Abstract
Computational design of proteins with altered ligand specificity is an emerging method for the creation of new biosensing systems. In this work, we investigated the outcome of site-directed mutagenesis on the Escherichia coli ribose binding protein (RBP), which is frequently used as a design scaffold for computational searches. A ribose biosensor was first constructed whereby an environmentally sensitive fluorescent probe was covalently attached to RBP at position S265C. This protein conjugate displayed a 54% decrease in emission intensity upon the addition of saturating ribose concentrations and exhibited an apparent dissociation constant (Kd) of 3.4 μM. Site-directed mutants within the RBP binding pocket were created and examined for ribose binding ability and overall structural stability. Because as many as 12 mutations are needed to alter ligand specificity in RBP, we measured the effect of single and multiple alanine mutations on stability and signal transduction potential of the ribose biosensor. Single alanine mutations had significant impact on both stability and signaling. Mutations of N190A and F214A each produced melting temperatures >8°C below those observed for the wild-type protein. Residue Q235, located in the hinge region of RBP, appeared to be a hot spot for global protein stability as well. Additional single alanine mutations demonstrated as much as 200-fold increase in apparent Kd but retained overall protein stability. The data collected from this study may be incorporated into design algorithms to help create more stable biosensors and optimize signal transduction properties for a variety of important analytes.
Keywords: biosensor, periplasmic binding proteins, ribose binding protein, fluorescence
Protein engineering is an emerging field that utilizes natural or designed polypeptide scaffolds for molecular recognition processes (Goodchild et al. 2006; Hosse et al. 2006). Although nature has provided biomolecules that are able to bind a number of potential analytical targets, there are many compounds of interest for which a binding molecule does not exist. Synthetic chemical warfare agents may be one of the most notable categories for which this is true (Igbal et al. 2000). To meet this need, stochastic and deterministic computational algorithms have been written to alter or to redesign the specificity of naturally occurring polypetides for the interaction with non-natural ligands for the purpose of chemical detection (DeGrado et al. 1999; Looger and Hellinga 2001; Rohl et al. 2004). Computational design of scaffolds and binders for many different chemical agents is an area of highly active research (Lee et al. 2000; Looger et al. 2003; Allert et al. 2004; Medintz et al. 2005).
The Escherichia coli periplasmic binding proteins (PBPs) are one family of scaffolds used for the design of fluorescent protein biosensors (Higgins 1992; Dwyer and Hellinga 2004). This family of structurally similar proteins is characterized by a single polypeptide chain folded into two domains connected by a hinge region that forms a cleft for substrate binding (Higgins 1992). In the presence of ligand, these proteins undergo a hinge-bending motion almost completely occluding the ligand from solvent (Quiocho and Ledvina 1996). Using X-ray crystallographic data for the open and closed forms, cysteine codons are engineered into the genetic code at specific locations that allow for the covalent modification of environmentally sensitive fluorophores (Marvin et al. 1997; Dattelbaum and Lakowicz 2001; deLorimier et al. 2002; Salins et al. 2004; Rizk et al. 2006). The ability to make quantitative measurements using other techniques (Tolosa et al. 2003; Wells 2006), including FRET (Deuschle et al. 2005; Medintz and Deschamps 2006) and electrochemistry (Benson et al. 2001), have also been explored using PBPs.
Although the main goal of protein design is to build a biomolecule with high specificity for the desired target, the overall stability of the newly designed scaffolds merits additional consideration during their construction. The wild-type PBPs are highly stable, making them attractive targets for protein engineering. However, the production of practical biosensors using this family necessitates that their stability be investigated following the high number of point mutations needed for building new analytical specificity. This work begins to analyze the contribution of individual amino acids toward the overall stability of PBP scaffolds. As a model system, we chose the Escherichia coli ribose binding protein (RBP) to investigate the effect of such large-scale site-directed mutation on protein stability. When using RBP as a design scaffold, there are at least 12 amino acids commonly mutated within the binding pocket of RBP to transform this protein from binding the sugar ribose to some other analyte (Looger et al. 2003; Allert et al. 2004; Dwyer et al. 2004). We have created mutations at each of these sites, singly and in tandem, to determine the effect on both RBP thermal stability and on the fluorescent allosteric signaling mechanism used to report ligand binding. The incorporation of these data into computational models may aid in the design of additional field-ready biosensors specific for many analytical targets.
