Abstract
The crystal structure of the flavodoxin-like protein WrbA with oxidized FMN bound reveals a close relationship to mammalian NAD(P)H:quinone oxidoreductase, Nqo1. Structural comparison of WrbA, flavodoxin, and Nqo1 indicates how the twisted open-sheet fold of flavodoxins is elaborated to form multimers that extend catalytic function from one-electron transfer between protein partners using FMN to two-electron reduction of xenobiotics using FAD. The structure suggests a novel physiological role for WrbA and Nqo1.
Keywords: soluble quinones, membrane quinones, peripheral membrane proteins, menaquinone, vitamin K, shikimate, chemotherapeutics
The 21 kDa protein WrbA from Escherichia coli is the founding member of a family of flavodoxin-like proteins conserved from bacteria to higher plants that are implicated in cellular responses to altered redox conditions and other kinds of stress (Grandori and Carey 1994) via their two-electron NAD(P)H:quinone oxidoreductase (Nqo) activity (Patridge and Ferry 2006). Nqo enzymes from bacteria to mammals are thought to shunt quinones from one-electron reduction pathways, thereby preventing redox cycling that can produce reactive oxygen species (Ernster 1987). Nqos are specific for FAD as redox cofactor but are characteristically unspecific for both electron donors (NADH or NADPH) and electron acceptors. Soluble mammalian Nqos can metabolically activate a wide range of electrophilic xenobiotics and are notable as both pro-oxidant and antioxidant enzymes, an apparent structure–function paradox that has been ascribed to the varying chemistries of their substrates (Cadenas 1995). Targeted drug therapies aim to exploit the naturally induced overexpression of Nqos in tumor cells to activate antitumor chemotherapeutics (Ernster 1987; Cadenas 1995; Li et al. 1995). The crystal structure of oxidized tetrameric E. coli WrbA (Wolfová et al. 2007) rationalizes functional distinctions with dimeric Nqos and monomeric flavodoxins and suggests a novel function for WrbAs and Nqos.
Results and Discussion
Overall fold
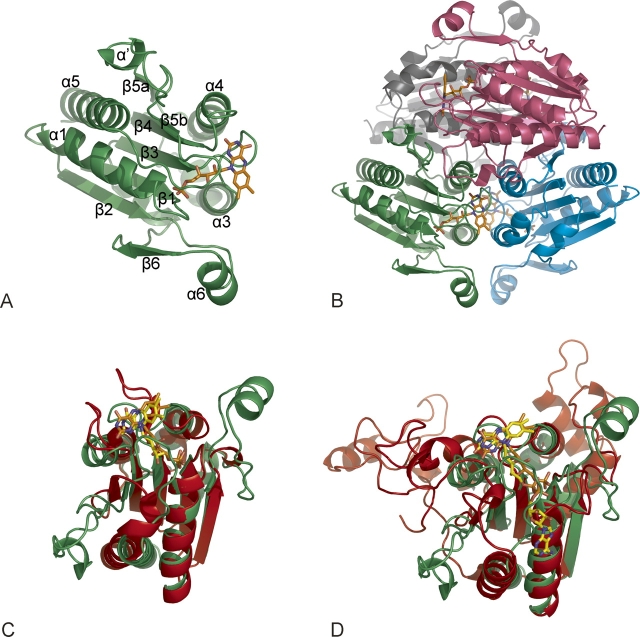
WrbA monomers (Fig. 1A) adopt the canonical αβα sandwich fold of the flavodoxins with FMN bound at the C-terminal end of the parallel 5-stranded sheet (Simondsen and Tollin 1980), but with characteristic flanking insertions and the ability to form tetramers (Fig. 1B) without a dedicated multimerization domain (Grandori and Carey 1994). Helix α' is part of a conserved sequence (Grandori and Carey 1994; Patridge and Ferry 2006) interrupting β5 that is typical of long-chain but not short-chain flavodoxins (Fig. 1C). Helix α6 is part of a highly variable subdomain inserted after β2 in Nqos (Fig. 1D) and some flavodoxins. A DALI search identifies several oxidoreductases as relatives of the WrbA fold, including mammalian 1QRD (Li et al. 1995), the prototypical Nqo1 (Ernster 1987; Cadenas 1995). The WrbA tetramer assembles as a dimer of dimers with 222 symmetry. Antiparallel monomers pair as in 1QRD, with each cofactor facing the last two turns of helix α3 in the other subunit. Dimers pair burying all four α4-helices at the equator with the FMNs facing outside and the α6-helices at the poles. The sequence including α4 is the most highly conserved region of WrbAs, but not flavodoxins. In 1QRD tetramerization is blocked by a C-terminal subdomain missing from WrbA that extends from each monomer toward the other and provides residues that contact the adenine riboside of the other monomer's NADH. This subdomain also packs against the distal half of bound dicoumarol (Asher et al. 2006), the defining inhibitor of the Nqos (Ernster 1987; Cadenas 1995), which does not inhibit WrbA (T. Gustavsson and J. Carey, unpubl.).
