemical characterization of Methanospirillum hungatii. Appl Environ Microbiol. 1977 Oct;34(4):371–376. doi: 10.1128/aem.34.4.371-376.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Hainline B. E., Rajagopalan K. V. Characterization of the molybdenum cofactor of sulfite oxidase, xanthine, oxidase, and nitrate reductase. Identification of a pteridine as a structural component. J Biol Chem. 1980 Mar 10;255(5):1783–1786. [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Methanococcus vannielii: culture and effects of selenium and tungsten on growth. J Bacteriol. 1977 Jun;130(3):1404–1406. doi: 10.1128/jb.130.3.1404-1406.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Reconstitution of a formate-NADP+ oxidoreductase from formate dehydrogenase and a 5-deazaflavin-linked NADP+ reductase isolated from Methanococcus vannielii. J Biol Chem. 1980 Feb 10;255(3):1049–1053. [PubMed] [Google Scholar]
- Jones J. B., Stadtman T. C. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem. 1981 Jan 25;256(2):656–663. [PubMed] [Google Scholar]
- Kearny J. J., Sagers R. D. Formate dehydrogenase from Clostridium acidiurici. J Bacteriol. 1972 Jan;109(1):152–161. doi: 10.1128/jb.109.1.152-161.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger A., Winkler E., Innerhofer A., Hackenberg H., Schägger H. The formate dehydrogenase involved in electron transport from formate to fumarate in Vibrio succinogenes. Eur J Biochem. 1979 Mar;94(2):465–475. doi: 10.1111/j.1432-1033.1979.tb12914.x. [DOI] [PubMed] [Google Scholar]
- Leonhardt U., Andreesen J. R. Some properties of formate dehydrogenase, accumulation and incorporation of 185W-tungsten into proteins of Clostridium formicoaceticum. Arch Microbiol. 1977 Dec 15;115(3):277–284. doi: 10.1007/BF00446453. [DOI] [PubMed] [Google Scholar]
- McKellar R. C., Shaw K. M., Sprott G. D. Isolation and characterization of a FAD-dependent NADH diaphorase from Methanospirillum hungatei strain GP1. Can J Biochem. 1981 Feb;59(2):83–91. doi: 10.1139/o81-013. [DOI] [PubMed] [Google Scholar]
- Schauer N. L., Ferry J. G. Metabolism of formate in Methanobacterium formicicum. J Bacteriol. 1980 Jun;142(3):800–807. doi: 10.1128/jb.142.3.800-807.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer P. A., Thauer R. K. Purification and properties of reduced ferredoxin: CO2 oxidoreductase from Clostridium pasteurianum, a molybdenum iron-sulfur-protein. Eur J Biochem. 1978 Apr;85(1):125–135. doi: 10.1111/j.1432-1033.1978.tb12220.x. [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Fuchs G., Käufer B. Reduced ferredoxin: CO2 oxidoreductase from Clostridium pasteurianum. Effect of ligands to transition metals on the activity and the stability of the enzyme. Hoppe Seylers Z Physiol Chem. 1975 Jun;356(6):653–662. doi: 10.1515/bchm2.1975.356.s1.653. [DOI] [PubMed] [Google Scholar]
- Tzing S. F., Bryant M. P., Wolfe R. S. Factor 420-dependent pyridine nucleotide-linked formate metabolism of Methanobacterium ruminantium. J Bacteriol. 1975 Jan;121(1):192–196. doi: 10.1128/jb.121.1.192-196.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R., Andreesen J. R. Differentiation between Clostridium acidiurici and Clostridium cylindrosporum on the basis of specific metal requirements for formate dehydrogenase formation. Arch Microbiol. 1977 Sep 28;114(3):219–224. doi: 10.1007/BF00446865. [DOI] [PubMed] [Google Scholar]
- Yagi T. Purification and properties of cytochrome c-553, an electron acceptor for formate dehydrogenase of Desulfovibrio vulgaris, Miyazaki. Biochim Biophys Acta. 1979 Oct 10;548(1):96–105. doi: 10.1016/0005-2728(79)90190-7. [DOI] [PubMed] [Google Scholar]



