Abstract
The Xist gene is expressed exclusively from the inactive X chromosome and plays a central role in regulating X chromosome inactivation. Here we describe experiments aimed at defining the extent of the active chromatin domain of the expressed Xist allele. By using an allele-specific general DNaseI sensitivity assay we show that there is preferential digestion of the expressed allele at sites within the transcribed locus but not in flanking sites located up to 70 kb 5′. A putative proximal boundary for the Xist domain is located within 10 kb upstream of promoter P1. Chromatin in the expressed domain was found to be acetylated at H4 in XX somatic cells but also in XY cells, where Xist is never expressed. A single clear exception to this was the Xist promoter, which is acetylated only in XX cells. These observations concur with the view that H4 acetylation may not be a general marker of active chromatin domains and further support data implicating local promoter acetylation as being of primary functional significance in vivo.
In female mammals, dosage compensation is achieved by the transcriptional silencing of one of the two X chromosomes, a process known as X inactivation (1). Features of the inactive X chromosome (Xi) that distinguish it from its active counterpart (Xa) are that it is condensed during interphase, replicates late in S phase, and is underacetylated at histones H3 and H4 (reviewed in ref. 2). More recently it has been shown that chromatin on Xi is highly enriched for the variant histone macroH2A (3).
The classically defined X inactivation center (Xic) is required both for initiation of X inactivation in early development and for propagation of the inactivation signal in cis (reviewed in ref. 4). Initiation of random X inactivation involves determining how many (counting) and which (choosing) X chromosome to inactivate. It has been suggested that this is achieved by cells blocking a single Xic and thus marking that chromosome as the active X chromosome. X inactivation then proceeds in cis from unblocked Xics at the onset of cellular differentiation (5).
The X inactive specific transcript (Xist) gene, originally identified as a candidate for the Xic (6–9), produces a large RNA with no apparent protein coding potential (10, 11). Xist RNA “coats” the inactive X chromosome domain in the interphase nucleus, suggesting that it provides the primary signal for in cis propagation of X inactivation (11, 12). A requirement for Xist in propagation of X inactivation in cis has been demonstrated by using targeted deletion of transcribed regions (13, 14). Importantly, the counting function of the Xic was unaffected in these experiments. A subsequent gene-targeting experiment indicated that sequences distal to Xist are important in counting (15). Analysis of XY embryonic stem (ES) cells bearing a 450-kb Xist yeast artificial chromosome transgene demonstrated that Xist is sufficient for X inactivation (16) and that transgenic loci recapitulate both counting and propagation functions (16, 17). This result was subsequently shown by using a much smaller 35-kb transgenic Xist construct that encompasses Xist, 9 kb of upstream sequence, and 6 kb of downstream sequence (18).
It has recently been shown that developmental up-regulation of Xist on the inactive Xi allele is attributable to RNA stabilization (19, 20) and that this in turn results from a developmentally regulated promoter switch (21). An upstream promoter (P0) transcribes an unstable isoform of Xist RNA before X inactivation. As cells differentiate, there is a switch to downstream promoters (P1, P2) and cis accumulation of stable Xist RNA on Xi. The Xa allele continues to transcribe unstable RNA from P0 for a short period and is then transcriptionally silenced.
Here we describe experiments directed toward defining the chromatin domain structure of the Xist locus by assaying both general nuclease sensitivity of Xi and Xa Xist alleles and by determining histone acetylation levels in XY and XX cells. The study was undertaken to identify boundaries at which there is a transition from the expressed Xist locus to flanking silent inactive X chromatin. Such boundaries could theoretically be important for insulating the Xist locus. In addition, we wished to define the probable maximal region encompassing regulatory elements required for Xist expression in XX somatic cells. Results from the DNase1 sensitivity assay indicate that the expressed locus lies in a relatively compact domain with a proximal limit located within 10 kb upstream of promoter P1. Analysis of H4 acetylation levels is consistent with acetylation/deacetylation playing a role in regulating initiation of transcription but not in defining the active chromatin domain.
MATERIALS AND METHODS
Chromatin Acetylation Assay.
Chromatin acetylation assays were carried out as described (22). In all experiments, we used an antibody mixture with specificity to highly acetylated H4 isoforms.
The following modifications were used to isolate nuclei from mouse tissue; thymus from 6- to 8-week-old BALB/c mice were removed into ice-cold PBS containing 5 mM sodium butyrate (Na butyrate) and gently teased apart to release cells. Cell pellets were resuspended in 10 ml of ice-cold 0.32 M sucrose/5 mM MgCl2/10 mM Tris⋅HCl, pH 7.5/0.2 mM PMSF/5 mM Na butyrate and the nuclei released by Dounce homogenization. Homogenates were centrifuged at 1,000 × g (10 minutes, 4°C), and the pellet was resuspended in 5 ml of ice-cold 2.2 M sucrose/5 mM MgCl2/10 mM Tris⋅HCl, pH 7.5/0.2 mM PMSF/5 mM Na butyrate and layered onto 5 ml of the same sucrose solution. Nuclei were pelleted at 50,000 × g (1 hr, 4°C) and resuspended in 1 ml of digestion buffer (0.32 M sucrose/50 mM Tris⋅HCl, pH 7.5/4 mM MgCl2/1 mM CaCl2/5 mM Na butyrate/0.1 mM PMSF).
