Abstract
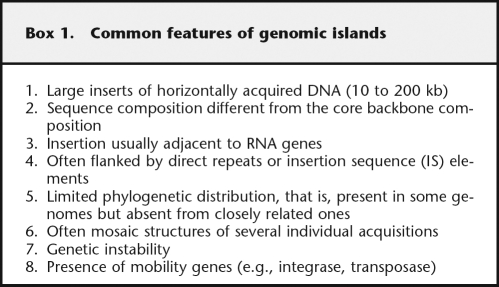
Large inserts of horizontally acquired DNA that contain functionally related genes with limited phylogenetic distribution are often referred to as genomic islands (GIs), and structural definitions of these islands, based on common features, have been proposed. Although a large number of mobile elements fall well within the GI definition, there are several concerns about the structural consensus for GIs: The current GI definition was put forward 10 yr ago when only 12 complete bacterial genomes were available, a large number of GIs deviate from that definition, and in silico predictions assuming a full/partial GI structural model bias the sampling of the GI structural space toward “well-structured” GIs. In this study, the structural features of genomic regions are sampled by a hypothesis-free, bottom-up search, and these are exploited in a machine learning approach with the aim of explicitly quantifying and modeling the contribution of each feature to the GI structure. Performing a whole-genome-based comparative analysis between 37 strains of three different genera and 12 outgroup genomes, 668 genomic regions were sampled and used to train structural GI models. The data show that, overall, GIs from the three different genera fall into distinct, genus-specific structural families. However, decreasing the taxa resolution, by studying GI structures across different genus boundaries, provides models that converge on a fairly similar GI structure, further suggesting that GIs can be seen as a superfamily of mobile elements, with core and variable structural features, rather than a well-defined family.
Horizontally acquired DNA sequences that contain functionally related genes with limited phylogenetic distribution, that is, present in some bacterial genomes while being absent from closely related ones, are often referred to as genomic islands (GIs). The location of those mobile elements often correlates with distinct structural features such as tRNA genes, direct repeats (DRs), and mobility genes, which has led to a definition of the GI structure that includes these features (Box 1; Hacker et al. 1997; Hacker and Kaper 2000; Schmidt and Hensel 2004).
Some of the GI-associated features are shared by other genomic elements such as integrated plasmids, bacteriophages, extracellular polysaccharide biosynthesis loci (Zhang et al. 1997; Hacker and Kaper 2000), and other gene clusters under specific constraints; these may or may not be recently horizontally acquired. However, GIs usually differ from bacteriophages and plasmids in the lack of autonomous replication origins (Schmidt and Hensel 2004).
GIs are also present in Gram-positive bacteria, but they can differ structurally from those present in Gram-negative bacteria; overall they do not exhibit specific junction sites (e.g., DRs), they are rarely inserted adjacent to RNA loci, and they are often stably integrated in the host genome because of the lack of mobility genes (Hacker et al. 1997).
Insertion of GIs into the bacterial chromosome is often a site-specific event. About 75% of GIs currently known have been inserted at the 3′-end of a tRNA locus (Hacker and Kaper 2002; Williams 2002). Other genes, though, may also act as insertion sites for GIs, for example, the cag pathogenicity island (PAI) has been inserted within the glr (glutamate racemase) gene of Helicobacter pylori (Censini et al. 1996). Often GIs are flanked at their boundaries by DRs with an average length of ∼20 bp (Kaper and Hacker 1999; Schmidt and Hensel 2004).
Several Web-based suites exploit the GI structural definition (Box 1) with the aim of implementing and automating the in silico prediction of genomic regions that share some or all of the GI-related signatures; those regions are subsequently annotated as novel GIs. For example Islander (Mantri and Williams 2004) and IslandPath (Hsiao et al. 2003), two Web-based suites, combine and overlap several GI-related features trying to predict genomic regions as close as possible to the GI structural definition.
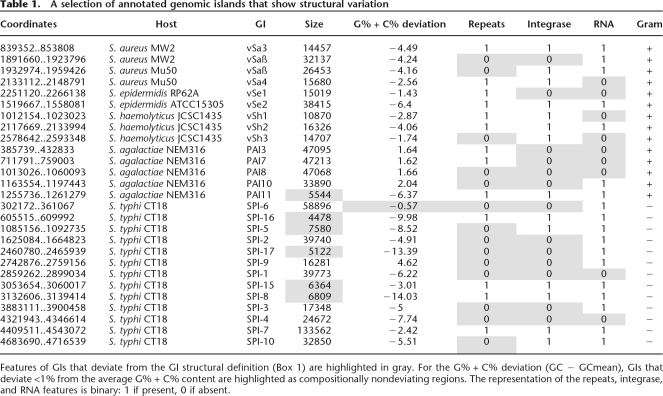
Although a large number of mobile elements fall well within the GI definition, there are several concerns about the structural consensus of GIs: Firstly, the current definition of the GI structure was put forward 10 yr ago (Hacker et al. 1997) when only 12 complete bacterial genomes were available; currently (May 2007) there are 558 complete published genomes and 1144 ongoing, enabling a more realistic sampling of the GI structural space for any potential structural variation to be captured. Secondly, there is a large number of GIs that deviate strongly from the GI definition (see Table 1). Thirdly, in silico prediction methods that assume a full or partial structure similar to the GI structural definition or search for GIs with some level of similarity to already known GI structures bias the sampling of the GI structural space toward “well-structured” GIs.
Table 1.
A selection of annotated genomic islands that show structural variation
Features of GIs that deviate from the GI structural definition (Box 1) are highlighted in gray. For the G% + C% deviation (GC − GCmean), GIs that deviate <1% from the average G% + C% content are highlighted as compositionally nondeviating regions. The representation of the repeats, integrase, and RNA features is binary: 1 if present, 0 if absent.
A fundamental property of GIs, independent of any a priori structural definition, is their horizontal origin: GIs are horizontally acquired mobile elements of limited phylogenetic distribution. Based on this concept, the search of the GI structural space is feasible in a hypothesis-free framework without the need to make any a priori assumptions about the GI structure that rely on previously seen examples of GIs.
The aim of this analysis is to study the structural variation of GIs and revisit the GI definition, taking into account only the fundamental property of GIs, that is, their horizontal origin. Instead of exploiting a top-down approach searching for GIs that follow the GI structural definition, we reverse this framework by pursuing a hypothesis-free, bottom-up search: In a first step GIs are defined as genomic regions with limited phylogenetic distribution consistent with recent acquisition (as identified by maximum parsimony), and in a second step those regions are structurally annotated. In a third step, the structural features sampled from this hypothesis-free search are exploited in a machine learning approach with the aim of explicitly quantifying and modeling their contribution to the GI structural definition.
A similar approach of a hypothesis-free identification of GIs, defined as genomic regions with limited phylogenetic distribution, was applied in eight Streptococcus agalactiae strains (Tettelin et al. 2005). Gene loss and gene gain are two distinct mechanisms that can both lead to limited phylogenetic distribution of a DNA sequence. However, Tettelin et al. (2005) did not apply any restriction (e.g., maximum parsimony) in order to differentiate gene gain from gene loss and defined as putative GIs any region (>5 kb) that was absent from at least one of the eight reference genomes.
In the present study, we focus on three different bacterial genera—Salmonella, Staphylococcus, and Streptococcus—for four major reasons: There are enough (>10) sequenced genomes for each genus, this collection of strains covers both Gram-negative and Gram-positive groups and has both commensal and pathogenic representatives, and HGT plays a key role in the evolution of these three lineages (Lawrence and Ochman 1997; Broker and Spellerberg 2004; Towers et al. 2004; Tettelin et al. 2005; Rosini et al. 2006; Waterhouse and Russell 2006; Novick and Subedi 2007; Vernikos et al. 2007).
Results
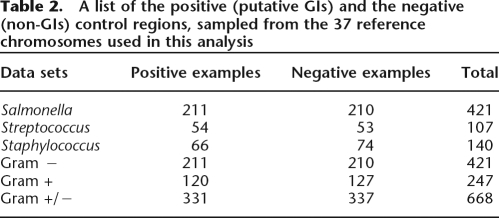
Implementing a whole-genome-based comparative analysis between 37 reference strains of three different genera and 12 outgroup genomes, a training set of 668 regions was built (Table 2). This training set, that includes both putative GIs (differentiated from gene loss events by a maximum parsimony approach) and randomly sampled regions (non-GIs), was used to study the structural variation of GIs and quantify the contribution of each feature to a GI structural model. As a starting point, GI structural models for each genus were built implementing a machine learning method, that of the Relevance Vector Machine (RVM) (Tipping 2001). In addition, in order to capture potential genus-specific signatures as well as to evaluate the ability of the RVM models to make generalizations on unseen data from different lineages, cross-genus GI models were built using different mixtures of training and test data sets. Overall, 11 structural GI models were built and analyzed (Table 3); the structural details of each model are discussed in detail in the following sections.
Table 2.
A list of the positive (putative GIs) and the negative (non-GIs) control regions, sampled from the 37 reference chromosomes used in this analysis
Table 3.
