Abstract
Functional chaperone cooperation between Hsp70 (DnaK) and Hsp104 (ClpB) was demonstrated in vitro. In a eubacterium Thermus thermophilus, DnaK and DnaJ exist as a stable trigonal ring complex (TDnaK⋅J complex) and the dnaK gene cluster contains a clpB gene. When substrate proteins were heated at high temperature, none of the chaperones protected them from heat inactivation, but the TDnaK⋅J complex could suppress the aggregation of proteins in an ATP- and TGrpE-dependent manner. Subsequent incubation of these heated preparations at moderate temperature after addition of TClpB resulted in the efficient reactivation of the proteins. Reactivation was also observed, even though the yield was low, if the substrate protein alone was heated and incubated at moderate temperature with the TDnaK⋅J complex, TGrpE, TClpB, and ATP. Thus, all these components were necessary for the reactivation. Further, we found that TGroEL/ES could not substitute TClpB.
Many molecular chaperones are heat shock proteins, and they help cells to survive the heat shock period. Among them, the Hsp70 family plays a crucial role (1, 2). The bacterial Hsp70 homolog is DnaK, and Escherichia coli DnaK, together with its cochaperones, DnaJ and GrpE, exhibits in vitro chaperone activity in recovery of firefly luciferase and E. coli RNA polymerase from a heat-inactivated state (3, 4). Binding and release of substrate proteins are regulated by the ATPase activity of DnaK, which is under tight control with DnaJ and GrpE (5, 6). DnaJ accelerates the rate of ATP hydrolysis of DnaK, whereas GrpE promotes the nucleotide exchange of ATP and ADP (7). DnaJ and GrpE are indispensable for the chaperone activity of DnaK in vivo and in vitro, and these three proteins constitute the DnaK⋅J-GrpE set (2).
Hsp104, originally identified as a factor for thermotolerance, is another molecular chaperone involved in recovery of the cell from heat-induced damage (8, 9). As a member of the Hsp100/Clp family, Hsp104 has two nucleotide-binding domains, each displaying one set of consensus ATP-binding sequence motifs (10), and it forms a homohexameric complex (11). In yeast, it has been demonstrated that Hsp104 plays a crucial role for recovery in the period after, rather than during, the heat shock to clear the aggregation in the cell (9). Interaction of Hsp104 with the yeast prion-like protein (Sup35) was also reported (12). The bacterial homolog of Hsp104 is ClpB, a protein found ubiquitously in various bacteria (13). E. coli ClpB is a σ32 (heat shock σ factor)-dependent heat shock protein (14, 15), and the purified ClpB has casein-activated ATPase activity (16). E. coli mutants lacking clpB exhibit growth defects at 44°C and undergo a more rapid death at 50°C than the wild type (15). Overproduction of other chaperones cannot compensate for the deleterious effect of the clpB null mutants at 50°C (17). ClpA, another Hsp100/Clp member in E. coli that is constitutively expressed, is involved in ATP-dependent protein degradation through an association with the unrelated protease ClpP. In the absence of ClpP, ClpA can display the chaperone function of the DnaK⋅J-GrpE set in activating the plasmid P1 initiator protein RepA; disassembly of inactive RepA dimers into active monomers (18, 19). These results implicate a unique chaperone activity of ClpB. However, the functional character of ClpB has been defined only to a small extent (20), and its putative chaperone activity has not been demonstrated.
Various molecular chaperones in the cell are thought to cooperate to assist protein folding. The functional relationship between Hsp70 and Hsp104 in yeast has been reported; overexpression of Hsp70 can partially suppress the phenotype of a HSP104 deletion (21), and inversely, deletion of HSP78 (a member of Hsp100/Clp family in yeast mitochondria) results in synthetic lethality on nonfermentable carbon sources in mutant strains carrying temperature-sensitive alleles of SSC1 (a member of the Hsp70 family in yeast mitochondria) (22, 23). These in vivo observations support the model of chaperone cooperation of the Hsp70 set and Hsp104. Indeed, Glover and Lindquist have demonstrated recently that yeast Hsp104, together with Hsp70 and Hsp40, could reactivate proteins that had been chemically denatured and allowed to aggregate (24).
We have studied chaperones of a thermophilic eubacteria, Thermus thermophilus. Unlike in other bacteria, DnaK and DnaJ are purified only as a trigonal ring complex from T. thermophilus (25). This TDnaK⋅J complex1 (the prefix T designates proteins of T. thermophilus hereafter) comprises three copies of each TDnaK, TdnaJ, and TDafA (26). TDafA is a newly found small protein necessary for the in vivo and in vitro assembly of TDnaK and TDnaJ. The TDnaK⋅J complex is stable during interaction with ATP, TGrpE, and a model denatured protein, reduced carboxymethylated α-lactalbumin (25, 26). TDnaJ alone is unstable at above 65°C, and it probably exists only in the TDnaK⋅J complex in the cell of T. thermophilus, whose optimum growth temperature is 75°C. The TDnaK⋅J complex can bind a model denatured protein, but so far folding-assisting activity has not been observed (25). TGrpE is isolated as a homodimer (27). Like in several other bacteria, the genes of the components of the TDnaK⋅J-GrpE set (TDnaK⋅J complex and TGrpE) in T. thermophilus are located in a single dnaK gene cluster (26), but unlike in other bacteria, this cluster also contains a gene for TClpB. The presence of TClpB gene in the dnaK gene cluster has prompted us to test the functional cooperation between the TDnaK⋅J-GrpE set and TClpB. As expected, TClpB cooperates with the TDnaK⋅J-GrpE set in the recovery of several kinds of heat-inactivated proteins.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids.
