Abstract
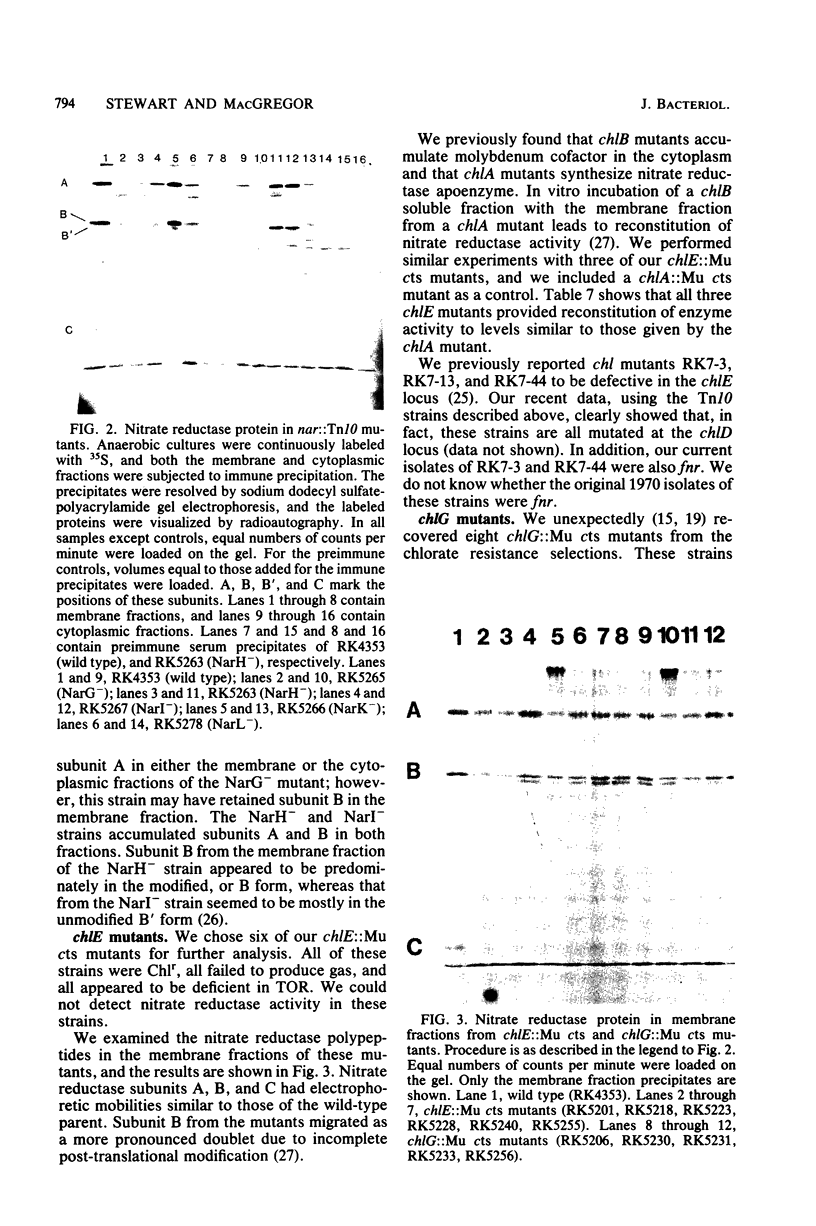
We examined the properties of mutants of E. coli which are defective with respect to nitrate reductase activity. chlE::Mu cts and chlG::Mu cts mutants were all chlorate resistant, and the strains that we examined all synthesized nitrate reductase apoenzyme. We concluded that the chlE and chlG loci, like the chlA, chlB, and chlD loci, are involved in the synthesis of insertion of molybdenum cofactor. We identified at least four distinct phenotypic classes of chlC::Tn10 mutants, all of which were fully or partially sensitive to chlorate. Two of these classes may represent lesions in the structural genes for nitrate reductase subunits A and C. Two other classes may be altered in the regulation of the expression of nitrate reductase or other anaerobic enzymes. We propose the mnemonic nar for naming individual genes within the chlC locus.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amy N. K. Identification of the molybdenum cofactor in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1981 Oct;148(1):274–282. doi: 10.1128/jb.148.1.274-282.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B. Linkage map of Escherichia coli K-12, edition 6. Microbiol Rev. 1980 Mar;44(1):1–56. doi: 10.1128/mr.44.1.1-56.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett E. L., Jackson C. E., Fukumoto H. T., Chang G. W. Formate dehydrogenase mutants of Salmonella typhimurium: a new medium for their isolation and new mutant classes. Mol Gen Genet. 1979;177(1):95–101. doi: 10.1007/BF00267258. [DOI] [PubMed] [Google Scholar]
- Bonnefoy-Orth V., Lepelletier M., Pascal M. C., Chippaux M. Nitrate reductase and cytochrome bnitrate reductase structural genes as parts of the nitrate reductase operon. Mol Gen Genet. 1981;181(4):535–540. doi: 10.1007/BF00428749. [DOI] [PubMed] [Google Scholar]
- Casse F., Chippaux M., Pascal M. C. Isolation from Salmonella typhimurium LT2 of mutants lacking specifically nitrate reductase activity and mapping of the chl-C gene. Mol Gen Genet. 1973 Aug 17;124(3):247–251. doi: 10.1007/BF00293095. [DOI] [PubMed] [Google Scholar]
- Casse F., Pascal M. C., Chippaux M., Ratouchniak J. Mapping of the chl-B gene in Salmonella typhimurium LT2. Mol Gen Genet. 1972;119(1):67–70. doi: 10.1007/BF00270445. [DOI] [PubMed] [Google Scholar]
- Clark D., Cronan J. E., Jr Escherichia coli mutants with altered control of alcohol dehydrogenase and nitrate reductase. J Bacteriol. 1980 Jan;141(1):177–183. doi: 10.1128/jb.141.1.177-183.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMoss J. A. Role of the chlC gene in formation of the formate-nitrate reductase pathway in Escherichia coli. J Bacteriol. 1978 Feb;133(2):626–630. doi: 10.1128/jb.133.2.626-630.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubourdieu M., Andrade E., Puig J. Molybdenum and chlorate resistant mutants in Escherichia coli K12. Biochem Biophys Res Commun. 1976 Jun 7;70(3):766–773. doi: 10.1016/0006-291x(76)90658-6. [DOI] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The role of a novel cytochrome b-containing nitrate reductase and quinone in the in vitro reconstruction of formate-nitrate reductase activity of E. coli. Biochem Biophys Res Commun. 1974 Dec 23;61(4):1234–1241. doi: 10.1016/s0006-291x(74)80416-x. [DOI] [PubMed] [Google Scholar]
