Abstract
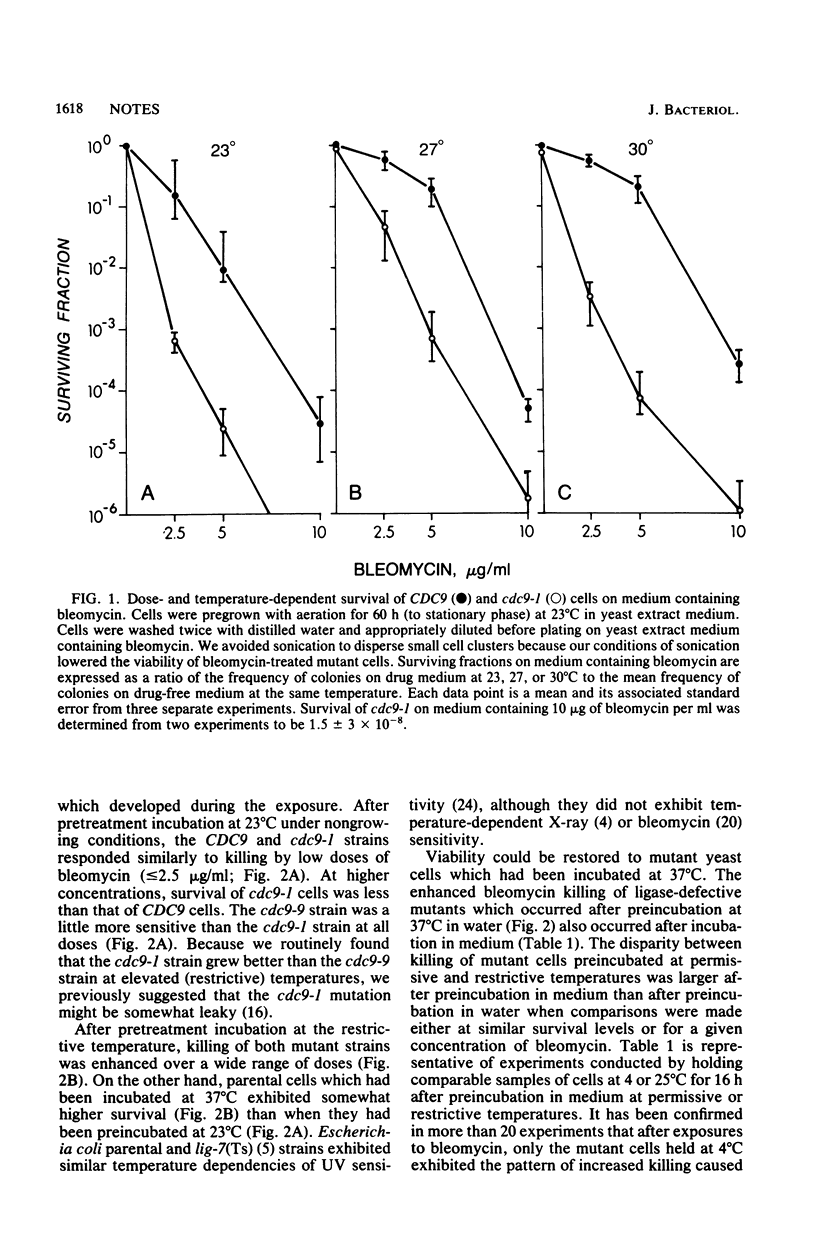
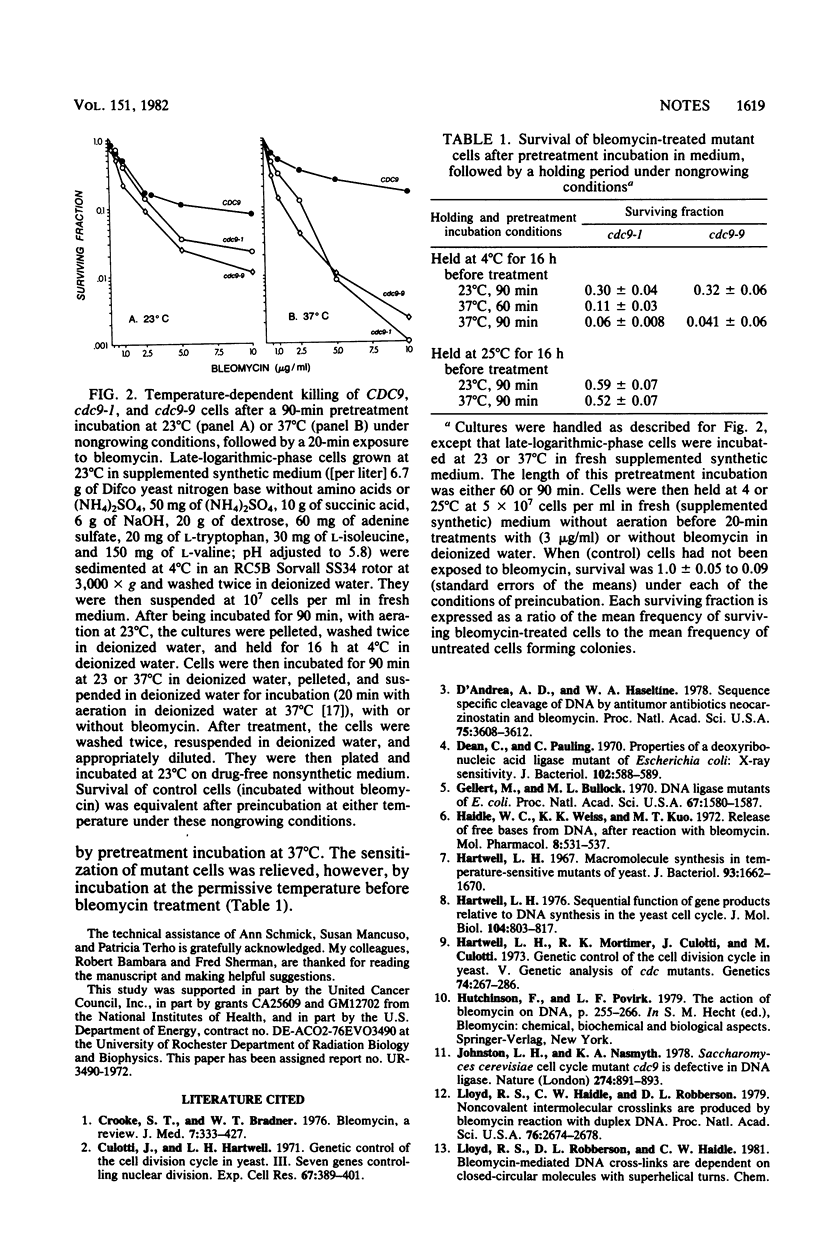
Conditional ligase-deficient mutants of Saccharomyces cerevisiae were more sensitive than their parental (CDC9) strain to dose-dependent killing by bleomycin, even when mutant cells were pregrown and exposed to the antibiotic at permissive temperatures. Pretreatment incubation at the restrictive temperature (37 degrees C) under growing or nongrowing conditions enhanced bleomycin killing of both cdc9-1 and cdc9-9 mutants. This sensitization could be relieved by incubation at the permissive temperature before treatment.
Full text
PDF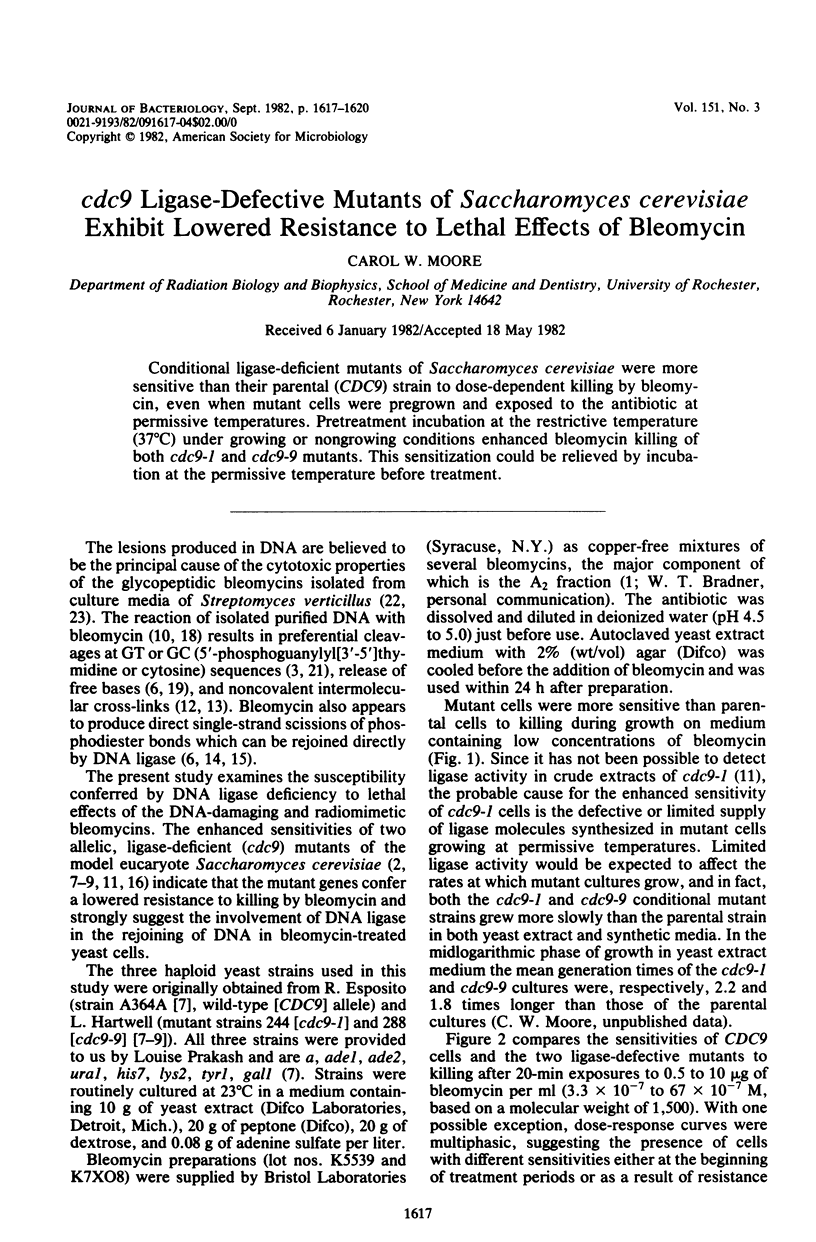

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Crooke S. T., Bradner W. T. Bleomycin, a review. J Med. 1976;7(5):333–428. [PubMed] [Google Scholar]
- Culotti J., Hartwell L. H. Genetic control of the cell division cycle in yeast. 3. Seven genes controlling nuclear division. Exp Cell Res. 1971 Aug;67(2):389–401. doi: 10.1016/0014-4827(71)90424-1. [DOI] [PubMed] [Google Scholar]
- D'Andrea A. D., Haseltine W. A. Sequence specific cleavage of DNA by the antitumor antibiotics neocarzinostatin and bleomycin. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3608–3612. doi: 10.1073/pnas.75.8.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C., Pauling C. Properties of a deoxyribonucleic acid ligase mutant of Escherichia coli: x-ray sensitivity. J Bacteriol. 