T. thermophilus strain HB8 (ATCC 27634) was used as a source of genomic DNA. E. coli strains JM109, CJ236, and BL21 (DE3) were used. Plasmid pUC118 and pET21c (Novagen) and helper phage M13KO7 were used. T. thermophilus was grown as described previously (25).
Cloning and Expression of T.clpB.
Chromosomal DNA from T. thermophilus was isolated as described previously (28). A 4.5-kbp KpnI fragment was cloned that contained the downstream region of the previously cloned 5.5-kbp fragment containing genes of the TDnaK·J-GrpE set and part of T.clpB gene. To express TClpB, NspI-PstI fragments encoding the C-terminal region of TClpB were subcloned into SphI-PstI sites of pUC118 (pMCBS1). A NdeI site was introduced to the translational initiation region of T.clpB gene in pMDJ1–1 (26) by the method of Kunkel et al. (29). The T.clpB gene carrying a NdeI site was digested with NdeI-NspI, and the region of the T.clpB gene corresponding to the C-terminus of TClpB was digested with SphI-EcoRI in pMCBS1. The obtained fragments were ligated into the corresponding NdeI-EcoRI sites in pET21c, and it was named the pMCB1.
Isolation of Proteins.
The TDnaK⋅J complex and TGrpE were expressed in E. coli carrying pMKJ8 (TDnaK⋅J complex) and pMGE3 (TGrpE), respectively, and purified as described previously (26, 27). TClpB was expressed in E. coli BL21 (DE3) carrying pMCB1. The cells were harvested, disrupted by a French press, and centrifuged at 100,000 × g for 40 min at 4°C. The supernatant was applied to a DE-52 cellulose column (Whatman). The column was eluted with a 20- to 250-mM linear gradient of NaCl in Buffer A. The TClpB fractions were pooled and solid ammonium sulfate was added to 400 mM. The solution was applied to a Butyl-Toyopearl column (Tosoh, Tokyo) and eluted with a 400- to 0-mM linear reverse gradient of ammonium sulfate. The TClpB fractions were pooled, concentrated, and applied on a Sepharose CL-6B equilibrated with Buffer A containing 100 mM Na2SO4. The peak fractions were pooled and stored in 2.8 M ammonium sulfate at 4°C. Throughout this paper, concentrations of substrate proteins are expressed as protomer and those of T. thermophilus chaperones, as trigonal complex (TDnaK⋅J complex), dimer (TGrpE), hexamer (TClpB), and TGroEL14-TGroES7 complex (TGroEL/ES).
ATPase Assay of TClpB.
ATPase activities were assayed in a 100-ml reaction mixture containing 25 mM Tris⋅HCl, pH 7.5, 5 mM MgCl2, 150 mM KCl, 1 mM ATP, 5 mg TClpB, and 0.1 mg/ml casein when added. The reaction was initiated by addition of ATP, terminated after a 10-min incubation by addition of 25 ml of 20% perchloric acid, and released Pi was measured by malachite green assay (30, 31).
Aggregation of Denatured Lactate Dehydrogenase (LDH).
LDH from Bacillus stearothermophilus (30 mM) was labeled with sulfosuccinimidyl-6-(biotinamido) hexanoate (Pierce, 150 mM) in 100 mM potassium phosphate, pH 7.0, at 37°C for 60 min, and biotinylated LDH was separated from free reagents by gel filtration. Biotinylated LDH (0.2 μM, 0.68 biotin/LDH protomer) was incubated at 73°C for 30 min in 50 mM 3-morpholinopropanesulfonic acid (MOPS)-NaOH buffer, pH 7.5, containing 5 mM MgCl2, 150 mM KCl, 1 mM DTT, and, when indicated, chaperones (0.4 μM TDnaK⋅J complex/0.8 μM TgrpE/0.4 μM TClpB)/3 mM ATP). The incubated solutions were immediately centrifuged at 19,000 × g for 10 min at 4°C. Supernatant and the pellet suspended in the original volume of the buffer were loaded to polyacrylamide gel (13%) electrophoresis in 0.1% sodium dodecylsulfate. Electrophoresed proteins were blotted to poly(vinylidene difluoride) membrane, and biotinylated LDH was detected by alkaline phosphatase-conjugated streptavidin (Vector Laboratories).
Reactivation of Heat-Inactivated Proteins.