DNA Probes.
GPT1 is a 1-kb XbaI fragment located 3 kb downstream of Xist exon VII. MP1 is a 500-bp PCR fragment spanning the P1 promoter. HP is a 1-kb HindIII–PstI fragment 2 kb upstream of P1; HH1.5 is a 1.5-kb HindIII fragment 3 kb upstream; 2.1(2)P is a 2-kb PstI fragment 17 kb upstream; E(2.3-1) is a 1-kb EcoRI fragment 20 kb upstream; 19E(2) is 2-kb EcoRI fragment 40 kb upstream, and E55(13) and E55(16) are EcoRI subclones (700 bp and 2 kb) located ≈70 kb upstream. 33H(2) is a 2-kb HindIII genomic fragment located between the E55(16) probe and DXCrc318. Other probes are as described; Xist cDNA clones mXist1, w5i and w7d (8, 10); NM18B (23); Cdx4 exon 3 region (24); Tsx (25); actin cDNA clone (26); R198 mouse minor satellite probe (27); DXSmh141 probe (28).
DNase1 Sensitivity Assay.
Livers of adult mice were removed and immediately Dounce-homogenized in 20–40 ml of ice-cold buffer S [buffer A (15 mM Tris⋅HCl, pH 7.5/15 mM NaCl/60 mM KCl/0.5 mM spermidine/0.15 mM spermine) containing 0.3 M sucrose/0.5 mM EGTA/2 mM EDTA/0.5 mM 2-mercaptoethanol] to make a single-cell suspension. Further homogenization using a tighter fitting pestle was carried out to release nuclei. The homogenate was centrifuged at 1,000 × g, (10 min, 4°C), and the pellet was gently resuspended in 5 ml of buffer S. An additional 5 ml of buffer S was added, followed by 10 ml of buffer S containing 0.2% (vol/vol) Triton X-100 and finally, 200 μl of 0.1 M PMSF. The nuclei were mixed and permeabilized on ice for 1–3 min before pelleting at 1,000 × g (10 min, 4°C). The pellet was then washed in 20 ml of buffer S followed by a 5-min spin to give a clean white pellet. Finally the nuclei were resuspended in 3 ml of buffer E (buffer A containing 0.5 mM EGTA/0.5 mM 2-mercaptoethanol).
DNase I digestions were carried out on ice with 1.2 × 107 nuclei and a range of enzyme concentrations. Buffer E and MgCl2 (to 5 mM) was added to each digest to give a volume of 480 μl. Digestion was initiated on addition of nuclei, and after 10 min was stopped by the addition of 20 μl of 0.5 M EDTA (pH 8). Each digest was then transferred to tubes containing 4.5 ml of lysis solution (20 mM EDTA/20 mM Tris⋅HCl, pH 8/1.1% SDS/proteinase K at 200 μg/ml) and incubated at 50°C overnight. The next day, the DNA was extracted with phenol/chloroform, ethanol precipitated, and resuspended in 0.5 ml of TE (10 mM Tris⋅HCl, pH8/1 mM EDTA, pH 8).
Single-Nucleotide Primer Extension (SNuPE) Assay.