A list of 11 structural GI models, built based on different training sets
Training sets include (1) 421 Salmonella regions, (2) 107 Streptococcus regions, (3) 140 Staphylococcus regions (including two regions overlapping rRNA operons), (4) 138 Staphylococcus regions (no rRNA operons), (5) 245 Staphylococcus–Streptococcus regions, (6) 559 Salmonella–Staphylococcus regions, (7) 528 Salmonella–Streptococcus regions, (8) 666 Salmonella–Staphylococcus–Streptococcus regions. Training sets 9–11 include three subsets of ∼140 different Salmonella-specific regions combined with the Staphylococcus- and Streptococcus-specific regions. Each model, expressed through function Si, is the weighted sum of eight basis functions (structural features): The Interpolated Variable Order Motif (IVOM) score that measures both low- and high-order compositional deviation from the backbone composition and is expressed as the relative entropy between the query and the genome-backbone (variable order) compositional distribution, the insertion point (INSP) of each genomic region; two states were (binary) evaluated: insertion point within a CDS locus (disrupting the corresponding CDS) or insertion within an intergenic part of the chromosome, the size (SIZE) of each genomic region, the gene density (DENS = number of genes per kilobase) of each region, presence or absence (binary) of direct/inverted repeats (REPEATS) flanking the boundaries of each genomic region, presence or absence (binary) of integrase and/or integrase-like (INT) protein domains, presence or absence (binary) of phage-related protein domains (PHAGE), presence or absence (binary) of non-coding RNA (RNA) in the proximity of each region.
GI structural models
Each GI model (Table 3) is the weighted sum of K basis functions, where K denotes the number of features used to describe a GI structure. In this analysis, eight structural features were used:
IVOM: The Interpolated Variable Order Motif score that measures both low- and high-order compositional deviation from the backbone composition (Vernikos and Parkhill 2006).
INTEGRASE: Presence or absence (binary) of integrase and/or integrase-like protein domains.
PHAGE: Presence or absence (binary) of phage-related protein domains.
SIZE: The size (in base pairs) of each genomic region.
RNA: Presence or absence (binary) of non-coding RNA in the proximity of each region.
DENSITY: The gene density (number of genes per kilobase) of each region.
REPEATS: Presence or absence (binary) of DRs or inverted repeats (IRs) flanking the boundaries of each genomic region.
INSP: The insertion point of each genomic region; two states were evaluated: insertion point within a coding sequence (CDS) locus (disrupting the corresponding CDS) or insertion within an intergenic part of the chromosome.
Each feature is evaluated during the training process of the RVM, and its overall contribution to the structural model is expressed by the corresponding feature weight. For example, a feature frequently related to GI structures (but absent from randomly sampled regions) receives typically higher weight (i.e., contributes more to the model) compared to a feature found equally frequently both in GIs and non-GIs; in the latter case, the feature weight will be lower or even zero (i.e., feature ignored).
In the following section, the contribution of each structural feature to the corresponding GI model is evaluated through a function (R) that quantifies the relative feature importance, rather than the actual feature weight (W). Briefly, the importance R of each feature is expressed as the product of the corresponding weight W and the corresponding standard deviation (SD) of the feature values in the training set. We prefer to assess the feature contribution to the model, through the R rather than the W value, because R takes into account the variability of the data set, normalizing the values with the corresponding SD. Consider, for example, two different structural features; the values of the first feature in the training set have higher dispersion relative to the values of the second feature. If both features have comparable W values, then the first feature will be more important than the second one, meaning that, because of its variability, it is more informative than the second feature. Based on that, it is not unusual for some features to have a very high value of W but a low value of R.
Genus-specific
Salmonella
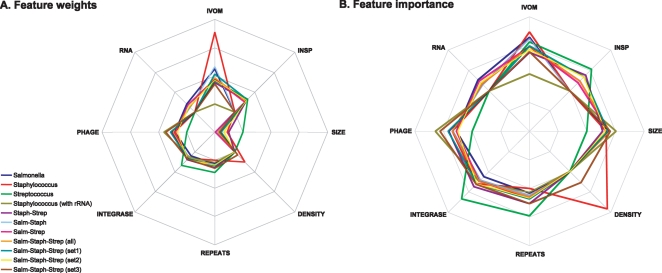
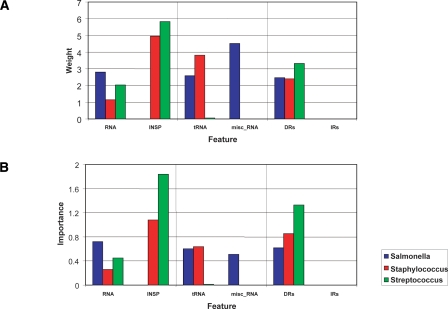
Using 211 positive (putative GIs) and 210 negative (randomly sampled) examples (Table 2; Supplemental Table 1), a model that describes the structure of GIs present in the Salmonella lineage was built (Fig. 1; Table 3). Overall under this model, the most “important” (informative) features are IVOM (RIVOM = 0.65), SIZE (RSIZE = 0.38), PHAGE (RPHAGE = 0.27), RNA (RRNA = 0.26), INTEGRASE (RINT = 0.13), and REPEATS (RREPEATS = 0.085); in this model, the DENSITY and INSP features were ignored. Note that the SIZE feature received a negative weight (WSIZE = −4.956); the same applies for all the other GI models apart from the one built based on the Streptococcus data set (see below) in which the SIZE feature is completely ignored (WSIZE = 0).
Figure 1.
Radar diagram illustrating (A) the feature weight and (B) “importance” of the eight structural features under different GI models, based on 11 training data sets. Features: (IVOM) feature composition; (INSP) insertion point; (SIZE) the size of each region; (DENSITY) gene density; (REPEATS) repeats flanking each region; (INTEGRASE) integrase-like protein domains; (PHAGE) phage-related protein domains; (RNA) non-coding RNAs. Each apex in the octagon-like diagram corresponds to one of the eight structural features, while the height of the plot at the corresponding apex is indicative of the actual (A) feature weight or (B) importance.
In order to investigate further the structural variation of GIs, in terms of preference for insertion within a specific locus and for different type of repeats flanking their boundaries, the RNA feature was further subdivided into tRNA and misc_RNA (any kind of non-coding RNA apart from tRNA) features; the same applies for the REPEATS feature that was further divided into DRs and IRs. The relative “importance” of those six structural features was evaluated pairwise: (RNA, INSP), (tRNA, misc_RNA), and (DRs, IRs) (Fig. 2). The results show that for GIs present in Salmonella chromosomes, insertion within an RNA (RRNA = 0.72) rather than a CDS locus (RINSP = 0.0) is the most informative feature when classifying unknown regions as GIs. In the case of RNA locus, insertion of GIs within a tRNA (RtRNA = 0.60) is slightly more informative than insertion within a misc_RNA locus (RmiscRNA = 0.51). In terms of type of repeats flanking the boundaries of GIs, DRs (RDRs = 0.63) rather than IRs (RIRs = 0.0) is the most informative feature.
Figure 2.
Bar chart illustrating (A) the feature weight and (B) “importance” of six structural features (evaluated pairwise), under three different dual-featured GI models, trained on Salmonella, Staphylococcus, and Streptococcus-specific regions, respectively. Features: [RNA, INSP], [tRNA, misc_RNA], [DRs, IRs].
Staphylococcus
The model that describes the structure of GIs present in Staphylococcus genomes was built based on 66 putative GIs and 74 randomly sampled regions (Table 2; Supplemental Table 1). Overall under this model, the most predictive informative structural features are PHAGE (RPHAGE = 0.65), SIZE (RSIZE = 0.51), INTEGRASE (RINT = 0.25), and REPEATS (RREPEATS = 0.07); the remaining features were ignored. Two randomly sampled regions had the two highest IVOM scores in this data set of 140 examples. These two regions (Staph.Epid_RP62.non.12 and Staph.MRSA252.non.21 in Supplemental Table 1) overlap with two rRNA operons. rRNA operons often deviate compositionally from the genome backbone composition mainly because of specific, well-preserved functional constraints rather than their horizontal origin (Vernikos and Parkhill 2006; Vernikos et al. 2007). Excluding those two regions and repeating the training, the GI model assigned weights to previously ignored features and modified each weight overall: DENSITY (RDENS = 0.92), IVOM (RIVOM = 0.74), PHAGE (RPHAGE = 0.35), SIZE (RSIZE = 0.34), INTEGRASE (RINT = 0.30); the rest of the features were ignored (Fig. 1; Table 3).
When GI models are trained (pairwise) only on selected structural features, insertion within a CDS locus (RINSP = 1.1) is more informative than insertion within an RNA locus (RRNA = 0.26). Between the different types of non-coding RNAs, insertion within a tRNA (RtRNA = 0.64) rather than a misc_RNA (RmiscRNA = 0.0) is the most informative feature. In terms of type of repeats, again DRs is the most informative feature (RDRs = 0.85, RIRs = 0.0) (Fig. 2). It is worth noting that under these three partial GI models, some previously ignored (under the full GI model above) structural features, that is, RNA, INSP, and REPEATS, are now informative predictors, further suggesting that those features were redundant predictors under the full model in which all eight features were evaluated.
Streptococcus
The training set for the Streptococcus genus consists of 54 and 53 positive and negative control examples, respectively (Table 2; Supplemental Table 1). Under this model, the most informative GI structural features are INTEGRASE (RINT = 0.67), IVOM (RIVOM = 0.56), INSP (RINSP = 0.53), and REPEATS (RREPEATS = 0.48). The remaining four features were ignored (Fig. 1; Table 3), giving the highest sparsity GI model that exploits only four (of the eight) basis functions.