LDH (Unitika, Osaka), glucose-6-phosphate dehydrogenase (G6PDH, Unitika) and α-glucosidase (Sigma) from B. stearothermophilus were used as substrate proteins. LDH (0.2 mM) was dissolved in 300 ml of the reaction mixture (50 mM MOPS-NaOH, pH 7.5/5 mM MgCl2/150 mM KCl/1 mM DTT) containing chaperones (0.4 μM TDnaK⋅J complex/0.8 μM TGrpE/0.4 μM TClpB) and ATP (3 mM), as indicated. The concentration of TGroEL/ES was 0.4 μM when used. The reaction mixture was incubated at 73°C for 30 min (inactivation period) and subsequently incubated at 55°C (reactivation period). An aliquot was taken out at indicated times in the reactivation period, and LDH activity was assayed at 55°C by the absorbance at 340 nm in the assay solution (100 mM potassium phosphate, pH 6.0/0.2 mM NADH/20 mM sodium pyruvate) (32). Varied temperatures of the reactivation period (50°C and 60°C) were also tested, but 55°C turned out to be the optimum temperature to give the best recovery yield. Measurements of inactivation and reactivation of α-glucosidase and G6PDH were carried out in 250 μl of the reaction mixture (50 mM MOPS-NaOH, pH 7.5/5 mM MgCl2/150 mM KCl) containing α-glucosidase (0.2 μM) or G6PDH (0.2 μM). The reaction mixture was incubated at 73°C for 20 min (α-glucosidase) or 10 min (G6PDH) (inactivation period) and subsequently incubated at 55°C for 1 hr (reactivation period). Recovery of activity from the heat-inactivated state was assessed by comparing activities of aliquots taken from the reaction mixtures immediately and 1 hr after initiation of the reactivation period. α-glucosidase activity was assayed by absorbance at 405 nm in the assay solution (50 mM sodium phosphate, pH 6.8/2 mM p-nitrophenyl-α-d-glucopyranoside) and G6PDH activity by absorbance at 340 nm in the assay solution (100 mM Tris⋅HCl, pH 8.8/40 mM MgCl2/1 mM NADP+/3 mM glucose 6-phosphate) at 55 °C (33). The chaperones (0.4 μM TDnaK⋅J complex/0.8 μM TGrpE/0.4 μM TClpB) and ATP (3 mM) were added as indicated.
RESULTS
A ClpB Homolog Is Encoded in the T. thermophilus dnaK Gene Cluster.
A dnaK gene cluster of T. thermophilus contains dnaK-grpE-dnaJ-dafA genes, and in addition we found another ORF downstream of dafA (26). The ORF encodes a protein with 854 residues, and the amino acid sequence shows overall similarity to that of E. coli ClpB (56.4% identity) (14, 15) and yeast Hsp104 (44.1% identity) (10). These two proteins belong to the Hsp104/ClpB subfamily in Hsp100/Clp family (34). Also found in the ORF are the consensus sequence motifs commonly found in members of the Hsp104/ClpB subfamily; two sets of Walker’s ATP-binding motif A (198GEPGVGKT205, 595GPTGVGKT602) and B (266ILFID270, 663VILFD667) (35), and the consensus I-V sequences (34). Therefore, the product of this ORF was T. thermophilus ClpB (TClpB), and the dnaK region in T. thermophilus contains five genes in the order dnaK-grpE-dnaJ-dafA-clpB.
TClpB Exhibits a Casein-Stimulated ATPase Activity.
TClpB exhibited an optimum ATPase activity (270 nmol/min/mg) at 70°C, a growth optimum temperature of T. thermophilus, and, like E. coli ClpB (16), this ATPase activity was stimulated by 1.6-fold by addition of a model denatured protein, casein. Stimulation by casein was observed at a wide temperature range (30≈85°C). BSA and RNaseA did not have stimulation effect. ATPase activity of TClpB was not affected by the presence of the TDnaK⋅J-GrpE set.
The TDnaK⋅J-GrpE Set Can Suppress Heat Aggregation.
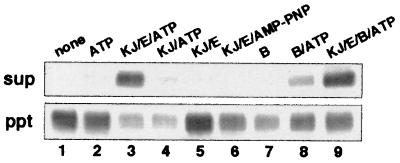
Thermophilic LDH incubated at 73°C for 30 min with indicated components was separated into the supernatant (soluble) fraction and the precipitate (aggregate) fraction by high-speed centrifugation, and each fraction was analyzed with SDS/PAGE (Fig. 1). LDH was biotinylated for detection. Compared with nonbiotinylated LDH, LDH with biotin label had similar enzymatic activity (97%) and was similarly inactivated at 73°C, that is, >90% of the activity was lost after 30 min. When LDH, alone or with ATP, was heated at 73°C for 30 min, it formed large aggregates and was precipitated (Fig. 1, lanes 1, 2). When LDH was heated in the presence of the TdnaK·J-GrpE set + ATP, nearly 80% of LDH remained in the soluble fraction (lane 3). TGrpE and ATP were indispensable for the prevention of aggregation (lanes 4, 5).‖ Adenosine 5′-[β,γ–imido]triphosphate was not able to substitute ATP (lane 6). These results indicate that the TDnaK⋅J-GrpE set can prevent the heat aggregation of other proteins in an ATP-, TGrpE-dependent manner. TClpB did not protect LDH from heat aggregation, but TClpB + ATP had a slight protective effect (lanes 7 and 8). When LDH was heated in the presence of the TDnaK⋅J-GrpE set + TClpB + ATP, a fraction slightly over half of LDH remained in the soluble fraction, and the rest aggregated (lane 9). When TDnaK⋅J-GrpE are present together, TClpB appears to interfere the aggregation-preventing function of the TDnaK⋅J-GrpE set.