SNuPE assays were performed as described (13, 29). Novel polymorphisms were identified by sequencing of PCR products and were subsequently checked in trial SNuPE reactions. Optimal conditions were established as follows; U2af1-rs1-flanking primers GAT CAG ACA TAC TCG GAT A and TGT GGT ACG GCC AGC CTA TG (270-bp product), SNuPE primer TAA CTG CAC AGG CCA GCT GT at 94°C 1 min; 65°C 1 min and 72°C 1 min (polymorphism; C57BL6, G; Mus spretus, A), Xist exon I-flanking primers CAA GGT GGA TTG ACT GTG A and GAG TTA CTT GAA CAT CCT CC (900-bp product), SNuPE primer TCT GTG GAA GAT CAG TGC A, 94°C 1 min; 56°C 1 min and 72°C 1 min (polymorphism; Mus domesticus, A; PGK, G). Xist exon V-flanking primers ACG ATC CCT AGG TGG AGA TG and GCA TGA GTA GGG TAC AGT (400-bp product), SNuPE primer GGT TCT CTC AGA AGC TAG GA, 94°C 1 min; 56°C 1 min and 72°C (polymorphism M. domesticus, A; PGK, G). Xist promoter P1-flanking primers CAT GGC TGG AGC AAG and TAT GGA GTC ACC AGG TTC CCA G (400-bp product), SNuPE primer GGT CCA ATA GAT GTC AGA 94°C 1 min; 62°C 1 min and 72°C 1 min (polymorphism M. domesticus, A; PGK, C). Xist 10-kb upstream-flanking primers GTA GAC CAG ACT GGG AAT CAG AAA and CAA GTA GGC CAA TCA ATA CC (400-bp product), SNuPE primer GTA AGT TCC AGA TCA GCC, 94°C 1 min; 50°C 1 min and 72°C 1 min (polymorphism M. domesticus, A; PGK, G). Xist 20-kb upstream-flanking primers TTC GGA TCT TCC TCT CCT ATA CAG and GTC CCT CAT CCT GCT GGT TT (300-bp product), SNuPE primer AAT GGA CAG AAG GGG TTA, 94°C 1 min; 58°C 1 min and 72°C 1 min (polymorphism M. domesticus, A; PGK, G). Xist 70-kb upstream-flanking primers TCG GAT CAG CCT GAC TGG TTG and GAG TGG GTT CTC TTA CAG (300-bp product), SNuPE primer TAC AAT ACA TGC TTC CTG G, 94°C 1 min; 58°C 1 min and 72°C 1 min (polymorphism M. domesticus, A; PGK, C). Pgk-1 SNuPE was carried out by using the previously described SNuPE primer and conditions (29) on a 700-bp PCR product generated from genomic DNA by using the flanking primers TTA AAG CTG AGC CCG GCC AAA A and GTC AGT TCC ATA CCA CTA AAC.
SNuPE reactions were quantitated on a PhosphorImager. Data was calculated as mean values for duplicate loadings after normalization for differences in specific activity of nucleotides by using F1 genomic DNA as a control. For newly described polymorphisms, we tested artificial mixes of genomic DNA (ranging from 1:9 to 9:1) to ensure linearity.
RESULTS
Allele-Specific DNaseI Sensitivity Assay.
Active chromatin domains are defined as regions encompassing genes that exhibit general sensitivity to nucleases relative to flanking sequences (30). To assess the extent of the active domain of the expressed Xist locus, we established an assay based on single-nucleotide primer extension (SNuPE) (29). The rationale behind the assay is to PCR-amplify regions spanning single-base polymorphisms from genomic DNA isolated from DNaseI-treated nuclei and to then use SNuPE to quantitate allelic ratios. If a given allele is more DNaseI-sensitive within the region defined by flanking primers, then it should be proportionately underrepresented in the resultant PCR products. This approach provides distinct advantages compared with methods that are based on measuring the relative rate of disappearance of polymorphic restriction-length fragments (31–34). Specifically, the SNuPE assay requires only single-base allelic differences, which can be identified more readily than polymorphic restriction-length fragments. In addition, fragment length differences do not need to be accounted for in assessing relative nuclease sensitivity.
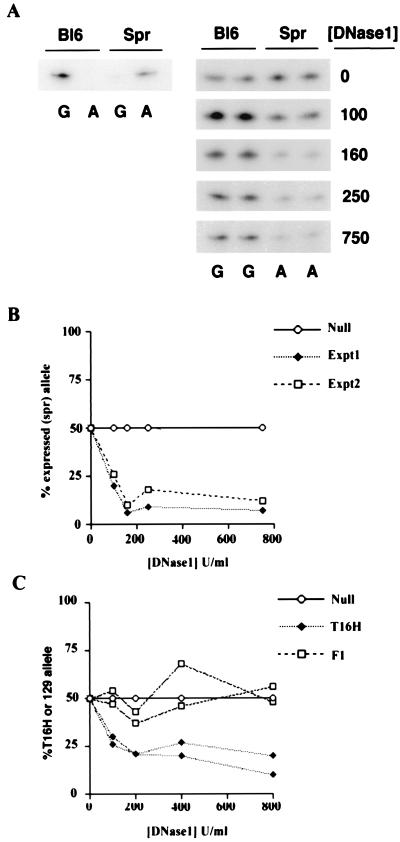
To test the assay system, we first analyzed a region of the imprinted U2af1-rs1 locus for which general DNase1 sensitivity assays based on digestion of polymorphic restriction fragments demonstrated ≈10-fold greater sensitivity of the paternal (expressed) allele relative to the maternal (silent) allele (34). SNuPE analysis was carried out by using a polymorphism between C57BL/6 (Bl6) and M. spretus located within the 5′-untranslated region of U2af1-rs1. A representative experiment analyzing DNase1-treated nuclei from (BL6 × M. spretus) F1 mice is illustrated in Fig. 1A. Relative levels of the expressed allele (M. spretus) are seen to decrease rapidly with increasing concentrations of DNase1. Quantitation of the above data and a second independent experiment is illustrated in Fig. 1B. In the concentration range of 160–750 units/ml DNase1, there is an ≈10-fold difference in the levels of the two alleles, consistent with previously reported data (34).
