Abstract
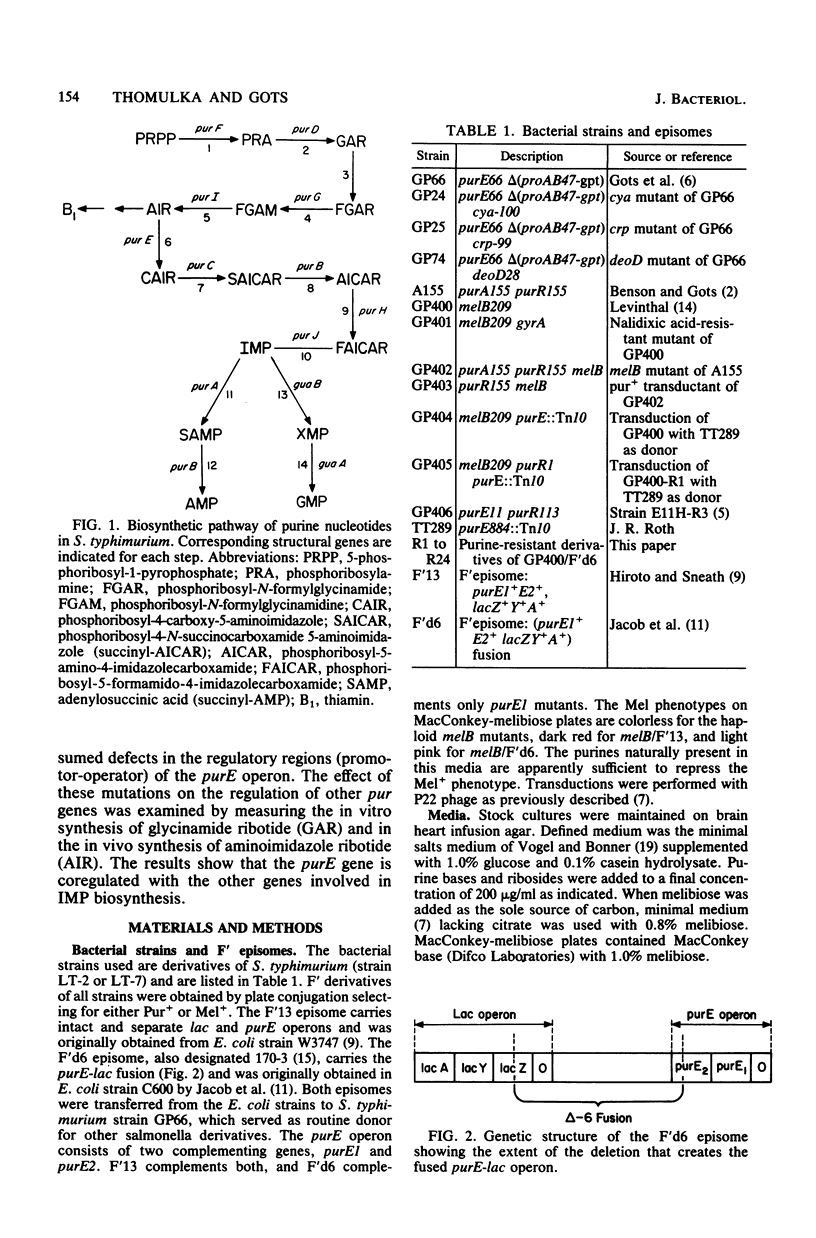
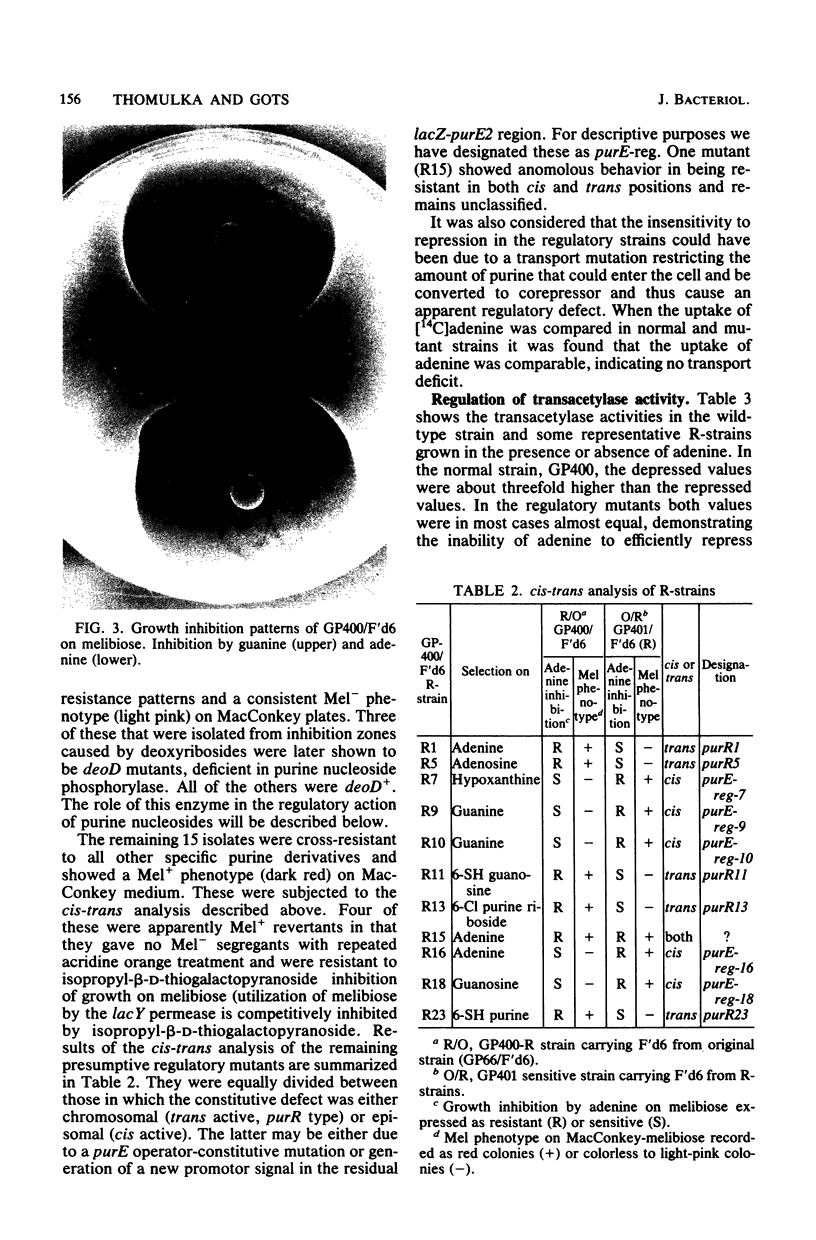
Expression of the purE operon of Salmonella typhimurium was analyzed by using an Escherichia coli F' episome containing a purE-lac fusion. The fusion removes the lacOP and part of the lacZ genes of the lac operon and places the intact lacY and lacA genes under control of the purE operon as shown by inhibition of growth on melibiose (lacY) and repression of thiogalactoside transacetylase (lacA) by various purines. Two classes of regulatory-deficient mutants were found among those resistant to inhibition by purines. One class was trans active (chromosomal) and corresponded to previously described purR mutants involving a deficient cytoplasmic repressor substance. These were also altered in the expression of the purF, purD, purG amd purI genes as evidenced by loss of repressibility of the synthesis of glycinamide ribotide and aminoimidazole ribotide. The other class was cis active (episomal), specific for only purE expression, and thus corresponded to an altered purE operon signal (operator or promoter). The metabolic requirements for the expression of purE were also monitored by measuring repression of the transacetylase in strains with various genetically altered metabolic backgrounds. Repression by guanine required an intact guanine phosphorbosyltransferase (gpt) and repression by adenine and all nucleosides required purine nucleoside phosphorylase (deoD). Synthesis of cyclic AMP (cya) and its receptor protein (crp) were no longer required for the expression of the lac genes under purE control.