Figure 1.
Effect of the TDnaK⋅J-GrpE set and TClpB on aggregation of LDH. LDH, biotinylated for detection, was incubated in the presence of indicated components at 73°C for 30 min. The solutions were centrifuged, and supernatant (sup) and precipitate (ppt) were analyzed by polyacrylamide gel electrophoresis in the presence of sodium dodecylsulfate. Electrophoresed proteins were blotted to the membrane, and biotinylated LDH was detected by alkaline phosphatase-conjugated streptavidin. KJ, E, and B represent TDnaK⋅J complex, TGrpE and TClpB, respectively.
The TDnaK⋅J-GrpE Set Alone Cannot Reactivate Heat-Inactivated LDH.
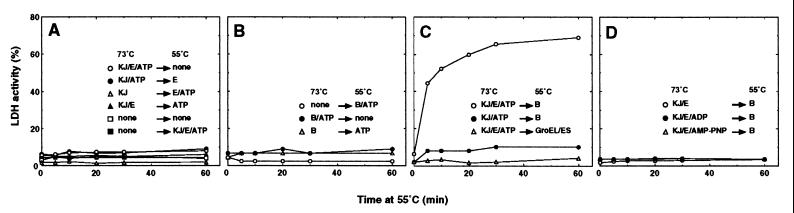
We examined whether the TDnaK⋅J-GrpE set (TDnaK⋅J complex and TGrpE) could mediate reactivation of heat-inactivated LDH. Experiments consisted of two steps: the first incubation at 73°C for 30 min (inactivation) was immediately followed by the second incubation at 55°C (reactivation). Indicated chaperones and nucleotides were added from the beginning of the 73°C incubation or at the time of temperature shift to 55°C. The values at time zero in Fig. 2A–D correspond to the residual LDH activities after the 30-min incubation at 73°C. LDH activities were measured at indicated times after the start of the 55°C incubation. It should be noted that components contained in the solution at the first 73°C incubation step remained present throughout the second 55°C incubation step. We confirmed that the TDnaK⋅J complex and TGrpE themselves were stable and active during incubation at 73°C for 30 min and at 55°C for 60 min. In the case when ATP was added, its concentration (3 mM) was high enough to ensure that a significant amount (>2 mM) of ATP remained unhydrolyzed after the incubations with TDnaK⋅J-GrpE and TClpB at 73°C for 30 min and at 55°C for 60 min.
Figure 2.
Effect of chaperones on reactivation of heat-inactivated LDH. Indicated components were added to the solutions containing LDH (final concentration, 0.2 μM), and the solutions were incubated at 73°C for 30 min. Then the temperature was shifted down to 55°C, and other indicated components were immediately supplemented to the solutions (0 min in abscissa). Incubation at 55°C continued, and LDH activities were measured at the indicated times. Final concentrations of added components were 0.4 μM (TDnaK⋅J complex), 0.8 μM (TGrpE), 0.4 μM (TClpB), 0.4 μM (TGroEL/ES), and 3 mM (nucleotides). Note that components added at 73°C remained present after the temperature shift. The KJ, E, and B represent the TDnaK⋅J complex, TGrpE, and TClpB, respectively. The orders of addition are shown. (A) Effect of the TDnaK⋅J complex and TGrpE. (B) Effect of TClpB. (C) Effect of TDnaK⋅J complex, TGrpE, TClpB, and TGroEL/ES. (D) Effect of nucleotides.
LDH lost the catalytic activity down to 6% or less of that of native LDH after the first incubation at 73°C for 30 min. Spontaneous recovery of the activity of the heat-inactivated LDH was not observed at 55°C, at which temperature the native LDH was stable and active (Fig. 2A). The TDnaK⋅J-GrpE set + ATP, or any other combinations of the TDnaK⋅J complex, TGrpE, and ATP, did not show a protective effect on LDH from heat inactivation at 73°C (see values at 0 min in Fig. 2A). Recovery of the LDH activity at 55°C was not observed in any experiments with all possible combinations of the TDnaK⋅J-GrpE set and ATP (Fig. 2A). Therefore, even though the TDnaK⋅J-GrpE set + ATP prevented the aggregation of LDH (Fig. 1, lane 3), the reactivation of the remaining soluble fraction was not observed. The molar ratio of the LDH:TDnaK⋅J complex:TGrpE was 1:2:4 (monomer base ratio, 1:6:8) in the above experiments, and we got the same result with double the amount of chaperones. We conclude that the TDnaK⋅J-GrpE set by itself can neither prevent LDH from heat inactivation nor mediate recovery of the activity of heat-inactivated LDH.