1970 May;102(2):588–589. doi: 10.1128/jb.102.2.588-589.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gellert M., Bullock M. L. DNA ligase mutants of Escherichia coli. Proc Natl Acad Sci U S A. 1970 Nov;67(3):1580–1587. doi: 10.1073/pnas.67.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidle C. W., Weiss K. K., Kuo M. T. Release of free bases from deoxyribonucleic acid after reaction with bleomycin. Mol Pharmacol. 1972 Sep;8(5):531–537. [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976 Jul 15;104(4):803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Johnston L. H., Nasmyth K. A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978 Aug 31;274(5674):891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- Lloyd R. S., Haidle C. W., Robberson D. L. Noncovalent intermolecular crosslinks are produced by bleomycin reaction with duplex DNA. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2674–2678. doi: 10.1073/pnas.76.6.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W. Ligase-deficient yeast cells exhibit defective DNA rejoining and enhanced gamma ray sensitivity. J Bacteriol. 1982 Jun;150(3):1227–1233. doi: 10.1128/jb.150.3.1227-1233.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W. Modulation of bleomycin cytotoxicity. Antimicrob Agents Chemother. 1982 Apr;21(4):595–600. doi: 10.1128/aac.21.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller W. E., Yamazaki Z., Breter H. J., Zahn R. K. Action of bleomycin on DNA and RNA. Eur J Biochem. 1972 Dec 18;31(3):518–525. doi: 10.1111/j.1432-1033.1972.tb02560.x. [DOI] [PubMed] [Google Scholar]
- Müller W. E., Zahn R. K. Bleomycin, an antibiotic that removes thymine from double-stranded DNA. Prog Nucleic Acid Res Mol Biol. 1977;20:21–57. doi: 10.1016/s0079-6603(08)60469-9. [DOI] [PubMed] [Google Scholar]
- Ross S. L., Sharma S., Moses R. E. DNA polymerase III-dependent repair synthesis in response to bleomycin in toluene-treated Escherichia coli. Mol Gen Genet. 1980;179(3):595–605. doi: 10.1007/BF00271750. [DOI] [PubMed] [Google Scholar]
- Tien Kuo M., Haidle C. W., Inners L. D. Characterization of bleomycin-resistant DNA. Biophys J. 1973 Dec;13(12):1296–1306. doi: 10.1016/s0006-3495(73)86063-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vig B. K., Lewis R. Genetic toxicology of bleomycin. Mutat Res. 1978;55(2):121–145. doi: 10.1016/0165-1110(78)90019-2. [DOI] [PubMed] [Google Scholar]
- Youngs D. A., Smith K. C. The involvement of polynucleotide ligase in the repair of UV-induced DNA damage in Escherichia coli K-12 cells. Mol Gen Genet. 1977 Mar 28;152(1):37–41. doi: 10.1007/BF00264937. [DOI] [PubMed] [Google Scholar]