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALPERS D. H., APPEL S. H., TOMKINS G. M. A SPECTROPHOTOMETRIC ASSAY FOR THIOGALACTOSIDE TRANSACETYLASE. J Biol Chem. 1965 Jan;240:10–13. [PubMed] [Google Scholar]
- Benson C. E., Gots J. S. Occurrence of a regulatory deficiency in purine biosynthesis among pur A mutants of Salmonella typhimurium. Mol Gen Genet. 1976 Apr 23;145(1):31–36. doi: 10.1007/BF00331554. [DOI] [PubMed] [Google Scholar]
- Gots J. S., Benson C. E., Jochimsen B., Koduri K. R. Microbial models and regulatory elements in the control of purine metabolism. Ciba Found Symp. 1977;(48):23–41. doi: 10.1002/9780470720301.ch3. [DOI] [PubMed] [Google Scholar]
- Gots J. S., Benson C. E., Shumas S. R. Genetic separation of hypoxanthine and guanine-xanthine phosphoribosyltransferase activities by deletion mutations in Salmonella typhimurium. J Bacteriol. 1972 Nov;112(2):910–916. doi: 10.1128/jb.112.2.910-916.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gots J. S., Dalal F. R., Shumas S. R. Genetic eparation of the inosinic acid cyclohydrolase-transformylase complex of Salmonella typhimurium. J Bacteriol. 1969 Aug;99(2):441–449. doi: 10.1128/jb.99.2.441-449.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gots J. S., Gollub E. G. SEQUENTIAL BLOCKADE IN ADENINE BIOSYNTHESIS BY GENETIC LOSS OF AN APPARENT BIFUNCTIONAL DEACYLASE. Proc Natl Acad Sci U S A. 1957 Sep 15;43(9):826–834. doi: 10.1073/pnas.43.9.