TClpB Alone Cannot Reactivate Heat-Inactivated LDH.
We then examined whether TClpB could mediate reactivation of heat-inactivated LDH (Fig. 2B). When LDH was incubated in the solution containing the TClpB and ATP at 73°C for 30 min, LDH lost 95% of its activity. Subsequent incubation at 55°C for 1 hr did not cause recovery of the LDH activity. TClpB itself was stable during incubation at 73°C for 30 min and at 55°C for 60 min. Therefore, TClpB could neither protect LDH from heat inactivation at 73°C nor mediate recovery of the LDH activity at 55°C. Addition of TClpB at 55°C, instead of at 73°C, did not change the result; no recovery of LDH activity was observed (Fig. 2B).
Cooperation of the TDnaK⋅J-GrpE Set and TClpB Is Required for Reactivation of Heat-Inactivated LDH.
Next the combination of the TDnaK⋅J-GrpE set and TClpB was tested (Fig. 2C). We heated LDH at 73°C for 30 min in the presence of the TDnaK⋅J-GrpE set + ATP and then added TClpB to the solution at the time of the temperature shift to 55°C, expecting the postheat shock effect of TClpB as implicated for yeast Hsp104 (9, 34). The result was remarkable: heat-inactivated LDH recovered the activity up to 70% of that of unheated LDH. Recovery of the activity at 55°C proceeded rather slowly, and it took 30 min to reach a maximum final yield. The yield of the recovery was decreased by prolonged exposure of LDH to 73°C over 30 min; for example, the 45- and 60-min incubations at 73°C resulted in 57% and 46% final recovery yields at 55°C, respectively. This decrease was not caused by loss of function of the TDnaK⋅J complex at 73°C, because the same degree of the yields was obtained in experiments where the TDnaK⋅J complex preheated for 30 min at 73°C was used instead of the unheated one. Therefore, the inactivated but chaperone-manageable state undergoes transition to a chaperone-unmanageable state with time at high temperatures. On the contrary, the yield was not changed by the delayed (by at least 30 min) addition of TClpB after the temperature was shifted to 55°C, indicating that the chaperone-manageable state is rather stable at moderate temperature. Recovery was only marginal when TGrpE was omitted. Thus, all of the three chaperone components used in these experiments, the TDnaK⋅J complex, TGrpE, and TClpB are essential for the reactivation of heat-inactivated LDH. Interestingly, the complex of chaperonin and cochaperonin from T. thermophilus (32) (TGroEL/ES) could not replace TClpB to mediate reactivation of the heat-inactivated LDH. The recovery of LDH activity depended on ATP hydrolysis, because no recovery was observed when adenosine 5′-[β,γ–imido]triphosphate or ADP was added instead of ATP (Fig. 2D).
Order of Addition of Chaperones.
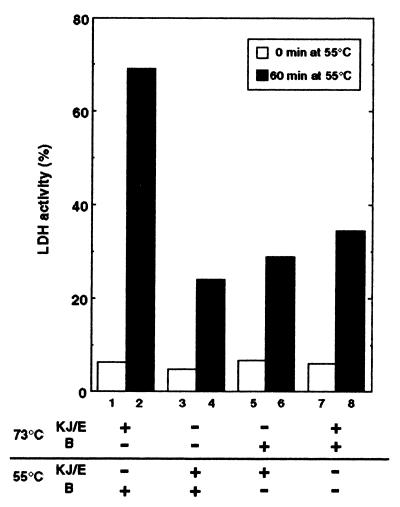
We then changed the order of addition of the TDnaK⋅J-GrpE set and TClpB (Fig. 3). A small but significant activity (25%) was recovered in the experiment where none of the chaperones were added at 73°C, and the TDnaK⋅J-GrpE set and TClpB were added together at 55°C (Fig. 3, bar 4). In this and other experiments shown in Fig. 3, ATP was always included from the 73°C incubation step. However, the same degree of recovery (38%) was observed in the experiment where ATP was added not at 73°C, but at 55°C together with all other components. Therefore, the presence of chaperones in the heat-inactivation step is not prerequisite for the subsequent reactivation at moderate temperature, even if the yield of reactivation is low. The presence of TClpB at 73°C did not improve significantly the yield of recovered LDH at 55°C (29%) (Fig. 3, bar 6). As described in the above paragraph, the presence of TDnaK⋅J-GrpE set at 73°C was necessary to achieve the high yield recovery (69%) at the subsequent 55°C incubation with TClpB (bar 2). Intermediate yield of the recovery (55%) was observed when the TDnaK⋅J-GrpE set (and ATP) was added 10 min after the start of the 73°C incubation. However, the high yield shown in bar 2 was reduced to 35% by the copresence of TClpB with the TDnaK⋅J-GrpE set at 73°C, even though both TClpB and the TDnaK⋅J-GrpE set were present at the 55°C incubation step (bar 8). TClpB retained its function after the 30-min incubation at 73°C because, in the experiment of bar 2, replacement of intact TClpB by the TClpB preheated at 73°C for 30 min did not reduce the high recovery yield (not shown). TClpB apparently has an inhibitory effect on the function of the TDnaK⋅J-GrpE set at 73 °C
Figure 3.