Figure 1.
WrbA bridges flavodoxins and oxidoreductases. (A) Overall fold. WrbA monomer, ribbon; FMN, skeletal (orange carbon atoms and atomic colors). Secondary structure numbering follows the convention introduced for Nqo1 (Li et al. 1995). Crystallization and diffraction analysis are described by Wolfová et al. (2007). (B) WrbA tetramer. View along the diagonal between crystallographic axes a and b, with each subunit a unique color. (C) Overlay with flavodoxin. Least-squares 3D alignment of WrbA monomer (rotated 90° from panel A) with short-chain flavodoxin (PDB ID 1J8Q; protein, red; FMN, yellow carbons and atomic colors). (D) Overlay with Nqo1. Mammalian Nqo1 monomer (PDB ID 1QRD; protein, red; FAD, yellow carbons and atomic colors). C-terminal subdomain at upper left.
Overlay of 1QRD and WrbA monomers (Fig. 1D) indicates that cofactor selectivity is influenced by residues in a short insertion after β5b that is characteristically conserved in WrbAs (Grandori and Carey 1994; Patridge and Ferry 2006). The adenine riboside portion of FAD is replaced by this irregular segment, which lies in a groove on the WrbA surface along the N-terminus of α1 at the same depth as the cofactor, with the sequence 171RQP173 occupying the position of the adenine and ribose rings. However, FAD substitutes for FMN in WrbA oxidoreductase activity (T. Gustavsson and J. Carey, unpubl.), suggesting this segment can move to accommodate adenine riboside.
Active Site
The WrbA FMN-binding site is in the same relative location as that of flavodoxins (Fig. 1C), but the disposition and redox properties of FMN are entirely different. Two protruding loops enclose the flavodoxin isoalloxazine in a narrow crevice lined by aromatic residues, leaving the dimethylbenzene ring edge exposed as an electron conduit (Simondsen and Tollin 1980). The flavodoxin pocket promotes the single-electron transfers required by its partner proteins by constraining the fully reduced flavoquinol to an unfavorable planar conformation without positive charges nearby, dramatically destabilizing it relative to the semiquinone (Simondsen and Tollin 1980). The flavodoxin crevice is missing in WrbA, and FMN is presented on a broad, flat surface consistent with assembly of its active sites at subunit interfaces. A planar isoalloxazine model was well-accommodated by the electron density, consistent with the oxidized state of the cofactor implied by the intense yellow color of the protein crystals (Wolfová et al. 2007).
As in flavodoxin, FMN is anchored by its ribityl-phosphate tail to conserved residues of the β1–α2 segment. Arg78 and Phe79 surround the dimethylbenzene ring (Fig. 2A), consistent with their predicted homology to flavodoxin crevice residues (Grandori and Carey 1994) despite the novelties of positive charge and topology with both residues on the same loop. All remaining residues of each FMN pocket are contributed by second (Trp97 and His132) and third (Tyr 142 and Phe 148) monomers. Thus, the topological difference in FMN-binding residues is linked to WrbA's assembly state, leading to cooperation among subunits for FMN binding that supports function as an obligate tetramer. The redox-active uracil end of isoalloxazine points directly toward Tyr142 (Oγ–FMN N3 ∼ 3.66 Å) and His132 (Nδ1–FMN O4 ∼ 2.86 Å), suggesting these residues correspond to the charge-relay pair of 1QRD (Tyr155, His161) that facilitates hydride transfer, promoting two-electron reduction (Li et al. 1995).