In terms of pairwise evaluation of selected structural features (Fig. 2), GIs present in Streptococcus genomes follow the same pattern of insertion point preference with the Staphylococcus GIs, that is, insertion within a CDS locus (RINSP = 1.84) is more informative than insertion within an RNA locus (RRNA = 0.45); the same applies for the type of non-coding RNAs (RtRNA = 0.013, RmiscRNA = 0.0) and the type of repeats (RDRs = 1.33, RIRs = 0.0).
Cross-genus
Staphylococcus–Streptococcus
Combining 138 Staphylococcus and 107 Streptococcus genomic regions, a data set of 245 (Gram-positive) examples was built in order to study the structural variation of GIs across genus/species boundaries. In this cross-genus GI model, the most informative features are PHAGE (RPHAGE = 0.41), INSP (RINSP = 0.39), IVOM (RIVOM = 0.374), INTEGRASE (RINT = 0.37), SIZE (RSIZE = 0.272), and REPEATS (RREPEATS = 0.270); the remaining structural features were ignored (Fig. 1; Table 3; Supplemental Fig. S2a).
Salmonella–Staphylococcus
A cross-genus data set of 421 Salmonella- and 138 Staphylococcus-specific regions was built and used to train a GI structural model; under this model, the most informative features are IVOM (RIVOM = 0.62), SIZE (RSIZE = 0.40), PHAGE (RPHAGE = 0.34), INTEGRASE (RINT = 0.21), RNA (RRNA = 0.15), and REPEATS (RREPEATS = 0.12). The remaining features were ignored (Fig. 1; Table 3; Supplemental Fig. S2b).
Salmonella–Streptococcus
Combining the Salmonella- and Streptococcus-specific regions, a data set of 528 examples was built. Under this cross-genus GI model, the most informative structural features are IVOM (RIVOM = 0.48), SIZE (RSIZE = 0.39), PHAGE (RPHAGE = 0.28), INTEGRASE (RINT = 0.25), RNA (RRNA = 0.24), INSP (RINSP = 0.20), and REPEATS (RREPEATS = 0.16) (Fig. 1; Table 3; Supplemental Fig. S2c).
Salmonella–Staphylococcus–Streptococcus
In order to study the structural variation of GIs across the three genera, taking into account the difference in the dimensionality of the three genus-specific data sets (421 Salmonella-, 138 Staphylococcus-, and 107 Streptococcus-specific regions), we followed two different approaches: In the first approach, a training set (N = 666) was built combining the full Salmonella and the other two genus-specific data sets; in the second approach, the Salmonella data set was split into three subsets (N ≈ 140 each), each of which was combined with the full Staphylococcus and Streptococcus data sets giving three training sets (namely, set1, set2, and set3) of ∼385 examples each; in each set, the three different genera contribute approximately the same number of examples.
Training the RVM on the full (N = 666) cross-genus data set (all), the most informative GI structural features are IVOM (RIVOM = 0.48), SIZE (RSIZE = 0.39), PHAGE (RPHAGE = 0.35), INTEGRASE (RINT = 0.25), INSP (RINSP = 0.24), RNA (RRNA = 0.17), and REPEATS (RREPEATS = 0.15) (Fig. 1; Table 3; Supplemental Fig. S3a).
Using each of the three smaller data sets (sets 1–3) to train the RVM, the most informative features under the three GI models are (for each model, respectively) IVOM (RIVOM = 0.49, 0.43, 0.39), PHAGE (RPHAGE = 0.42, 0.25, 0.56), SIZE (RSIZE = 0.37, 0.32, 0.41), INTEGRASE (RINT = 0.29, 0.30, 0.31), INSP (RINSP = 0.34, 0.34, 0.0), REPEATS (RREPEATS = 0.19, 0.17, 0.27), DENSITY (RDENS = 0.0, 0.0, 0.26), and RNA (RRNA = 0.0, 0.19, 0.0) (see Fig. 1; Table 3; Supplemental Fig. S3). Based on the four RVM trainings (all, set1, set2, and set3), the four models that capture the structural variation of GIs across the three genera have converged over fairly similar GI structures, with the exception of genus-specific features, that is, the RNA feature for Salmonella, the INSP feature for Streptococcus, and the DENSITY feature for Staphylococcus (see Supplemental Fig. S3e and Discussion).
Prediction accuracy—benchmarking
In order to evaluate the prediction accuracy of the RVM classifier, each data set was split into five smaller subsets of approximately the same size, and the RVM was trained on four-fifths of the data set and tested on the remaining one-fifth; this process was repeated five times (for each data set), classifying non-overlapping test sets each time (fivefold cross-validation). Moreover, in order to evaluate further the generalization properties of each GI structural model, we performed six “genus-blind” cross-validations, training a model only on examples of one genus and testing it on examples of the other two. This blind test was performed in order to investigate how different genus-specific models would perform in classifying regions from unknown taxa. In order to estimate the relative accuracy and generalization properties of each model, we performed a receiver operating characteristic (ROC) curve analysis. The area under the ROC curve (AUC) is a measure of the accuracy of a given classifier; the closer the AUC is to 1, the more accurate the classifier (see Supplemental Material for details).
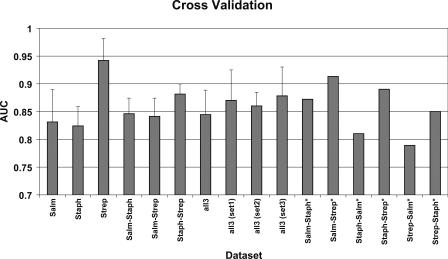
Overall, throughout the 10 fivefold cross-validations, the different GI models made good generalizations on unseen data, classifying with high accuracy (AUC: 0.82–0.94) unknown examples (GIs and non-GIs) (Fig. 3; Supplemental Fig. S4). Between the three different genus-specific GI models, the Streptococcus (Strep) model is the most accurate, followed by the Salmonella (Salm) and the Staphylococcus (Staph) models (AUC: 0.94, 0.83, and 0.82, respectively).
Figure 3.
A bar chart illustrating the average performance of the RVM classifier, under different training and test data sets. Each data set is split into five subsets of approximately equal size; four of the five subsets are used to train an RVM model, while the omitted subset is used to test the performance of this model. This process is repeated five times on non-overlapping test sets (fivefold cross-validation). The performance of the RVM models was evaluated through the receiver operating characteristic (ROC) curve. The area under the ROC curve (AUC) is a measure of the model’s accuracy: The closer the curve follows the left-hand and the top border of the ROC space, the more accurate the classification model. A perfect classifier would give an AUC of 1, while a classifier that makes a random guess would give an AUC of 0.5. The average value and ±1 SD of the AUC over the five subsets of the fivefold cross-validation is calculated for the first 10 data sets. The AUC values for the last six data sets (with the asterisk) summarize the performance of the RVM, when trained on the whole data set of the first genus and tested on the whole data set of the second genus, for example, for the Salm-Strep* data set, the 421 Salmonella-specific regions were used to train a GI model that was tested on the 107 Streptococcus-specific regions.
Between the three different GI models, trained on a mixture of examples from two different genera, the Staph-Strep (Gram-positive) model is the most accurate, followed by the Salm-Staph and the Salm-Strep models (AUC: 0.881, 0.846 and 0.841, respectively). Overall, the Salm-Staph model performs better than the corresponding two genus-specific Salm and Staph models (Fig. 3); similarly the Salm-Strep and Staph-Strep models are overall more accurate than the Salm and Staph models, respectively.
GI models trained on a mixture of examples from all the three genera show fairly similar performance (AUC: 0.84–0.88). More specifically, the three GI models trained on data sets in which the three genera are equally represented (i.e., set1, set2, and set3) perform equally well (AUC: 0.87, 0.86, 0.88) and slightly better than the model trained on all (N = 666) examples (AUC: 0.84), underlining the increased sparsity property of the RVM method.
The evaluation of the three genus-specific GI models, under a “genus-blind” cross-validation framework, indicates that the RVM classifier can very accurately predict unseen examples from close or distantly related genera that are not included in the training set (Fig. 3). More specifically, using the Salm model to classify Staphylococcus- and Streptococcus-specific regions can be overall more (AUC: 0.87 vs. 0.82) or similarly (AUC: 0.91 vs. 0.94) accurate compared to the corresponding genus-specific models, respectively. The Staph model shows high accuracy (AUC: 0.81 and 0.89) in classifying Salmonella- and Streptococcus-specific regions, respectively; overall, this model is slightly less accurate than the corresponding genus-specific models (AUC: 0.83 and 0.94, respectively). Similar conclusions can be drawn for the performance (AUC: 0.79 and 0.85) of the Strep model when classifying Salmonella- and Staphylococcus-specific regions, respectively. Again this model is more accurate in classifying Staphylococcus-specific regions than the Staph model (AUC: 0.85 and 0.82, respectively), but is less accurate in classifying Salmonella-specific regions than the Salm model (AUC: 0.79 and 0.83, respectively).
Discussion
The aim of this analysis was to study the structural variation of GIs, quantifying and modeling the “importance” of genetic features that can be informative when classifying GIs and non-GI regions, enabling a quantitative rather than a descriptive definition of the actual GI structure to be proposed. The basic principle behind this analysis is a hypothesis-free framework, in which no a priori assumptions are made about the GI structure.