Results
Construction of a ribose sensing fluorescent protein
The periplasmic space of gram-negative bacteria is host to a variety of soluble binding proteins which help bacteria obtain nutrients from the environment. These periplasmic binding proteins (PBP) typically undergo large conformational changes upon interaction with a specific ligand (Fig. 1). The ability to detect ligand binding to a specific PBP is commonly performed using environmentally sensitive fluorophores covalently attached at specific sites on the protein. Many groups have published work describing the rational placement of fluorophores using both inductive and computational methods (Gilardi et al. 1994; Brune et al. 1998; Marvin and Hellinga 1998; Dattelbaum and Lakowicz 2001; deLorimier et al. 2002; Shrestha et al. 2002; Thomas et al. 2006). Choosing the site of fluorophore attachment as well as the identity of the fluorophore is consistently an empirical exercise for each binding protein under study. In order to measure the interaction between RBP and ribose, we constructed a single cysteine mutant of RBP as described previously (deLorimier et al. 2002). Wild-type RBP does not contain any Cys residues; hence, introduction of an L265C modification allows for the covalent and site-specific modification of RBP with a thiol-reactive fluorophore. This mutant was constructed as a translational fusion with an N-terminal His-Tag for affinity purification.
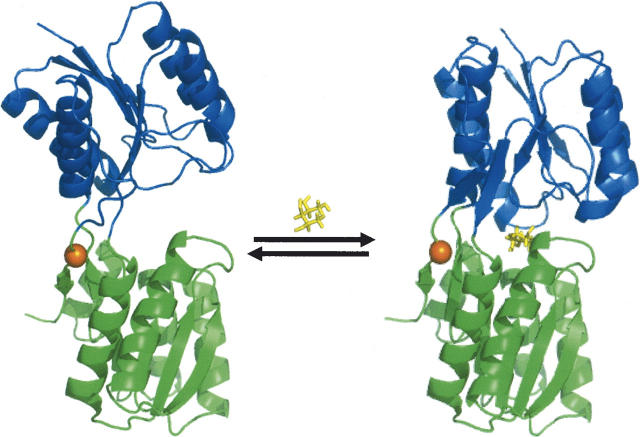
Figure 1.
Conformational change observed for the E. coli ribose binding protein in the absence (1URP) and presence (2DRI) of ribose. The alpha carbon of residue L265C, which serves as the site of covalent attachment for ABD-F, is marked by a sphere in the hinge region. Molecular rendering performed using PyMOL (DeLano 2002).
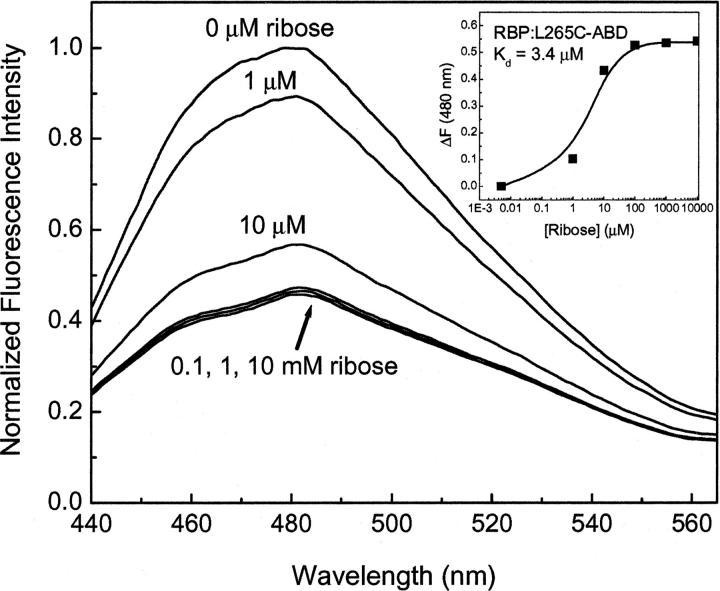
A number of commercially available fluorescent probes were investigated for their ability to signal ribose binding to RBP, including fluorescein iodoacetamide, acrylodan, ABD-F, IANBD ester, and IANBD amide. Also used for this purpose were JPW4095 and JPW4044 (Millard et al. 2004), provided as a kind gift from Leslie Lowe and colleagues (University of Connecticut). Most of these probes yielded little or no change in fluorescence upon ribose binding (data not shown). However, covalent modification of RBP L265C with ABD-F displayed a 54% decrease in fluorescence emission upon titration of the conjugated protein with ribose (Fig. 2). The peak of fluorescence emission recorded for each measurement was then used to calculate a dissociation constant of 3.4 μM for ribose interaction with ABD-labeled RBP (Fig. 2, insert). This number is consistent with that previously reported using other environmentally sensitive probes for signal transduction of ribose binding (deLorimier et al. 2002).
Figure 2.
Normalized fluorescence emission spectra for RBP:L265C-ABD in the presence of increasing amounts of ribose. Measurements were performed in 5 mM phosphate buffer (pH 7.0) at room temperature.