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmeyer J., Neuhard J. Metabolism of exogenous purine bases and nucleosides by Salmonella typhimurium. J Bacteriol. 1971 Apr;106(1):14–24. doi: 10.1128/jb.106.1.14-24.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Levine R. A., Taylor M. W. Selection for purine regulatory mutants in an E. coli hypoxanthine phosphoribosyl transferase-guanine phosphoribosyl transferase double mutant. Mol Gen Genet. 1981;181(3):313–318. doi: 10.1007/BF00425604. [DOI] [PubMed] [Google Scholar]
- Levinthal M. Biochemical studies of melibiose metabolism in wild type and mel mutant strains of Salmonella typhimurium. J Bacteriol. 1971 Mar;105(3):1047–1052. doi: 10.1128/jb.105.3.1047-1052.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low K. B. Escherichia coli K-12 F-prime factors, old and new. Bacteriol Rev. 1972 Dec;36(4):587–607. doi: 10.1128/br.36.4.587-607.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson B. C., Jargiello P., Blank J., Hoffee P. A. Genetic regulation of ribonucleoside and deoxyribonucleoside catabolism in Salmonella typhimurium. J Bacteriol. 1970 Jun;102(3):628–635. doi: 10.1128/jb.102.3.628-635.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Hartman P. E. Linkage map of Salmonella typhimurium, edition V. Microbiol Rev. 1978 Jun;42(2):471–519. doi: 10.1128/mr.42.2.471-519.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Westby C. A., Gots J. S. Genetic blocks and unique features in the biosynthesis of 5'-phosphoribosyl-N-formylglycinamide in Salmonella typhimurium. J Biol Chem. 1969 Apr 25;244(8):2095–2102. [PubMed] [Google Scholar]