Figure 1.
Allele-specific DNase1 sensitivity assay. (A) Left, autoradiograph illustrating SNuPE reactions detecting polymorphism between Bl6 and M. spretus (Spr) U2af-rs1 alleles (G and A, respectively). Right, autoradiographs illustrating relative levels of alleles in (C57BL6 × M. spretus) F1 DNA prepared from nuclei treated with various concentrations of DNase1. SNuPE reactions were loaded in duplicate. (B) Quantitation of data shown above (Expt1) and an independent experiment (Expt2) showing expressed allele as % of total signal. The result expected if both alleles exhibit equal DNase1 sensitivity (Null) is included for illustrative purposes. (C) Quantitation of two independent allele-specific DNase1 sensitivity assays for the Pgk-1 locus. Data is shown for (PGK × T16H) F1 female (T16H), and for control (PGK × 129) F1 female (F1). Data points in B and C are mean values of duplicate loadings.
We went on to assess relative DNaseI sensitivity of expressed and silent alleles of the X chromosome-linked Pgk-1 gene on Xa and Xi. To do this, we analyzed DNaseI-treated nuclei from XX female T(X;16)16H (T16H) × C3H.pgk1a (PGK) F1 females. T16H causes complete nonrandom X inactivation of the PGK strain X chromosome. As a control, we analyzed nuclei from (129 × PGK) F1 female animals, which undergo normal random X inactivation. For SNuPE analysis, we used a defined polymorphism between PGK and standard laboratory strains (29). The results obtained from independent experiments by using separate nuclear preparations are illustrated in Fig. 1C. As anticipated, the expressed (T16H) allele exhibited greater DNaseI sensitivity compared with the silent (PGK) allele, and this difference was not detectable in the control (129 × PGK) F1 nuclei. The allelic sensitivity difference was ≈2–3 fold, less than that seen for U2af1-rs1. Importantly, the result was highly reproducible in independent determinations.
DNaseI Sensitivity of the Xist Domain.
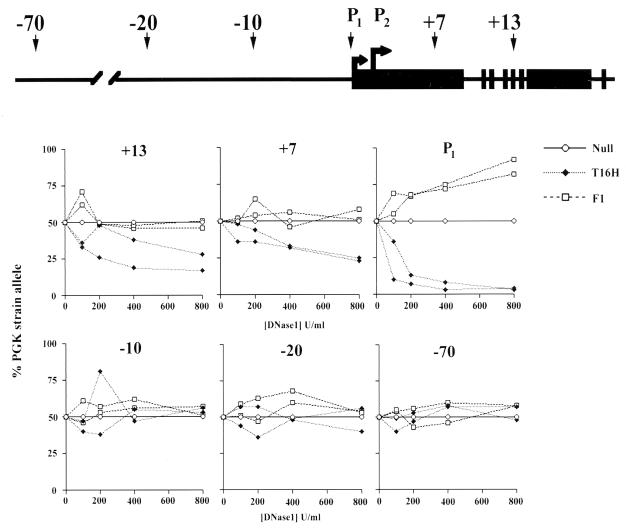
We went on to identify SNuPE polymorphisms between PGK and T16H within the Xist locus and flanking regions. Results from two independent determinations are illustrated in Fig. 2. Polymorphisms located in exon V (+13) and exon I (+7) of the gene revealed greater sensitivity of the expressed (PGK strain) allele. In control (129 × PGK) F1 female animals, both alleles exhibit similar levels of DNaseI sensitivity. The difference in sensitivity of expressed and silent alleles was similar to that seen at the Pgk-1 locus, i.e., ≈2–3 fold and again was reproducible in independent experiments.
Figure 2.
Allele-specific sensitivity of expressed and silent Xist loci. The location of polymorphisms within the transcribed locus and at various sites upstream are shown on the diagram above. DNase1-sensitivity assay results from two independent experiments are shown for each polymorphism. Both (PGK × T16H) F1 female (T16H) and (PGK × 129) F1 female controls (F1) were analyzed. Data points are mean values calculated from duplicate loadings. Null represents expected result if allelic sensitivities are equivalent.
We also analyzed a polymorphism close to promoter P1. In this case, we observed a very strong difference in sensitivity (Fig. 2C). This likely reflects the fact that the polymorphism is in close proximity to a DNase1 hypersensitive site located over promoter P1 (35). In control (129 × PGK) F1 animals we observed the opposite effect (i.e., greater sensitivity of the 129 allele), albeit to a lesser extent. The reason for this may be that the large differences in allelic sensitivity amplify skewed X inactivation patterns attributable to different Xce alleles on 129 and PGK-derived X chromosomes. Theoretically, this effect should be more apparent where there are large differences in allelic sensitivity, e.g., DNase1 hypersensitive sites, compared with regions with relatively mild general DNase1 sensitivity differences.