Single alanine mutations affect RBP stability
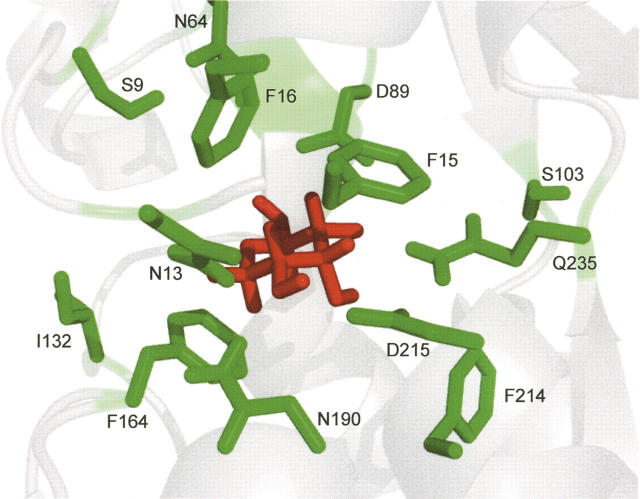
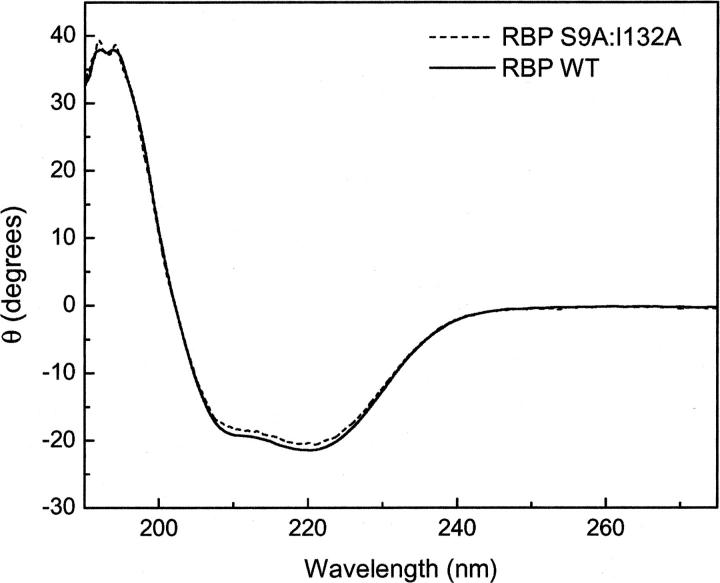
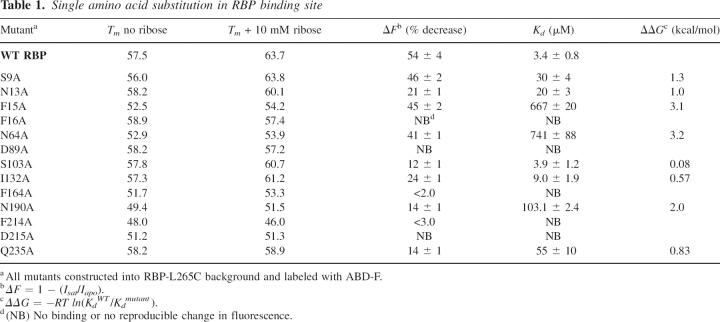
In order to examine the contribution to overall stability of individual amino acids in the ribose binding pocket, we constructed 13 individual alanine mutations in RBP L265C (Fig. 3). These particular residues were chosen based on their proximity to ribose in the X-ray crystal structure of RBP as well as from previous computational work using RBP as a design scaffold. To confirm proper folding of each mutant, wavelength circular dichroism (CD) spectra were measured. All mutant CD spectra were nearly identical to those observed for the wild-type protein, which is consistent with little or no change to overall RBP structure, as expected (Fig. 4). While global RBP structure appeared unchanged, the temperature of protein unfolding showed great variations, even with single alanine mutations. The data summarized in Table 1 include thermal unfolding measurements for WT RBP (RBP with the L265C mutation) in the presence and absence of ribose. CD data for WT RBP indicate an increase in melting temperature from 57.5°C to 63.7°C upon the addition of ribose. In contrast, only four of the single alanine mutants (S9A, N13A, S103A, and I132A) displayed a significant increase in stability in the presence of ribose similar to the wild-type protein with melting temperatures above 60°C. Many of the single alanine mutations resulted in a greater than expected decrease in overall protein stability as analyzed by melting temperature. In particular, F214A and N190A produced the most significant destabilization of the global protein structure. Mutation at these two sites resulted in a decrease of melting temperature to 49°C and 48°C, respectively, demonstrating almost no change upon the addition of ribose. These two residues are located in the lower RBP domain in close proximity to each other with an inter alpha-carbon distance of ∼4 Å (Fig. 3).
Figure 3.