Figure 2.
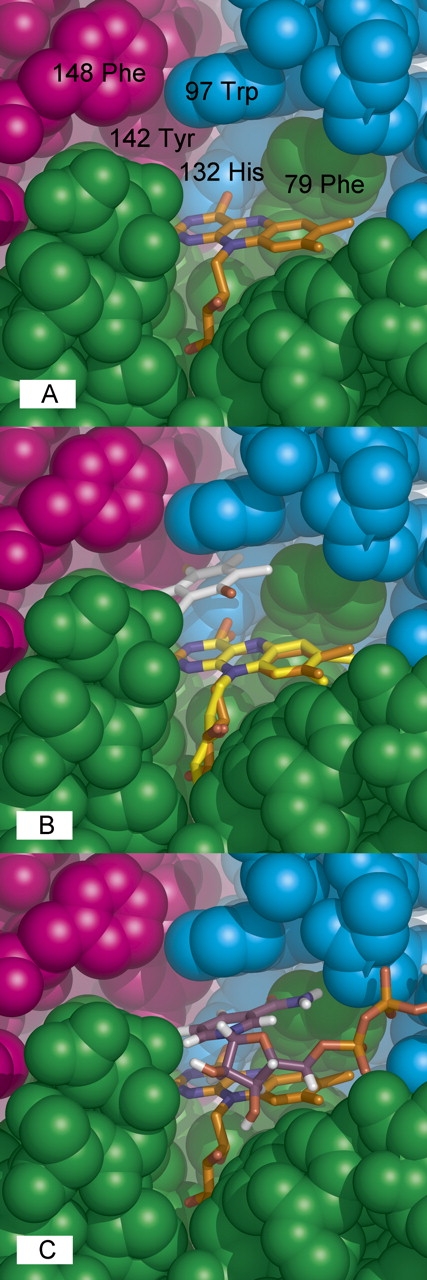
WrbA active site. (A) Key residues. Space-filling representation with residues numbered and chain colors as in Figure 1B. (B) Quinone binding. The WrbA active site was overlaid with that of 1QRD with bound duroquinone (Li et al. 1995) by superposition of the common flavin atoms from isoalloxazine through phosphate. (C) NADH binding. The program FlexX (Rarey et al. 1996) was used to dock NADH into the WrbA tetramer (nicotinamide and adenine riboside carbons, purple; pyrophosphate, orange; other colors atomic).
Overlay with the 1QRD structure containing bound tetramethyl-1,4-benzoquinone (Li et al. 1995) indicates that WrbA accommodates quinones in a highly similar, cavernous active site (Fig. 2B). NADH binding by WrbA examined with docking (Fig. 2C) shows that the nicotinamide ring is easily accommodated in a position similar to that found in 1QRD (Li et al. 1995). The promiscuity of both proteins for electrophilic substrates in the reductive half-reaction may reflect the requirement for an active site large enough to accommodate the adjacent ribose of nicotinamide in the oxidative half-reaction. However, the relevant oxidation state of FMN for quinone binding is the reduced state; in this state the isoalloxazine ring system displays butterfly bending of ∼18°–28° about the N5–N10 centerline (Walsh and Miller 2003) that would presumably narrow the space available for quinones and help to displace oxidized nicotinamide. In any case, the active site is not large enough for simultaneous binding of flavin, nicotinamide, and quinone; thus, WrbA is predicted to share the characteristic ping-pong kinetic mechanism of the Nqos (Ernster 1987; Cadenas 1995).