Effect of order of additions of the TDnaK⋅J-GrpE set and TClpB on the recovery of LDH activity. The order of addition of chaperones are indicated. For all experiments, ATP (3 mM) was added from the beginning of the 73°C incubation. The LDH activities at 0 min (blank bars) and 60 min (black bars) after the temperature shift to 55°C were measured.
Other Proteins Are Also Rescued from the Heat-Inactivated State by Cooperation of the Two Chaperones.
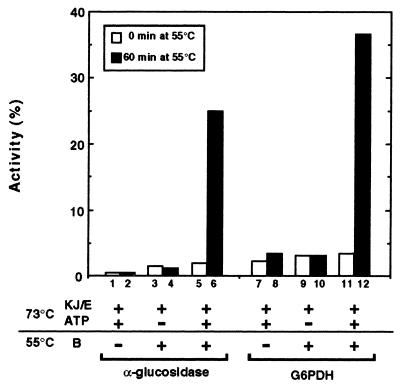
To know whether TDnaK⋅J-GrpE set- and TClpB-dependent reactivation of heat-inactivated protein is universal or specific only to LDH, we have examined two other proteins as substrates, thermophilic α-glucosidase (a monomer of the 65 kDa subunit) and G6PDH (a homotetramer of the 53-kDa subunit). These proteins lost their enzymatic activities after incubation at 73°C for 20 min (α-glucosidase) or 10 min (G6PDH). The enzymes inactivated at 73°C did not recover activity spontaneously at 55°C where the native enzymes were stable and active (not shown). The TDnaK⋅J-GrpE set ± ATP were able neither to protect the enzymes from inactivation at 73°C (Fig. 4, bars 1, 3, 5, 7, 9, 11) nor to help the enzymes to recover activity at 55°C (Fig. 4, bars 2, 4, 8, 10). Like the case of LDH, the enzymes recovered activities, up to 25% (α-glucosidase) and 37% (G6PDH), when they were heated at 73°C with the TDnaK⋅J-GrpE set + ATP and subsequently incubated at 55°C for 60 min with further addition of TClpB (Fig. 4, bars 6, 12). ATP was indispensable for the recovery (Fig. 4, bars 4, 10). Again, similar to LDH, as the incubation period at 73°C increased, the yield of the recovery decreased. For example, α-glucosidase and G6PDH heated for 30 min at 73°C with the TDnaK⋅J-GrpE set + ATP recovered 17% and 18%, respectively, of their activities after the subsequent 60-min incubation at 55°C with TClpB. Thus, cooperation of the TDnaK⋅J-GrpE set and TClpB to rescue proteins from the heat-inactivated state is not restricted to LDH.
Figure 4.
Reactivation of heat-inactivated α-glucosidase and G6PDH mediated by the TDnaK⋅J-GrpE set and TClpB. α-glucosidase (0.2 μM) and G6PDH (0.2 μM) were incubated at 73°C for 20 min (α-glucosidase) or 10 min (G6PDH) and then subsequently incubated at 55°C. The activities of these enzymes were measured at 0 min and 60 min after the temperature shift to 55°C. Activities of α-glucosidase and G6PDH are expressed as percent of those before heat inactivation.
DISCUSSION
The cooperation of the Hsp70 chaperone family and Hsp104 family in thermotolerance of eukaryotic cells has been indicated from in vivo experiments of yeast (9, 21). This contention gained support by the recent in vitro demonstration of reactivation of chemically denatured proteins, which were supposed to mimic the heat-denatured partly aggregated state by cooperative action of purified yeast chaperones Hsp70, Hsp40, and Hsp104 (24). This report provides another support for this cooperation: recovery of heat-inactivated enzymes by the cooperative action of the TDnaK (Hsp70), DnaJ (Hsp40), GrpE, and TClpB (Hsp104) was directly demonstrated. We also show that the cooperation could be functional not only in eukaryotes but also in eubacteria. T. thermophilus is different from E. coli in that TDnaK is isolated as a trigonal complex with TDnaJ and TDafA. Nonetheless, the general importance of this cooperation in the eubacterial kingdom is suggested by the fact that hsp104/clpB genes, or closely related genes, as well as hsp70/dnaK genes, are found in all of the eubacteria whose genomes have been determined so far.
When proteins are exposed at denaturing temperatures, they are at first denatured with concomitant loss of biological activity and then start to form aggregation. Aggregation undergoes various stages with time, and finally insoluble large aggregates are formed. Neither the TDnaK⋅J-GrpE set nor TClpB can protect other proteins from heat inactivation. However, analogous to yeast counterparts (24, 36), the TDnaK⋅J-GrpE set can suppress the aggregation of heat-inactivated proteins by using energy of ATP hydrolysis (Fig. 1). TClpB by itself has a small effect in blocking the aggregation, but it apparently inhibits in some degree the aggregation-prevention function of the TDnaK⋅J-GrpE set. Also, TClpB has an inhibitory effect on the final recovery yield when, in addition to the TDnaK⋅J-GrpE set, it is added to the LDH solution in the inactivation period (Fig. 3). During incubation at 73°C, inactivation and reactivation of LDH might cycle in the solution containing the TDnaK⋅J-GrpE set + TClpB + ATP. However, in each cycle, a small fraction of inactivated LDH could fall into aggregates before they are reactivated. Another explanation is that TClpB competes for inactivated LDH, and then the TClpB-bound LDH becomes permanently inactivated.