Amino acids involved in RBP binding to D-ribose and mutated to alanine (shown in green; ribose ligand is shown in red). Figure generated with PyMOL (DeLano 2002).
Figure 4.
Representative circular dichroism wavelength scans for WT RBP and RBP S9A. Measurements were performed in 5 mM phosphate buffer (pH 7.0) at room temperature.
Table 1.
Single amino acid substitution in RBP binding site
Two separate peptide segments comprise the hinge region that connects the upper and lower domains of RBP (Fig. 1). S103A is found on one of these segments and Q235A on the other. While S103A produced a melting temperature similar to the wild type, Q325A demonstrated no change in melting temperature upon interaction with ribose. This mutation may have locked the RBP into a stable yet open structure even after binding to ribose. Examples of ligand binding to the open confirmation of PBPs have been seen for the structurally similar maltose binding protein as well (Duan et al. 2001; Duan and Quiocho 2002).
Ribose sensing is significantly altered with binding pocket mutations
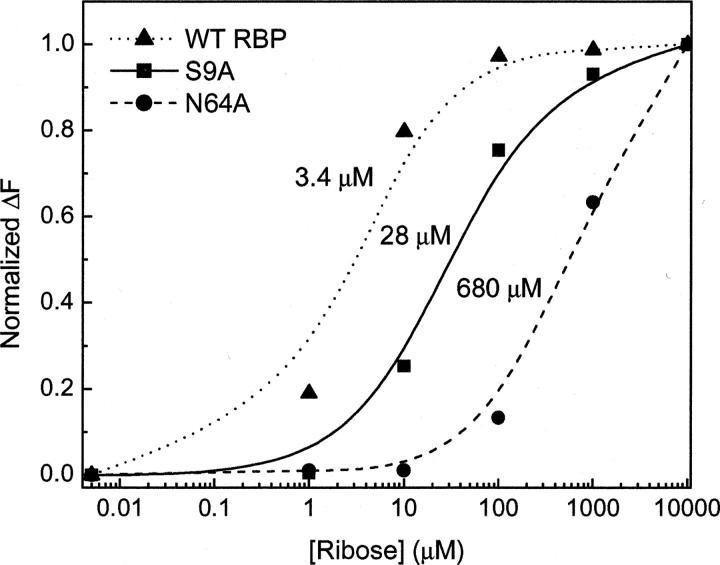
While mutations to amino acids located in the RBP binding pocket were expected to alter Kd, extreme changes in ΔF were not expected for most single alanine mutations. Figure 5 shows the apparent dissociation constants obtained for the wild-type RBP and two single alanine mutants. Although the actual change in fluorescence intensity does not vary greatly (Table 1), there was a 250-fold variation in binding constant among these mutants. To determine if specificity for ribose binding was altered, RBP N64A, which displayed an increased Kd value, was screened for cross reactivity with structurally similar sugars. There were no observable changes in fluorescence emission upon the addition of 10 mM fructose, glucose, or potassium gluconate to either the wild-type RBP or RBP N64A construct (data not shown). These data demonstrate that simple mutations within the ligand binding pocket may be useful for altering the dynamic range for analyte sensing while retaining sugar specificity.
Figure 5.
Relative binding constants for WT RBP and for two single alanine mutants. Data from normalized fluorescence emission were replotted and were analyzed using the nonlinear curve fitting algorithm in Origin 6.0.
We hypothesized that single alanine mutations would not disrupt the signal transducing properties for ribose sensing. However, we found that the majority of single alanine mutations caused significant decreases in ΔF (Table 1). Eight of the 13 mutants demonstrated less than 15% change in fluorescence intensity (down from 54% in the wild type) upon binding ribose. Five of these mutations (F16A, D89A, F164A, F214A, and D215A) produced almost no measurable change in intensity upon the addition of ribose. While it is not surprising that these results suggest that ligand binding is a complex process governed by many intermolecular forces, subtle alterations of these forces may have significant impact on not only ribose binding ability but also on signal transducing potential. This latter realization has implications in the sensitivity one can expect when building biosensors using this protein as a scaffold.