In contrast to the above, polymorphisms upstream of P1 (located at approximately −10, −20, and −70 kb) showed no significant difference in relative allelic sensitivity. Thus, analysis of general DNaseI sensitivity suggests that the domain of the expressed Xist locus on the inactive X chromosome extends no farther than 10 kb upstream. We were unable to define usable SNuPE polymorphisms in the region downstream of Xist and therefore cannot determine the 3′ boundary of the Xist domain by using this approach (but see Discussion).
Acetylation Analysis of the Xist Locus.
We went on to analyze patterns of histone H4 acetylation at the Xist locus by using chromatin immunoprecipitation as described by O’Neill and Turner (22). To distinguish the contribution of the two alleles in XX cells, we compared results for XX and XY cells. As males have only an active X (silent Xist), the acetylation status of the expressed allele in XX cells can be indirectly inferred.
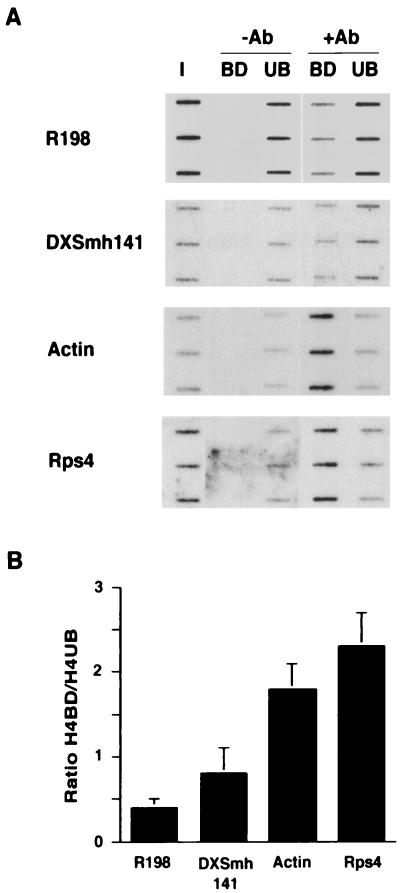
Control experiments were carried out by using probes for heterochromatic and genic regions (Fig. 3A). The R198 probe, derived from a minor satellite repeat within M. musculus centromeres (27), hybridizes predominantly to the antibody unbound (hypoacetylated) chromatin fraction. This result is consistent with previously published cytogenetic and immunoprecipitation analyses (22, 36). The DXSmh141 probe, corresponding to a long complex repeat unit sequence island located in the Giemsa-positive A3 band on the mouse X chromosome (28, 37), also hybridizes predominantly to the antibody unbound (hypoacetylated) chromatin fraction. This observation concurs with cytogenetic analysis of histone H4 acetylation levels that suggest that late-replicating Giemsa-negative bands are hypoacetylated (36). In contrast to heterochromatin probes, genic probes for the Actin gene and the X linked Rps4 gene both hybridize predominantly to the antibody-bound (acetylated) chromatin fraction. Fig. 3B illustrates quantified data from three independent determinations expressed as the ratio of bound to unbound signal. A ratio of <1 was obtained for the heterochromatin probes whereas genic probes give a ratio of >1.5.
Figure 3.
Histone acetylation assay. (A) Hybridization of heterochromatin and genic probes to slot blots with triplicate loading of ≈100 ng of input DNA (I) or DNA from antibody-bound (BD) and -unbound (UB) chromatin fractions without (−Ab) or with (+Ab) antibody to hyperacetylated histone H4 isoforms. (B) Ratio of signal for H4-bound/H4-unbound fractions calculated from three independent determinations. Slot blot data was quantified by using PhosphorImager analysis.