The best recovery of LDH activity was obtained for the LDH, previously heated with the TDnaK⋅J-GrpE set, in which the soluble fraction is maximal (Fig. 1 and Fig. 2C). Therefore, the heat-inactivated soluble proteins appear to be the favorite substrates for the cooperative action of the TDnaK⋅J-GrpE set and TClpB. Because the soluble fraction, a supernatant of the centrifugation (19,000 × g, 10 min) may contain denatured monomers and small aggregates, the cooperative system of the TDnaK⋅J-GrpE set and TClpB may have the ability to rescue both of them. Moreover, this cooperative system can probably rescue proteins contained in even larger aggregates. This contention is indicated from the observation that LDH heated at 73°C in the absence of any chaperones can recover a small but significant activity at 55°C by the TDnaK⋅J-GrpE set and TClpB (Fig. 3). Under this condition, almost all LDH in the heated solution could be precipitated when centrifuged (Fig. 1). As proposed by Glover and Lindquist for yeast Hsp104 (24), we suggest that TClpB and the TDnaK⋅J-GrpE set function cooperatively to disaggregate and assist refolding of proteins.
The individual roles of TClpB and the TDnaK⋅J-GrpE set in the reactivation of heat-inactivated proteins are not clear. In our experiments, chaperones contained in the solution at the first 73°C incubation step were not removed at the time of temperature shift and attempts to reisolate the folding-competent proteins have been unsuccessful. However, two observations are worth noting. First, to recover activity of LDH heated alone, both TClpB and the TDnaK⋅J-GrpE set were required in the reactivation period (Fig. 3). Besides prevention of aggregation, the TDnaK⋅J-GrpE set appears to have another function, mediation of refolding, which needs TClpB as a partner. One plausible explanation of the necessity for both chaperones in the reactivation period is that a putative refolding intermediate is directly transferred between TClpB and the TDnaK⋅J-GrpE set. This explanation is consistent with the second observation that TGroEL/ES could not replace TClpB in the reactivation reaction (Fig. 2C). In spite of the fact that TGroEL/ES by itself has the ability at moderate temperatures to assist refolding of nonnative LDH (32), the TDnaK⋅J-GrpE set chooses only TClpB as a specific partner. Discrimination of the TClpB from TGroEL/ES is explained if the TDnaK⋅J-GrpE set can transfer the nonnative LDH only to TClpB (but not to TGroEL/ES). This explanation assumes the physical interaction of the TDnaK⋅J-GrpE set and TClpB. Glover and Lindquist reported that Hsp104-mediated refolding is tightly linked to the presence of the Hsp70/40 and cannot be separated into sequential reactions, and that a weak physical interaction exists between yeast Hsp40 and Hsp104 (24). Cooperation between Hsp70/Hsp40 and Hsp104 has been established, but further studies are required to clarify the role of each chaperone.
Acknowledgments
We thank Ms. N. Kato for technical assistance of DNA sequencing and Dr. H. Taguchi for a kind gift of purified TGroEL/ES. K.M. was the recipient of a research fellowship of the Japanese Society for the Promotion of Science for Young Scientists. This work was supported by a Grant-in-Aid for Scientific Research on Priority Area (no. 09276101) from the Ministry of Education, Science, Sports and Culture of Japan.
ABBREVIATIONS
- LDH
lactate dehydrogenase
- G6PDH
glucose-6-phosphate dehydrogenase
- TDnaK⋅J complex
the trigonal complex of DnaK3DnaJ3DafA3 of T. thermophilus
- TDnaK⋅J-GrpE set
TDnaK⋅J complex + TgrpE
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AB012390).
For lane 4, repeated experiments showed that the supernatant band of LDH was always very faint and barely visible, but the band intensity of the precipitation fraction was sometimes weaker than the control one, e.g., lane 2. The exact reason for this has not been known, but we assume that the size of the aggregate formed in the absence of TGrpE could be small, and the weakly packed precipitate was lost during subsequent procedures.