Multiple alanine mutations may be tolerated by the RBP scaffold
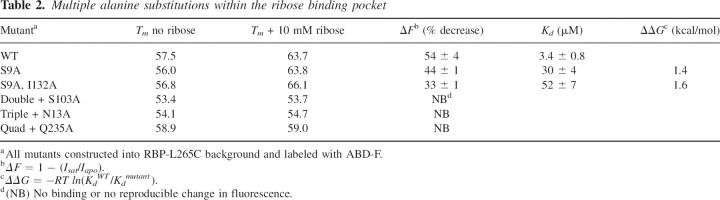
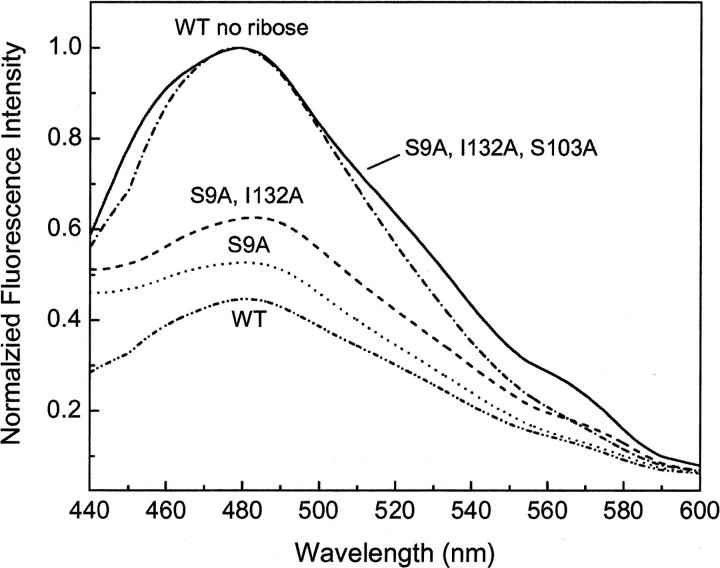
While single alanine mutations may yield information about specific locations within the ribose binding pocket, construction of computationally designed proteins typically involves as many as 13 simultaneous mutations. As a result, we examined the tolerance of RBP to multiple alanine mutations within the ribose binding pocket (Table 2). We started with amino acids on the outside of the binding pocket and added additional mutations reaching toward the hinge at the back of the binding pocket, resulting in a quintuple alanine mutant. The ability to bind ribose quickly diminishes as the number of mutations is increased. Changes in fluorescence intensity in response to ribose were observed for the first two mutations, but no change was observed with additional alanine insertions (Fig. 6). The decrease in ribose-dependent fluorescence was accompanied by an increase in Kd (Table 2). Additionally, an increase in destabilization was observed as alanine residues were added further into the binding pocket. Interestingly, mutation Q235A inserted into a quadruple alanine background showed an ∼5°C increase in protein stability. These data indicate that, while multiple mutations are in general detrimental to overall protein stability, there may be some sites that can aid in designing more stable scaffolds for the design process.
Table 2.
Multiple alanine substitutions within the ribose binding pocket
Figure 6.
Normalized fluorescence emission spectra for RBP containing multiple alanine mutations within the ribose binding pocket. Emission spectra for WT and all mutants are shown in the presence of ribose. The spectra for the triple-, quad-, and quintuple-alanine RBP mutants in the presence of ribose overlap the WT no ribose spectra. Measurements were performed in 5 mM phosphate buffer (pH 7.0) at room temperature.
Discussion
Many successful examples of using RBP and other periplasmic binding proteins as scaffolds for the design of new ligand binders exist in the literature (Looger et al. 2003; Allert et al. 2004; Dwyer et al. 2004). While these proteins are known to be quite stable, the large number of localized mutations necessary to alter ligand specificity may cause significant destabilization and reduction in signal transducing ability. In order to design highly specific binding molecules, one does not necessarily want to give up sensitivity. The limits on the interplay between specificity and sensitivity in biosensor development are a focus of this project.
Using site-directed mutagenesis, we investigated the possibility of incorporating experimentally determined constraints back into design algorithms in order to produce more stable biosensing systems. The constructed fluorescent protein biosensors show a large decrease in fluorescence intensity in response to ribose. Alteration of specific residues to alanine within the RBP binding pocket results in the ability to detect ribose over several orders of magnitude (Fig. 5). To examine the effective shelf life of the RBP biosensors, fluorescence response was monitored periodically over the past 5 mo for wild-type RBP and for RBP Q235A stored at 4°C. Both of these systems retained consistent ribose-dependent fluorescence changes over this time period. In previous work mutating a few of the amino acids in the RBP binding pocket using a genetically encoded ribose biosensor, Lager et al. (2003) observed similar changes in binding constant. The energetic contribution of the alanine mutations to the binding constant is seen using ΔΔG (Table 1). For example, mutation of N64A is found to contribute over 3 kcal/mol to the binding of ribose by RBP. This significant change in binding free energy shows that careful consideration is needed for introducing site-specific mutations into an RBP scaffold.