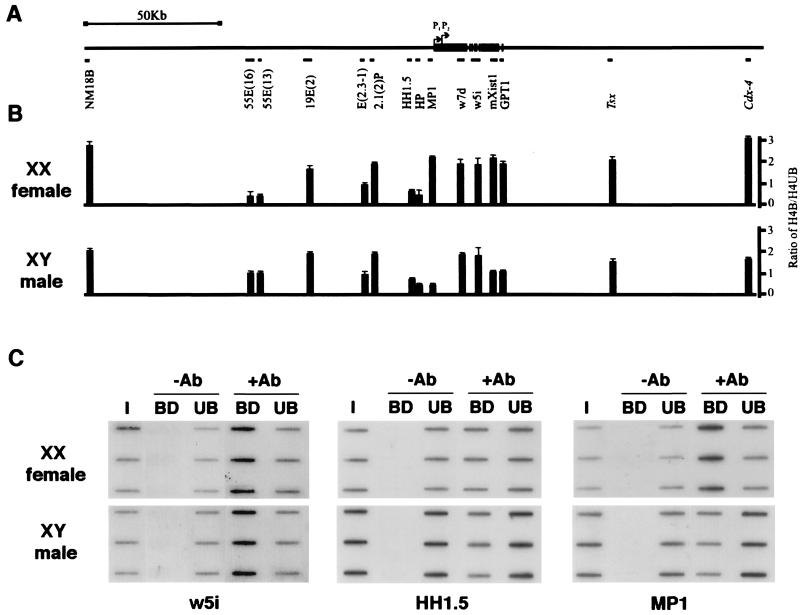
We next analyzed a series of probes derived from across the Xist locus and in flanking 3′ and 5′ regions (Fig. 4A). Probes were hybridized to immunoprecipitated chromatin from XX female and XY male animals. Quantified results obtained from at least two independent determinations are shown in Fig. 4B. Selected examples are illustrated in Fig. 4C.
Figure 4.
Chromatin acetylation profile of the Xist locus. (A) Diagram illustrating the location of single-copy probes relative to the Xist gene. (B) Ratio of H4-bound/H4-unbound chromatin in XY male and XX female cells. Each value represents the mean of at least two independent experiments. (C) Examples of immunoprecipitations of XX female and XY male chromatin probed with w5i, MP1, and HH1.5 showing triplicate loadings of input DNA (I), and antibody-bound (BD) and -unbound (UB) fractions either in the absence (−) or presence (+) of specific antibody (Ab).
In XX female cells, probes within the Xist domain detected similar levels of acetylation compared with the control Actin and Rps4 genic probes shown in Fig. 3B. However, much of the domain was also acetylated to this level in XY somatic cells where Xist is never expressed (Fig. 4B, and see for example, w5i in Fig. 4C). Thus, acetylated chromatin is not specifically associated with the domain of the expressed locus. Acetylation patterns in flanking regions were variable but generally similar in male and female cells. Probes HP, HH1.5, 55E(16), and 55E(13) hybridize to hypoacetylated chromatin, most notably in XX cells (Fig. 4B and for example HH1.5 in Fig. 4C). The low levels of acetylation are similar to constitutive heterochromatin (Fig. 3B). Probes NM18B, 19E(2), 2.1(2)P, Tsx, and Cdx4, on the other hand, detect acetylated chromatin both in XY and XX cells (Fig. 4B). The NM18B probe corresponds to a CpG-rich island (23), but an associated gene has not been identified to date. Both Tsx and Cdx4 probes correspond to genes that are not expressed in the cell type analyzed (24, 25). There are no known genes associated with the other loci. Thus, as is the case for the Xist domain, acetylation patterns do not appear to correlate with the domains of active transcription units.
Importantly, and in contrast to the aforementioned results, the Xist promoter probe MP1 hybridized to acetylated chromatin in XX cells but to hypoacetylated chromatin in XY cells (see Fig. 4 B and C). Thus, in this localized region, there appears to be a good correlation between H4 acetylation and expression. Presumably, the acetylated chromatin in XX cells derives specifically from the expressed allele, although this has not been formally demonstrated.
DISCUSSION
One aim of this study was to identify putative boundaries between the expressed Xist locus and inactive X chromatin. Our analysis indicates that the upstream boundary lies within 10 kb of promoter P1. Based on this observation, we suggest that regulatory elements involved in maintaining expression from somatic cell promoters P1 (and also the recently identified promoter P2; ref. 21), lie within or downstream of this region. This conclusion is consistent with transgenic studies, which indicate that sequences within 9 kb upstream and 5 kb downstream of Xist are sufficient to establish appropriate expression patterns (18).
We were unable to extend our analysis to define a boundary downstream of Xist. Previous studies have shown that the Brx locus, located 65 kb downstream, is subject to normal X inactivation (38). On this basis, we would anticipate a transition from the expressed Xist domain and the silent Brx domain somewhere within this span. Interestingly, deletion of the 65-kb span in ES cells does not affect cis-inactivation of Brx (15). This result suggests that at least in the distal region, a defined boundary with insulator properties is not required to separate the expressed Xist locus and cis-inactivated material (15).
Our analysis revealed heterogeneity in general nuclease sensitivity differences ranging from 2- to 3-fold for the Xist and Pgk-1 loci to ≈10-fold for U2af1-rs1. The significance of this is not clear, although it may reflect a unique chromatin configuration for the inactive X chromosome. Previous studies have also revealed relatively mild nuclease-sensitivity differences for active and inactive X chromosome alleles (31–33).