Implementing a machine learning-oriented approach, genomic regions (both GIs and randomly sampled regions) from 37 chromosomes of three different genera were exploited in order to build genus-specific as well as cross-genus GI structural models. Overall, the three genus-specific GI models show both core and variable structural features with distinct genus-specific signatures. For example, the IVOM and INT features are informative in all three GI models; on the other hand, the RNA, INSP, and DENSITY features are Salmonella-, Streptococcus-, and Staphylococcus-specific features, respectively (Fig. 1; Table 3). Moreover, in the Strep model apart from the INSP feature, the INT and REPEATS features contribute more to the overall structural model compared to the other two genus-specific models (Fig. 1), while the SIZE and PHAGE features seem to be informative only in the Salm and Staph structural GI models.
Care should be taken when interpreting the “importance” of each of the eight structural features. In this analysis, GI models are built evaluating how informative each feature is, taking into account cross-feature relationships and information redundancy. Mapping the eight features in a high-dimensional space enables cross-feature relationships to be captured: If some features contain information present already in other features (redundant information), then for the sake of model sparsity, those features (basis functions) will be ignored by setting their weight to zero value. That, however, does not necessarily mean that those features may not be informative when seen on their own, that is, in single-featured GI models (Supplemental Fig. S6). Therefore, it is more intuitive to interpret the importance of each feature as it is relative (in combination with the rest of the features) rather than its absolute importance under a GI model. For example, in the Strep model, the PHAGE feature is ignored when building a model evaluating all the eight features. However, when the PHAGE feature is evaluated in a single-featured model, it turns out to be the second most informative feature (Supplemental Fig. S6); this observation is in line with previous studies showing the impact of bacteriophage elements in the evolution of Streptococci (Broudy et al. 2001; Banks et al. 2003; Fischetti 2007). Perhaps some of the information in the PHAGE feature is already present in some other features (e.g., phage integrase protein domains of the INTEGRASE feature) making the PHAGE feature a redundant predictor under a multifeatured GI model.
The same observation applies for the SIZE feature. In a multifeatured model, SIZE is a very informative feature for the Salm and Staph models; however, in a single-featured model (i.e., evaluated on its own), the SIZE feature is ignored in all three genera models (Supplemental Fig. S6). This further suggests that in multifeatured models, some structural features correlate with the SIZE feature. Moreover, throughout this analysis, the SIZE feature received a negative weight in all GI models apart from the Strep model. Generally, during the training process some features may correlate positively or even negatively (e.g., the SIZE feature) with class membership. This does not necessarily suggest that true GIs are always of small size, but rather that the SIZE feature is negatively correlated with some other features. This observation becomes much clearer in the case of the Strep model in which both the SIZE and the PHAGE features received a weight of zero. However, in the other 10 models, the same two features received a negative and a positive weight, respectively (Table 3). Perhaps the SIZE feature is inversely correlated with the PHAGE feature, suggesting that GIs of phage origin are on average larger than GIs of different origin. Indeed, for the Salmonella and the Staphylococcus data set, the average size of GIs of phage origin is significantly larger than the size of GIs of different origin (p-value = 1.17 × 10−7 and 1 × 10−5, respectively). In order for the reverse correlation of the SIZE and some features to be captured in the model, the SIZE feature has to have a negative weight.
The fact that in the Strep GI model, three structural features (i.e., INTEGRASE, REPEATS, and INSP) are unusually highly informative (relative to the other two genus-specific models), while at the same time, those three features are frequently involved in the mobilization of genomic DNA (i.e., integration/excision), leaves open the possibility of a GI model that is capturing a distinct Streptococcus-specific mechanism of genetic element mobilization, via integration preferably within CDS loci. It is worth noting that the Strep GI model shows the highest sparsity exploiting only half of the basis functions (four out of the eight structural features), compared to the Staph (five out of eight) and the Salm (six out of eight) GI models, proposing a much simpler structural model, in order to describe GIs in the Streptococcus lineage (Table 3); this observation is in line with the outstanding classification accuracy of the Strep GI model (AUC: 0.94−) (Fig. 3).
The distinct structural feature with the highest contribution to the Staph GI model, while being ignored in the other two genus-specific models, is the DENSITY feature (Fig. 1; Supplemental Fig. S1). Overall, the average gene density of GIs present in Staphylococcus genomes is significantly (p-value = 1.4 × 10−6) higher than that of randomly sampled regions; in Salmonella and Streptococcus lineages, this feature is less informative when predicting GIs (p-value = 1.7 × 10−3 and 1.3 × 10−2, respectively). Again, it is possible that this genus-specific GI model is capturing the underlying origin of GIs present in Staphylococcus genomes, suggesting chromosomes of higher gene density than that characterizing the Staphylococcus lineage as the potential source of those GIs, one obvious possibility being bacteriophage genomes.
Increasing further the resolution within certain GI structural features (i.e., insertion within a CDS or RNA locus, tRNA, or misc_RNA and DRs or IRs), training the RVM pairwise only on those selected features, the genus-specific signatures of each model become more evident (Fig. 2). For the prediction of GIs in the Salmonella lineage, integration within a non-coding RNA locus is much more informative than within a CDS locus. The opposite observation can be made for the Staphylococcus and Streptococcus models. In the case of non-coding RNA, insertion within a tRNA or a misc_RNA locus is almost equally informative for the prediction of Salmonella GIs, while in Staphylococcus and Streptococcus lineages, insertion within a tRNA locus is much and slightly more informative than insertion within a misc_RNA, respectively. In all three genera, the predominant type of repeats associated with GIs is DRs.
Although the three genus-specific GI structural models show distinct signatures, suggesting well-defined GI families with core and variable regions, when the RVM training takes place on a mixture of cross-genus examples, the various GI models converge over fairly similar GI structures (Fig. 1; Supplemental Figs. S2–S3). This observation further supports the idea that GIs overall represent a superfamily of mobile elements with significant structural variation, rather than a well-defined family when looking across genus boundaries. When the predictive accuracy and generalization properties of the cross-genus models are evaluated, many of those models perform overall equally well or better compared to the corresponding genus-specific models (Fig. 3). This observation perhaps suggests that in some cases the RVM method has overfitted slightly on a subset of a genus-specific training data set, misclassifying the remaining subset; when more training examples from other genera are included in the training data set, models with much lower degrees of overfitting are trained.
Between the cross-genus GI models, trained on a mixture of two different genera examples, the Staph-Strep model shows the highest accuracy compared to the Salm-Staph and Salm-Strep models. Perhaps this cross-genus GI model is capturing structural properties of GIs found in Gram-positive bacteria that are less or not informative for the prediction of GIs in Gram-negative bacteria (Hacker et al. 1997).
Even when the cross-validation is based on a GI model that is trained on a genus-specific data set and tested on examples of a different genus, the prediction accuracy remains remarkably high, further supporting the concept of the GI superfamily. For example, the accuracy of the model trained on Salmonella examples and tested on Streptococcus examples is very similar to that of the Streptococcus-specific model (Fig. 3). Moreover, the genus-specific GI model with the highest sparsity, that is, the Strep model, discriminates GIs remarkably well from randomly sampled regions when tested on examples from the other two genera (Supplemental Fig. S5).
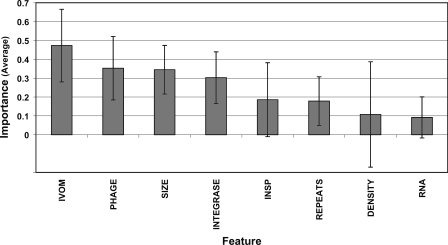
Overall, the evaluation of the eight structural features across the 11 training data sets shows that the IVOM, PHAGE, SIZE, and INTEGRASE features are, on average, the most informative ones, followed by the INSP, REPEATS, DENSITY, and RNA features (Fig. 4). It seems that the four most informative structural features are important predictors when classifying GIs from any of the three genera, suggesting that there are core features of a superfamily of mobile elements, whereas the other four, less-informative features are capturing genus-specific properties of GIs (being informative only when predicting GIs from a single genus), suggesting these may be variable features of distinct genus-specific GI families.
Figure 4.
Bar chart illustrating the average “importance,” across 11 structural GI models, of the eight structural features evaluated in this analysis. The eight features have been sorted (in decreasing order) based on their average importance. Error bars show ±1 SD.
The analysis carried out in this study forms the first attempt to quantify the actual GI structure in a probabilistic framework taking into account the contribution of all the informative structural features. Instead of vaguely describing putative GIs, we can explicitly quantify our level of confidence that they fit an empirically derived structure. This probabilistic scoring framework enables a systematic description of GI elements, which can be ranked based on their underlying structural information and subsequently classified into distinct structural families.
Although this methodology provides some new insights about the structural variation of GIs, there are some limitations that have to be taken into account:
(1) The RVM method shows increased sparsity, providing simple models that can very accurately capture the underlying structural variation in some cases (e.g., the Strep model). On the other hand, the RVM method overfitted twice, to some extent, to the Staphylococcus data set: firstly, the two Staph models (with and without the two rRNA operons) show significantly different weights, and secondly the Staph model models the Staphylococcus data set more poorly than any of the other two genus-specific models (Salm and Strep), perhaps overfitting to the DENSITY feature. To test whether this is, indeed, the case for the Staph model, we removed the DENSITY feature from the training and test data sets and repeated the cross-validation using the three models (Salm, Staph, Strep), re-evaluating their performance on the Staphylococcus data set. The data support the suggestion that the poorer performance of the Staph model on the Staphylococcus data set, relative to the other two genus-specific models, is due to overfitting of the model to 20% of the data set that had examples with significantly higher gene density than the rest of the data set. The new Staph model outperforms the other two models when tested on the Staphylococcus data set; more specifically, the AUC before and after the removal of the DENSITY feature for the three models is as follows, respectively: (Staph = 0.824, 0.875), (Salm = 0.872, 0.865), (Strep = 0.850, 0.850).