The amino acid mutations described here result in loss of signaling, loss of stability, both, or neither. Peristeric residues (e.g., I132A and S9A) that are along the binding cleft but not directly involved in ligand binding produce almost no change to either signaling or thermal stability. Mutation of endosteric residues that directly contact the ribose ligand and are located further into the binding cleft appear to have a greater impact on stability and signaling but not necessarily both (e.g., D89). Residues N190 and F214 seem to act as loci for both stability and signaling. Thermal unfolding data for residue Q235, which is located in the hinge region, indicate the presence of a hot spot for global protein stability (Table 1). Indeed, a mutation of Q235V retained ribose binding ability and resulted in an increase in overall RBP stability to 68°C in the presence of ribose (data not shown), which is higher than that found for the wild-type protein. Taken together, these data further support the important role residue 235 may have in overall protein stability. Because of the increase in stability for this mutant, the design process could be altered so that this mutation is fixed during the search algorithm. While this may limit the number of possible ligand poses used during the computational search, a significant benefit to signal transduction and stability may be realized.
While a substantial gain in stability is observed following ligand binding and hinge bending conformational changes, RBP is optimized for its role in E. coli nutrient acquisition and not necessarily for biosensor design. We show that RBP is able to absorb at least five alanine mutations simultaneously within the binding pocket without too much loss in stability (Table 2). Increasing the stability of the starting scaffold is one way to create more stable biosensing systems. In addition to point mutation described here, homologous proteins from thermophilic organisms offer attractive alternatives (Cuneo et al. 2006; deLorimier et al. 2006). However, because ligand binding is measured as a function of global conformational changes, these proteins may require higher incubation temperatures to function properly. Because of the known structural homology between RBP and other such binding proteins, these results may serve as a model to help influence the computational design of other members of this structural superfamily.
Materials and methods
Preparation of RBP mutants
Primers for the E. coli rbsB gene, encoding the ribose binding protein, and a His10-C-terminal tag were used to clone into the NdeI–EcoRI site of pET21a (Novagen, Inc.). For insertion of point mutations, overlapping PCR site-directed mutagenesis was performed on this construct as previously described (Ho et al. 1989). All constructs were confirmed by DNA sequencing (VCU Nucleic Acid Research Facility). RBP mutants were expressed from E. coli BL21(DE3) pLysS and purified using Ni-NTA affinity chromatography (GE Biosciences) according to manufacturer-recommended protocols. Protein purity was checked using a Bruker Omniflex MALDI-TOF as well as SDS-PAGE. Purified RBP mutants were applied to an Econo-Pac 10 DG gel filtration column (Bio-Rad) to equilibrate the samples in 5 mM phosphate buffer (pH 7.0) for storage at 4°C.
Preparation of protein conjugates
In a typical labeling procedure, 10–50 μM protein in 5 mM phosphate buffer (pH 7.0) was incubated with a threefold molar excess of thiol-reactive fluorophore in the dark, at 4°C overnight. After an initial screening of the available environmentally sensitive probes (see list below) using RBP:L265C, ABD-F was used to covalently modify all RBP mutants. Unreacted fluorophore was removed from labeled protein conjugates using Econo-Pac 10 DG columns pre-equilibrated with 5 mM phosphate buffer (pH 7.0). For simplicity, RBP:L265C will be designated WT RBP. 60
Fluorescein-iodoacetamide, acrylodan, NBD ester, and NBD amide were purchased from Invitrogen/Molecular Probes. ABD-F was purchased from AnaSpec, Inc. JPW4095 and JPW4044 were kind gifts of Leslie Lowe at the University of Connecticut. All dyes tested were dissolved in dry DMSO and used without further purification.
Steady-state fluorescence
Fluorescence measurements were performed using a Varian Cary Eclipse spectrofluorometer equipped with a plate reader and thin film polarizers set to magic angle (0° excitation and 54.7° emission). Titrations of protein conjugates (1 μM) in 5 mM phosphate buffer (pH 7.0) were performed using a 96-well plate reader while increasing ribose concentrations from 0 to 10 mM ribose and using an excitation wavelength of 380 nm. Fluorescence intensity at the emission maximum as a function of ribose concentration was used to determine binding constants by fitting data to a single binding isotherm as previously described (deLorimier et al. 2002). All data analysis was performed using the nonlinear curve fitting procedures employed in the Origin 6.0 software package. A minimum of three experimental trials were averaged for calculation of binding constants. Unless otherwise stated, all measurements were conducted at room temperature.
Circular dichroism
All CD measurements were performed using a Jasco J-720 spectrometer equipped with a thermal peltier temperature control module, and data were analyzed using the spectra manager software (ver. 5.1.0.0) supplied with the instrument. All measurements were performed using stirred proteins solutions of ∼0.5 μM in 5 mM phosphate buffer (pH 7.0) with or without the addition of 10 mM ribose.
Acknowledgments
The authors thank the Thomas F. Jeffress and Kate Miller Jeffress Memorial Trust for their generous support of this research. B.D. thanks the Howard Hughes Medical Institute for funding support.