Based on both cytogenetic evidence (36) and immunoprecipitation analysis (22), it has been suggested that histone acetylation acts as a marker by which the genome is partitioned into coding and noncoding regions. Our data demonstrating hypoacetylation of the DXSmh141 repeat sequence island located in a G dark interstitial chromosome band lends further support to this view. Nevertheless, it remains unclear how acetylation levels vary at the level of individual genes. Analysis of chromatin acetylation at the chicken β-globin locus demonstrated hyperacetylation comapping with the domain of the active locus as defined by general DNase1 sensitivity (39). In contrast to this result, O’Neill and Turner (22) have demonstrated that acetylation levels are similar at a number of loci regardless of expression status. Our results indicate that for Xist, and also at flanking loci, acetylation levels do not correlate with active chromatin domains and are therefore in agreement with findings in the latter study. However, it should be taken into account that some of the flanking probes used are not fully characterized in terms of being genic or nongenic and also that the probes used do not provide universal coverage of the entire region of interest.
Although we did not find a general correlation between acetylation levels and expression status, we did observe a correlation specifically within the promoter region of Xist. This result is consistent with a number of biochemical studies that have highlighted the likely importance of histone acetylation in the initiation of transcription (reviewed in ref. 40). A recent study of Gcn5p-related histone hyperacetylation of target genes in yeast has provided clear in vivo evidence that promoter-specific acetylation is linked to transcription initiation (41). Conversely, it is possible that hypoacetylation of the silent P1 in XY cells is functionally more relevant. In this respect, it is interesting to note that the P1 region is known to be methylated on the silent Xist allele (42) and that recent evidence has linked methylation with the recruitment of histone deacetylase complexes (43, 44).
Results presented here indicate that the Xist promoters could provide a useful in vivo model to study promoter acetylation. An analysis of acetylation levels of Xist in ES cells is the subject of an independent study (45). In further studies, it will be of interest to determine acetylation levels of individual alleles in XX somatic cells, and to this end it should be possible to adapt the SNuPE assay to assess the allelic contribution in immunoprecipitated H4 acetylated chromatin. In addition, it will be interesting to assess acetylation levels at the recently described promoter P2.
Acknowledgments
We thank Robert Feil for help and materials used in analyzing DNase1 sensitivity of U2af-rs1 and members of the X Inactivation Group and R. Festenstein for helpful discussions. This work was supported by the Medical Research Council, United Kingdom and by the Wellcome Trust (L.O’N. and B.T.).
ABBREVIATIONS
- Xi
X inactive
- Xa
X active, Xic, X inactivation center
- SNuPE
single-nucleotide primer extension
References
- 1.Lyon M. Nature (London) 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 2.Heard E, Clerc P, Avner P. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 3.Costanzi C, Pehrson J R. Nature (London) 1998;393:599–601. doi: 10.1038/31275. [DOI] [PubMed] [Google Scholar]
- 4.Rastan S, Brown S D. Genet Res. 1990;56:99–106. doi: 10.1017/s0016672300035163. [DOI] [PubMed] [Google Scholar]
- 5.Rastan S. J Embryol Exp Morphol. 1983;78:1–22. [PubMed] [Google Scholar]
- 6.Brown C J, Ballabio A, Rupert J L, Lafreniere R G, Grompe M, Tonlorenzi R, Willard H F. Nature (London) 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 7.Brown C J, Lafreniere R G, Powers V E, Sebastio G, Ballabio A, Pettigrew A L, Ledbetter D H, Levy E, Craig I W, Willard H F. Nature (London) 1991;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- 8.Brockdorff N, Ashworth A, Kay G F, Cooper P, Smith S, McCabe V M, Norris D P, Penny G D, Patel D, Rastan S. Nature (London) 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 9.Borsani G, Tonlorenzi R, Simmler M C, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. Nature (London) 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 10.Brockdorff N, Ashworth A, Kay G F, McCabe V M, Norris D P, Cooper P J, Swift S, Rastan S. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 11.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 12.Clemson C M, McNeil J A, Willard H F, Lawrence J B. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Nature (London) 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 14.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 15.Clerc P, Avner P. Nat Genet. 1998;19:249–253. doi: 10.1038/924. [DOI] [PubMed] [Google Scholar]