Functional hypothesis
Both WrbA and Nqo1 are typically purified as soluble proteins (Ernster 1987). Although reduction of soluble electrophiles is an undisputed biochemical property of Nqos and WrbAs in vitro and in vivo (Ernster 1987; Cadenas 1995; Li et al. 1995; Laskowski et al. 2002; Asher et al. 2006; Patridge and Ferry 2006), many such compounds could react spontaneously at the reducing potential of mammalian or bacterial cytosol. A surface feature of WrbA suggests it may yet prove to have a physiological role involving membrane interaction. Helix α6 faces the middle two turns of α3 in the same subunit without contacting it, forming a protrusion and a hydrophobic channel to the active site. Irregular segments connecting α6 could provide flexible docking at the membrane, permitting long-chain quinones to reach reduced flavin. 1QRD presents a more complex α6 insertion than WrbA (Li et al. 1995); its association with the membrane would orient Nqo1 dimers with their C-terminal subdomains facing the cytosol, suggesting that proteins proposed to interact there (Asher et al. 2006) could be functional partners. A ping-pong kinetic mechanism could suggest that membrane localization is transitory.
The suggestion that both WrbA and the Nqos may have physiological roles involving membrane binding is consistent with a large number of observations that have not yet been reconciled. E. coli WrbA is regulated by phosphorylated ArcA (Liu and De Wulf 2004), the global sensor of redox state in the energy-linked quinone pool (Georgellis et al. 2001). WrbA is among a group of specific proteins that accumulate as cytoplasmic aggregates when membrane protein fusions are overexpressed in E. coli (Wagner et al. 2007), apparently due to saturation of the cytoplasmic membrane protein translocation apparatus; levels of respiratory chain complexes in the cytoplasmic membrane are also strongly reduced, and the Arc system is activated. WrbA may be linked to tryptophan biosynthesis (Yang et al. 1993), which lies on one branch of the shikimate pathway with menaquinone biosynthesis on the other. The phylogenetic distribution of WrbAs (Grandori and Carey 1994; Patridge and Ferry 2006) is the same as the distribution of the shikimate pathway. Nqo1 is thought to be a physiologically relevant target of the coumarol anticoagulants (Ernster 1987; Cadenas 1995; Li et al. 1995; Asher et al. 2006), consistent with its largely unexplored role in menaquinone-dependent protein carboxylation (Wallin et al. 1987). An uncharacterized hydrophobic fraction of Nqo1 remains associated with membranes (Ernster 1987). Soluble Nqo1 can reduce vitamin K or CoQ10 in vesicles (Martius et al. 1975; Beyer et al. 1996) and requires coactivators (e.g., BSA, Triton) (Ernster 1987) that may mimic the membrane. Yeast WrbA presents a palmitoylation signal (Grandori and Carey 1994) and is membrane-localized (G. Petsko, pers. comm.). Thus it appears possible that overexpression of Nqos in tumor cells, and of both proteins in response to xenobiotics or oxidative stress (Ernster 1987; Grandori and Carey 1994; Cadenas 1995; Li et al. 1995; Georgellis et al. 2001; Laskowski et al. 2002; Liu and De Wulf 2004; Asher et al. 2006; Patridge and Ferry 2006), may be related only indirectly to reduction of soluble electrophiles.
Acknowledgments
J.C. thanks A. Bocarsly and T. Silhavy (Princeton University) for useful discussions. This work was supported by grants NSF INT 03-09049 to J.C., Kontakt ME640 to I.K.S., LC06010 to R.E. and I.K.S., and LC06077 to J.B. Additional support from the Academy of Sciences of the Czech Republic (AV0Z60870520) and the Institutional Research Concept (MSM6007665808) is acknowledged.
Footnotes
Reprint requests to: Jannette Carey, Chemistry Department, Princeton University, Washington Road and William Street, Princeton, NJ 08544-1009, USA; e-mail: jcarey@princeton.edu; fax: (609) 258-6746.
Article and publication are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073018907.