(2) The RVM method, as implemented in the present study, gave an error margin of 10%–20%. Possible sources of this error margin include significant structural intersection of the GIs and the randomly sampled regions; some randomly sampled regions were sampled close to classical GI-related structural features (e.g., tRNA) simply by chance, while a few GIs lack most (or all) of the classical GI-related features (since no a priori structural assumptions were made). Moreover, the phylogenetic sample used in the present study strongly affects the validity of the training data sets; overall, 11–13 strains and four outgroups were analyzed for each reference genus. Regions of limited phylogenetic distribution (under a maximum parsimony evaluation) were defined as GIs, while inter-GI chromosomal regions were randomly sampled. Under this framework, there are two possibilities to be taken into account: Firstly, some predicted GIs might not actually represent true GIs, if the phylogenetic resolution is further increased, that is, including more reference strains and more distantly related outgroups. Secondly, some randomly sampled regions might have been sampled over “ancient” GIs that were acquired prior to the divergence of the reference and the outgroup lineages. Consequently, care should be taken when interpreting the results of this analysis; the parameters of the RVM models and the validity of the actual training data sets directly affect the conclusions drawn about the structural variation of GIs. These conclusions are specific only for the three data sets analyzed, the structural annotation methodology and the machine learning method implemented in this study.
The species sample used in this analysis is inevitably small in the context of a wide, representative sampling of the GI structural space. However, it forms a proof of concept showing that the components of a GI structure can explicitly be quantified through a probabilistic framework. Under this concept, more species and many more structural components (e.g., the distance of GIs from the origin of replication oriC, their relative time of acquisition, number of pseudogenes per island, and coding strand bias) can be taken into account and evaluated, enabling the construction of more sophisticated and more detailed structural models.
Overall in this analysis, we showed that GIs tend to fall within structural families with well-defined signatures when looking within certain lineage boundaries, but when the taxa resolution decreases, that is, looking at GIs across different species, universally distributed structural GI components emerge. Perhaps overall, GIs should be seen as a superfamily of mobile elements with unifying and variable structural features rather than a single, well-defined family.
Methods
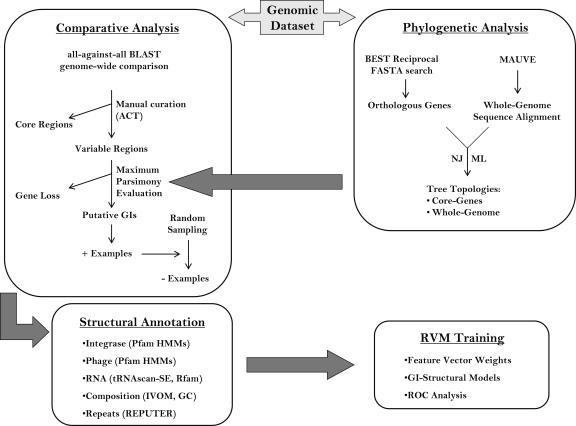
The methodology followed throughout this analysis is summarized as a flowchart in Figure 5 and described in the following sections.
Figure 5.
Flowchart summarizing the major steps in the methodology followed throughout this analysis: A phylogenetic analysis using both whole-genome sequence (if applicable) and the amino acid sequence of the core gene products was carried out enabling the construction of the reference tree topology for each genus. In a second step, a comparative analysis (genomewise) was performed between the chromosomes of each genus and the corresponding outgroups, leading to the identification of regions with limited phylogenetic distribution. In a third step, a maximum parsimony model (based on the reference tree topology) was applied in order to differentiate gene gain from gene loss events and exclude regions with limited phylogenetic distribution due to a gene loss event. The remaining regions formed the positive control data set (i.e., putative GIs) of this analysis. The negative control data set (i.e., non-GIs) was built implementing a random sampling approach, sampling regions only within the inter-GI parts of the chromosome; both positive and negative examples were annotated structurally. In a final step, the structural features of each region were used as input vectors to a machine learning method (RVM) leading to the construction of structural GI models.
Genomic data set
A list of all the 49 strains used in this comparative analysis is provided in Table 4. Throughout this analysis, we focused on the analysis of 37 reference bacterial strains from three different genera, namely, Salmonella, Staphylococcus, and Streptococcus. In order to differentiate a limited phylogenetic distribution pattern due to a gene gain or a gene loss event (under a maximum parsimony evaluation), 12 more distantly related bacterial strains that formed outgroups for the three reference genera were also included in this analysis. The 12 outgroup genomes were used only in the maximum parsimony evaluation of the predicted regions and do not form part of the actual data set for which the data were produced. Briefly, 11 Salmonella strains with four outgroups (Escherichia coli, Shigella), 13 Staphylococcus strains with four outgroups (Bacillus, Listeria), and 13 Streptococcus strains with four outgroups (Lactobacillus, Lactococcus, Enterococcus) were analyzed.
Table 4.
The list of 49 strains used in this comparative analysis
Best reciprocal FASTA
For each of the three genera, all genomes were (pairwise) compared against the other including the four outgroups. In order to infer the orthologous genes in each pair of genomes compared, we applied a best reciprocal FASTA method as implemented in Vernikos et al. (2007) (for details, see Supplemental material). Overall, 1952, 741, and 429 orthologous genes were identified in the Salmonella, Staphylococcus, and Streptococcus data sets (including the corresponding four outgroups), respectively (Supplemental Fig. S7).
Phylogenetic analysis
For the construction of the reference tree topology, modules of the PHYLIP package version 3.65 (Felsenstein 1989) were implemented. More specifically, for the whole-genome sequence alignments (Salmonella and Staphylococcus data sets), the DNADIST module with the gamma-based method for correcting the rate heterogeneity among sites was used. We also used the NEIGHBOR module, which implements the Neighbor-Joining (NJ) method (Saitou and Nei 1987), and the DNAML module, which implements the Maximum Likelihood (ML) method for DNA sequences (Felsenstein and Churchill 1996). For the construction of tree topologies using the amino acid sequence alignment of the core gene products for each genus (and the corresponding outgroups), we used the PROTDIST, NEIGHBOR, and PROML modules of PHYLIP. Different tree topologies for a given lineage were evaluated further through the PROML and TREE-PUZZLE (Schmidt et al. 2002) methods, exploiting the model with the highest number of parameters; for each genus, the tree topology with the highest likelihood was selected as the reference one. All the parameters were determined from the data using the TREE-PUZZLE software. Tree topologies were drawn using the TREEVIEW software (Page 1996).
Comparative analysis—GI detection
The genomic sequences of each genus and the corresponding outgroups were compared using a genome-wide, all-against-all BLAST (Altschul et al. 1997) comparison; the results were visualized through ACT (Carver et al. 2005) and manually inspected. Genomic regions (≥2 CDSs) of limited phylogenetic distribution that were present in some of the strains while being absent from the rest were processed further (at this stage, core genomic regions shared by all strains were excluded). In a second step, regions of limited phylogenetic distribution were analyzed applying a maximum parsimony model, in order to differentiate gene gain (HGT) from gene loss; the maximum parsimony model is based on the reference tree topology of each genus (Supplemental Figs. S8–S10). The comparative based approach followed to identify GIs assuming a parsimony model has been described previously (Vernikos et al. 2007). Briefly, a genomic region that shows limited phylogenetic distribution that can more likely be explained by a gene gain rather than a gene loss event, based on a reference tree topology, is considered to be a putative GI. For example, a region that is present in the Salmonella lineage and absent from E. coli MG1655 might well be either a true HGT in the former or deletion in the latter. However, if, for example, the same region is also present in E. coli EDL933 and E. coli CFT073, then we can infer more reliably that this event represents probably a deletion (in E. coli MG1655) rather a true HGT in the Salmonella lineage. Conversely, a sequence that is confined to one lineage is more likely to have been horizontally acquired than to have been deleted independently from multiple lineages (Lawrence and Ochman 1998). Genomic regions identified under this framework formed the positive control set of this analysis; overall, 331 putative GIs were sampled from the 37 reference chromosomes (Table 2; Supplemental Fig. S11).
Random sampling
For the construction of the negative control data set, that is, genomic regions that are not GIs, a random sampling approach was followed. For each genome with identified putative GIs, an equal number of non-GI regions was randomly sampled, sampling the size distribution of the corresponding genus-specific GIs (Supplemental Fig. S12). Overall, this analysis yielded 337 non-GIs, giving a total number of 668 training sets (Table 2; Supplemental Table 1). Random sampling was “forced” to occur only within inter-GI regions of each chromosome. Note that the results of the random sampling approach were manually curated, removing randomly sampled regions that had been already sampled from other chromosomes of different strains of the same genus; the manual curation filtered out any redundancy in the training set that could possibly affect the training and evaluation process. For these reasons, the numbers of positive and negative examples for each genus are slightly different.
Machine learning
In order to build structural models of GIs, eight features were taken into account: The IVOM score (relative entropy), insertion point (1 if within a CDS locus, 0 otherwise), size of each region (in base pairs), gene density (genes/kilobase), repeats (binary: 1 if present, 0 otherwise), phage-related protein domains (binary), integrase(-like) protein domains (binary), and non-coding RNA (binary). Furthermore, the RNA feature was further divided into tRNA and misc_RNA subcategories; the same applies for the repeats feature, which was further divided into DRs and IRs subcategories.