References
- 1.Hartl F U. Nature (London) 1996;381:571–580. doi: 10.1038/381571a0. [DOI] [PubMed] [Google Scholar]
- 2.Bukau B, Horwich A L. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 3.Schröder H, Langer T, Hartl F U, Bukau B. EMBO J. 1993;12:4137–4144. doi: 10.1002/j.1460-2075.1993.tb06097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ziemienowicz A, Skowyra D, Zeilstra-Ryalls J, Fayet O, Georgopoulos C, Zylicz M. J Biol Chem. 1993;268:25425–25431. [PubMed] [Google Scholar]
- 5.Szabo A, Langer T, Schröder H, Flanagan J, Bukau B, Hartl F U. Proc Natl Acad Sci USA. 1994;91:10345–10349. doi: 10.1073/pnas.91.22.10345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarty J S, Buchberger A, Reinstein J, Bukau B. J Mol Biol. 1995;249:126–137. doi: 10.1006/jmbi.1995.0284. [DOI] [PubMed] [Google Scholar]
- 7.Liberek K, Marszalek J, Ang D, Georgopoulos C. Proc Natl Acad Sci USA. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sanchez Y, Lindquist S L. Science. 1990;248:1112–1115. doi: 10.1126/science.2188365. [DOI] [PubMed] [Google Scholar]
- 9.Parsell D A, Kowal A S, Singer M A, Lindquist S. Nature (London) 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 10.Parsell D A, Sanchez Y, Stitzel J D, Lindquist S. Science. 1990;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- 11.Parsell D A, Kowal A S, Lindquist S. J Biol Chem. 1994;269:4480–4487. [PubMed] [Google Scholar]
- 12.Schirmer E C, Lindquist S. Proc Natl Acad Sci USA. 1997;94:13932–13937. doi: 10.1073/pnas.94.25.13932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottesman S, Squires C, Pichersky E, Carrington M, Hobbs M, Mattick J S, Dalrymple B, Kuramitsu H, Shiroza T, Foster T, et al. Proc Natl Acad Sci USA. 1990;87:3513–3517. doi: 10.1073/pnas.87.9.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitagawa M, Wada C, Yoshioka S, Yura T. J Bacteriol. 1991;173:4247–4253. doi: 10.1128/jb.173.14.4247-4253.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Squires C L, Pedersen S, Ross B M, Squires C. J Bacteriol. 1991;173:4254–4262. doi: 10.1128/jb.173.14.4254-4262.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo K M, Kim K I, Goldberg A L, Ha D B, Chung C H. J Biol Chem. 1992;267:20429–20434. [PubMed] [Google Scholar]
- 17.Thomas J G, Baneyx F. J Bacteriol. 1998;180:5165–5172. doi: 10.1128/jb.180.19.5165-5172.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi M R. Proc Natl Acad Sci USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pak M, Wickner S. Proc Natl Acad Sci USA. 1997;94:4901–4906. doi: 10.1073/pnas.94.10.4901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S K, Kim K I, Woo K M, Seol J H, Tanaka K, Ichihara A, Ha D B, Chung C H. J Biol Chem. 1993;268:20170–20174. [PubMed] [Google Scholar]
- 21.Sanchez Y, Parsell D A, Taulien J, Vogel J L, Craig E A, Lindquist S. J Bacteriol. 1993;175:6484–6491. doi: 10.1128/jb.175.20.6484-6491.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moczko M, Schönfisch B, Voos W, Pfanner N, Rassow J. J Mol Biol. 1995;254:538–543. doi: 10.1006/jmbi.1995.0636. [DOI] [PubMed] [Google Scholar]
- 23.Schmitt M, Neupert W, Langer T. EMBO J. 1995;14:3434–3444. doi: 10.1002/j.1460-2075.1995.tb07349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glover J R, Lindquist S. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 25.Motohashi K, Taguchi H, Ishii N, Yoshida M. J Biol Chem. 1994;269:27074–27079. [PubMed] [Google Scholar]
- 26.Motohashi K, Yohda M, Endo I, Yoshida M. J Biol Chem. 1996;271:17343–17348. doi: 10.1074/jbc.271.29.17343. [DOI] [PubMed] [Google Scholar]
- 27.Motohashi K, Yohda M, Odaka M, Yoshida M. FEBS Lett. 1997;412:633–636. doi: 10.1016/s0014-5793(97)00847-8. [DOI] [PubMed] [Google Scholar]
- 28.Amada K, Yohda M, Odaka M, Endo I, Ishii N, Taguchi H, Yoshida M. J Biochem. 1995;118:347–354. doi: 10.1093/oxfordjournals.jbchem.a124913. [DOI] [PubMed] [Google Scholar]
- 29.Kunkel T A, Bebenek K, McClary J. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 30.Baykov A A, Evtushenko O A, Avaeva S M. Anal Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 31.Geladopoulos T P, Sotiroudis T G, Evangelopoulos A E. Anal Biochem. 1991;192:112–116. doi: 10.1016/0003-2697(91)90194-x. [DOI] [PubMed] [Google Scholar]
- 32.Taguchi H, Yoshida M. J Biol Chem. 1993;268:5371–5375. [PubMed] [Google Scholar]
- 33.Okuno H, Nagata K, Nakajima H. J Appl Biochem. 1985;7:192–201. [PubMed] [Google Scholar]
- 34.Schirmer E C, Glover J R, Singer M A, Lindquist S. Trends Biochem Sci. 1996;21:289–296. [PubMed] [Google Scholar]
- 35.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cyr D M. FEBS Lett. 1995;359:129–132. doi: 10.1016/0014-5793(95)00024-4. [DOI] [PubMed] [Google Scholar]