Footnotes
Reprint requests to: Jonathan D. Dattelbaum, Department of Chemistry, University of Richmond, Gottwald Center for the Sciences, 28 Westhampton Way, Richmond, VA 23173, USA; e-mail: jdattelb@richmond.edu; fax: (804) 287-1897.
Abbreviations: PBP, bacterial periplasmic binding protein; RBP, Escherichia coli ribose binding protein; ABD-F, [4-Fluoro-7-aminosulfonylbenzofurazan]; CD, circular dichroism.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062595707.
References
- Allert M., Rizk, S.S., Looger, L.L., and Hellinga, H.W. 2004. Computational design of receptors for an organophosphate surrogate of the nerve agent soman. Proc. Natl. Acad. Sci. 101: 7907–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson D.E., Conrad, D.W., de Lorimier, R.M., Trammell, S.A., and Hellinga, H.W. 2001. Design of bioelectronic interfaces by exploiting hinge-bending motions in proteins. Science 293: 1641–1644. [DOI] [PubMed] [Google Scholar]
- Brune M., Hunter, J.L., Howell, S.A., Martin, S.R., Hazlett, T.L., Corrie, J.E., and Webb, M.R. 1998. Mechanism of inorganic phosphate interaction with phosphate binding protein from Escherichia coli . Biochemistry 37: 10370–10380. [DOI] [PubMed] [Google Scholar]
- Cuneo M.J., Changela, A., Warren, J.J., Beese, L.S., and Hellinga, H.W. 2006. The crystal structure of a thermophilic glucose binding protein reveals adaptations that interconvert mono and di-saccharide binding sites. J. Mol. Biol. 362: 259–270. [DOI] [PubMed] [Google Scholar]
- Dattelbaum J.D. and Lakowicz, J.R. 2001. Optical determination of glutamine using a genetically engineered protein. Anal. Biochem. 291: 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGrado W.F., Summa, C.M., Pavone, V., Nastri, F., and Lombardi, A. 1999. Denovo design and structural characterization of proteins and metalloproteins. Annu. Rev. Biochem. 68: 779–819. [DOI] [PubMed] [Google Scholar]
- DeLano W.L.. 2002. The PyMOL Molecular Graphics System. Delano Scientific, San Carlos, CA.
- deLorimier R.M., Smith, J.J., Dwyer, M.A., Looger, L.L., Sali, K.M., Paavola, C.D., Rizk, S.S., Sadigov, S., Conrad, D.W., and Loew, L., et al. 2002. Construction of a fluorescent biosensor family. Protein Sci. 11: 2655–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deLorimier R.M., Tian, Y., and Hellinga, H.W. 2006. Binding and signaling of surface-immobilized reagentless fluorescent biosensors derived from periplasmic binding proteins. Protein Sci. 15: 1936–1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuschle K., Okumoto, S., Fehr, M., Looger, L.L., Koszhukh, L., and Frommer, W.B. 2005. Construction and optimization of a family of genetically encoded metabolite sensors by semirational protein engineering. Protein Sci. 14: 2304–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X. and Quiocho, F.A. 2002. Structural evidence for a dominant role of nonpolar interactions in the binding of a transport/chemosensory receptor to its highly polar ligands. Biochemistry 41: 706–712. [DOI] [PubMed] [Google Scholar]
- Duan X., Hall, J.A., Nikaido, H., and Quiocho, F.A. 2001. Crystal structures of the maltodextrin/maltose-binding protein complexed with reduced oligosaccharides: Flexibility of tertiary structure and ligand binding. J. Mol. Biol. 306: 1115–1126. [DOI] [PubMed] [Google Scholar]
- Dwyer M.A. and Hellinga, H.W. 2004. Periplasmic binding proteins: A versatile superfamily for protein engineering. Curr. Opin. Struct. Biol. 14: 495–504. [DOI] [PubMed] [Google Scholar]
- Dwyer M.A., Looger, L.L., and Hellinga, H.W. 2004. Computational design of a biologically active enzyme. Science 304: 1967–1971. [DOI] [PubMed] [Google Scholar]
- Gilardi G., Zhou, L.Q., Hibbert, L., and Cass, A.E. 1994. Engineering the maltose binding protein for reagentless fluorescence sensing. Anal. Chem. 66: 3840–3847. [DOI] [PubMed] [Google Scholar]
- Goodchild S., Love, T., Hopkins, N., and Mayers, C. 2006. Engineering antibodies for biosensor technologies. Adv. Appl. Microbiol. 58: 185–226. [PubMed] [Google Scholar]
- Higgins C.F.. 1992. ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 8: 67–113. [DOI] [PubMed] [Google Scholar]
- Ho S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77: 51–59. [DOI] [PubMed] [Google Scholar]