The aim of the machine learning in this analysis is dual: GI structural models will be trained in order to quantify (i.e., assign weights) the relative contribution of each feature to the GI structure, and in a second step, the derived models will be used to classify previously unseen examples (GIs and non-GIs), enabling evaluation of the generalization properties of each model and capturing of any potential variation in the GI structure. For this purpose, 668 training sets were used to train 11 GI models using a Biojava (http://www.biojava.org) implementation of the Relevance Vector Machine (RVM) (Tipping 2001).
The RVM is a method for sparse, Bayesian-based learning with applications in classification and regression analysis. RVM is a model of identical functional form to the well-known Support Vector Machine (SVM) that nonetheless overcomes a few of the disadvantages of the latter (Tipping 2001). RVM models exploit overall fewer basis functions relative to an SVM model, offering the advantage of increased sparsity, building simpler models with better generalization properties on unseen data. The RVM method has been previously applied in detecting binding sites in human protein-coding sequences (Down et al. 2006), in the identification of transcriptional start sites in mammalian DNA (Down and Hubbard 2002), and in a vertebrate gene finding method (Carter and Durbin 2006). For further details about technical aspects of the RVM method, refer to the Supplemental Material.
Further analysis
Details about the multiple sequence alignments, the structural annotation of GIs, the ROC curve analysis, and the cross-validation carried out in this study can be found in the Supplemental Material.
Acknowledgments
The authors thank Thomas Down and David Carter for providing the source code for the RVM training implementation and for making valuable suggestions regarding the RVM training; and Nicholas Thomson and Matthew Holden for discussion on aspects of the comparative analysis. This work was supported by the Wellcome Trust, and G.S.V. is supported by a Wellcome Trust Sanger Institute Ph.D. studentship.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.7004508
References
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang J., Zhang Z., Miller W., Lipman D.J., Zhang Z., Miller W., Lipman D.J., Miller W., Lipman D.J., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks D.J., Lei B., Musser J.M., Lei B., Musser J.M., Musser J.M. Prophage induction and expression of prophage-encoded virulence factors in group A Streptococcus serotype M3 strain MGAS315. Infect. Immun. 2003;71:7079–7086. doi: 10.1128/IAI.71.12.7079-7086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres S.B., Richter E.W., Nagiec M.J., Sumby P., Porcella S.F., DeLeo F.R., Musser J.M., Richter E.W., Nagiec M.J., Sumby P., Porcella S.F., DeLeo F.R., Musser J.M., Nagiec M.J., Sumby P., Porcella S.F., DeLeo F.R., Musser J.M., Sumby P., Porcella S.F., DeLeo F.R., Musser J.M., Porcella S.F., DeLeo F.R., Musser J.M., DeLeo F.R., Musser J.M., Musser J.M. Molecular genetic anatomy of inter- and intraserotype variation in the human bacterial pathogen group A Streptococcus. Proc. Natl. Acad. Sci. 2006;103:7059–7064. doi: 10.1073/pnas.0510279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner F.R., Plunkett G., Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Plunkett G., Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., Glasner J.D., Rode C.K., Mayhew G.F., Rode C.K., Mayhew G.F., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bolotin A., Wincker P., Mauger S., Jaillon O., Malarme K., Weissenbach J., Ehrlich S.D., Sorokin A., Wincker P., Mauger S., Jaillon O., Malarme K., Weissenbach J., Ehrlich S.D., Sorokin A., Mauger S., Jaillon O., Malarme K., Weissenbach J., Ehrlich S.D., Sorokin A., Jaillon O., Malarme K., Weissenbach J., Ehrlich S.D., Sorokin A., Malarme K., Weissenbach J., Ehrlich S.D., Sorokin A., Weissenbach J., Ehrlich S.D., Sorokin A., Ehrlich S.D., Sorokin A., Sorokin A. The complete genome sequence of the lactic acid bacterium Lactococcus lactis ssp. lactis IL1403. Genome Res. 2001;11:731–753. doi: 10.1101/gr.169701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A., Quinquis B., Renault P., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Quinquis B., Renault P., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Renault P., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Sorokin A., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Ehrlich S.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Lapidus A., Goltsman E., Mazur M., Pusch G.D., Goltsman E., Mazur M., Pusch G.D., Mazur M., Pusch G.D., Pusch G.D., et al. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 2004;22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broker G., Spellerberg B., Spellerberg B. Surface proteins of Streptococcus agalactiae and horizontal gene transfer. Int. J. Med. Microbiol. 2004;294:169–175. doi: 10.1016/j.ijmm.2004.06.018. [DOI] [PubMed] [Google Scholar]
- Broudy T.B., Pancholi V., Fischetti V.A., Pancholi V., Fischetti V.A., Fischetti V.A. Induction of lysogenic bacteriophage and phage-associated toxin from Group A Streptococci during coculture with human pharyngeal cells. Infect. Immun. 2001;69:1440–1443. doi: 10.1128/IAI.69.3.1440-1443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter D., Durbin R., Durbin R. Vertebrate gene finding from multiple-species alignments using a two-level strategy. Genome Biol. 2006;7:S6.1–S6.12. doi: 10.1186/gb-2006-7-s1-s6. (Suppl 1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver T.J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rajandream M.A., Barrell B.G., Parkhill J., Barrell B.G., Parkhill J., Parkhill J. ACT: The Artemis comparison tool. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- Censini S., Lange C., Xiang Z., Crabtree J.E., Ghiara P., Borodovsky M., Rappuoli R., Covacci A., Lange C., Xiang Z., Crabtree J.E., Ghiara P., Borodovsky M., Rappuoli R., Covacci A., Xiang Z., Crabtree J.E., Ghiara P., Borodovsky M., Rappuoli R., Covacci A., Crabtree J.E., Ghiara P., Borodovsky M., Rappuoli R., Covacci A., Ghiara P., Borodovsky M., Rappuoli R., Covacci A., Borodovsky M., Rappuoli R., Covacci A., Rappuoli R., Covacci A., Covacci A. cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. 1996;93:14648–14653. doi: 10.1073/pnas.93.25.14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Liou S.R., Plunkett G., Mayhew G.F., Rose D.J., Burland V., Kodoyianni V., Schwartz D.C., Blattner F.R., Liou S.R., Plunkett G., Mayhew G.F., Rose D.J., Burland V., Kodoyianni V., Schwartz D.C., Blattner F.R., Plunkett G., Mayhew G.F., Rose D.J., Burland V., Kodoyianni V., Schwartz D.C., Blattner F.R., Mayhew G.F., Rose D.J., Burland V., Kodoyianni V., Schwartz D.C., Blattner F.R., Rose D.J., Burland V., Kodoyianni V., Schwartz D.C., Blattner F.R., Burland V., Kodoyianni V., Schwartz D.C., Blattner F.R., Kodoyianni V., Schwartz D.C., Blattner F.R., Schwartz D.C., Blattner F.R., Blattner F.R. Comparative genomics of Salmonella enterica serovar Typhi strains Ty2 and CT18. J. Bacteriol. 2003;185:2330–2337. doi: 10.1128/JB.185.7.2330-2337.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep B.A., Gill S.R., Chang R.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Gill S.R., Chang R.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Chang R.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Phan T.H., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Chen J.H., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Davidson M.G., Lin F., Lin J., Carleton H.A., Mongodin E.F., Lin F., Lin J., Carleton H.A., Mongodin E.F., Lin J., Carleton H.A., Mongodin E.F., Carleton H.A., Mongodin E.F., Mongodin E.F., et al. Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet. 2006;367:731–739. doi: 10.1016/S0140-6736(06)68231-7. [DOI] [PubMed] [Google Scholar]
- Down T.A., Hubbard T.J., Hubbard T.J. Computational detection and location of transcription start sites in mammalian genomic DNA. Genome Res. 2002;12:458–461. doi: 10.1101/gr.216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down T., Leong B., Hubbard T.J., Leong B., Hubbard T.J., Hubbard T.J. A machine learning strategy to identify candidate binding sites in human protein-coding sequence. BMC Bioinformatics. 2006;7:419. doi: 10.1186/1471-2105-7-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Felsenstein J., Churchill G.A., Churchill G.A. A Hidden Markov Model approach to variation among sites in rate of evolution. Mol. Biol. Evol. 1996;13:93–104. doi: 10.1093/oxfordjournals.molbev.a025575. [DOI] [PubMed] [Google Scholar]
- Fischetti V.A. In vivo acquisition of prophage in Streptococcus pyogenes. Trends Microbiol. 2007;15:297–300. doi: 10.1016/j.tim.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Glaser P., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Frangeul L., Buchrieser C., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Buchrieser C., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Rusniok C., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Amend A., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Baquero F., Berche P., Bloecker H., Brandt P., Chakraborty T., Berche P., Bloecker H., Brandt P., Chakraborty T., Bloecker H., Brandt P., Chakraborty T., Brandt P., Chakraborty T., Chakraborty T., et al. Comparative genomics of Listeria species. Science. 2001;294:849–852. doi: 10.1126/science.1063447. [DOI] [PubMed] [Google Scholar]