References
- Asher G., Dym, O., Tsvetkov, P., Adler, J., and Shaul, Y. 2006. The crystal structure of NAD(P)H:quinone oxidoreductase 1 in complex with its potent inhibitor dicoumarol. Biochemistry 45: 6372–6378. [DOI] [PubMed] [Google Scholar]
- Beyer R.E., Segura-Aguilar, J., DiBernardo, S., Cavazzoni, M., Fato, R., Fiorentini, D., Galli, M.C., Setti, M., Landi, L., and Lenaz, G. 1996. The role of DT-diaphorase in the maintenance of the reduced antioxidant form of coenzyme-Q in membrane systems. Proc. Natl. Acad. Sci. 93: 2528–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas E. 1995. Antioxidant and prooxidant functions of DT-diaphorase in quinone metabolism. Biochem. Pharmacol. 49: 127–140. [DOI] [PubMed] [Google Scholar]
- Ernster L. 1987. DT-Diaphorase: A historical review. Chem. Scr. 27A: 1–13. [Google Scholar]
- Georgellis D., Kwon, O., and Lin, E.C. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292: 2314–2316. [DOI] [PubMed] [Google Scholar]
- Grandori R. and Carey, J. 1994. Six new candidate members of the α/β twisted open-sheet family detected by sequence similarity to flavodoxin. Protein Sci. 3: 2185–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski M.J., Dreher, K.A., Gehring, M.A., Abel, S., Gensler, A.L., and Sussex, I.M. 2002. FQR1, a novel primary auxin-response gene, encodes a flavin mononucleotide-binding quinone reductase. Plant Physiol. 128: 578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Bianchet, M.A., Talalay, P., and Amzel, L.M. 1995. The three-dimensional structure of NAD(P)H:quinone reductase, a flavoprotein involved in cancer chemoprotection and chemotherapy: Mechanism of the two-electron reduction. Proc. Natl. Acad. Sci. 92: 8846–8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X. and De Wulf, P. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 279: 12588–12597. [DOI] [PubMed] [Google Scholar]
- Martius C., Ganser, R., and Viviani, A. 1975. The enzymatic reduction of K-vitamins incorporated in the membrane of liposomes. FEBS Lett. 59: 13–14. [DOI] [PubMed] [Google Scholar]
- Patridge E.V. and Ferry, J.G. 2006. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 188: 3498–3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rarey M., Kramer, B., Lengauer, T., and Klebe, G. 1996. A fast flexible docking method using an incremental construction algorithm. J. Mol. Bio. 261: 470–489. [DOI] [PubMed] [Google Scholar]
- Simondsen R.P. and Tollin, G. 1980. Structure–function relations in flavodoxins. Mol. Cell. Biochem. 33: 13–24. [DOI] [PubMed] [Google Scholar]
- Wagner S., Baars, L., Ytterberg, A.J., Klussmeier, A., Wagner, C.S., Nord, O., Nygren, P.-Å., van Wijk, K.J., and de Gier, J.-W. 2007. Consequences of membrane protein overexpression in Escherichia coli . Mol. Cell. Proteomics http://www.mcponline.org/cgi/reprint/M600431-MCP200v1. [DOI] [PubMed]
- Wallin R., Rannels, S.R., and Martin, L.F. 1987. DT-Diaphorase and vitamin K-dependent carboxylase in liver and lung microsomes and in macrophages and type II epithelial cells isolated from rat lung. Chem. Scr. 27A: 193–202. [Google Scholar]
- Walsh J.D. and Miller, A.-F. 2003. Flavin reduction potential tuning by substitution and bending. THEOCHEM 623: 185–195. [Google Scholar]
- Wolfová J., Mesters, J.R., Brynda, J., Grandori, R., Natalello, A., Carey, J., and Kutá Smatanová, I. 2007. Crystallization and preliminary diffraction analysis of E. coli WrbA in complex with its cofactor flavin mononucleotide. Acta Crystallograph. F 63: 571–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Ni, L., and Somerville, R.L. 1993. A stationary-phase protein of Escherichia coli that affects the mode of association between the trp repressor protein and operator-bearing DNA. Proc. Natl. Acad. Sci. 90: 5796–5800. [DOI] [PMC free article] [PubMed] [Google Scholar]