- Hosse R.J., Rothe, A., and Power, B.E. 2006. A new generation of protein display scaffolds for molecular recognition. Protein Sci. 15: 14–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igbal S.S., Mayo, M.W., Bruno, J.G., Bronk, B.V., Batt, C.A., and Chambers, J.P. 2000. A review of molecular recognition technologies for detection of biological threat agents. Biosens. Bioelectron. 15: 549–578. [DOI] [PubMed] [Google Scholar]
- Lager I., Fehr, M., Frommer, W.B., and Lalonde, S. 2003. Development of a fluorescent nanosensor for ribose. FEBS Lett. 533: 85–89. [DOI] [PubMed] [Google Scholar]
- Lee W.E., Thompson, H.G., Hall, J.G., and Bader, D.E. 2000. Rapid detection and identification of biological and chemical agents by immunoassay, gene probe assay and enzyme inhibition using a silicon-based biosensor. Biosens. Bioelectron. 14: 795–804. [DOI] [PubMed] [Google Scholar]
- Looger L.L. and Hellinga, H.W. 2001. Generalized dead-end elimination algorithms make large-scale protein side-chain structure prediction tractable: Implications for protein design and structural genomics. J. Mol. Biol. 307: 429–445. [DOI] [PubMed] [Google Scholar]
- Looger L.L., Dwyer, M.A., Smith, J.J., and Hellinga, H.W. 2003. Computational design of receptor and sensor proteins with novel functions. Nature 423: 185–190. [DOI] [PubMed] [Google Scholar]
- Marvin J.S. and Hellinga, H.W. 1998. Engineering biosensors by introducing fluorescent allosteric signal transducers: Construction of a novel glucose sensor. J. Am. Chem. Soc. 120: 7–11. [Google Scholar]
- Marvin J.S., Corcoran, E.E., Hattangadi, N.A., Zhang, J.V., Gere, S.A., and Hellinga, H.W. 1997. The rational design of allosteric interactions in a monomeric protein and its applications to the construction of biosensors. Proc. Natl. Acad. Sci. 94: 4366–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medintz I.L. and Deschamps, J.R. 2006. Maltose-binding protein: A versatile platform for prototyping biosensing. Curr. Opin. Biotechnol. 17: 17–27. [DOI] [PubMed] [Google Scholar]
- Medintz I.L., Goldman, E.R., Lassman, M.E., Hayhurst, A., Kusterbeck, A.W., and Deschamps, J.R. 2005. Self-assembled TNT biosensor based on modular multifunctional surface-tethered components. Anal. Chem. 77: 365–372. [DOI] [PubMed] [Google Scholar]
- Millard A.C., Jin, L., Wei, J.P., Wuskell, A., Lewis, A., and Loew, L.M. 2004. Sensitivity of second harmonic generation from styryl dyes to transmembrane potential. Biophys. J. 86: 1169–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho F.A. and Ledvina, P.S. 1996. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: Variation of common themes. Mol. Microbiol. 20: 17–25. [DOI] [PubMed] [Google Scholar]
- Rizk S.S., Cuneo, M.J., and Hellinga, H.W. 2006. Identification of cognate ligands for the Escherichia coli phnD protein product and engineering of a reagentless fluorescent biosensor for phosphonates. Protein Sci. 15: 1745–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohl C.A., Strauss, C.E.M., Misura, K.M.S., and Baker, D. 2004. Protein structure prediction using rosetta. Methods Enzymol. 383: 66–93. [DOI] [PubMed] [Google Scholar]
- Salins L.L., Deo, S.K., and Daunert, S. 2004. Phosphate binding protein as the biorecognition element in a biosensor for phosphate. Sens. Actuators B Chem. 97: 81–89. [DOI] [PubMed] [Google Scholar]
- Shrestha S., Salins, L.L., Mark Ensor, C., and Daunert, S. 2002. Rationally designed fluorescently labeled sulfate-binding protein mutants: evaluation in the development of a sensing system for sulfate. Biotechnol. Bioeng. 78: 517–526. [DOI] [PubMed] [Google Scholar]
- Thomas K.J., Sherman, D.B., Amiss, T.J., Andalus, S.A., and Pitner, B. 2006. A long-wavelength fluorescent glucose biosensor based on bioconjugates of galactose/glucose binding protein and Nile red derivatives. Diabetes Technol. Ther. 8: 261–268. [DOI] [PubMed] [Google Scholar]
- Tolosa L., Ge, X., and Rao, G. 2003. Reagentless optical sensing of glutamine using a dual-emitting glutamine-binding protein. Anal. Biochem. 314: 199–205. [DOI] [PubMed] [Google Scholar]
- Wells M.. 2006. Advances in optical detection strategies for reporter signal measurements. Curr. Opin. Biotechnol. 17: 28–33. [DOI] [PubMed] [Google Scholar]