- Glaser P., Rusniok C., Buchrieser C., Chevalier F., Frangeul L., Msadek T., Zouine M., Couve E., Lalioui L., Poyart C., Rusniok C., Buchrieser C., Chevalier F., Frangeul L., Msadek T., Zouine M., Couve E., Lalioui L., Poyart C., Buchrieser C., Chevalier F., Frangeul L., Msadek T., Zouine M., Couve E., Lalioui L., Poyart C., Chevalier F., Frangeul L., Msadek T., Zouine M., Couve E., Lalioui L., Poyart C., Frangeul L., Msadek T., Zouine M., Couve E., Lalioui L., Poyart C., Msadek T., Zouine M., Couve E., Lalioui L., Poyart C., Zouine M., Couve E., Lalioui L., Poyart C., Couve E., Lalioui L., Poyart C., Lalioui L., Poyart C., Poyart C., et al. Genome sequence of Streptococcus agalactiae, a pathogen causing invasive neonatal disease. Mol. Microbiol. 2002;45:1499–1513. doi: 10.1046/j.1365-2958.2002.03126.x. [DOI] [PubMed] [Google Scholar]
- Hacker J., Kaper J.B., Kaper J.B. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- Hacker J., Kaper J.B., Kaper J.B., editors. Pathogenicity islands and the evolution of pathogenic microbes. Springer; New York: 2002. Vol. 1. [Google Scholar]
- Hacker J., Blum-Oehler G., Muhldorfer I., Tschape H., Blum-Oehler G., Muhldorfer I., Tschape H., Muhldorfer I., Tschape H., Tschape H. Pathogenicity islands of virulent bacteria: Structure, function and impact on microbial evolution. Mol. Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- Herron-Olson L., Fitzgerald J.R., Musser J.M., Kapur V., Fitzgerald J.R., Musser J.M., Kapur V., Musser J.M., Kapur V., Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE. 2007;2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M.T., Feil E.J., Lindsay J.A., Peacock S.J., Day N.P., Enright M.C., Foster T.J., Moore C.E., Hurst L., Atkin R., Feil E.J., Lindsay J.A., Peacock S.J., Day N.P., Enright M.C., Foster T.J., Moore C.E., Hurst L., Atkin R., Lindsay J.A., Peacock S.J., Day N.P., Enright M.C., Foster T.J., Moore C.E., Hurst L., Atkin R., Peacock S.J., Day N.P., Enright M.C., Foster T.J., Moore C.E., Hurst L., Atkin R., Day N.P., Enright M.C., Foster T.J., Moore C.E., Hurst L., Atkin R., Enright M.C., Foster T.J., Moore C.E., Hurst L., Atkin R., Foster T.J., Moore C.E., Hurst L., Atkin R., Moore C.E., Hurst L., Atkin R., Hurst L., Atkin R., Atkin R., et al. Complete genomes of two clinical Staphylococcus aureus strains: Evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins J., Alborn W.E., Arnold J., Blaszczak L.C., Burgett S., DeHoff B.S., Estrem S.T., Fritz L., Fu D.J., Fuller W., Alborn W.E., Arnold J., Blaszczak L.C., Burgett S., DeHoff B.S., Estrem S.T., Fritz L., Fu D.J., Fuller W., Arnold J., Blaszczak L.C., Burgett S., DeHoff B.S., Estrem S.T., Fritz L., Fu D.J., Fuller W., Blaszczak L.C., Burgett S., DeHoff B.S., Estrem S.T., Fritz L., Fu D.J., Fuller W., Burgett S., DeHoff B.S., Estrem S.T., Fritz L., Fu D.J., Fuller W., DeHoff B.S., Estrem S.T., Fritz L., Fu D.J., Fuller W., Estrem S.T., Fritz L., Fu D.J., Fuller W., Fritz L., Fu D.J., Fuller W., Fu D.J., Fuller W., Fuller W., et al. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 2001;183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao W., Wan I., Jones S.J., Brinkman F.S., Wan I., Jones S.J., Brinkman F.S., Jones S.J., Brinkman F.S., Brinkman F.S. IslandPath: Aiding detection of genomic islands in prokaryotes. Bioinformatics. 2003;19:418–420. doi: 10.1093/bioinformatics/btg004. [DOI] [PubMed] [Google Scholar]
- Jin Q., Yuan Z., Xu J., Wang Y., Shen Y., Lu W., Wang J., Liu H., Yang J., Yang F., Yuan Z., Xu J., Wang Y., Shen Y., Lu W., Wang J., Liu H., Yang J., Yang F., Xu J., Wang Y., Shen Y., Lu W., Wang J., Liu H., Yang J., Yang F., Wang Y., Shen Y., Lu W., Wang J., Liu H., Yang J., Yang F., Shen Y., Lu W., Wang J., Liu H., Yang J., Yang F., Lu W., Wang J., Liu H., Yang J., Yang F., Wang J., Liu H., Yang J., Yang F., Liu H., Yang J., Yang F., Yang J., Yang F., Yang F., et al. Genome sequence of Shigella flexneri 2a: Insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 2002;30:4432–4441. doi: 10.1093/nar/gkf566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaper J.B., Hacker J., Hacker J. Pathogenicity islands and other mobile virulence elements. American Society for Microbiology Press; Washington, DC: 1999. [Google Scholar]
- Kleerebezem M., Boekhorst J., van Kranenburg R., Molenaar D., Kuipers O.P., Leer R., Tarchini R., Peters S.A., Sandbrink H.M., Fiers M.W., Boekhorst J., van Kranenburg R., Molenaar D., Kuipers O.P., Leer R., Tarchini R., Peters S.A., Sandbrink H.M., Fiers M.W., van Kranenburg R., Molenaar D., Kuipers O.P., Leer R., Tarchini R., Peters S.A., Sandbrink H.M., Fiers M.W., Molenaar D., Kuipers O.P., Leer R., Tarchini R., Peters S.A., Sandbrink H.M., Fiers M.W., Kuipers O.P., Leer R., Tarchini R., Peters S.A., Sandbrink H.M., Fiers M.W., Leer R., Tarchini R., Peters S.A., Sandbrink H.M., Fiers M.W., Tarchini R., Peters S.A., Sandbrink H.M., Fiers M.W., Peters S.A., Sandbrink H.M., Fiers M.W., Sandbrink H.M., Fiers M.W., Fiers M.W., et al. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Ogasawara N., Moszer I., Albertini A.M., Alloni G., Azevedo V., Bertero M.G., Bessieres P., Bolotin A., Borchert S., Ogasawara N., Moszer I., Albertini A.M., Alloni G., Azevedo V., Bertero M.G., Bessieres P., Bolotin A., Borchert S., Moszer I., Albertini A.M., Alloni G., Azevedo V., Bertero M.G., Bessieres P., Bolotin A., Borchert S., Albertini A.M., Alloni G., Azevedo V., Bertero M.G., Bessieres P., Bolotin A., Borchert S., Alloni G., Azevedo V., Bertero M.G., Bessieres P., Bolotin A., Borchert S., Azevedo V., Bertero M.G., Bessieres P., Bolotin A., Borchert S., Bertero M.G., Bessieres P., Bolotin A., Borchert S., Bessieres P., Bolotin A., Borchert S., Bolotin A., Borchert S., Borchert S., et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- Lawrence J., Ochman H., Ochman H. Amelioration of bacterial genomes: Rates of change and exchange. J. Mol. Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Lawrence J., Ochman H., Ochman H. Molecular archaeology of the Escherichia coli genome. Proc. Natl. Acad. Sci. 1998;95:9413–9417. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantri Y., Williams K.P., Williams K.P. Islander: A database of integrative islands in prokaryotic genomes, the associated integrases and their DNA site specificities. Nucleic Acids Res. 2004;32:D55–D58. doi: 10.1093/nar/gkh059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Sanderson K.E., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Spieth J., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Clifton S.W., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Latreille P., Courtney L., Porwollik S., Ali J., Dante M., Du F., Courtney L., Porwollik S., Ali J., Dante M., Du F., Porwollik S., Ali J., Dante M., Du F., Ali J., Dante M., Du F., Dante M., Du F., Du F., et al. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- McClelland M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Sanderson K.E., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Clifton S.W., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Latreille P., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Porwollik S., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Sabo A., Meyer R., Bieri T., Ozersky P., McLellan M., Meyer R., Bieri T., Ozersky P., McLellan M., Bieri T., Ozersky P., McLellan M., Ozersky P., McLellan M., McLellan M., et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat. Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- McGillivary G., Tomaras A.P., Rhodes E.R., Actis L.A., Tomaras A.P., Rhodes E.R., Actis L.A., Rhodes E.R., Actis L.A., Actis L.A. Cloning and sequencing of a genomic island found in the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Infect. Immun. 2005;73:1927–1938. doi: 10.1128/IAI.73.4.1927-1938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R.P., Subedi A., Subedi A. The SaPIs: Mobile pathogenicity islands of Staphylococcus. Chem. Immunol. Allergy. 2007;93:42–57. doi: 10.1159/000100857. [DOI] [PubMed] [Google Scholar]
- Page R.D. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Parkhill J., Dougan G., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Dougan G., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., James K.D., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Thomson N.R., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Pickard D., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Wain J., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Churcher C., Mungall K.L., Bentley S.D., Holden M.T., Mungall K.L., Bentley S.D., Holden M.T., Bentley S.D., Holden M.T., Holden M.T., et al. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature. 2001;413:848–852. doi: 10.1038/35101607. [DOI] [PubMed] [Google Scholar]