- 16.Lee J T, Strauss W M, Dausman J A, Jaenisch R. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 17.Lee J T, Jaenisch R. Nature (London) 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 18.Herzing L B, Romer J T, Horn J M, Ashworth A. Nature (London) 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 19.Panning B, Dausman J, Jaenisch R. Cell. 1997;90:907–916. doi: 10.1016/s0092-8674(00)80355-4. [DOI] [PubMed] [Google Scholar]
- 20.Sheardown S A, Duthie S M, Johnston C M, Newall A E T, Formstone E J, Arkell R M, Nesterova T B, Alghisi G-C, Rastan S, Brockdorff N. Cell. 1997;91:99–107. doi: 10.1016/s0092-8674(01)80012-x. [DOI] [PubMed] [Google Scholar]
- 21.Johnston C M, Nesterova T B, Formstone E J, Newall A E T, Duthie S M, Sheardown S A, Brockdorff N. Cell. 1998;94:809–817. doi: 10.1016/s0092-8674(00)81739-0. [DOI] [PubMed] [Google Scholar]
- 22.O’Neill L P, Turner B M. EMBO J. 1995;14:3946–3957. doi: 10.1002/j.1460-2075.1995.tb00066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cooper P, Keer J T, McCabe V M, Hamvas R M, Brown S D, Rastan S, Brockdorff N. Genomics. 1993;15:570–575. doi: 10.1006/geno.1993.1109. [DOI] [PubMed] [Google Scholar]
- 24.Horn J M, Ashworth A. Hum Mol Genet. 1995;4:1041–1047. doi: 10.1093/hmg/4.6.1041. [DOI] [PubMed] [Google Scholar]
- 25.Simmler M C, Cunningham D B, Clerc P, Vermat T, Caudron B, Cruaud C, Pawlak A, Szpirer C, Weissenbach J, Claverie J M, Avner P. Hum Mol Genet. 1996;5:1713–1726. doi: 10.1093/hmg/5.11.1713. [DOI] [PubMed] [Google Scholar]
- 26.Minty A J, Alonso S, Caravatti M, Buckingham M E. Cell. 1982;30:185–192. doi: 10.1016/0092-8674(82)90024-1. [DOI] [PubMed] [Google Scholar]
- 27.Kipling D, Wilson H E, Mitchell A R, Taylor B A, Cooke H J. Chromosoma. 1994;103:46–55. doi: 10.1007/BF00364725. [DOI] [PubMed] [Google Scholar]
- 28.Mileham P, Brown S D M. Mamm Genome. 1996;7:253–261. doi: 10.1007/s003359900077. [DOI] [PubMed] [Google Scholar]
- 29.Singer Sam J, Lebon J M, Dai A, Riggs A D. PCR Methods Appl. 1992;1:160–163. doi: 10.1101/gr.1.3.160. [DOI] [PubMed] [Google Scholar]
- 30.Stalder J, Larsen A, Engel J D, Dolan M, Groudine M, Weintraub H. Cell. 1980;20:451–460. doi: 10.1016/0092-8674(80)90631-5. [DOI] [PubMed] [Google Scholar]
- 31.Riley D E, Canfield T K, Gartler S M. Nucleic Acids Res. 1984;12:1829–1845. doi: 10.1093/nar/12.4.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang T P, Caskey C T. Mol Cell Biol. 1987;7:2994–2998. doi: 10.1128/mcb.7.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerem B S, Goitein R, Richler C, Marcus M, Cedar H. Nature (London) 1983;304:88–90. doi: 10.1038/304088a0. [DOI] [PubMed] [Google Scholar]
- 34.Feil R, Boyano M, Allen N D, Kelsey G. J Biol Chem. 1997;272:20893–20906. doi: 10.1074/jbc.272.33.20893. [DOI] [PubMed] [Google Scholar]
- 35.Sheardown S A, Newall A E T, Norris D P, Rastan S, Brockdorff N. Gene. 1997;203:159–168. doi: 10.1016/s0378-1119(97)00507-6. [DOI] [PubMed] [Google Scholar]
- 36.Jeppesen P, Turner B M. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 37.Nasir J, Fisher E M, Brockdorff N, Disteche C M, Lyon M F, Brown SD. Proc Natl Acad Sci USA. 1990;87:399–403. doi: 10.1073/pnas.87.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmler M C, Heard E, Rougelle C, Cruaud C, Weissenbach J, Avner P. Mamm Genome. 1997;8:760–766. doi: 10.1007/s003359900561. [DOI] [PubMed] [Google Scholar]
- 39.Hebbes T R, Clayton A L, Thorne A W, Crane Robinson C. EMBO J. 1994;13:1823–1830. doi: 10.1002/j.1460-2075.1994.tb06451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Struhl K. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- 41.Kuo M-H, Zhou J, Jambeck P, Churchill M E A, Allis C D. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norris D P, Patel D, Kay G F, Penny G D, Brockdorff N, Sheardown S A, Rastan S. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 43.Nan X S, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Nature (London) 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 44.Jones P L, Veenstra G J C, Wade P A, Vermaak D, Kass S U, Landsberger N, Wolffe A P. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 45.O’Neill, L. P., Keohane, A. M., Lavender, J. S., McCabe., V., Heard, E., Avner, P., Brockdorff, N. & Turner, B. M. (1999) EMBO J., in press. [DOI] [PMC free article] [PubMed]