- Forget P. The bacterial nitrate reductases. Solubilization, purification and properties of the enzyme A of Escherichia coli K 12. Eur J Biochem. 1974 Mar 1;42(2):325–332. doi: 10.1111/j.1432-1033.1974.tb03343.x. [DOI] [PubMed] [Google Scholar]
- Glaser J. H., DeMoss J. A. Comparison of nitrate reductase mutants of Escherichia coli selected by alternative procedures. Mol Gen Genet. 1972;116(1):1–10. doi: 10.1007/BF00334254. [DOI] [PubMed] [Google Scholar]
- Guest J. R. Biochemical and genetic studies with nitrate reductase C-gene mutants of Escherichia coli. Mol Gen Genet. 1969;105(4):285–297. doi: 10.1007/BF00277583. [DOI] [PubMed] [Google Scholar]
- Howe M. M., Bade E. G. Molecular biology of bacteriophage mu. Science. 1975 Nov 14;190(4215):624–632. doi: 10.1126/science.1103291. [DOI] [PubMed] [Google Scholar]
- Jones R. W., Garland P. B. Sites and specificity of the reaction of bipyridylium compounds with anaerobic respiratory enzymes of Escherichia coli. Effects of permeability barriers imposed by the cytoplasmic membrane. Biochem J. 1977 Apr 15;164(1):199–211. doi: 10.1042/bj1640199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Lambden P. R., Guest J. R. Mutants of Escherichia coli K12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976 Dec;97(2):145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- Lester R. L., DeMoss J. A. Effects of molybdate and selenite on formate and nitrate metabolism in Escherichia coli. J Bacteriol. 1971 Mar;105(3):1006–1014. doi: 10.1128/jb.105.3.1006-1014.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H. Anaerobic cytochrome b1 in Escherichia coli: association with and regulation of nitrate reductase. J Bacteriol. 1975 Mar;121(3):1111–1116. doi: 10.1128/jb.121.3.1111-1116.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., McElhaney G. E. New mechanism for post-translational processing during assembly of a cytoplasmic membrane protein? J Bacteriol. 1981 Nov;148(2):551–558. doi: 10.1128/jb.148.2.551-558.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A., Normansell D. E. Purification and properties of nitrate reductase from Escherichia coli K12. J Biol Chem. 1974 Aug 25;249(16):5321–5327. [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A. Reconstitution of nitrate reductase activity and formation of membrane particles from cytoplasmic extracts of chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1973 Jun;114(3):1164–1176. doi: 10.1128/jb.114.3.1164-1176.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H. Synthesis of nitrate reductase components in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1975 Mar;121(3):1117–1121. doi: 10.1128/jb.121.3.1117-1121.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman B. M., Cole J. A. The chromosomal location and pleiotropic effects of mutations of the nirA+ gene of Escherichia coli K12: the essential role of nirA+ in nitrite reduction and in other anaerobic redox reactions. J Gen Microbiol. 1978 May;106(1):1–12. doi: 10.1099/00221287-106-1-1. [DOI] [PubMed] [Google Scholar]