- Paulsen I.T., Banerjei L., Myers G.S., Nelson K.E., Seshadri R., Read T.D., Fouts D.E., Eisen J.A., Gill S.R., Heidelberg J.F., Banerjei L., Myers G.S., Nelson K.E., Seshadri R., Read T.D., Fouts D.E., Eisen J.A., Gill S.R., Heidelberg J.F., Myers G.S., Nelson K.E., Seshadri R., Read T.D., Fouts D.E., Eisen J.A., Gill S.R., Heidelberg J.F., Nelson K.E., Seshadri R., Read T.D., Fouts D.E., Eisen J.A., Gill S.R., Heidelberg J.F., Seshadri R., Read T.D., Fouts D.E., Eisen J.A., Gill S.R., Heidelberg J.F., Read T.D., Fouts D.E., Eisen J.A., Gill S.R., Heidelberg J.F., Fouts D.E., Eisen J.A., Gill S.R., Heidelberg J.F., Eisen J.A., Gill S.R., Heidelberg J.F., Gill S.R., Heidelberg J.F., Heidelberg J.F., et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Perna N.T., Plunkett G., Burland V., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., Plunkett G., Burland V., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., Burland V., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., Mau B., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., Glasner J.D., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., Rose D.J., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., Mayhew G.F., Evans P.S., Gregor J., Kirkpatrick H.A., Evans P.S., Gregor J., Kirkpatrick H.A., Gregor J., Kirkpatrick H.A., Kirkpatrick H.A., et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–533. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- Pridmore R.D., Berger B., Desiere F., Vilanova D., Barretto C., Pittet A.C., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., Berger B., Desiere F., Vilanova D., Barretto C., Pittet A.C., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., Desiere F., Vilanova D., Barretto C., Pittet A.C., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., Vilanova D., Barretto C., Pittet A.C., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., Barretto C., Pittet A.C., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., Pittet A.C., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., Zwahlen M.C., Rouvet M., Altermann E., Barrangou R., Rouvet M., Altermann E., Barrangou R., Altermann E., Barrangou R., Barrangou R., et al. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsoniiNCC 533. Proc. Natl. Acad. Sci. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden A.E., Mota L.J., Munter S., Shorte S.L., Holden D.W., Mota L.J., Munter S., Shorte S.L., Holden D.W., Munter S., Shorte S.L., Holden D.W., Shorte S.L., Holden D.W., Holden D.W. The SPI-2 type III secretion system restricts motility of Salmonella-containing vacuoles. Cell Microbiol. 2007;9:2517–2529. doi: 10.1111/j.1462-5822.2007.00977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosini R., Rinaudo C.D., Soriani M., Lauer P., Mora M., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Rinaudo C.D., Soriani M., Lauer P., Mora M., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Soriani M., Lauer P., Mora M., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Lauer P., Mora M., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Mora M., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Maione D., Taddei A., Santi I., Ghezzo C., Brettoni C., Taddei A., Santi I., Ghezzo C., Brettoni C., Santi I., Ghezzo C., Brettoni C., Ghezzo C., Brettoni C., Brettoni C., et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Schmidt H., Hensel M., Hensel M. Pathogenicity islands in bacterial pathogenesis. Clin. Microbiol. Rev. 2004;17:14–56. doi: 10.1128/CMR.17.1.14-56.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H.A., Strimmer K., Vingron M., von Haeseler A., Strimmer K., Vingron M., von Haeseler A., Vingron M., von Haeseler A., von Haeseler A. TREE-PUZZLE: Maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Takeuchi F., Watanabe S., Baba T., Yuzawa H., Ito T., Morimoto Y., Kuroda M., Cui L., Takahashi M., Ankai A., Watanabe S., Baba T., Yuzawa H., Ito T., Morimoto Y., Kuroda M., Cui L., Takahashi M., Ankai A., Baba T., Yuzawa H., Ito T., Morimoto Y., Kuroda M., Cui L., Takahashi M., Ankai A., Yuzawa H., Ito T., Morimoto Y., Kuroda M., Cui L., Takahashi M., Ankai A., Ito T., Morimoto Y., Kuroda M., Cui L., Takahashi M., Ankai A., Morimoto Y., Kuroda M., Cui L., Takahashi M., Ankai A., Kuroda M., Cui L., Takahashi M., Ankai A., Cui L., Takahashi M., Ankai A., Takahashi M., Ankai A., Ankai A., et al. Whole-genome sequencing of Staphylococcus haemolyticus uncovers the extreme plasticity of its genome and the evolution of human-colonizing Staphylococcal species. J. Bacteriol. 2005;187:7292–7308. doi: 10.1128/JB.187.21.7292-7308.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Nelson K.E., Paulsen I.T., Eisen J.A., Read T.D., Peterson S., Heidelberg J., DeBoy R.T., Haft D.H., Dodson R.J., Nelson K.E., Paulsen I.T., Eisen J.A., Read T.D., Peterson S., Heidelberg J., DeBoy R.T., Haft D.H., Dodson R.J., Paulsen I.T., Eisen J.A., Read T.D., Peterson S., Heidelberg J., DeBoy R.T., Haft D.H., Dodson R.J., Eisen J.A., Read T.D., Peterson S., Heidelberg J., DeBoy R.T., Haft D.H., Dodson R.J., Read T.D., Peterson S., Heidelberg J., DeBoy R.T., Haft D.H., Dodson R.J., Peterson S., Heidelberg J., DeBoy R.T., Haft D.H., Dodson R.J., Heidelberg J., DeBoy R.T., Haft D.H., Dodson R.J., DeBoy R.T., Haft D.H., Dodson R.J., Haft D.H., Dodson R.J., Dodson R.J., et al. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science. 2001;293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- Tettelin H., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., Masignani V., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., Cieslewicz M.J., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., Donati C., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., Medini D., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., Ward N.L., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., Angiuoli S.V., Crabtree J., Jones A.L., Durkin A.S., Crabtree J., Jones A.L., Durkin A.S., Jones A.L., Durkin A.S., Durkin A.S., et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome.”. Proc. Natl. Acad. Sci. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipping M.E. Sparse Bayesian learning and the relevance vector machine. J. Mach. Learn. Res. 2001;1:211–244. [Google Scholar]
- Towers R.J., Gal D., McMillan D., Sriprakash K.S., Currie B.J., Walker M.J., Chhatwal G.S., Fagan P.K., Gal D., McMillan D., Sriprakash K.S., Currie B.J., Walker M.J., Chhatwal G.S., Fagan P.K., McMillan D., Sriprakash K.S., Currie B.J., Walker M.J., Chhatwal G.S., Fagan P.K., Sriprakash K.S., Currie B.J., Walker M.J., Chhatwal G.S., Fagan P.K., Currie B.J., Walker M.J., Chhatwal G.S., Fagan P.K., Walker M.J., Chhatwal G.S., Fagan P.K., Chhatwal G.S., Fagan P.K., Fagan P.K. Fibronectin-binding protein gene recombination and horizontal transfer between group A and G Streptococci. J. Clin. Microbiol. 2004;42:5357–5361. doi: 10.1128/JCM.42.11.5357-5361.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos G.S., Parkhill J., Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- Vernikos G.S., Thomson N.R., Parkhill J., Thomson N.R., Parkhill J., Parkhill J. Genetic flux over time in the Salmonella lineage. Genome Biol. 2007;8:R100. doi: 10.1186/gb-2007-8-6-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse J.C., Russell R.R., Russell R.R. Dispensable genes and foreign DNA in Streptococcus mutans. Microbiol. 2006;152:1777–1788. doi: 10.1099/mic.0.28647-0. [DOI] [PubMed] [Google Scholar]
- Welch R.A., Burland V., Plunkett G., Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Burland V., Plunkett G., Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Plunkett G., Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Redford P., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Roesch P., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Rasko D., Buckles E.L., Liou S.R., Boutin A., Hackett J., Buckles E.L., Liou S.R., Boutin A., Hackett J., Liou S.R., Boutin A., Hackett J., Boutin A., Hackett J., Hackett J., et al. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. 2002;99:17020–17024. doi: 10.1073/pnas.252529799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K.P. Integration sites for genetic elements in prokaryotic tRNA and tmRNA genes: Sublocation preference of integrase subfamilies. Nucleic Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Radziejewska-Lebrecht J., Krajewska-Pietrasik D., Toivanen P., Skurnik M., Radziejewska-Lebrecht J., Krajewska-Pietrasik D., Toivanen P., Skurnik M., Krajewska-Pietrasik D., Toivanen P., Skurnik M., Toivanen P., Skurnik M., Skurnik M. Molecular and chemical characterization of the lipopolysaccharide O-antigen and its role in the virulence of Yersinia enterocolitica serotype O:8. Mol. Microbiol. 1997;23:63–76. doi: 10.1046/j.1365-2958.1997.1871558.x. [DOI] [PubMed] [Google Scholar]
- Zhang Y.Q., Ren S.X., Li H.L., Wang Y.X., Fu G., Yang J., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., Ren S.X., Li H.L., Wang Y.X., Fu G., Yang J., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., Li H.L., Wang Y.X., Fu G., Yang J., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., Wang Y.X., Fu G., Yang J., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., Fu G., Yang J., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., Yang J., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., Qin Z.Q., Miao Y.G., Wang W.Y., Chen R.S., Miao Y.G., Wang W.Y., Chen R.S., Wang W.Y., Chen R.S., Chen R.S., et al. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228) Mol. Microbiol. 2003;49:1577–1593. doi: 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]