- Oltmann L. F., Claassen V. P., Kastelein P., Reijnders W. N., Stouthamer A. H. Influence of tungstate on the formation and activities of four reductases in Proteus mirabilis: identification of two new molybdo-enzymes: chlorate reductase and tetrathionate reductase. FEBS Lett. 1979 Oct 1;106(1):43–46. doi: 10.1016/0014-5793(79)80691-2. [DOI] [PubMed] [Google Scholar]
- Pascal M. C., Chippaux M., Abou-Jaoudé A., Blaschkowski H. P., Knappe J. Mutants of Escherichia coli K12 with defects in anaerobic pyruvate metabolism. J Gen Microbiol. 1981 May;124(1):35–42. doi: 10.1099/00221287-124-1-35. [DOI] [PubMed] [Google Scholar]
- Pugsley A. P., Schnaitman C. A. Identification of three genes controlling production of new outer membrane pore proteins in Escherichia coli K-12. J Bacteriol. 1978 Sep;135(3):1118–1129. doi: 10.1128/jb.135.3.1118-1129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve E. C., Doherty P. Linkage relationships of two genes causing partial resistance to chloramphenicol in Escherichia coli. J Bacteriol. 1968 Oct;96(4):1450–1451. doi: 10.1128/jb.96.4.1450-1451.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimokawa O., Ishimoto M. Purification and some properties of inducible tertiary amine N-oxide reductase from Escherichia coli. J Biochem. 1979 Dec;86(6):1709–1717. doi: 10.1093/oxfordjournals.jbchem.a132691. [DOI] [PubMed] [Google Scholar]
- Showe M. K., DeMoss J. A. Localization and regulation of synthesis of nitrate reductase in Escherichia coli. J Bacteriol. 1968 Apr;95(4):1305–1313. doi: 10.1128/jb.95.4.1305-1313.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperl G. T., DeMoss J. A. chlD gene function in molybdate activation of nitrate reductase. J Bacteriol. 1975 Jun;122(3):1230–1238. doi: 10.1128/jb.122.3.1230-1238.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables W. A. Genetic studies with nitrate reductase-less mutants of Escherichia coli. I. Fine structure analysis of the narA, narB and narE loci. Mol Gen Genet. 1972;114(3):223–231. doi: 10.1007/BF01788891. [DOI] [PubMed] [Google Scholar]
- Venables W. A., Guest J. R. Transduction of nitrate reductase loci of Escherichia coli by phages P-1 and lambda. Mol Gen Genet. 1968;103(2):127–140. doi: 10.1007/BF00427140. [DOI] [PubMed] [Google Scholar]
- Wimpenny J. W., Cole J. A. The regulation of metabolism in facultative bacteria. 3. The effect of nitrate. Biochim Biophys Acta. 1967 Oct 9;148(1):233–242. doi: 10.1016/0304-4165(67)90298-x. [DOI] [PubMed] [Google Scholar]
- del Campillo-Campbell A., Campbell A. Molybdenum cofactor requirement for biotin sulfoxide reduction in Escherichia coli. J Bacteriol. 1982 Feb;149(2):469–478. doi: 10.1128/jb.149.2.469-478.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]